We reported the distribution and genetic composition of 66 HML-6 elements. We analyzed the phylogeny of the HML-6 sequences and identified two main clusters. We provided the first description of a Rec domain within the env sequence of 23 HML-6 elements. A Rec domain was also predicted within the ERVK3-1 transcript sequence, revealing its expression in various healthy tissues. Evidence about the context of insertion and colocalization of 19 HML-6 elements with functional human genes are also reported, including the sequence 16p11.2, whose 5′ long terminal repeat overlapped the exon of one transcript variant of a cellular zinc finger upregulated and involved in hepatocellular carcinoma. The present work provides the first complete overview of the HML-6 elements in GRCh37(hg19), describing the structure, phylogeny, and genomic context of insertion of each locus. This information allows a better understanding of the genetics of one of the most expressed HERV groups in the human genome.
KEYWORDS: endogenous retrovirus, HERV, HERV-K-MEL, HML-6, RetroTector, bioinformatics
ABSTRACT
Eight percent of the human genome is composed of human endogenous retroviruses (HERVs), remnants of ancestral germ line infections by exogenous retroviruses, which have been vertically transmitted as Mendelian characters. The HML-6 group, a member of the class II betaretrovirus-like viruses, includes several proviral loci with an increased transcriptional activity in cancer and at least two elements that are known for retaining an intact open reading frame and for encoding small proteins such as ERVK3-1, which is expressed in various healthy tissues, and HERV-K-MEL, a small Env peptide expressed in samples of cutaneous and ocular melanoma but not in normal tissues.
IMPORTANCE We reported the distribution and genetic composition of 66 HML-6 elements. We analyzed the phylogeny of the HML-6 sequences and identified two main clusters. We provided the first description of a Rec domain within the env sequence of 23 HML-6 elements. A Rec domain was also predicted within the ERVK3-1 transcript sequence, revealing its expression in various healthy tissues. Evidence about the context of insertion and colocalization of 19 HML-6 elements with functional human genes are also reported, including the sequence 16p11.2, whose 5′ long terminal repeat overlapped the exon of one transcript variant of a cellular zinc finger upregulated and involved in hepatocellular carcinoma. The present work provides the first complete overview of the HML-6 elements in GRCh37(hg19), describing the structure, phylogeny, and genomic context of insertion of each locus. This information allows a better understanding of the genetics of one of the most expressed HERV groups in the human genome.
INTRODUCTION
About 8% of the human genome consists of human endogenous retroviruses (HERVs) (1), resulted from the occasional integration of exogenous retroviruses into the human germ line, which occurred mostly more than 30 million years ago. These proviruses were vertically transmitted to the offspring and then fixed in the genome of the population during the evolution (2, 3), accumulating, over time, several mutations that in most cases compromised their coding capability.
Nevertheless, some important examples of HERV involvement in human biology have been demonstrated (2, 4–6), such as the retroviral protein Syncytin-1, a functional envelope (Env) protein coded by an HERV-W provirus that is involved in trophoblastic-cell fusion during pregnancy (7, 8). However, in the majority of studies, only general HERV groups and not individual HERV loci have been investigated in detail for their correlation to diseases (9), especially due to the lack, until recently, of any information regarding the HERV characterization and classification at the genomic level (10). For this reason, the putative role of HERVs as etiological agents, contributing factors, or markers of diseases has been repetitively suggested (4, 9, 11, 12), but it still remains to be better investigated and definitively established in the majority of cases (2, 4, 5, 9).
Recently, a comprehensive collection and classification of HERV sequences has been provided, revealing the presence of a totality of 3,173 most complete proviral elements in the human genome assembly GRCh37/hg19 (13). This study noted the previous HERV subdivision in classes I (Gamma- and Epsilon-like), II (Beta-like), and III (Spuma-like) (10, 13), depending on the similarity to exogenous retroviruses, and included all the HERV elements in 39 groups, revealing a high number of recombination events that took place over time among different HERVs (13).
Between the HERV groups, the human MMTV-like (HML) supergroup of class II is one of the most investigated, mainly due to the fact it includes some of the youngest and best conserved elements, belonging to the HML-2 clade (14). This supergroup consists of 10 clades (HML-1 to HML-10) that are related to the exogenous mouse mammary tumor virus (MMTV) (13). Accordingly, the genome of HML elements is composed of four main retroviral genes (gag, pro, pol, and env) flanked by two long terminal repeats (LTRs), and it shows several beta-retroviral typical features, including the primer-binding site (PBS) for lysine (K) tRNA (3, 13). In addition, it has been reported that some HML-2 elements are able to encode an mRNA nuclear export protein, Rec, coded from a doubly spliced transcript which is a functional homolog of the retroviral regulatory proteins MMTV Rem (15), HIV Rev, and HTLV Rex (16, 17). The HML-2 rec accessory gene can be present in two forms: the first with a full-length sequence (characteristic of type 2 HML-2 elements) and the second with a 292-bp deletion that codes for a smaller protein, NP9 (associated with type 1 HML-2 sequences) (17, 18). Recently, the Rec domain has also been found within the genomes of some HML-10 elements (19).
In addition to HML-2 and HML-10, HML-6 is also a highly investigated HML clade. The earliest studies about this group collected 10 sequences identified by using a PCR approach with HML-6-specific primers (20, 21). A first phylogenetic characterization of these elements indicated that the HML-6 subgroup was a heterogeneous but distinct group of elements belonging to the HERV-K superfamily, with a PBS for lysine tRNA (20, 21). The HML-6 betaretroviral features were also detected: two zinc fingers in gag and both dUTPase and G-patch domains in pro (20, 21). Notably, the dUTPase tree showed a different phylogeny from the one of the other genes, and further analysis concerning the presence of dUTPase in the various HERV-K subgroups demonstrated that the HML-6 dUTPase sequences appear to be more related to the MMTV dUTPases than to those of the other HML members (20, 22). According to RetroTector (ReTe) analysis, this subgroup includes 48 canonical elements and additional 17 noncanonical elements coming from recombination events (13), and the internal sequences are flanked by two LTRs identified among four types (LTR3, LTR3A, LTR3B, and LTR3B_v) by RepBase.
A first important study reported an extensive transcriptional activity of HML-6 elements through retrovirus-specific microarray (23). HML-6 transcripts were found in all the 19 healthy tissues analyzed (23). Of note, an HML-6 element in locus 19q13.43b that contains an intact open reading frame (ORF) was reported to encode a small transcript, ERVK3-1, expressed in various healthy tissues (ENSG00000142396), and gave support to the hypothesis of an extensive HML-6 expression activity (23). Subsequently, besides physiological expression, HML-6 sequences were reported to be of particular interest due to either the selective activation or the increased activity of several proviral loci in malignant mammary gland tissue from patients with human breast cancer (24). Other examples of HML-6 expression in cancer were also reported in cutaneous and ocular melanoma cells, in which a small peptide from an HML-6 env gene, namely, HERV-K-MEL, was detected in tumor tissues but not in normal tissues (25). Although these findings raised the possibility of an HML-6 contribution to diseases, the causal relationship between HML-6 expression and cancer is still not clear, and further expression studies of the individual HML-6 loci are needed to clarify their potential contribution to human pathogenesis.
The present study aims to provide for the first time a comprehensive structural characterization of the HML-6 group members, which can be useful to direct further and more detailed studies of expression. The results show a high heterogeneity between the HML-6 elements due to the presence of two distinct subgroups. Moreover, the analysis of their genomic context of insertion provides updated information on the HML-6 presence in the human genome, which is essential to understand their potential role in physiological and pathological contexts and to focus further expression analyses on specific loci of interest.
RESULTS
Collection of 66 HML-6 loci in the human genome sequence.
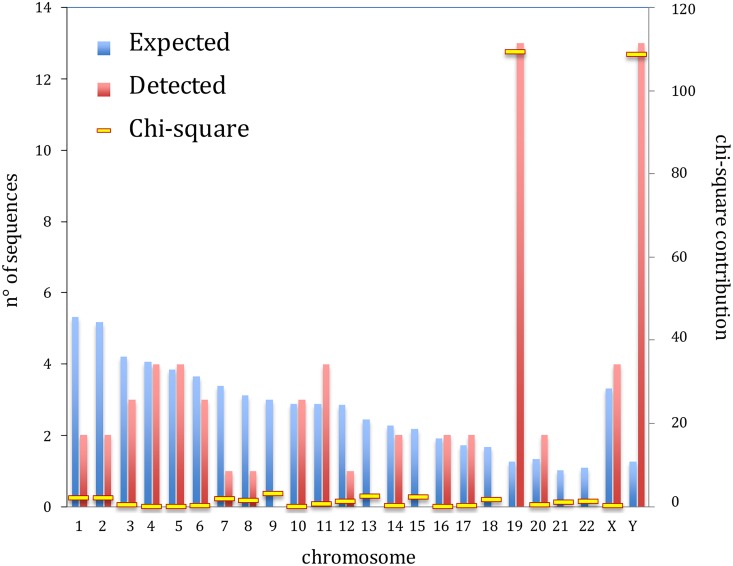
We collected the HML-6 sequences provided by Vargiu et al. (13), in which ReTe analysis of the genome assembly hg19 allowed to identify and classify all the most intact HERV proviruses in our genome. Moreover, we compared these coordinates and sequences to those obtained with a genome browser BLAT search in genome assembly hg19, using as a query the LTR3A-HERVK3-LTR3A consensus sequence assembled from Dfam data set. In particular, two HML-2 sequences—in locus 10q11.21 and 10q25.1—were detected only by ReTe and provided in Vargiu et al., but we were not able to find them by BLAT searching. Similarly, two sequences in locus 5q13.2, showing 100% identity and flanked by an identical region, were both detected by BLAT searching, even if only one sequence in locus 5q13.2 was previously reported by Vargiu et al. (13). Therefore, through this integrated search approach, we obtained the genomic coordinates of 66 HML-6 sequences (Table 1 ; see also File S1 in the supplemental material for the coordinates in hg38). We named the HML-6 elements in conformity with their genomic localization, and in the case of presence of multiple sequences within the same genomic locus we unequivocally indicated the sequence order with alphabetical letters. When we analyzed the distribution of the HML-6 insertion, almost all chromosomes showed an apparent random distribution of HML-6 loci, in the sense that the number of sequences was approximately proportional to the chromosome size. The exceptions were chromosome 19 and chromosome Y, in which we detected more HML-6 proviruses than expected. An overall chi-square test including all chromosomes indicated a nonrandom distribution (P < 0.0001) of HML-6 loci, with a very prominent contribution of chi-square values calculated for the chromosomes 19 and Y (Fig. 1). To confirm this finding, we also performed all comparisons between each pair of chromosomes and, owing to the large number of tests involved (n = 276), filtered P values by using the Benjamini-Hochberg procedure (26) to maintain a cumulative probability of false discoveries in all tests lower than 5%. The data confirmed a significant enrichment on chromosomes 19 and Y (see Fig. S1 in the supplemental material).
TABLE 1.
HML-6 proviral sequences and their localization in the human genome GRCh37/hg19 assembly
| Locus | Chromosome | Strand | Position |
LTR type | Length (bp) | Subtype | |
|---|---|---|---|---|---|---|---|
| Start | End | ||||||
| 1p21.1 | 1 | + | 103298830 | 103306681 | LTR3 | 7,851 | 1a |
| 1q25.2 | 1 | – | 179406261 | 179412404 | LTR3A | 6,143 | 1a |
| 2q14.23 | 2 | – | 128372842 | 128376247 | LTR3B_v | 3,405 | 2 |
| 2q22.1 | 2 | – | 136829388 | 136834831 | LTR3A | 5,443 | 1a |
| 3p25.1 | 3 | + | 14266558 | 14271762 | LTR3A | 5,204 | 1a |
| 3p21.31a | 3 | – | 46087646 | 46095966 | LTR3A | 8,320 | 1a |
| 3p21.31b | 3 | + | 46468034 | 46475121 | LTR3A | 7,087 | 1a |
| 4p14 | 4 | – | 39540876 | 39545998 | LTR3B | 5,122 | 1b |
| 4q13.2 | 4 | – | 69610304 | 69616956 | LTR3B | 6,652 | 1b |
| 4q13.3 | 4 | + | 71418184 | 71420406 | LTR3B_v | 2,222 | 2 |
| 4q21.1 | 4 | – | 78313436 | 78321358 | LTR3 | 7,922 | 1a |
| 5p14.1 | 5 | + | 24649773 | 24654829 | LTR3A | 5,056 | 1a |
| 5q13.2a | 5 | + | 69641005 | 69643229 | Only 3′LTR3B_v | 2,224 | 2 |
| 5q13.2b | 5 | + | 69958435 | 69960659 | Only 3′LTR3B_v | 2,224 | 2 |
| 5q13.2c | 5 | + | 70867724 | 70874228 | LTR3A | 6,504 | 1a |
| 6p22.2 | 6 | – | 26288250 | 26296494 | LTR3B | 8,244 | 1b |
| 6p21.32a | 6 | – | 32443272 | 32447375 | Only 5′LTR3 | 4,103 | 1a |
| 6p21.32b | 6 | – | 32527497 | 32535122 | Only 5′LTR3 | 7,625 | 1a |
| 7q36.1 | 7 | – | 150279386 | 150283313 | LTR3B_v | 3,927 | 2 |
| 8q11.1 | 8 | + | 47395171 | 47403000 | LTR3A | 7,829 | 1a |
| 10q11.21a | 10 | – | 43788582 | 43796372 | LTR3B_v | 7,790 | 2 |
| 10q11.21b | 10 | + | 45774424 | 45782353 | LTR3 | 7,929 | 1a |
| 10q25.1 | 10 | + | 110488980 | 110492726 | LTR3B_v | 3,746 | 2 |
| 11p15.4 | 11 | – | 7920872 | 7927779 | LTR3A | 6,907 | 1a |
| 11q12.3a | 11 | – | 61817251 | 61823827 | LTR3A | 6,576 | 1a |
| 11q12.3b | 11 | + | 62019229 | 62021808 | Only 5′LTR3 | 2,579 | 1a |
| 11q23.2 | 11 | – | 112795351 | 112800806 | LTR3A | 5,455 | 1a |
| 12q24.12 | 12 | – | 112253979 | 112263658 | LTR3A | 9,679 | 1a |
| 14q12 | 14 | + | 28879914 | 28890437 | Only 3′LTR3A | 10,523 | 1a |
| 14q24.2 | 14 | + | 70278180 | 70282740 | LTR3A | 4,560 | 1a |
| 16p11.2 | 16 | – | 30627018 | 30635602 | LTR3B | 8,584 | 1b |
| 16p11.1 | 16 | + | 34750975 | 34758850 | LTR3B | 7,875 | 1b |
| 17q21.31 | 17 | + | 41949365 | 41952180 | LTR3B_v | 2,815 | 2 |
| 17q25.1 | 17 | + | 72580505 | 72583070 | Only 5′LTR3B | 2,565 | 1b |
| 19p13.2a | 19 | – | 9618707 | 9625736 | LTR3 | 7,029 | 1a |
| 19p13.2b | 19 | – | 11964097 | 11971799 | LTR3 | 7,702 | 1a |
| 19p12a | 19 | + | 21416592 | 21420867 | LTR3B_v | 4,275 | 2 |
| 19p12b | 19 | – | 21968952 | 21975023 | Only 3′LTR3 | 6,071 | 1a |
| 19p12c | 19 | + | 22376043 | 22379350 | Only 3′LTR3 | 3,307 | 1a |
| 19p12d | 19 | + | 24047631 | 24054030 | LTR3 | 6,399 | 1a |
| 19q13.41a | 19 | – | 52307949 | 52315192 | LTR3A | 7,243 | 1a |
| 19q13.41b | 19 | + | 52479404 | 52484683 | LTR3A | 5,279 | 1a |
| 19q13.41c | 19 | – | 52913436 | 52917986 | --- | 4,550 | 1a |
| 19q13.41d | 19 | – | 52978909 | 52981951 | LTR3A | 3,042 | 1a |
| 19q13.41e | 19 | – | 53487788 | 53492829 | Only 3′LTR3B | 5,041 | 1b |
| 19q13.43a | 19 | + | 58023984 | 58029856 | LTR3B | 5,872 | 1b |
| 19q13.43b | 19 | + | 58817037 | 58826633 | LTR3B | 9,596 | 1b |
| 20p13 | 20 | – | 1377446 | 1383348 | LTR3A | 5,902 | 1a |
| 20p11.21 | 20 | + | 25374769 | 25383907 | LTR3A | 9,138 | 1a |
| Xp11.22 | X | + | 53188296 | 53193008 | LTR3 | 4,712 | 1a |
| Xp11.21 | X | – | 57129414 | 57135829 | LTR3A | 6,415 | 1a |
| Xq13.2 | X | + | 73397834 | 73402327 | Only 3′LTR3A | 4,493 | 1a |
| Xq27.1 | X | – | 140290665 | 140293656 | Only 5′LTR3B | 2,991 | 1b |
| Yq11.221 | Y | + | 19443452 | 19448989 | LTR3A | 5,537 | 1a |
| Yq11.222a | Y | + | 19958329 | 19963018 | Only 3′LTR3B | 4,689 | 1b |
| Yq11.222b | Y | + | 20051008 | 20055589 | Only 3′LTR3B | 4,581 | 1b |
| Yq11.222c | Y | – | 20074149 | 20078731 | Only 3′LTR3B | 4,582 | 1b |
| Yq11.222d | Y | – | 20216759 | 20221448 | Only 3′LTR3B | 4,689 | 1b |
| Yq11.223a | Y | – | 25964947 | 25969633 | Only 3′LTR3B | 4,686 | 1b |
| Yq11.223b | Y | + | 26162248 | 26166935 | Only 3′LTR3B | 4,687 | 1b |
| Yq11.23a | Y | – | 26263056 | 26267731 | Only 3′LTR3B | 4,675 | 1b |
| Yq11.23b | Y | – | 26277752 | 26282358 | Only 3′LTR3B | 4,606 | 1b |
| Yq11.23c | Y | + | 27680077 | 27684680 | Only 3′LTR3B | 4,603 | 1b |
| Yq11.23d | Y | – | 27694700 | 27699373 | Only 3′LTR3B | 4,673 | 1b |
| Yq11.23e | Y | + | 27795496 | 27800183 | Only 3′LTR3B | 4,687 | 1b |
| Yq11.23f | Y | + | 27992754 | 27997440 | Only 3′LTR3B | 4,686 | 1b |
FIG 1.
Histogram of expected and detected chromosomal distributions of HML-6 proviruses. The enrichment in chromosomes 19 and Y is particularly clear, as also highlighted by their contributions to the chi-square distribution.
Phylogenetic analyses and subtype classification of HML-6 proviral internal sequences.
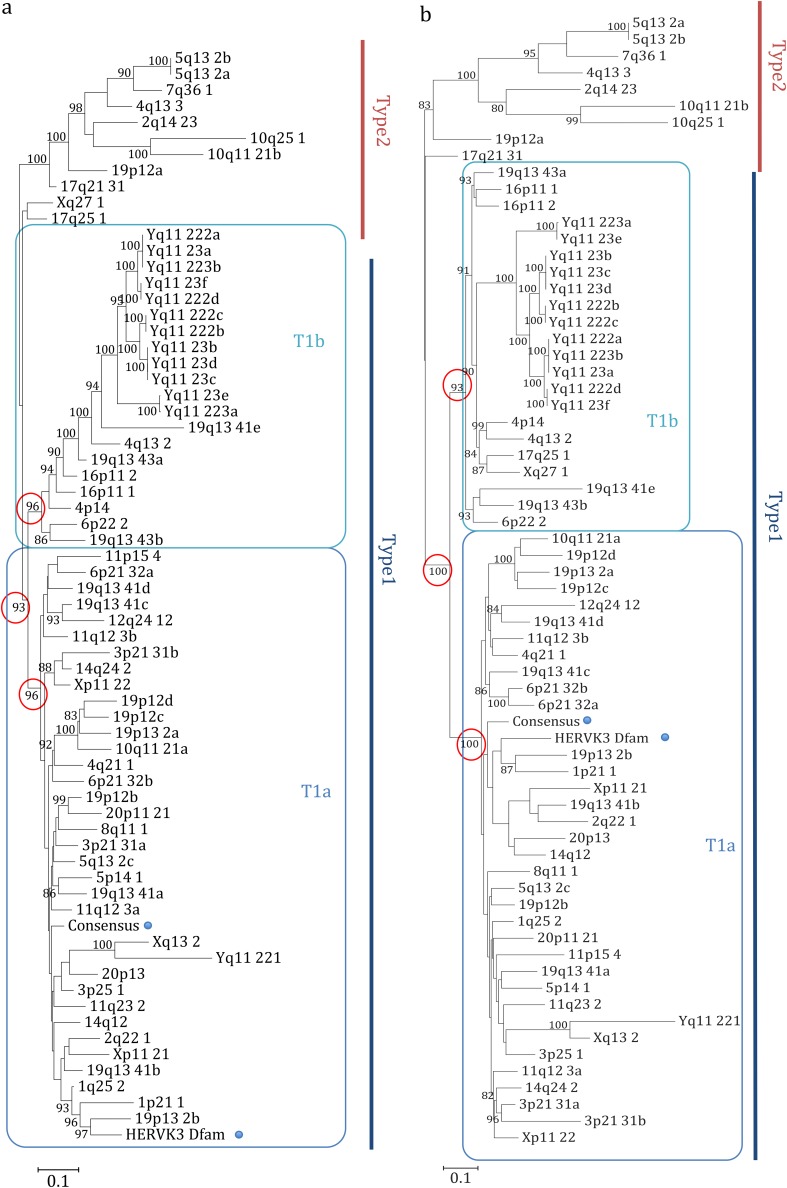
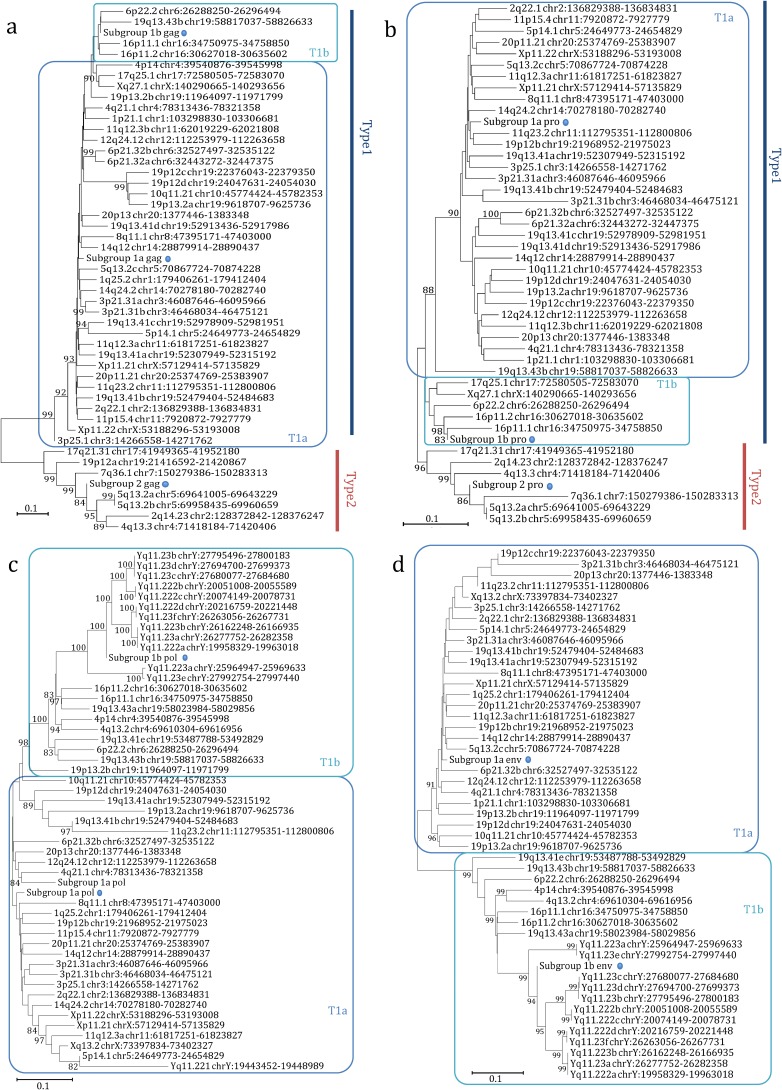
In order to characterize the structure of each single provirus, we created a consensus multiple sequence alignment of (i) all the 66 HML-6 internal sequences and (ii) the consensus sequence HERVK3 from Dfam. Next, we performed a neighbor-joining (NJ) analysis of the created consensus alignment with the Kimura model test. The resulting tree revealed the presence of two main clusters that we named type 1 and 2, including 55 and 11 elements, respectively (Fig. 2a). Moreover, type 1 elements showed an additional internal subdivision in two further clusters of 35 and 20 elements that we named type 1a and type 1b, respectively. To better understand the meaning of this phylogenetic information, we implemented the analysis by creating a maximum-likelihood (ML) tree, selecting the K80 model for the phylogenetic analysis (Fig. 2b). Also, in this case, we obtained a similar result, as we were able to identify the two main clusters with type 1 and 2 HML-6 proviruses and the type 1 subdivision in type 1a and 1b. Loci Xq27.1 and 17q25.1 were assigned to type 2 cluster by the NJ analysis but to the type 1b cluster by the ML analysis. Subsequently, phylogeny and evolutionary relations of the HML-6 group was investigated with respect to the others HML elements. We generated a majority-rule consensus sequence for the gag gene of each subgroup, selecting this gene due to the fact that it was the most conserved within the HML-6 group. Then, we performed a NJ analysis of amino acid sequences of Gag, by using our translated consensus and the Gag consensus sequences of the other HML groups (13), whereas the Gag sequences of the exogenous betaretroviruses MMTV, Mason-Pfizer monkey virus (MPMV), and Jaagsiekte sheep retrovirus (JSRV) and the Gag sequence of ZAM Drosophila endogenous retrovirus were used as outgroups. This analysis confirmed that all analyzed sequences belonged indeed to the HML-6 group (Fig. S2). Interestingly, all the HML-6 Gag consensus sequences grouped together outside the HML clade, suggesting that HML-6 may represent an intermediate group between the HERV-K and the other MMTV-related clades, in agreement with what has been reported in other works (22, 27, 28).
FIG 2.
Phylogenetic analysis of the internal sequences. The phylogeny was investigated by using the NJ method and the Kimura two-parameter model (a) and by performing an ML analysis and a K80 model test (b) of a consensus alignment of internal sequences. The two intragroup clusters (types 1 and 2) are indicated by blue and red lines, respectively, whereas the additional distribution of type 1 elements in two subtypes (1a and 1b) is indicated by squares.
Structural characterization of HML-6 proviral sequences.
The Dfam assembled HML-6 consensus sequence shows a typical proviral genome structure, in which 5′LTRs and 3′LTRs flank the gag, pro, pol, and env genes encoding the structural proteins and the essential enzymes (21). The gag gene (positions 205 to 1850) encodes the matrix (MA), capsid (CA) and nucleocapsid (NC) elements; the pro gene (1666 to 2621) encodes the protease (Pro) enzyme; the pol gene (2578 to 5276) determines the production of reverse transcriptase (RT) and integrase (IN); and the env gene (5225 to 7166) encodes the surface (SU) and transmembrane (TM) proteins. In addition, the analysis of conserved domains allowed identifying a predicted Rec domain between pol and env (5272 to 5445).
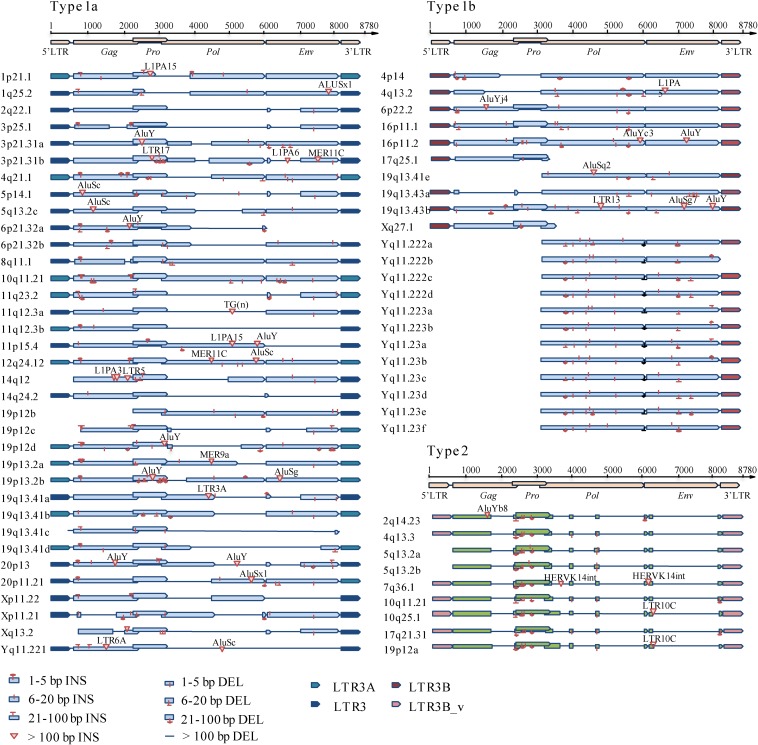
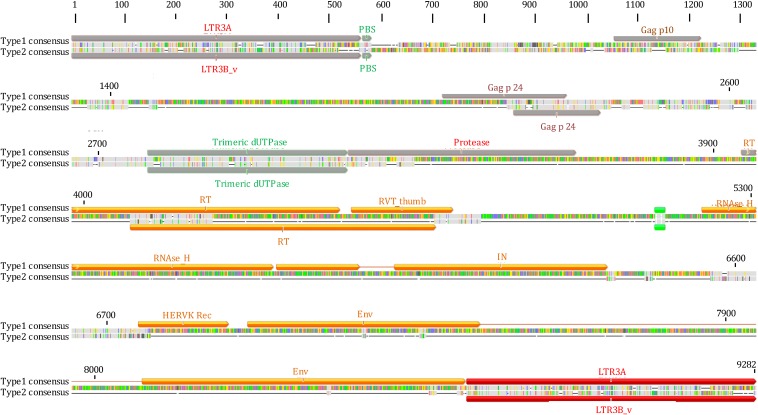
We hence attempted to define the structural characteristics of the HML-6 proviral types, annotating all the insertions and deletions within the internal sequences with respect to the consensus (Fig. 3). In general, compared to the consensus sequence, which was 7,166 bp in length excluding the LTRs, the overall average length of 4,918 bp was below the expectation: in fact, only nine elements maintained a complete structure, whereas the majority of sequences were incomplete due to the lack of large viral portions (Fig. 3). We hence annotated these variations, observing that a number of them are shared between elements of the same subgroup: (i) 51% of subgroup 1a elements lacked nucleotides (nt) 2727 to 4060 within the pol gene (RT portion), whereas the 57% lacked nt 5176 to 6185 within the env gene (Rec and SU portion); (ii) the gag and pro genes were completely missing in 69% of subgroup 1b elements and, moreover, a deletion of nt 5236 to 5333 (Rec portion) within the env gene was also found in 69% of these sequences; (iii) the viral portions between nt 717 and 1000, 2239 and 2919, 3078 and 3492, 3592 and 5071, and 5249 and 7120 were deleted in all the subgroup 2 elements, resulting in the partial deletion of the gag (p17 portion) and pro genes and in the complete deletion of the pol and env genes. We consequently summarized a consensus structure for each subgroup (Fig. 4 and see File S2 in the supplemental material).
FIG 3.
Structural characterization of 66 HML-6 sequences. Nucleotide insertions and deletions of each HML-6 nucleotide sequence are annotated by comparison to the HML-6 consensus sequence from Dfam.
FIG 4.
HML-6 type 1 and type 2 consensus sequences. Type 2 elements show common nucleotide deletions partially or totally corresponding to pro, pol, and env protein domains, as shown in the alignments of type 1 and 2 consensus sequences.
In addition, we annotated all minor insertions and deletions in order to define not only the subgroup overall identity but also the singularity of each HML-6 sequence (Fig. 3, see Data Set S1 in the supplemental material). Such a detailed structural characterization may hence provide a specific background for the structural investigation of single HML-6 loci and the unequivocal match to their eventual expression products.
Phylogenetic analysis of individual HML-6 retroviral genes.
To further verify the previous phylogenetic and structural studies, we performed NJ analyses for the individual HML-6 gag, pro, pol, and env genes that confirmed the presence of two main proviral types (1 and 2), as well as the additional subdivision within type 1 (1a and 1b) for all genes (Fig. 5).
FIG 5.
Phylogenetic analysis of the HML-6 nucleotide sequences of gag (a), pro (b), pol (c), and env (d) genes. Blue and red lines indicate genes of type 1 and 2 proviruses, while type 1a and 1b clusters are indicated by squares. The three intragroup consensus sequences of the genes are also included in the analysis and indicated by a dot. The evolutionary relationship has been ascertained by using the NJ method and the Kimura two-parameter model; the phylogenetic tree was built by using the bootstrap method with 1,000 replicates.
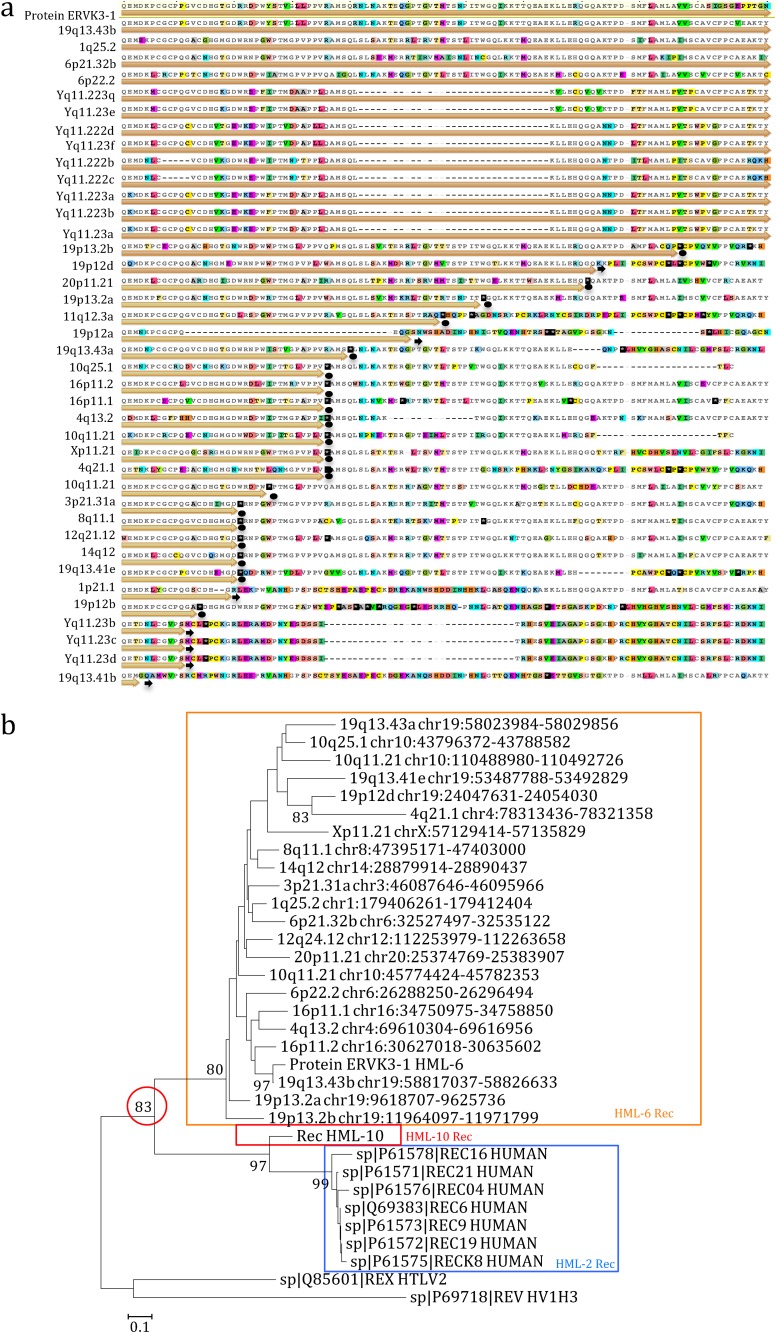
Moreover, we inspected the characteristics of the newly identified HML-6 Rec putative domain. First, we used the ERVK3-1 rec nucleotide sequence as a reference for a multiple alignment, finding that 39 of the 66 identified HML-6 elements included the rec sequence within their env gene. Then, we created a multiple alignment of the predicted Rec amino acid sequences, finding a full-length Rec putative domain within 23 HML-6 loci, while 16 loci showed an incomplete Rec domain due to the presence of several deletions (Fig. 6a). We hence built a consensus sequence from the multiple alignment (File S2 in the supplemental material), a NJ phylogenetic tree of 23 full-length HML-6 Rec amino acid sequences (Fig. 6b). We included the following as reference sequences: (i) seven HML-2 Rec amino acid sequences reported in UniProt; (ii) the recently described HML-10 Rec consensus amino acid sequence (19); (iii) the amino acid sequence of the functional homologue HIV-1 Rev; and (iv) the amino acid sequence of the functional homologue HTLV-1 Rex (see Materials and Methods for the corresponding accession numbers). Remarkably, the Rec NJ tree showed a high phylogenetic relationship between HML-6 and HML-2 Rec putative proteins. Second, to investigate their possible relevance, we also analyzed the integrity of the ORFs compared to the ERVK3-1 Rec amino acid sequence (Fig. 6a), observing that 4 of 23 ORFs have a predicted intact coding structure devoid of premature stop codons and frameshifts. Third, we focused on the four HML-6 Rec putative amino acid sequences with predicted intact ORFs, searching the functional domains involved into the nuclear localization signal (NLS) and nuclear export signal (NES) as described for the HML-2 and HML-10 Rec domains (17, 19). The results showed that the HML-6 Rec putative domain harbors a conserved NES domain, as reported for the HML-2 and HML-10 Rec, but not the NLS domains reported for the HML-2 Rec, as well as for HIV Rev and HTLV Rex (data not shown). In addition, we investigated the presence of a putative Rec responsive element (RcRe) by searching within the env sequence or within the LTR similarities with the reported HML-2 RcRe and the HIV RRE. This analysis did not allow us to find any putative HML-6 RcRe element; however, we cannot exclude the presence of a RcRe element characterized by different structures.
FIG 6.
Multiple alignment and phylogenetic relationships of HML-6 Rec domains. (a) Multiple alignment of the HML-6 Rec amino acid sequences with the protein ERVK3-1 used as reference. The colors in the sequences show disagreements in the alignments; black lines represent the deletion. ORFs are indicated by orange arrows, eventually stopping in correspondence of stop codons (black dots) or frameshift mutation (black arrows). (b) Relationship between the four best-preserved HML-6 Rec domains and the known HML-2 and HML-10 Rec domains is shown in a phylogenetic tree. This relationship was ascertained by using the NJ method and the Kimura two-parameter model with 1,000 bootstrap replicates.
Characterization and phylogenetic analysis of LTRs and time of insertion.
The HML-6 group has been associated with four different types of LTRs—LTR3, LTR3A, LTR3B, and LTR3B_v—according to the RepeatMasker annotations. We hence attempted to characterize the structure and phylogeny of LTR sequences, inspecting in particular whether the different LTR types were associated with specific proviral types.
First, we performed a nucleotide sequence comparison between the four LTR types, observing that LTR3A appeared to be a 3′-end extension variant of LTR3 (46 nt longer), while LTR3B_v seemed to be a 3′-end extension variant of LTR3B (62 nt longer) (Fig. S3). Second, to better identify the differences between LTRs, to have a comparison and to verify the RepeatMasker annotations, we built an NJ tree of all HML-6 proviral LTRs, which showed the presence of three clusters of sequences, one including LTR3A and LTR3, one including LTR3B, and one including LTR3B_v elements (Fig. 7). Hence, we sought to determine whether there was any association between the type of LTR and the different HML-6 types. Interestingly, the results showed that type 1a elements were associated with only LTR3 and LTR3A, type 1b sequences only occurred with LTR3B, and LTR3B_v was only related to type 2 members (Table 1 and Fig. 3). The presence of LTR3B associated with loci Xq27.1 and 17q25.1, which were assigned to type 2 and type 1b clusters by the NJ and ML analyses, respectively, allowed us to identify them as type 1b HML-6 loci. We characterized the LTRs identifying the most conserved structures in betaretroviral LTRs. The polyadenylation signal was clearly present at positions 303 to 308 as an AATAAA box, and we found a putative GT/CT area immediately after this motif. We did not found any evidences of TATA box structure (Fig. S3).
FIG 7.
Phylogenetic characterization of HML-6 LTRs. The evolutionary distance between the LTRs is shown in a phylogenetic tree that includes all HML-6 LTRs and the LTR Dfam consensus sequences (black dots). The tree shows three different clusters of sequences showing LTR3A and LTR3 (green line), LTR3B (light blue line), and LTR3B_v (orange line) elements.
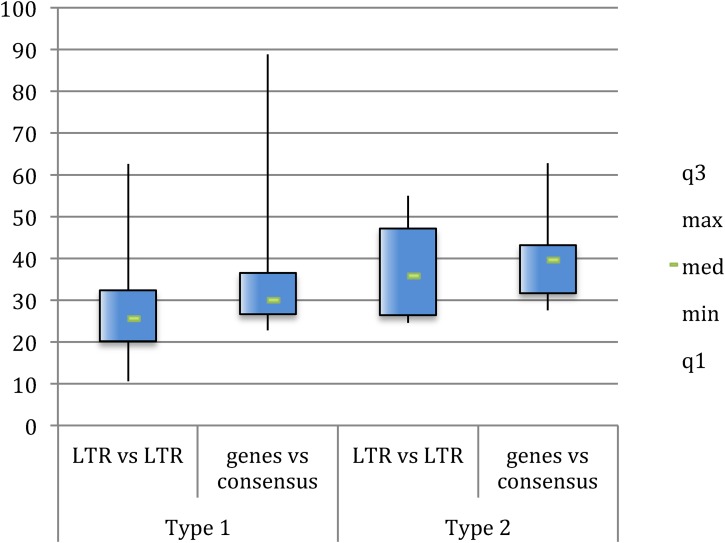
Next, we collected the solitary LTRs by using the LTR3A, LTR3, LTR3B, and LTR3B_v as consensus sequences for a BLAT search on human assembly hg19, identifying 385 mostly intact LTRs, whose coordinates are reported in File S1. It is well known that the 5′LTRs and 3′LTRs of the same provirus are identical at the time of integration (29) and that they independently accumulate random substitutions comparably to the internal proviral sequences and the host genome, allowing us to assess the provirus time of integration according to the nucleotide divergence between LTRs. However, due to deletions and rearrangements, in many instances only one (or none) provirus-associated LTR is available, impairing the estimation of the time of insertion. Hence, we recently implemented the calculation of time of insertion with the use of multiple divergence data between individual genic portions and their consensus (30). Considering a mutation rate in humans of 0,002/nucleotide/million years (31), we estimated the evolutionary age of each HML-6 sequence by calculating nucleotide divergences both between the 5′LTR and 3′LTR of each provirus and between nt 150 to 350 portions of the gag, pol, and env genes and a generated consensus for each subgroup (Fig. 8 and see Data Set S2 in the supplemental material).
FIG 8.
Time of integration of HML-6 elements. The time of integration was computationally evaluated by using divergences both between 5′LTRs and 3′LTRs and between 150- to 350-nt portions of the gag, pro, pol, and env genes and a generated consensus.
While the combination of both LTR-based and consensus-based divergence calculation was possible for only 16 elements (Data Set S2), the two methods in combination allowed us to estimate the time of integration of an overall 54 HML-6 proviruses (82%). The results showed that type 2 elements were probably acquired around 35 to 40 million years ago, while type 1 elements were probably acquired around 25 to 30 million years ago, possibly suggesting the existence of two waves of HML-6 viral insertions.
Genomic context of insertion.
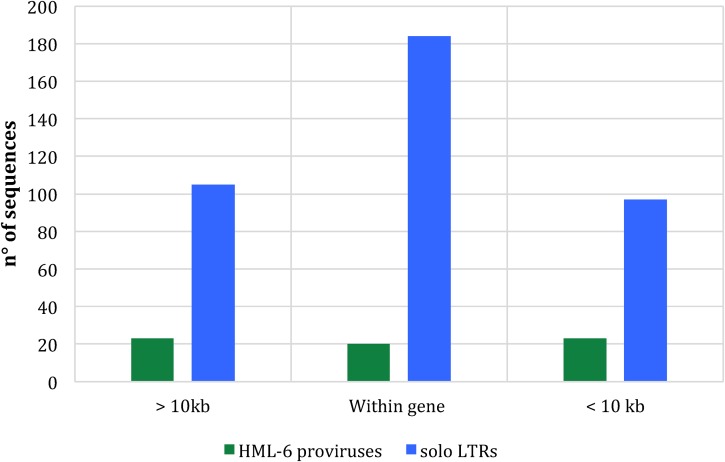
The impact of HERVs on the human genome largely depends on their context of integration, since proviral insertions in proximity or within human genes are able to influence their expression, both in sense and in antisense orientations, depending on (i) the regulatory activity of the LTRs; (ii) the possible insertion of retroviral splice donor and splice acceptor within the human genes; and (iii) the regulatory activity of antisense transcripts (32–34). For this reason, resulting from a negative selection pressure, HERVs were mainly inserted into intergenic regions, whereas the majority of intragenic insertions occurred in the antisense direction to gene transcription (32, 33). Therefore, we analyzed the context of integration of all 66 HML-6 elements, attempting to design a map of the elements that could be useful to understand their potential effects on human health through further investigations of the genes involved. We found only 19 sequences (representing about 30% of the HML-6 elements) included into intragenic regions: 11 elements were inserted within coding genes, mainly into introns (9/11), and 8 elements into processed or unprocessed pseudogenes (Table 2). Interestingly, 7 of the 11 host genes that include HML-6 within their sequences encode zinc finger proteins and may be involved in transcriptional regulation. In addition, the elements integrated into coding regions showed a prevalent antisense orientation with respect to the enclosing genes and seem to be mostly integrated into intronic portions. However, HML-6 loci 16p11.2 and 19q13.41c were inserted into the exons (FLJ90415 and ZNF528, respectively) and, in the case of 16p11.2, showed the same sense orientation of the surrounding gene. We hence focused the analysis on locus 16p11.2, finding that the 5′LTR of this sequence overlapped with the first exon of one of the processed transcripts of the ZNF689 gene (Gencode ID ENST00000566673.1), in agreement to the Ensembl and Gencode annotations (35, 36). Importantly, the ZNF689 gene has been reported to code for a zinc finger protein involved in suppressing the apoptosis of hepatocellular carcinoma cells and to be overexpressed in hepatocellular carcinoma (HCC) (37). We confirm that the sequence 6p21.32b, also known as HERVK3I, is located within the intron 1 of DRB2 and DRB6 pseudogenes, in the major histocompatibility complex (MHC) region, as already reported by Doxiadis et al. (33). In addition to that, we extended the analysis of the context of insertion to the HML-6 solitary LTRs we detected. Interestingly, we observed that a large number of LTRs were integrated close to or within human genes. Indeed, 284 solo LTRs were included within the sequence of the genes, and 97 solo LTRs were integrated within a 10-kb window of distance from the nearest-neighbor gene (Fig. 9).
TABLE 2.
HML-6 genomic context of insertion into human coding and noncoding genes
| Sequence | Type | Gene name | Gene type | Description |
|---|---|---|---|---|
| 1p21.1(+) | 1a | RP5-936J12.1(–) | Known lincRNA | |
| 1q25.2(–) | 1a | AXDND1(+) | Known protein coding | Axonemal dynein light chain domain containing protein 1 |
| 2q14.23(–) | 2 | MYO7B(+) | Known protein coding | Myosin VIIB |
| 4p14(–) | 1b | UGDH-AS1(+) | Known antisense | UGDH antisense RNA 1 |
| 4q21.1(–) | 1a | CCNG2(+) | Known protein coding | Cyclin G2 |
| 6p21.32b(–) | 1a | HLA-DRB6(–) | Known transcribed unprocessed pseudogene | Major histocompatibility complex, class II, DR β6 (pseudogene) |
| 11p15.4(–) | 1a | RP11-494M8.4(–) | Known lincRNA | |
| 11q12.3b(+) | 1a | RP11-703H8.9(–) | Known antisense | |
| 14q12(+) | 1a | CTD-2591A6.2(+) | Known lincRNA | |
| 16p11.2(–) | 1b | FLJ90415(–) | Known protein coding | Zinc finger protein 689 |
| 17q25.1(+) | 1b | CD300D(–); FLJ31882(+) | Known protein coding; known antisense | Immune receptor expressed on myeloid cells 1 |
| 19p13.2a(–) | 1a | CTC-543D15.3(+) | Known lincRNA | |
| 19p13.2b(–) | 1a | DKFZp571K0837(+) | Known protein coding | Zinc finger protein 439 |
| 19p12c(+) | 1a | ZNF676(–) | Known protein coding | Zinc finger protein 676 |
| 19q13.41a(–) | 1a | FPR3(+) | Known protein coding | Formyl peptide receptor 3 |
| 19q13.41b(+) | 1a | ZNF350(–) | Known protein coding | Zinc finger protein 350 |
| 19q13.41c(–) | 1a | ZNF528(+) | Known protein coding | Zinc finger protein 528 |
| 19q13.41d(–) | 1a | ZNF578 (+) | Known protein coding | Zinc finger protein 578 |
| 19q13.41e(–) | 1b | ZNF702P(–) | Known transcribed processed pseudogene | Zinc finger protein 702 |
| 19q13.43b(+) | 1b | ERVK3-1 | Known protein coding | Endogenous retrovirus group K3 member 1 |
FIG 9.
HML-6 provirus and solo LTR distance ranges from the nearest-neighbor gene. The histogram clearly shows a pattern of HML-6 element distribution close to the human genes.
PBS and betaretroviral structural features.
Medstrand et al. identified the PBS of the HML-6 sequences to be complementary to lysine tRNA and consequently included the subgroup in the HERVK clade (20). Given that such a classification was based on a limited number of HML-6 members at the time, we aimed to expand that analysis, including all 66 HML-6 sequences collected, to examine possible variations within the subtypes. We found that 38 HML-6 proviruses conserved the PBS regions, 20 of which maintained a well-preserved PBS sequence. As expected, all of these PBSs were predicted to recognize lysine tRNA (Fig. S4).
As further analysis, we searched other typical betaretrovirus markers, such as two zinc fingers in gag, as well as dUTPase and G-patch in pro (13, 20). The gag zinc fingers have a conserved composition (Cys-X2-Cys-X4-His-X4-Cys) and were found at positions 1483 to 1533 (zf-CCHC motif) and 1586 to 1678 (zf-CCHC_5 motif). We found at least one zinc finger motif in >51% of the HML-6 sequences and in >13% of the sequences present in both zf-CCHC and zf-CCHC_5 motifs. In particular, we found 19 loci with a zf-CCHC motif and 24 loci with a zf-CCHC_5 motif. The pro dUTPase and G-patch domains have also been detected: we observed a trimeric dUTPase conserved domain (positions 1768 to 1800) within 20 sequences and a G-patch domain (positions 2479 to 2604) within 12 sequences. Interestingly, we also found that the type 1 and type 2 HML-6 dUTPase sequences differed from each other in the first 80 amino acid residues (Fig. S4).
DISCUSSION
The HML-6 group, member of the class II betaretrovirus-like HERVs, includes several proviral loci with established transcriptional activity in physiological conditions and an increased transcriptional activity in some human cancers (20, 21, 23, 25). In particular, two HML-6 transcripts were already shown to contain intact ORFs: (i) ERVK3-1 is expressed in various healthy tissues (ENSG00000142396), and (ii) HERV-K-MEL encodes a small Env peptide in cutaneous and ocular melanoma cells but not in normal tissues (25). Nevertheless, due to the absence of a comprehensive description of the HML-6 group at the genomic level, the specific contribution of the individual HML-6 loci to human transcriptome, such as their role in human physiological and pathological conditions, is overall still unclear. In the present study, we analyzed in great detail the distribution, genetic composition, and phylogeny of all 66 HML-6 elements retrieved in human genome assembly hg19, providing a complete characterization of the HML-6 group. Overall, based on their chromosomal distribution, HML-6 proviruses showed a random integration pattern, with the only exception of sequences in chromosomes 19 and Y, with a higher number of integrations than expected. Such a pattern of distribution is in agreement with those observed in other HML groups, such as HML-5 and HML-10, and with HERVs in general (13, 19, 38).
In order to better characterize the group, we analyzed the sequences of the PBS; this has been used for the first classification of the group and was expected to be complementary to lysine tRNA, as reported for other HML members (13). Even if, in general, the value of the PBS as a phylogenetic marker is not totally consistent, given the occurrence of alternative PBS types for some HERV groups (30, 39), such an analysis corroborated the previous findings for the HML-6 elements (20), confirming that they harbor a type K PBS sequence.
The characterization of the HERVK (HML-6) consensus sequence confirmed a structure resembling the typical proviral genome, with the retroviral genes gag, pro, pol, and env flanked by 5′LTRs and 3′LTRs. It is worth noting that the structural analysis revealed that 23 HML-6 sequences present an Env Rec domain, whose presence has been reported here for the first time and has been confirmed through the phylogenetic analysis of Rec putative proteins. The Rec protein, a functional homologue of the retroviral regulatory proteins HIV Rev and HTLV Rex, was initially considered present in the sole HML-2 elements (16, 17), and HML2 Rec has been shown to interact with the promyelocytic leukemia zinc finger protein (PLZF), suggesting that Rec may contribute to germ cell tumor development (40, 41). Recently, the same domain has been also predicted within the sequence of five elements belonging to another HML subgroup, HML-10 (19). Similar to what was observed for HML-10, no evidence of NP9 protein domain was observed in HML-6, which is limited to the sole type 1 HML-2 group (18). Importantly, one of the identified Rec domain lies within the ERVK3-1 gene that has eight transcripts expressed in several tissues, including six transcripts predicted to be coding (28, 36). In addition, the overexpression of HML-2 Rec in a pluripotent cell line is sufficient to inhibit HIV infections (42). This information, together with the previously described HML-2 Rec implication in human pathology, suggests the need for further investigations of the role of this domain in HML-6 sequences.
Phylogenetic analysis of HML-6 internal sequences revealed the presence of two main clusters, which we named types 1 and 2, with type 1 showing an additional internal subdivision in two clusters: type 1a and type 1b. Phylogenetic analysis of the HML-6 Gag amino acid sequences allowed us to confirm the group division into type 1 and type 2 elements. We also confirmed that the HML-6 internal sequences are associated with four types of LTRs: LTR3, LTR3A, LTR3B, and LTR3B_v. These LTR types showed differences in sequence length and were grouped in three phylogenetic clusters, one including LTR3A and LTR3, one including LTR3B, and one including LTR3B_v elements. Indeed, analyses of solo LTRs is necessary for further structural characterizations and the creation of more representative consensus sequences. Interestingly, we observed that type 1a elements were associated only with LTR3 and LTR3A, and type 1b sequences were associated with LTR3B, whereas LTR3B_v was only related to type 2. The structural analysis showed the presence of a polyadenylation signal and a putative GT/CT-rich region in all the HML-6 LTR types. Target site duplications and T-rich regions are also present in retroviral classes and families of LTRs, and it has been proposed as a binding site for the cellular factor Sp1 (43, 44). Although the GT/CT-rich region is present in most HERVs, to the best of our knowledge, its functional role in HML-6 LTRs has not yet been elucidated. Interestingly, as also Benachenhou et al. (44) reported, the TATA box was absent, and it is possible to speculate on a role for the AATAAA motif as a TATA box. Finally, the structural and phylogenetic distances between type 1 and type 2 HML-6 elements seemed to reflect different times of insertion and may indicate two separate integration waves for the two types. These results also suggest that the integration of type 2 HML-6 occurred after the divergence between New World monkeys and Old World monkeys at the time of the Catarrhini primate speciation (about 40 millions years ago). In contrast, the integration of type 1 HML-6 elements seems to be specific for hominoid primates, as it was predicted to be occurred about 30 million years ago, after the divergence between Old World monkeys and hominoids. An open question is whether the two subsequent waves of integration for type 1 and type 2 elements can be linked to the fact that the acquisition of a retroviral element into the genome might be favored by the presence of a preexisting endogenous retrovirus through recombination events. Indeed, as recently reported in koalas, the presence of older HERVs facilitates the disruption, and thus the endogenization, of a coexisting exogenous species (45).
The analysis of the HML-6 genomic context of insertion and colocalization with functional genes and sequences putatively involved in disease showed that its pattern is comparable to the ones of other HERV elements (33), showing a higher HML-6 presence in intergenic regions, whereas the majority of sequences within intragenic regions resulted integrated in an antisense orientation. Interestingly, the HML-6 elements were often integrated within host genes coding for zinc finger proteins (7/11), that may be involved in transcriptional regulation. Indeed, we found that sequence 16p11.2 overlaps with a processed transcript of the zinc finger protein 689 gene, which is overexpressed in HCC (37). Anyway, colocalization with zinc fingers, which is particularly evident in chromosome 19, may be a consequence of the prevalence of zinc fingers in this chromosome.
Of particular interest was the finding of a large portion of solo LTRs close to human genes. Although we did not perform a complete phylogenetic and structural characterization of all solo HML-6 LTRs, the possible presence of polyadenylation signals and GT/CT-rich regions, observed in the proviral LTRs, may have an influence on the neighbor human gene expression, as already reported in other studies (46, 47).
In conclusion, the performed analysis gives complete and updated information on the HML-6 individual loci in the human genome GRCh37/hg19, essential to better understand the genetics of this group, including the possible contribution in physiological and pathological contexts, and its comprehensive transcriptional/translational analysis.
MATERIALS AND METHODS
The HML-6 sequences were collected from the human genome assembly GRCh37/hg19 both by employing RetroTector analysis on GRCh37/hg19 assembly and by retrieving chromosome coordinates in the UCSC Genome Browser database (48, 49) using assembled LTR3A-HERVK3-LTR3A consensus sequences from the Dfam database as a BLAT query (50). Elements obtained from both strategies were combined, and the identity of the HML-6 sequences was confirmed by multiple alignments with respect to the assembled HERVK3 consensus sequence. We estimated the expected distribution of HML-6 loci in each chromosome by using the formula: e = Cl*66/Tl, where e is the number of expected integration in the chromosome, Cl is the chromosome length, 66 is the total number of HML-6 loci in the human genome hg19, and Tl is the sum of all chromosome lengths.
Using the LTR3, LTR3A, LTRB, and LTR3B_v consensus sequences from Dfam as queries for a BLAT search, we collected the HML-6 solitary LTRs. The coordinates have been compared in order to exclude replicates. A consensus nucleotide alignment of the internal sequences has been created with MCoffee from the TCoffee package, v12.00.7fb08c2 (51). The integrity of each HML-6 element was analyzed compared to the assembled HERVK3 consensus sequence from the Dfam database (50). The genomic structure was further defined by using the RetroTector algorithm (52) in ReTe online. Additional multiple alignments were performed with MAFFT online, version 7 (53), for the inspection of LTRs composition with respect to the LTR3, LTR3A, LTRB, and LTR3B_v consensus sequences from Dfam. The obtained alignments were visualized using Geneious bioinformatics software, version 8.1.3 (54). The consensus sequences for type 1a, type 1b, and type 2 were generated from multiple alignments following the majority-based rule using Geneious bioinformatics software.
We selected the Kimura model (K80) as the more appropriate for analyze the HML-6 internal sequence evolution with JmodelTest, version 2.1.10 (55). Neighbor-joining phylogenetic trees were built with MEGA software (56), version 6.06, using pairwise deletion and the p-distance method with 1,000 bootstrap replications. Maximum-likelihood trees were built with PhyML 3.0 online (http://www.atgc-montpellier.fr/phyml/), selecting the K80 model and 100 bootstrap replication (57). The Gag amino acid sequences of the other HML consensus and of the exogenous retroviruses MPMV (P07567), MMTV, and JSRV (P31622) were included in the analysis as controls, as well as ZAM (O46144), used as an outgroup. Phylogenetic trees of Rec sequences were built with MEGA software (56), version 6.06, using pairwise deletion and the Poisson method. The HML-10 (19), HML-2 Rec (P61573, P61572, P61573, P61575, P61576, P61571, and P61578), HTLV-1 Rex (Q85601), and HIV-1 Rev (P69718) amino acid sequences were included in the analysis.
Estimation of time of integration.
Considering the HML-6 coevolution with the host genome and assuming a human genome substitution rate of 0.2% per nucleotide per million years, the time of integration of the HML-6 sequences (T) was estimated using the formula T = D/0.2, calculating the percentage of divergent nucleotides (D) between 150- and 350-nt portions of gag, pol, and env genes and a generated consensus for each type and subtype. The consensus sequences used in these analyses were generated with Geneious software from visually inspected multiple alignments and following the majority rule. The time of integration based on the 5′LTR versus the 3′LTR divergence was also evaluated, considering that each LTR of the same sequence accumulates mutations independently according to the formula T = D/0.2/2. Divergence values were estimated using MEGA 6.06 (56) using pairwise deletion and the Kimura two-parameter model and excluding CpG dinucleotides from the alignments. The final age of the sequences was expressed, when possible, as the average value obtained from all methods, excluding those with a standard deviation >25%.
PBS and betaretroviral structural features.
The PBS nucleotide sequences were analyzed and characterized through MAFFT multiple alignments compared to the PBS reference sequences kindly provided by J. Blomberg. Nucleocapsid zinc fingers, Pro dUTPases, and Pol G-patch amino acid motifs were aligned using the MUSCLE algorithm in MEGA (56). All analyses were visualized on the Geneious platform. The composition of PBS and structural features was represented using WebLogo (http://weblogo.berkeley.edu).
Genomic context.
The genomic context of the HML-6 elements was retrieved by analyzing their genomic coordinates on the Data Integrator tool in the UCSC Genome Browser (48, 49), selecting the “genes and genes prediction” track. Moreover, all the sequences were visualized using Genome Browser concurrently with the activation of GENECODE v24, RefSeq genes, ENCODE, and Gtex annotations. The distances between HERV proviruses/solitary LTRs and human genes have been computed by using the function “distance” from the package GenomicRanges, version 1.30.3, in RStudio (R, v3.4.4). Human gene coordinates were collected from GENCODE v24.
Search for conserved domains.
The conserved domains present in the sequences were identified by using NCBI conserved domain search software (58).
Supplementary Material
ACKNOWLEDGMENT
We thank Giacomo Diaz for valuable critical revisions of the statistics in the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00110-19.
REFERENCES
- 1.Hattori M. 2005. Finishing the euchromatic sequence of the human genome. Tanpakushitsu Kakusan Koso 50:162–168. [PubMed] [Google Scholar]
- 2.Jern P, Coffin JM. 2008. Effects of retroviruses on host genome function. Annu Rev Genet 42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 3.Bannert N, Kurth R. 2006. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet 7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- 4.Voisset C, Weiss RA, Griffiths DJ. 2008. Human RNA “rumor” viruses: the search for novel human retroviruses in chronic disease. Microbiol Mol Biol Rev 72:157–196. doi: 10.1128/MMBR.00033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grandi N, Tramontano E. 2018. HERV envelope proteins: physiological role and pathogenic potential in cancer and autoimmunity. Front Microbiol 9:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grandi N, Tramontano E. 2018. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front Immunol 9:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheynet RIE, Bouton O, Blond J, Lavillette D. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol 4:3321–3329. doi: 10.1128/JVI.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, Lavallie E, Tang X, Edouard P, Howes S, Keith JC Jr, McCoy JM. 2000. Syncytin is a captive retroviral envelope protein involved. Nature 403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 9.Grandi N. 2017. Integrations and their mobilization by L1 machinery: contribution to the human transcriptome and impact on the host physiopathology. Viruses 9:1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blomberg J, Benachenhou F, Blikstad V, Sperber G, Mayer J. 2009. Classification and nomenclature of endogenous retroviral sequences (ERVs): problems and recommendations. Gene 448:115–123. doi: 10.1016/j.gene.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Kassiotis G. 2014. Endogenous retroviruses and the development of cancer. J Immunol 192:1343–1349. doi: 10.4049/jimmunol.1302972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blikstad V, Benachenhou F, Sperber GO, Blomberg J. 2008. Endogenous retroviruses—evolution of human endogenous retroviral sequences: a conceptual account. Cell Mol Life Sci 65:3348–3365. doi: 10.1007/s00018-008-8495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vargiu L, Rodriguez-Tomé P, Sperber GO, Cadeddu M, Grandi N, Blikstad V, Tramontano E, Blomberg J. 2016. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 13:2–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. 2011. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mertz JA, Simper MS, Lozano MM, Payne SM, Dudley JP. 2005. Mouse mammary tumor virus encodes a self-regulatory RNA export protein and is a complex retrovirus. J Virol 79:14737–14747. doi: 10.1128/JVI.79.23.14737-14747.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magin C, Löwer R, Löwer J. 1999. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J Virol 73:9496–9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer J, Ehlhardt S, Seifert M, Sauter M, Müller-Lantzsch N, Mehraein Y, Zang KD, Meese E. 2004. Human endogenous retrovirus HERV-K(HML-2) proviruses with Rec protein coding capacity and transcriptional activity. Virology 322:190–198. doi: 10.1016/j.virol.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 18.Armbruester V, Sauter M, Roemer K, Best B, Hahn S, Nty A, Schmid A, Philipp S, Mueller A, Mueller-Lantzsch N. 2004. Np9 protein of human endogenous retrovirus K interacts with ligand of numb protein X Np9 protein of human endogenous retrovirus K interacts with ligand of numb protein X. J Virol 78:10310–10319. doi: 10.1128/JVI.78.19.10310-10319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grandi N, Cadeddu M, Pisano MP, Esposito F, Blomberg J, Tramontano E. 2017. Identification of a novel HERV-K(HML10): comprehensive characterization and comparative analysis in non-human primates provide insights about HML10 proviruses structure and diffusion. Mob DNA 8:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medstrand P, Mager DL, Yin H, Dietrich U, Blomberg J. 1997. Structure and genomic organization of a novel human endogenous retrovirus family: HERV-K (HML-6). J Gen Virol 78:1731–1744. doi: 10.1099/0022-1317-78-7-1731. [DOI] [PubMed] [Google Scholar]
- 21.Yin H, Medstrand P, Kristofferson A, Dietrich U, Åman P, Blomberg J. 1999. Characterization of human MMTV-like (HML) elements similar to a sequence that was highly expressed in a human breast cancer: further definition of the HML-6 group. Virology 256:22–35. doi: 10.1006/viro.1998.9587. [DOI] [PubMed] [Google Scholar]
- 22.Mayer J, Meese EU. 2003. Presence of dUTPase in the various human endogenous retrovirus K (HERV-K) families. J Mol Evol 57:642–649. doi: 10.1007/s00239-003-2514-6. [DOI] [PubMed] [Google Scholar]
- 23.Seifarth W, Frank O, Zeilfelder U, Spiess B, Greenwood AD, Hehlmann R, Leib-Mösch C. 2005. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J Virol 79:341–352. doi: 10.1128/JVI.79.1.341-352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank O, Verbeke C, Schwarz N, Mayer J, Fabarius A, Hehlmann R, Leib-Mösch C, Seifarth W. 2008. Variable transcriptional activity of endogenous retroviruses in human breast cancer. J Virol 82:1808–1818. doi: 10.1128/JVI.02115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiavetti F, Thonnard J, Colau D, Boon T, Coulie PG. 2002. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res 62:5510–5516. [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 27.Tristem M. 2000. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol 74:3715–3730. doi: 10.1128/jvi.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medstrand P, Blomberg J. 1993. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J Virol 67:6778–6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lebedev YB, Belonovitch OS, Zybrova NV, Khil PP, Kurdyukov SG, Vinogradova TV, Hunsmann G, Sverdlov ED. 2000. Differences in HERV-K LTR insertions in orthologous loci of humans and great apes. Gene 247:265–277. doi: 10.1016/S0378-1119(00)00062-7. [DOI] [PubMed] [Google Scholar]
- 30.Grandi N, Cadeddu M, Blomberg J, Tramontano E. 2016. Contribution of type W human endogenous retrovirus to the human genome: characterization of HERV-W proviral insertions and processed pseudogenes. Retrovirology 13:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson WE, Coffin JM. 1999. Constructing primate phylogenies from ancient retrovirus sequences. Proc Natl Acad Sci U S A 96:10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van de Lagemaat LN, Medstrand P, Mager DL. 2006. Multiple effects govern endogenous retrovirus survival patterns in human gene introns. Genome Biol 7:R86. doi: 10.1186/gb-2006-7-9-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doxiadis GGM, de Groot N, Bontrop RE. 2008. Impact of endogenous intronic retroviruses on major histocompatibility complex class II diversity and stability. J Virol 82:6667–6677. doi: 10.1128/JVI.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mack M, Bender K, Schneider PM. 2004. Detection of retroviral antisense transcripts and promoter activity of the HERV-K (C4) insertion in the MHC class III region. Immunogenetics 56:321–332. doi: 10.1007/s00251-004-0705-y. [DOI] [PubMed] [Google Scholar]
- 35.Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Fernandez Banet J, Billis K, García Girón C, Hourlier T, Howe K, Kähäri A, Kokocinski F, Martin FJ, Murphy DN, Nag R, Ruffier M, Schuster M, Tang YA, Vogel JH, White S, Zadissa A, Flicek P, Searle SMJ. 2016. The Ensembl gene annotation system. Database (Oxford) 2016:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigó R, Hubbard TJ. 2012. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res 22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shigematsu S, Fukuda S, Nakayama H, Inoue H, Hiasa Y, Onji M, Higashiyama S. 2011. ZNF689 suppresses apoptosis of hepatocellular carcinoma cells through the downregulation of Bcl-2 family members. Exp Cell Res 317:1851–1859. doi: 10.1016/j.yexcr.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 38.Lavie L, Medstrand P, Schempp W, Meese E, Mayer J. 2004. Human endogenous retrovirus family HERV-K (HML-5): status, evolution, and reconstruction of an ancient betaretrovirus in the human genome. J Virol 78:8788–8798. doi: 10.1128/JVI.78.16.8788-8798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jern P, Sperber GO, Blomberg J. 2005. Use of endogenous retroviral sequences (ERVs) and structural markers for retroviral phylogenetic inference and taxonomy. Retrovirology 2:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galli UM, Sauter M, Lecher B, Maurer S, Herbst H, Roemer K, Mueller-Lantzsch N. 2005. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene 24:3223–3228. doi: 10.1038/sj.onc.1208543. [DOI] [PubMed] [Google Scholar]
- 41.Boese A, Sauter M, Galli U, Best B, Herbst H, Mayer J, Kremmer E, Roemer K, Mueller-Lantzsch N. 2000. Human endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene 19:4328–4336. doi: 10.1038/sj.onc.1203794. [DOI] [PubMed] [Google Scholar]
- 42.Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, Pera RAR, Wysocka J. 2015. HHS public access. Nature 522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sjøttem EVA, Anderssen S, Johansen T. 1996. The promoter activity of long terminal repeats of the HERV-H family of human retrovirus-like elements is critically dependent on Sp1 family proteins interacting with a GC/GT box located immediately 3 J to the TATA box. J Virol 70:188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benachenhou F, Jern P, Oja M, Sperber G, Blikstad V, Somervuo P, Kaski S, Blomberg J. 2009. Evolutionary conservation of orthoretroviral long terminal repeats (LTRs) and ab initio detection of single LTRs in genomic data. PLoS One 4:e5179. doi: 10.1371/journal.pone.0005179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waugh CA, Hanger J, Loader J, King A, Hobbs M, Johnson R, Timms P. 2017. Infection with koala retrovirus subgroup B (KoRV-B), but not KoRV-A, is associated with chlamydial disease in free-ranging koalas (Phascolarctos cinereus). Sci Rep 7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST, Mohan GS, Li W, Ye L, Compans RW, Yang C, Bowley DR, Labrijn AF, Zwick MB, Burton DR, Weissenhorn W, Lee KH, Skehel JJ, Wiley DC, Bray M, Davis K, Geisbert T, Schmaljohn C, Huggins J, Shen Y, Maupetit J, Derreumaux P, Sircar A, Chaudhury S, Gray JJ, Williams C, Diseases I. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351:1083–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pi W, Zhu X, Wu M, Wang Y, Fulzele S, Eroglu A, Ling J, Tuan D. 2010. Long-range function of an intergenic retrotransposon. Proc Natl Acad Sci U S A 107:12992–12997. doi: 10.1073/pnas.1004139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hinrichs AS, Learned K, Lee BT, Li CH, Raney BJ, Rhead B, Rosenbloom KR, Sloan CA, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ. 2014. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res 42:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kent WJ, Sugnet CW, Furey TS, Roskin KM. 1976. The Human Genome Browser at UCSCW. J Med Chem 19:1228–1231.186604 [Google Scholar]
- 50.Hubley R, Finn RD, Clements J, Eddy SR, Jones TA, Bao W, Smit AFA, Wheeler TJ. 2016. The Dfam database of repetitive DNA families. Nucleic Acids Res 44:D81–D89. doi: 10.1093/nar/gkv1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wallace IM, Sullivan OO, Higgins DG, Notredame C. 2006. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res 34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sperber GO, Airola T, Jern P, Blomberg J. 2007. Automated recognition of retroviral sequences in genomic data: RetroTector©. Nucleic Acids Res 35:4964–4976. doi: 10.1093/nar/gkm515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Darriba D, Taboada GL, Doallo R, Posada D. 2015. Europe PMC funders group jModelTest 2: more models, new heuristics, and high-performance computing. Nat Methods 9:6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 58.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.