Zika virus (ZIKV) is an emerging vector-borne pathogen causing a febrile disease. ZIKV infection might also trigger Guillain-Barré syndrome, neuropathy, and myelitis. Vertical transmission of ZIKV can cause fetus demise, stillbirth, or severe congenital abnormalities and neurological complications. There is no vaccine or specific antiviral treatment against ZIKV. We used a genome-wide CRISPR activation screen, where genes are activated from their native promoters to identify host cell factors that protect cells from ZIKV infection or associated cell death. The results provide a better understanding of key host factors that protect cells from ZIKV infection and might assist in identifying novel antiviral targets.
KEYWORDS: CRISPR activation, genome-wide screen, interferon-stimulated genes, interferons, Zika virus
ABSTRACT
Zika virus (ZIKV) is an arthropod-borne emerging pathogen causing febrile illness. ZIKV is associated Guillain-Barré syndrome and other neurological complications. Infection during pregnancy is associated with pregnancy complications and developmental and neurological abnormalities collectively defined as congenital Zika syndrome. There is still no vaccine or specific treatment for ZIKV infection. To identify host factors that can rescue cells from ZIKV infection, we used a genome-scale CRISPR activation screen. Our highly ranking hits included a short list of interferon-stimulated genes (ISGs) previously reported to have antiviral activity. Validation of the screen results highlighted interferon lambda 2 (IFN-λ2) and interferon alpha-inducible protein 6 (IFI6) as genes providing high levels of protection from ZIKV. Activation of these genes had an effect on an early stage in viral infection. In addition, infected cells expressing single guide RNAs (sgRNAs) for both of these genes displayed lower levels of cell death than did the controls. Furthermore, the identified genes were significantly induced in ZIKV-infected placenta explants. Thus, these results highlight a set of ISGs directly relevant for rescuing cells from ZIKV infection or its associated cell death and substantiate CRISPR activation screens as a tool to identify host factors impeding pathogen infection.
IMPORTANCE Zika virus (ZIKV) is an emerging vector-borne pathogen causing a febrile disease. ZIKV infection might also trigger Guillain-Barré syndrome, neuropathy, and myelitis. Vertical transmission of ZIKV can cause fetus demise, stillbirth, or severe congenital abnormalities and neurological complications. There is no vaccine or specific antiviral treatment against ZIKV. We used a genome-wide CRISPR activation screen, where genes are activated from their native promoters to identify host cell factors that protect cells from ZIKV infection or associated cell death. The results provide a better understanding of key host factors that protect cells from ZIKV infection and might assist in identifying novel antiviral targets.
INTRODUCTION
Zika virus (ZIKV) is a positive-strand RNA virus belonging to the Flavivirus genus of the Flaviviridae family that also includes West Nile virus (WNV), Japanese encephalitis virus, yellow fever virus, and dengue virus (DENV). ZIKV is transmitted by Aedes mosquitoes and typically causes a mild self-resolving disease. ZIKV has gained international attention following its spread to the Western Hemisphere causing the 2015-2016 epidemic in Brazil. The main reason for this attention was the association between ZIKV infection, microcephaly, and Guillain-Barré syndrome (1). During 2015 to 2016, 1.5 million cases of ZIKV infection and 1,950 cases of confirmed infection-related microcephaly were reported in Brazil (2). Currently, there is still no approved vaccine or specific treatment for ZIKV infection. Identification of host factors capable of restricting ZIKV infection might assist in designing anti-ZIKV therapies.
Flaviviruses enter the host cells via receptor-mediated endocytosis. The positive-strand RNA genome is released from the endosome and is immediately translated in the endoplasmic reticulum (ER). The translated polyprotein is then cleaved by viral and host proteases to form the active viral proteins. These include 3 structural proteins (capsid, membrane, and envelope) and 7 nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The viral proteins localize to the ER, where they form a viral replication complex built on modified ER membranes. The virus replicates from a double-stranded RNA (dsRNA) intermediate transcribed by the viral RNA-dependent RNA polymerase. Virus assembly and budding occur at the ER, followed by transport of the virion to the Golgi, where it matures and is exocytosed (3).
ZIKV, like other RNA viruses, strongly depends on host factors due to the limited number of proteins it expresses. Several important studies identified key genes essential for ZIKV infection using loss-of-function genome-scale screens applying pooled clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 and/or RNA interference (RNAi) approaches (4–7). However, complementary screens aiming to identify genes that when overexpressed rescue cells from recurrent ZIKV infection were not reported. To identify such genes, we employed a CRISPR activation approach. This method uses single guide RNAs (sgRNAs) directed to various regions in endogenous promoters to recruit a catalytically inactive Cas9 (dCas9) fused to a transcription activator (8–14). Recruitment of the activator to these locations specifically activates the gene controlled by the targeted promoter. CRISPR activation libraries were developed by different groups, and these differ mainly in the number, location, and type of activators used (8–14). Although generally comparable and dependent on the specific conditions used, a synergistic activation mediator (SAM) seemed to consistently deliver high levels of gene induction compared to some of the other systems (15). Activation of genes from their native promoter has several advantages over traditional overexpression, including the activation of splicing variants and large transcripts. Here, CRISPR SAM was used to identify genes that when overexpressed rescue cells from ZIKV infection or ZIKV-induced cell death. Nine out of the 10 top-ranked genes were interferon-stimulated genes (ISGs) previously reported to have antiviral capabilities (16–20), confirming the validity of the screen. Of the validated top hits, interferon lambda 2 (IFN-λ2) and interferon alpha-inducible protein 6 (IFI6) showed the highest levels of protection from ZIKV infection.
Our results provide a list of ZIKV-specific ISGs, which adds basic information on how the host copes with ZIKV infection and might assist in identifying new antiviral targets.
RESULTS
CRISPR activation identifies host factors that rescue cells from ZIKV infection.
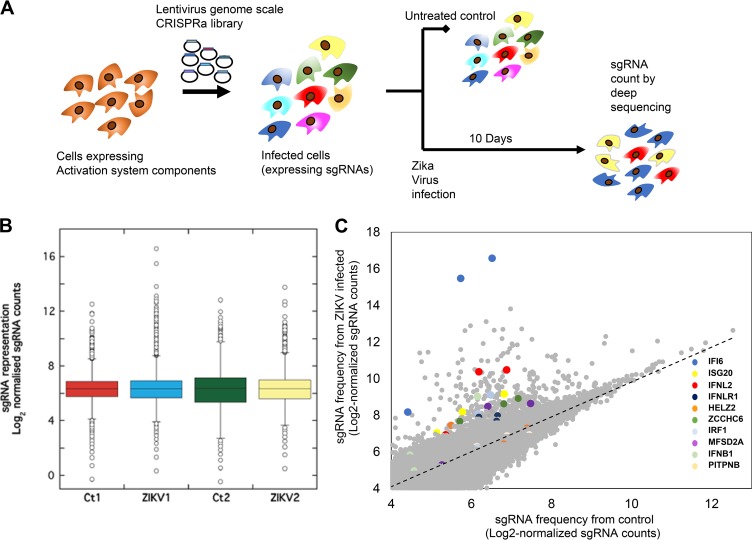
CRISPR activation was employed to identify genes rescuing cells from multiple cycles of ZIKV infection. Lentiviruses prepared from vectors carrying a pooled genome-scale sgRNA library were used to infect Huh7 hepatoma cells stably expressing the dCas9 and activation domains (12, 21). Huh7 cells were chosen due their high susceptibility and pronounced cytopathic effect following ZIKV infection (22). While the liver is not a major organ in ZIKV pathogenesis, high levels of ZIKV RNA were detected in the liver of mouse models (23, 24). Moreover, a limited number of studies reported jaundice and/or hepatic dysfunction in patients with ZIKV infection as well (25, 26). The Huh7 cells were infected with ZIKV MR776 at a low multiplicity of infection (MOI) (0.2). The surviving cells were collected 10 days postinfection. Genomic DNA prepared from these cells was used to amplify the sgRNA for next-generation sequencing (NGS) (Fig. 1A). We found that at 10 days postinfection, the sgRNA distributions were different between the infected cells and controls, with a subset of guides showing enrichment in infected cells (Fig. 1B). The pooled library used contained 3 sgRNAs for each gene in the RefSeq database (23,430 isoforms) (12). For most enriched genes, more than one sgRNA targeting the same gene was enriched in the infected cells (Fig. 1C). Two ranking algorithms were used to analyze the obtained results, “RNAi gene enrichment ranking” (RIGER) (27) and “model-based analysis of genome-wide CRISPR-Cas9 knockout” (MAGeCK) (28). The tools use different statistical models for the ranking of genes. While RIGER relies on a Kolmogorov-Smirnov-based statistic for ranking, MAGeCK uses a negative binomial model in combination with a robust ranking aggregation algorithm. We applied both methods to test our data for robustness against the statistical model used. The obtained top-ranking genes are shown in Table 1 . The order of the genes in the table is relative to the gene ranking results of MAGeCK applied to the first screen. Our six highest ranking genes are robust across the two screens performed and against the statistical method used for ranking. Moreover, as mentioned above, most of these genes are ISGs with known antiviral activity, further confirming these results (16–20).
FIG 1.
ZIKV CRISPR activation screen. (A) A scheme summarizing the screen conditions. (B) Box plots showing the distribution of sgRNA frequencies after control or ZIKV infection from n = 2 infection replicates. Multiple guides are seen to be enriched following infection, revealing guides that promote resistance to ZIKV. Boxes, 25th to 75th percentiles; whiskers, 1st to 99th percentiles. (C) Scatter plot showing enrichment of sgRNAs targeting the top candidate genes identified by RIGER and MAGeCK (colored dots) compared with other sgRNAs in the library (gray dots) after ZIKV infection.
TABLE 1.
Analysis of the CRISPR screena
| Gene IDb | Gene rank by algorithm |
|||
|---|---|---|---|---|
| MAGeCK1 | MAGeCK2 | RIGER 1 | RIGER 2 | |
| IFI6 | 1 | 1 | 1 | 1 |
| ISG20 | 2 | 2 | 2 | 26 |
| ZCCHC6 | 3 | 236 | 3 | 85 |
| IFN-λ2 | 4 | 3 | 9 | 64 |
| HELZ2 | 5 | 6 | 3 | 85 |
| IFN-λR1 | 6 | 4 | 6 | 40 |
| EGFR | 7 | 18,134 | 10,142 | 17,934 |
| IRF1 | 8 | 7 | 140 | 76 |
| MAVS | 9 | 127 | 847 | 19,704 |
| TRIM25 | 10 | 89 | 228 | 788 |
| MFSD2A | 14 | 3,966 | 149 | 10,188 |
| PITPNB | 24 | 927 | 647 | 16,783 |
A comparison of the 2 screen repeats and two different analysis algorithms used RIGER and MAGeCK. Values represent gene ranks assigned by the corresponding algorithm in the different repeats. Highest-ranking genes are sorted relative to the results of MAGeCK applied on the first screen. The gene ranks of the first six candidates are robust across the replicates and the algorithm used.
ID, identifier.
Functional validation of the CRISPR activation screen results.
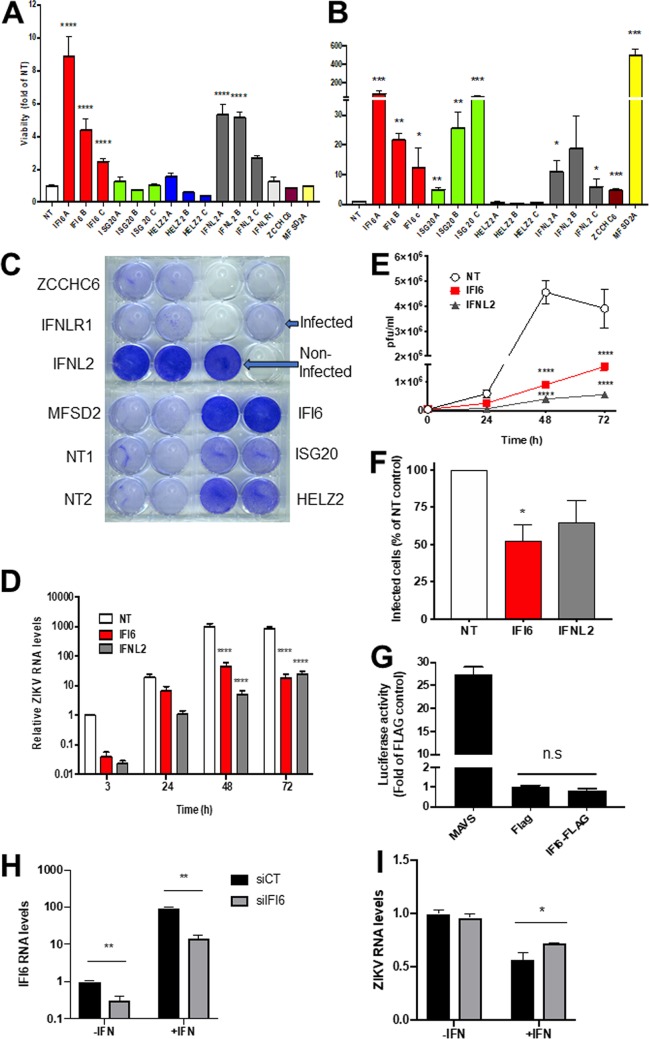
To validate the screen results, we recloned the top-ranking individual sgRNAs into the library backbone. Lentiviruses carrying these sgRNAs were prepared and used to create stable Huh7 cell lines. Cells expressing nontargeting sgRNAs served as controls. The obtained cell lines were infected with ZIKV, and viability was determined 7 days postinfection (Fig. 2A and C). Initially, we cloned the top-ranking sgRNA for each gene. Additional sgRNAs were cloned for genes showing a protective effect. Crystal violet staining of the infected wells shows significantly less cell death in the cells expressing interferon alpha-inducible protein 6 (IFI6) and interferon lambda 2 (IFN-λ2) (Fig. 2A and C). A smaller protective effect was observed for interferon-stimulated exonuclease gene 20 (ISG20) and helicase with zinc finger 2 (HELZ2). Quantification of the crystal violet staining indicated that for IFI6 and IFN-λ2, most sgRNAs targeting the same gene showed significant though varied levels of protection (Fig. 2A). The activation of the targeted genes by the sgRNAs was confirmed using real-time PCR (Fig. 2B). Overall, most of the targeted genes were significantly activated at levels ranging from 3- to >400-fold. Significant activation was observed for all genes showing a protective effect, except HELZ2. For IFI6, there was a clear correlation between the activation and protection levels. Such a correlation was not observed for the rest of the genes. Taken together, these results confirm a protective role from ZIKV infection by the activation of IFI6, IFN-λ2, and, to a lesser extent, ISG20. To further confirm the roles of IFI6 and IFN-λ2 in protection from ZIKV infection, we tested ZIKV RNA levels in infected CRISPR-activated cell lines expressing nontargeting (NT), IFI6, or IFN-λ2 sgRNAs. Activation of both genes significantly inhibited the accumulation of ZIKV RNA (Fig. 2D). The ZIKV RNA levels were >20-fold lower in cells expressing IFI6 sgRNAs than in cells expressing NT sgRNAs after 48 h and 47-fold lower at 72 h postinfection. In cells expressing IFN-λ2 sgRNAs, ZIKV RNA levels were 195-fold lower than in cells expressing NT sgRNAs after 48 h, and 34-fold lower at 72 h postinfection. Viral production from these cells was significantly inhibited in the IFI6- and IFN-λ2-activated cells as well (Fig. 2E). Virus production was reduced more than 5-fold in IFI6-activated cells and 11-fold in IFN-λ2-activated cells after 48 h. Although the effect was still significant, the reduction in virus production at 72 h postinfection diminished to 2.5-fold and 7-fold in IFI6- and IFN-λ2-activated cells, respectively. The use of flow cytometry analysis of ZIKV-infected Huh7 cells stably expressing NT, IFI6, and IFN-λ2 sgRNAs indicated that the activation of IFI6 inhibited infection by 48%, while IFN-λ2 inhibited infection by 35% (Fig. 2F). IFI6 activation itself conferred high levels of protection from ZIKV infection, comparable to that of IFN-λ2, which activates other ISGs. This fact led us to test if IFI6 is capable of activating the transcription of other ISGs using an ISG56 promoter reporter cell line (29). Mitochondrial antiviral-signaling (MAVS) protein was used as a positive control. IFI6 did not activate ISG56 promoter-dependent transcription (Fig. 2G), indicating that its antiviral activity is a direct effect. To determine the role of IFI6 in controlling ZIKV infection, we performed a knockdown experiment using small interfering RNAs (siRNAs) (Fig. 2H and I). In the presence of interferon alpha (IFN-α), IFI6 levels were induced by approximately 94-fold compared to nontreated control cells. IFI6 siRNA reduced this elevation by 4.3-fold (Fig. 2H; **, P < 0.01). A smaller but significant 0.7-fold reduction was observed in controls in the absence of IFN (**, P < 0.01). These results confirm the efficiency of these siRNAs.
FIG 2.
Functional validation of the ZIKV CRISPR activation screen. (A) Huh7 cell lines expressing the indicated sgRNAs were infected with ZIKV (MOI, 0.2) for 2 h. After 7 days, the cells were stained with crystal violet. The graph shows the quantification of the crystal violet staining by measurement of the absorbance at 570 nm. Bar graphs show mean values ± standard deviation (SD) from two independent experiments performed in duplicate. ****, P < 0.001, one-way analysis of variance (ANOVA), Dunnett’s multiple-comparison test. (B) Expression levels of the activated genes as determined by real-time PCR using gene-specific primers. Shown are mean values ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001, multiple two-tailed unpaired t tests. (C) Representative crystal violet staining results. (D) Time course of relative ZIKV RNA levels as determined by real-time PCR following infection (MOI, 1) of nontargeting (NT), IFI6, and IFN-λ2 CRISPR activated cell lines. ****, P < 0.001, two-way ANOVA with Tukey’s multiple-comparison test. (E) Time course of ZIKV production NT, IFI6, and IFN-λ2 CRISPR activated cell lines as determined using plaque assays. ****, P < 0.001, two-way ANOVA with Tukey’s multiple-comparison test. (F) Flow cytometry of infected Huh7 cells expressing NT or IFI6 sgRNAs at 48 h postinfection. The results are the mean values ± SD. *, P < 0.05, two-tailed unpaired t test. (G) ISG56-luciferase activity in cells transfected with MAVS, empty FLAG plasmid, or an IFI6-FLAG expressing plasmid. The results are the mean values ± SD. n.s., nonsignificant, two-tailed unpaired t test. (H) IFI6 mRNA levels in cells transfected with IFI6 or nontargeting control siRNAs in the presence or absence of IFN-α (1,000 U/ml, 18 h), **, P < 0.01, two-tailed unpaired t test. (I) IFI6 knockout slightly rescues ZIKV infection in IFN-α-treated cells. Huh7 cells were transected with the indicated siRNAs. The next day, the indicated samples were pretreated with IFN-α (for 2 h) and infected with ZIKV (MOI, 1). The cells were harvested for qPCR analysis at 24 h postinfection. *, P < 0.05, two-tailed unpaired t test.
IFN-α suppressed ZIKV RNA levels by 50%. ZIKV RNA levels were slightly but significantly (*, P < 0.05) elevated as a result of IFI6 downregulation compared to cells transfected with nontargeting siRNA cells (Fig. 2I). In the absence of IFN-α, ZIKV RNA levels were not significantly affected. These results indicate that IFI6 has a role in controlling ZIKV replication.
Subcellular localization of IFI6.
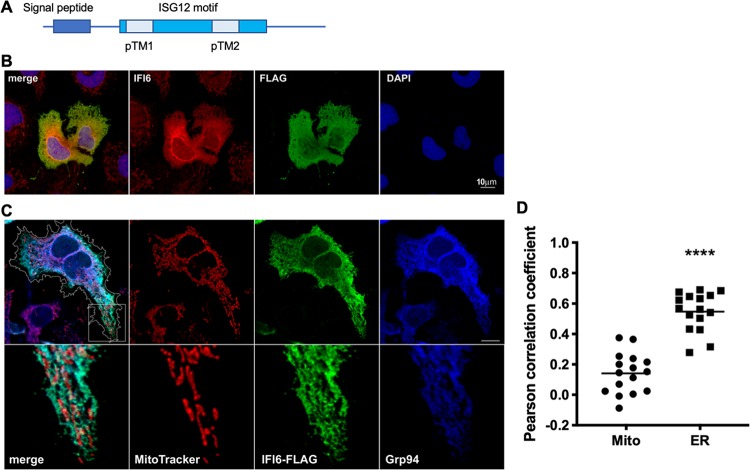
IFI6 is a member of the ISG12 family containing four related ISG12 genes, IFI6 and ISG12a (IFI27), ISG12b(IFI27L2), and ISG12c(IFI27L1) (30). The IFI6 gene encodes a small 130-amino-acid (aa)-long protein containing a predicted N-terminal signal peptide and two predicted transmembrane domains (Fig. 3A). IFI6 was previously reported to localize to the mitochondria and to inhibit apoptosis in DENV-infected cells (31, 32). To confirm the localization of IFI6, we constructed a C-terminal FLAG fusion protein. First, we transfected our IFI6-FLAG construct into Huh7 cells and stained the cells both with FLAG (red) and commercial IFI6-specific (green) antibodies (Fig. 3B). While a significant amount of the two signals seems to colocalize, IFI6 background staining could be observed in nonexpressing cells. Thus, we continued our analysis with the IFI6-FLAG (green) construct, which was further stained with both GRP94 (blue), a known ER marker (33), and MitoTracker (red, Fig. 3C and D). In our hands, IFI6 showed significant colocalization with GRP94 (Pearson correlation coefficient, 0.55 ± 0.03 versus 0.14 ± 0.03 for the mitochondria; n = 16), indicating that the majority of IFI6 localizes to the ER (Fig. 3D). This localization suggests that IFI6 might inhibit a stage in the viral life cycle that occurs at the ER.
FIG 3.
Immunofluorescence microscopy of cells expressing IFI6-FLAG. (A) A schematic representation of IFI6 structural domain organization. (B) Huh7 cells were transfected with IFI6-FLAG. After 24 h, the cells were stained with IFI6-specific and FLAG-specific antibodies. Scale bar = 10 μm. (C) Huh7 cells were transfected with IFI6-FLAG and after 24 h, the cells were stained with MitoTracker, fixed, and costained with anti-FLAG and anti-GRP94 antibodies. Bottom panel is 3.5-fold enlargement of the area labeled with a white square. The cells were visualized using a confocal microscope. The contour of the IFI6-expressing cell is outlined in white. Scale bar = 10 μm. (D) Graph indicates the Pearson correlation coefficients of IFI6 and MitoTracker or GRP94 localization in 16 cells. ****, P < 0.001, two-tailed, unpaired t test.
IFI6 and IFN-λ2 inhibit an early step in ZIKV and DENV infection.
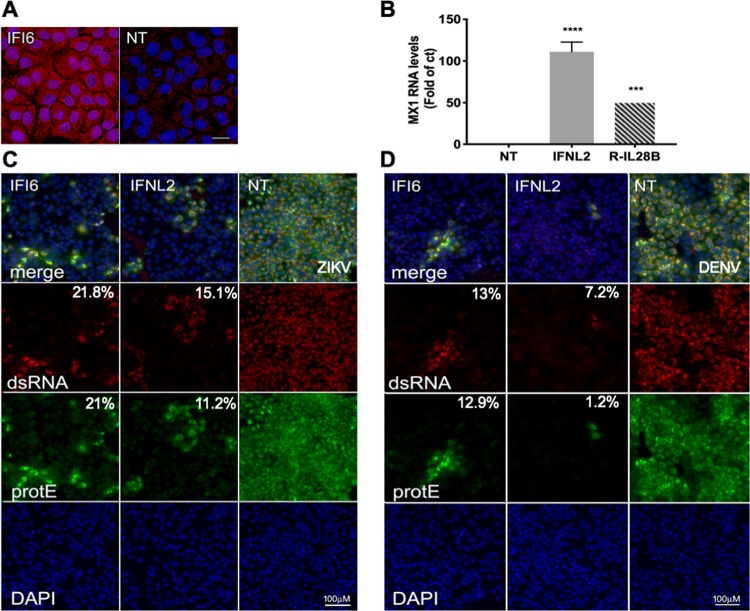
The overexpression of IFI6 at the protein level in our activated stable cell line, compared to a nontargeting control, was confirmed using the specific antibody mentioned above (Fig. 4A). In spite of the apparent high background, a significant overexpression could be observed. Fluorescence intensity was over 20-fold higher in the cells expressing IFI6 sgRNAs than in the NT controls. While we could not confirm the overexpression of IFN-λ2 at the protein level due to a lack of appropriate specific antibodies, we performed a functional assay. In this assay, medium from NT or IFN-λ2-activated cells was used to treat Huh7 cells for 18 h. Following treatment, the cells were harvested and tested for the expression of Myxovirus resistance 1 (Mx1), a known ISG that is strongly elevated in the presence of IFN-λ, using quantitative PCR (qPCR) (34). Mx1 levels were elevated by 100-fold compared to controls treated with medium from NT cells, indicating that IFN-λ2 is activated at the protein level and secreted (Fig. 4B).
FIG 4.
Immunofluorescence of IFI6- and IFN-λ2-expressing ZIKV-infected cells. (A) Huh7 cell lines expressing IFI6 or nontargeting sgRNAs were stained with an IFI6-specific antibody Scale bar = 20 μm. (B) Medium from Huh7 cell lines expressing IFN-λ2 or NT sgRNAs was used to treat Huh7 cells. After 18 h, the cells were harvested and analyzed by qPCR using Mx1 primers. Recombinant IL-28b (IFN-λ3, R-IL-28B) was used as a positive control (10 ng/μl). ***, P < 0.01; ****, P < 0.001, one-way ANOVA Dunnett’s multiple-comparison test. (C and D) Huh7 cell lines expressing IFI6, IFN-λ2, or nontargeting sgRNAs were infected with ZIKV (MOI, 10) (C) or DENV (MOI, 1) (D) for 2 h. After 24 h (ZIKV) or 48 h (DENV), the cells were fixed and stained with flavivirus protein E-specific antibody and an antibody recognizing dsRNA. DAPI was used for nucleus staining. Scale bar = 100 μm. The relative mean fluorescence intensity of each image compared to the control was quantified using Fiji and is presented below each image.
The phenotypic readout used in our screen and functional validation was cell viability following ZIKV infection. This readout cannot distinguish between genes preventing ZIKV infection and genes protecting cells from ZIKV-induced cell death. To distinguish between these two possibilities, we infected Huh7 stable cell lines expressing NT, IFI6, and IFN-λ2 sgRNAs with ZIKV (Fig. 4C) or DENV (Fig. 4D). The cells were fixed and stained with an antibody against flavivirus E protein and antibodies recognizing dsRNA. Nearly all the cells expressing the control nontargeting sgRNAs were stained with both protein E and dsRNA antibodies, indicating high levels of infection, while cells expressing the sgRNAs for IFI6 and IFN-λ2 showed considerably lower levels of infection. The fact that most of the sgRNAs expressing cells do not contain dsRNA indicates that their activation most likely inhibits a stage in viral infection prior to its formation. To further identify the mechanistic details of IFI6 antiviral activity, the effects of its activation on various steps in the flavivirus life cycle occurring in the ER were determined using an IFI6-activated cell line. To determine if IFI6 affects viral RNA translation, we used a DENV translation reporter containing a firefly luciferase gene flanked by the viral untranslated regions (35). In vitro-transcribed viral RNA was transfected into control and IFI6-activated cells. Luciferase levels were determined 1 and 4 h posttransfection. The luciferase levels were similar in control cells and in the IFI6-activated cell line, indicating that IFI6 does not affect viral RNA translation or stability. Processing of the viral polyprotein by host protease was tested using FLAG-tagged NS4B-2K. IFI6 activation did not affect host protease processing of this junction. In addition, the possible interaction between IFI6 and several viral replicase proteins that was tested using immunoprecipitation and did not yield any significant results. ISG12a, an IFI6-related protein, inhibits hepatitis C virus by degrading nonstructural protein NS5A (36). IFI6 activation, however, did not seem to affect the stability of several tested ZIKV proteins. Last, as flaviviruses modify host membranes and thus depend on host lipid stores for membrane proliferation, we tested the possible effect of IFI6 activation on lipid droplet (LD) formation. IFI6 activation did not affect LD number, size, or distribution.
IFI6 and IFN-λ2 activation inhibits cell death in ZIKV-infected cells.
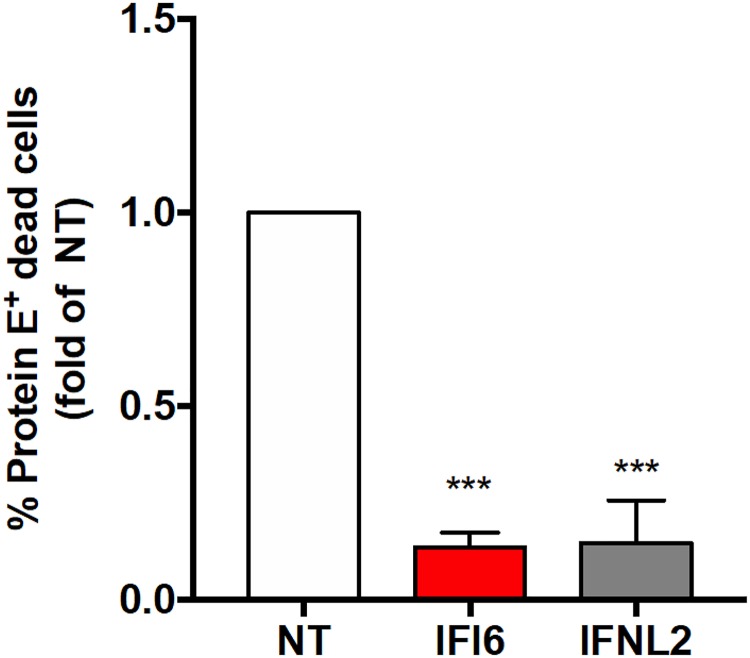
IFI6 was previously reported to inhibit apoptosis in DENV-infected cells (31, 32), in the context of parvovirus infection (37), and in cancer cells (38–40). Our results (Fig. 4) thus far agree with an effect of IFI6 and IFN-λ2 on an early stage in viral infection due to the fact that most of the infected cells expressing IFI6 or IFN-λ2 sgRNAs do not contain dsRNA at 24 h postinfection. This, however, does not rule out an additional later effect on cell death as well. To test this possibility, we infected Huh7 stable cell lines expressing NT, IFI6, and IFN-λ2 sgRNAs with ZIKV and analyzed the cells using live/dead staining flow cytometry at 72 h posttransfection. Both IFI6 and IFN-λ2 significantly inhibited infection-induced cell death (Fig. 5). While the number of infected cells varied in the 4 experiments we performed, in the cells expressing nontargeting sgRNAs, approximately 25% ± 8% of the infected cells were consistently dead in most experiments after 72 h. However, in the cells with activated IFI6 or IFN-λ2, rates of cell death were approximately 10-fold lower (Fig. 5). These results suggest that overexpression of both of these genes could inhibit infection-associated cell death.
FIG 5.
Quantification of cell death in ZIKV-infected IFI6-expressing cells. Huh7 cells stably expressing nontargeting (NT), IFI6, or IFN-λ2 sgRNAs were infected with ZIKV (MOI, 1). The cells were stained with Zombie Yellow, a dye discriminating between live and dead cells, fixed, and stained with anti-flavivirus protein E antibody at 72 h postinfection. The results are from 4 independent experiments. Values are mean fold changes of the NT control ± SD. ***, P < 0.001, two-tailed unpaired t test.
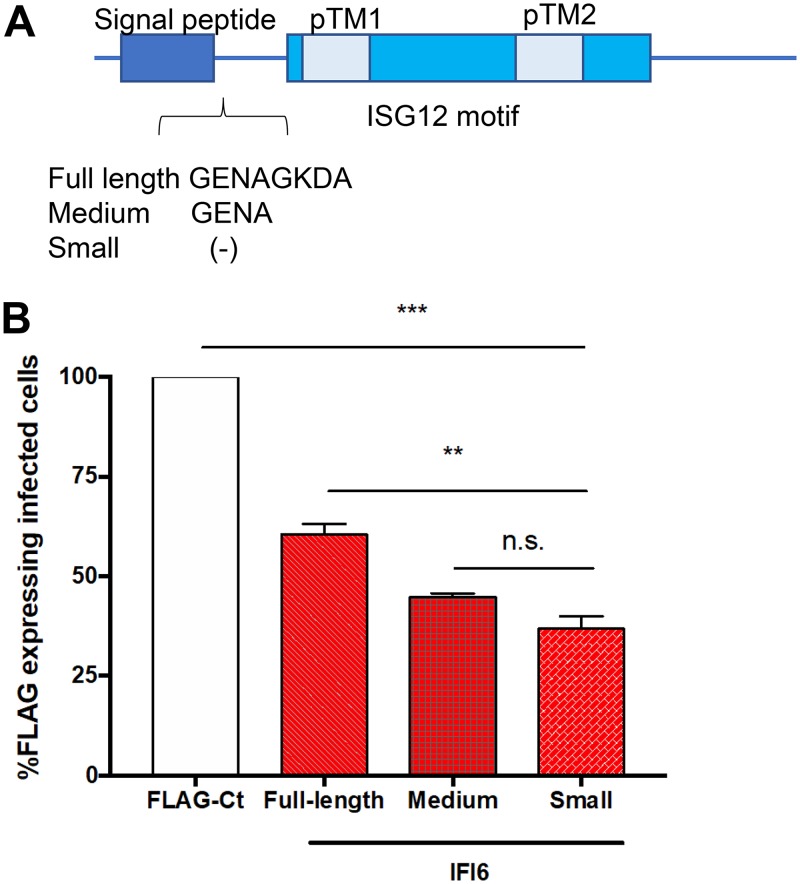
Antiviral activity of IFI6 splice variants.
IFI6 has three different splice variants, as follows: isoform A (UniProt P09912-1, small) is considered the canonical isoform, is shorter than the other isoforms, and lacks 8 aa at the N terminus of the protein immediately following the signal peptide (Fig. 6A); isoform C (UniProt P09912-3, full length) is the longest variant, containing all 8 aa; and isoform B (UniProt P09912-2, medium) has only the first 4 aa of the 8 aa at this location. Alternatively, spliced variants of the related ISG12a were previously reported to have distinct tissue expression and function (41, 42); thus, we tested if the IFI6 alternative splicing has functional consequences in terms of its antiviral activity. IFI6 was amplified from IFN-treated cells, the PCR products were cloned, and the percentages of the different isoforms were analyzed using specific primers. We found that the small isoform of IFI6 is the most abundant form, appearing in approximately 70% of our amplified clones, while the full-length isoform was detected in only 25% of the clones and the medium isoform in the remaining 5%. To test the possible differences in the antiviral activities of the different isoforms, we cloned them into an expression vector with a C-terminal FLAG tag. The plasmids were transfected into Huh7 cells, followed by ZIKV infection. The cells were analyzed by flow cytometry at 48 h postinfection (Fig. 6B). Although the differences between the variants were statistically significant, the difference in antiviral activities between the isoforms was approximately 20%. Thus, we do not believe that such a difference could have a significant measurable functional relevance in this system.
FIG 6.
Antiviral activity of IFI6 isoforms. (A) Schematic representation of IFI6 isoforms. (B) Plasmids expressing FLAG-tagged IFI6 isoforms or FLAG-tagged controls were transfected into Huh7 cells. After 24 h, the cells were infected with ZIKV (MOI, 1). After 48 h, the cells were fixed and stained with flavivirus protein E and FLAG antibodies and analyzed using flow cytometry. The results are presented as a percentage of the FLAG control (TBC1D20-R105A). The results are the mean values ± SD. **, P < 0.01; ***, P < 0.001, one-way ANOVA with Tukey’s multiple-comparison test.
IFI6, IFN-λ2, and ISG20 are induced following ZIKV infection of placenta explants.
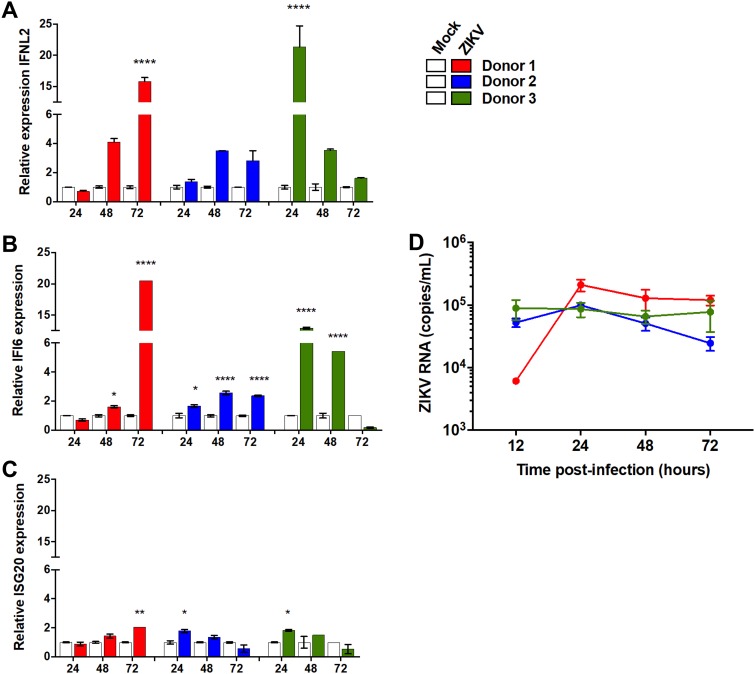
To test the relevance of the identified genes in disease relevant primary tissue, we tested the expression of these genes in ex vivo placenta explants following ZIKV infection. First trimester pregnancy samples were obtained from healthy women undergoing elective termination of pregnancy. Tissue explants from three independent donors were prepared from placenta and infected with ZIKV. Infected tissue was analyzed by real-time PCR for ZIKV RNA and ISG expression levels. All three identified genes were induced in all three donors; however, these inductions were highly varied in their magnitude and kinetics (Fig. 7). ISG20 showed the lowest levels of induction at ∼2-fold in all donors (Fig. 7C), while IFI6 was significantly induced in all three donors and at high levels, at >5-fold in 2/3 donors (Fig. 7B). IFN-λ2 showed a generally similar pattern (Fig. 7C). ZIKV RNA levels, however, did not significantly differ between the different donors (Fig. 7D). While a direct link between the induction of the expression of these proteins and ZIKV RNA levels could not be established, their induction suggests a potential role for these proteins in controlling viral infection.
FIG 7.
Expression of the identified ISG following in ZIKV-infected ex vivo placenta explants. Tissue explants were prepared from placenta samples from first trimester pregnancy terminations. Tissue explants were infected with ZIKV (MOI, 1; overnight). (A to C) RNA was produced at the indicated time points and analyzed using qRT-PCR with IFN-λ2 (A), IFI6 (B), or ISG20 (C). (D) The graph in shows the infection levels doing the course of the experiment. Shown are data from 3 different donors. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ***, P < 0.0001, two-way ANOVA with Sidak’s multiple-comparison test.
DISCUSSION
CRISPR activation was used to identify genes that upon activation rescue cells from ZIKV infection or ZIKV-induced cell death. Our results identify several genes conferring protection, all of which were previously associated with antiviral activity. Validation experiments show that activation of IFI6 and IFN-λ2 is sufficient to confer high levels of protection from multiple cycles of ZIKV infection, while ISG20 and HELZ2 showed low levels of protection when activated individually.
Interferon lambda 2 (IFN-λ2) was previously shown to reduce ZIKV infection (17, 43). Interestingly, IFN-λR1, a subunit of the IFN-λ2 receptor, was also ranked high in our screen. It failed, however, to confer protection when activated individually.
HELZ2 was recently shown to inhibit DENV infection by altering the host cell lipid metabolism (19). Its limited protective activity in our experiments was not consistent between the various sgRNAs. Furthermore, HELZ2 did not seem to be activated in cells expressing the relevant sgRNAs, as determined by real-time PCR.
Our top-ranking gene, IFI6, showed very high levels of protection from ZIKV and DENV infection. These were comparable or exceeded the protection conferred by IFN-λ2, which induces the activation of multiple ISGs. IFI6, on the other hand, does not appear to activate the transcription of other ISGs. Thus, the high levels of protection conferred by IFI6 seem to be caused by its own activity.
IFI6 was recently shown to restrict yellow fever virus infection using a CRISPR knockout screen (16). Interestingly, IFI6, STAT2, and IRF9 were the only ISGs identified in this screen. This work reported that IFI6 inhibits flaviviruses when expressed before infection by preventing the formation of the virus-induced membrane alterations on which the viral replication complex is formed (16). Part of our results are in line with this mechanism and support an antiviral role for IFI6 at an early stage in the viral life cycle occurring at the ER; this is suggested by the localization of the protein and the fact that there was no dsRNA formed in infected cells with activated IFI6. However, we have also confirmed a previously described reduction in cell death following infection of IFI6- or IFN-λ2-activated cells with ZIKV at later time points (31, 32). These results point to an interesting possibility that when expressed before infection, such as in cells surrounding the infected cell following paracrine interferon secretion, these genes can prevent infection. However, when expressed following infection, these genes might be involved in reducing infection-induced cell death.
IFI6 has three known splice isoforms. We found significant differences in the expression levels of the various variants, where the shortest variant was the most abundantly expressed (70%). However, despite these differences, there was only a 20% difference in the antiviral activity against ZIKV between the full-length and shorter forms. Thus, at least in this system, these variants do seem to play substantially different roles. IFI6 was also reported to inhibit apoptosis under various oncogenic conditions (38–40), and thus, these variants might be significant in other cell types or for other functions of this protein.
Both IFI6 and IFN-λ2 were significantly and strongly induced in placenta explants infected with ZIKV from three different donors. Although the time course of induction was different between the various donors, the significant induction of these genes suggests a potential role for these proteins in controlling viral infection in the placenta.
An effective IFN response is generally mediated by an additive effect of more than one ISG (44, 45). Thus, it will be reasonable to hypothesize that the ISGs identified in our screen most likely function together to achieve higher levels of protection from ZIKV infection. Accordingly, our screen identified a short list of ISGs providing protection from ZIKV. This list might have a diagnostic value for infection outcome. Alternately, this list might assist in the design of antiviral drug combinations.
A CRISPR activation screen to detect proteins rescuing cells from influenza virus infection was previously reported (46). This screen identified a glycosyltransferase that modified sialic acid and thus inhibits infection by influenza virus.
Interestingly, none of the top hits in this screen were IFN related. Furthermore, the reported screen did not detect known influenza virus restriction factors. The authors suggested that this is likely due to poor gene induction, provided by one sgRNA per cell in the SAM library (46). Although we used the same library, most of our top hits were ISGs (Table 1), indicating that the presence of one sgRNA per cell is most likely not the limiting factor. Our limited experience with this system suggests different possible explanations. One reason might be that activated ISGs do not completely block infection; thus, their individual activation might not be sufficient to prevent cell death. Alternatively, we observed significant differences in activation in cell lines from different origins. In our hands, a gene activated in one cell type, for example, 293T, might not be activated in another, e.g., Huh7 cells. This might be caused by differences in chromatin state, the availability of different essential activation factors, or other differences between the cell lines. CRISPR activation was recently used to identify genes inhibiting murine norovirus infection in HeLa cells using a different library (47). Several ISGs were identified among the genes rescuing cells from norovirus infection. Interestingly, although norovirus is a positive-strand RNA virus as well, there was no overlap with the ISGs identified in our screen. While all of these screens highlighted important specific host factors rescuing cells from infection, differences in the cell lines used for these screens should be considered. However, together these screens highlight CRISPR activation as a powerful and specific tool for the identification of host factors rescuing cells from viral infection.
MATERIALS AND METHODS
Cell culture, viruses, and transfections.
Vero, HEK293T, and Huh7 cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum and 1% (vol/vol) penicillin-streptomycin (Biological Industries, Bet-Haemek, Israel). The Huh7 cells stably expressing the MS2-P65-HSF1 activator plasmid (Addgene plasmid 89308) were grown in the presence of hygromycin (0.8 mg/ml). Lipofectamine 2000 (Invitrogen, Carlsbad, CA) or polyethyleneimine (PEI; Polysciences, Warrington, PA) were used for plasmid DNA transfections of subconfluent cells.
ZIKV MR766 was a kind gift from Leslie Lobel (Ben-Gurion University, Israel), and dengue virus 2 New Guinea C strain stocks were purchased from Culture Collections (Public Health England). Virus stocks were propagated in Vero cells, and the supernatant was harvested at 3 to 7 days postinfection. Viral titers were determined by plaque assays on Vero cells. Interferon alpha B2 was purchased from PBL (NJ; catalog no. 11115-1), and recombinant human interleukin 28B (IL-28B)/IFN-λ 3 protein was from R&D Systems (catalog no. 5259-IL).
sgRNA library amplification and lentivirus production.
A pooled library of lentiviral vectors (lentiSAMv2) containing genome-scale sgRNA was purchased from Addgene (plasmid 1000000078, a gift from Feng Zhang). The library was amplified according to the depositor’s protocol (21). sgRNA distribution was verified following amplification by NGS. HEK293T cells were used for lentivirus production. The cells were seeded at 40% confluence in 4 T225 flasks. The next day, cells were transfected using PEI with 3.4 μg pMD2.G, 6.8 μg psPAX2, and 13.6 μg of the amplified library (21). The supernatant was harvested at 2 days posttransfection, filtered, and stored at −80°C. The library titer was determined through transduction in Huh7 cells stably expressing the MS2-P65-HSF1 activator plasmid, as described previously (21).
CRISPR activation screen.
To transduce the Huh7 cells stably expressing the MS2-P65-HSF1 activator plasmid, the cells were seeded into 6-well plates at a density of 2.45 × 106 cells per well. The cells were transduced with lentiviral pseudoparticles at an MOI of 0.3. A total of 1.17 × 108 Huh7 cells were infected with the library plasmids to yield 3.5 × 107 transduced cells at an MOI of 0.3. Since the library contains 70,290 sgRNAs, this number is sufficient for infection of at least 500 cells with each sgRNA (12, 21). The cells were then spinoculated at 1,000 × g for containing 3% fetal bovine serum (FBS), 20 mM HEPES, and 4 μg ml−1 Polybrene. The next day, the cells were replated at low confluence in T175 flasks and selected with 6 μg/ml blasticidin for 7 days. Following selection, the cells were replated at 40% confluence and infected with ZIKV (MOI, 0.2) for 2 h. The cells were washed extensively and incubated for 10 days. During the incubation period the cells were washed and the medium was replaced every 3 days. Genomic DNA was prepared from the cells and used to amplify the sgRNA for NGS, as described previously (21). Samples were sequenced on an Illumina HiSeq 2000 platform. The screen was repeated twice.
NGS and screen hit analysis.
The resulting reads from the NGS experiments were analyzed with two different algorithms, RIGER (27) (utilizing the GENE-E framework) and MAGeCK v0.5.6 (28). In order to prepare the input for RIGER, we used the Python scripts available online (21). Key regions in our reads were detected with python –no-g counter_spacers.py, available in reference 12.
RIGER and MAGeCK were used with default parameters following the workflow given in reference 12. As the two methods yielded similar lists in terms of high-ranking genes, we continued using the list produced by MAGeCK. Raw sgRNA sequencing read numbers and the MAGeCK scored and ranked list of genes from the two screen repeats are added in the supplemental material (Data Sets S1 to S4).
Validation of screen hits.
To validate the screen hits, sgRNAs for the top-ranking genes were individually cloned into the library backbone plasmid (lentiSAMv2; Addgene). Lentiviruses produced from these plasmids were used to infect Huh7 cells stably expressing the MS2-P65-HSF1 activator plasmid. The cells were selected with 6 μg/ml blasticidin for 7 days to create stable cell lines. The obtained cell lines were infected with ZIKV (MOI, 0.2) for 2 h, followed by extensive washes. After 7 days, the cells were stained with crystal violet. To quantify the crystal violet staining, the dye was extracted using 10% acetic acid. The obtained solution was diluted and transferred to a 96-well plate. Absorbance at 570 nm was measured using a plate reader.
For qPCR experiments, total RNA was extracted from each cell line using Tri Reagent (Sigma, Rehovot, Israel). The obtained RNA was reverse transcribed using ExcelRT (SMOBIO). Real-time PCR was performed using Fast SYBR green mastermix (Roche) and the primers listed in Table 2. Hypoxanthine phosphoribosyltransferase 1 (HPRT) was used as a housekeeping control. The results are the fold change from nontargeting sgRNA-expressing cells calculated using the ΔΔCT method.
TABLE 2.
Primers used for real-time PCR
| Target gene | Primer sequence |
|
|---|---|---|
| Forward | Reverse | |
| IFI6 | AGCTGGTCTGCGATCCTGAATG | TTACCTATGACGACGCTGCTGC |
| ISG20 | AGTGAGCGCCTCCTACACAAG | ACCAGCTTGCCTTTCAGGAG |
| IFN-λ2 | ACACCCTGCACCATATCCTCTC | CGGAAGAGGTTGAAGGTGACAG |
| HELZ | GTGTCCTCCATCACCAAGAGC | TCCACAACGAAGCCCAGAAAC |
| MSD2A | ACCAAGTTTGCCTCTGGAGTG | ACTTGACACGTTCCGGCTG |
| ZCCHC6 | GTGTTAACTGAAGGAGAGCTGGC | TCTTTGGTTCAGGGCATCTTCC |
| HPRT | TGACACTGGCAAAACAATGCA | GGTCCTTTTCACCAGCAAGCT |
| ZIKV | ATGCCATTTGCTCGGCTGTG | GGTCGTTCTCCTCAATCCACAC |
| ZIKV | TTGGTCATGATACTGCTGATTGC | CCTTCCACAAAGTCCCTATTGC |
| Mx1 | CAGAGAGAAGGAGCTGGAAGAA | GCTGGCCTCCTGGTGATA |
Plaque assay.
Virus-containing medium from Zika-infected cells was collected at 2, 24, 48, and 72 h postinfection. Confluent Vero cells grown in 6-well plates were infected with serial dilutions (up to 1 × 10–6) of the virus-containing medium for 2 h, washed, and overlaid with 0.5% hydroxypropyl methylcellulose in DMEM. The cells were stained with crystal violet at 5 days postinfection, and the plaques were counted.
Luciferase reporter assay.
Reporter HEK293 cells stably expressing Toll-like receptor 3 (TLR3) and ISG56-luciferase (pGL3Basic [48]) were a gift to J. U. Jung. Subconfluent ISG56-Luc cells in 24 wells were transfected with the indicated plasmids using PEI. Luciferase activity was measured at 24 h posttransfection.
siRNA experiments.
Nontargeting control siRNA and siIFI6 (Dharmacon, SMARTpool catalog number L-003672-00-0005 for the IFI6 and D-001810-10-05 for the nontargeting pool) were repeatedly transfected (at 24 h and 48 h postplating) to Huh7 cells using Lipofectamine 2000 (Invitrogen). The cells were treated with IFN-α (human IFN alpha B2 [Alpha 8], 1,000 IU/ml) for 2 h and then infected with ZIKV at an MOI of 1. qPCR was used to quantify IFI6 and ZIKV levels at 24 h postinfection, and HPRT was used as a housekeeping control. The primer sequences are listed in Table 2. The graph shows representative results from three independent experiments.
Immunofluorescence microscopy.
Cells were cultured on coverslips, washed with PBS, fixed for 20 min with methanol at −20°C, and blocked with 1% bovine serum albumin (BSA) in PBS for 1 h. Immunolabeling was performed overnight with the IFI6 antibody (catalog no. ab192314, 1:100 dilution; Abcam), flavivirus group antigen antibody (4G2, 1:200 dilution; Novus, Littleton, CO), or J2 dsRNA antibody (1:200 dilution; Scion, Budapest, Hungary). For colocalization studies, MitoTracker Red (1:1,000 dilution; Thermo), anti-FLAG (mouse monoclonal, 1:500 dilution; Sigma-Aldrich), or anti-GRP94 (catalog no. ADI-SPA-850, 1:1,000 dilution; Enzo) was used. The secondary antibody was Alexa Fluor 488 goat anti-mouse or Alexa Fluor 555 goat anti-rabbit (1:2,000 dilution). 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes, Eugene, OR) was used for nucleus staining. Images were acquired using a Zeiss LSM800 confocal laser scanning microscope (Carl Zeiss MicroImaging, Jena, Germany) hooked to an inverted microscope. Relative fluorescent intensity was quantified using ImageJ or Fiji (49, 50). For colocalization analysis between GRP94 or MitoTracker and IFI6, immunofluorescent signals were analyzed for Pearson’s colocalization using the ZEN software (Zeiss). Analysis was performed on 16 images.
Plasmids.
IFI6 was amplified from the cDNA of IFN-α-treated Huh7 cells using the following primers: forward (FW), CAAGCTTATGCGGCAGAAGGCGGTATC, and reverse (RV), AGGTACCCTCCTCATCCTCCTCACTATC. The PCR product was ligated into the pJET1.2/Blunt donor plasmid (Thermo, Waltham, MA), verified by sequencing, and further cloned using Gibson Assembly into a modified pEF-IRES-puro vector encoding a C-terminal 3×FLAG tag (51). The percentages of full-length, medium, and small splice variants were calculated by performing colony PCR with distinguishing primers on pJet-IFI6-containing bacterial colonies. The FW primer used for detection of the full-length form of IFI6 is CAGGTGAGAATGCGGGTAAGG, and the FW primer for medium-length IFI6 is CAGGTGAGAATGCGGGTAAGA. Dengue virus translation reporter (52) was prepared from dengue virus strain 16681 luciferase replicon clone (53). The DENV open reading frame (ORF) was removed by PCR, leaving only luciferase reporter flanked by 5′ untranslated region (5′ UTR) and 3′ UTR, and the plasmid was religated. The primers used were FW, AAAGCAAAACTAACATGAAACAAGGCT, and RV, TTAGATAGATCTTTGTTCATTTTTGAGAACTCG. A capped in vitro-translated transcript was prepared as described previously (54). ZIKV PRVABC59 (ATCC VR-1843) NS4B-2K amplified from infected cells by reverse transcription-PCR (RT-PCR) and cloned into modified pEF-IRES-puro with a C-terminal 3×FLAG tag using Gibson Assembly. FLAG-tagged MAVS was expressed from a modified pEF-IRES-puro with a C-terminal 3×FLAG tag (51).
Flow cytometry analysis.
For live/dead cells and infection experiments, Huh7 cells expressing nontargeting or IFI6 sgRNAs were infected ZIKV at MOI of 1. The cells were stained for 30 min with Zombie Yellow (1:500 dilution; BioLegend) in PBS at 72 h postinfection. The cells were then fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton, blocked (1% FBS), and stained with anti-protein E (flavivirus group antigen antibody, 4G2, 1:200 dilution; Novus, Littleton, CO). The secondary antibody was anti-rabbit Alexa Fluor 488 (1:1,000 dilution; Thermo). For the experiment with the IFI6 splicing variants, Huh7 cells were transfected with IFI6-FLAG variants or a FLAG control (FLAG-TBC1D20 R105A [55] or FLAG-VP35 [56] a gift from L. Lobel). Since an empty FLAG plasmid cannot be detected using fluorescence-activated cell sorting (FACS), we used two different FLAG-tagged controls presumed to be inert in these experiments: an inactive mutant form of an ER-localized Rab-GAP FLAG-TBC1D20 R105A (55) or Ebola virus VP35. Similar results were obtained with the two proteins. The cells were infected at 24 h posttransfection with ZIKV at an MOI of 1. The cells were fixed and stained as described above at 48 h postinfection. The primary antibodies were anti-protein E and FLAG M2 (Sigma). The secondary antibodies were anti-rabbit Alexa Fluor 488 or anti-mouse Alexa Fluor 555 (1:1,000 dilution; Thermo). Cells were analyzed using the CytoFLEX flow cytometer (Beckman Coulter Life Sciences).
Ex vivo placenta explant cultures and ZIKV infection.
First trimester pregnancy (7 to 12 weeks gestation) samples were obtained from healthy women undergoing elective termination of pregnancy at Paule de Viguier Toulouse University Hospital, France. Tissue explants (0.3 cm2) were prepared from placenta and infected with the Asian strain of Zika virus. Tissue explants were infected overnight with 5.6 × 1010 RNA copies/ml (MOI, 1) in DMEM-F12 containing 2% FBS. After five washes in an excess volume of PBS, explants were cultured in 10% FBS–DMEM-F12 medium for up to 3 days. For quantitative reverse transcription-PCR (qRT-PCR) analysis, placenta tissue was immersed in liquid nitrogen and ground. Samples were transferred to new tubes and lysed in RNA lysis buffer. Total RNA was isolated using an RNA minikit (Qiagen), following the manufacturer’s instructions. Purified RNA was reverse transcribed using the SuperScript III first-strand synthesis system kit (Invitrogen). Placental gene expression was quantified by qRT-PCR using LightCycler 480 SYBR green I master (Roche). HPRT was used as a housekeeping control. The results are the fold change from time-matched mock controls calculated using the ΔΔCT method. qRT-PCR was performed in 96-well plates and run on a LightCycler 480 instrument. Total RNA was isolated from the supernatants of infected human placental explants using the QIAamp viral RNA minikit (Qiagen), following the manufacturer’s instructions. Quantification of viral RNA was performed by a qRT-PCR assay using ZIKV-specific primers (ZIKV 835 forward AND 911c reverse). A standard curve was obtained by amplification of a fragment of ZIKV E-envelope region and cloned into pCR-XL-2 plasmid (Invitrogen). A standard curve was generated using 10-fold serial dilutions of constructed ZIKV envelope plasmid and was used to quantify ZIKV RNA in supernatants. Viral RNA copies were extrapolated from the standard curve using the sample threshold cycle (CT) and represented as copies per milliliter of supernatant.
Ethics statement.
This study was approved by the South-West & Outmer II ethics committee and registered at the Ministry of Higher Education and Research (number DC-2016-2772). All participants provided prior written informed consent in agreement with the guidelines of the Declaration of Helsinki, with the experiments performed in accordance with approved guidelines.
Statistical analyses.
Statistical analyses were performed using Prism (GraphPad, San Diego, CA).
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Jung laboratory (University of Southern California) and Eran Bacharach (Tel Aviv University) for helpful conversations.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00211-19.
REFERENCES
- 1.Coyne CB, Lazear HM. 2016. Zika virus–reigniting the TORCH. Nat Rev Microbiol 14:707–715. doi: 10.1038/nrmicro.2016.125. [DOI] [PubMed] [Google Scholar]
- 2.de Oliveira WK, de Franca GVA, Carmo EH, Duncan BB, de Souza Kuchenbecker R, Schmidt MI. 2017. Infection-related microcephaly after the 2015 and 2016 Zika virus outbreaks in Brazil: a surveillance-based analysis. Lancet 390:861–870. doi: 10.1016/S0140-6736(17)31368-5. [DOI] [PubMed] [Google Scholar]
- 3.Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R. 2018. Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol 16:125–142. doi: 10.1038/nrmicro.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma H, Dang Y, Wu Y, Jia G, Anaya E, Zhang J, Abraham S, Choi JG, Shi G, Qi L, Manjunath N, Wu H. 2015. A CRISPR-based screen identifies genes essential for West-Nile-virus-induced cell death. Cell Rep 12:673–683. doi: 10.1016/j.celrep.2015.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, Carette JE. 2016. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savidis G, McDougall WM, Meraner P, Perreira JM, Portmann JM, Trincucci G, John SP, Aker AM, Renzette N, Robbins DR, Guo Z, Green S, Kowalik TF, Brass AL. 2016. Identification of Zika virus and dengue virus dependency factors using functional genomics. Cell Rep 16:232–246. doi: 10.1016/j.celrep.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 7.Zhang R, Miner JJ, Gorman MJ, Rausch K, Ramage H, White JP, Zuiani A, Zhang P, Fernandez E, Zhang Q, Dowd KA, Pierson TC, Cherry S, Diamond MS. 2016. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 535:164–168. doi: 10.1038/nature18625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. 2013. CRISPR RNA-guided activation of endogenous human genes. Nat Methods 10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. 2013. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. 2013. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods 10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, Iyer EPR, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, Church GM. 2015. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. 2015. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS. 2014. Genome-cale CRISPR-mediated control of gene repression and activation. Cell 159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. 2014. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chavez A, Tuttle M, Pruitt BW, Ewen-Campen B, Chari R, Ter-Ovanesyan D, Haque SJ, Cecchi RJ, Kowal EJK, Buchthal J, Housden BE, Perrimon N, Collins JJ, Church G. 2016. Comparison of Cas9 activators in multiple species. Nat Methods 13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson RB, Ohlson MB, Eitson JL, Kumar A, McDougal MB, Boys IN, Mar KB, De La Cruz-Rivera PC, Douglas C, Konopka G, Xing C, Schoggins JW. 2018. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat Microbiol doi: 10.1038/s41564-018-0244-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jagger BW, Miner JJ, Cao B, Arora N, Smith AM, Kovacs A, Mysorekar IU, Coyne CB, Diamond MS. 2017. Gestational stage and IFN-lambda signaling regulate ZIKV infection in utero. Cell Host Microbe 22:366–376.e3. doi: 10.1016/j.chom.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang D, Weidner JM, Qing M, Pan X-B, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi P-Y, Block TM, Guo J-T. 2010. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J Virol 84:8332. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fusco DN, Pratt H, Kandilas S, Cheon SS, Lin W, Cronkite DA, Basavappa M, Jeffrey KL, Anselmo A, Sadreyev R, Yapp C, Shi X, O'Sullivan JF, Gerszten RE, Tomaru T, Yoshino S, Satoh T, Chung RT. 2017. HELZ2 is an IFN effector mediating suppression of dengue virus. Front Microbiol 8:240. doi: 10.3389/fmicb.2017.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlin AF, Plummer EM, Vizcarra EA, Sheets N, Joo Y, Tang W, Day J, Greenbaum J, Glass CK, Diamond MS, Shresta S. 2017. An IRF-3-, IRF-5-, and IRF-7-independent pathway of dengue viral resistance utilizes IRF-1 to stimulate type I and II interferon responses. Cell Rep 21:1600–1612. doi: 10.1016/j.celrep.2017.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, Zhang F. 2017. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 12:828–863. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan JF, Yip CC, Tsang JO, Tee KM, Cai JP, Chik KK, Zhu Z, Chan CC, Choi GK, Sridhar S, Zhang AJ, Lu G, Chiu K, Lo AC, Tsao S, Kok KH, Jin DY, Chan KH, Yuen KY. 2016. Differential cell line susceptibility to the emerging Zika virus: implications for disease pathogenesis, non-vector-borne human transmission and animal reservoirs. Emerg Microbes Infect 5:e93. doi: 10.1038/emi.2016.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowall SD, Graham VA, Rayner E, Atkinson B, Hall G, Watson RJ, Bosworth A, Bonney LC, Kitchen S, Hewson R. 2016. A susceptible mouse model for Zika virus infection. PLoS Negl Trop Dis 10:e0004658. doi: 10.1371/journal.pntd.0004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zmurko J, Marques RE, Schols D, Verbeken E, Kaptein SJ, Neyts J. 2016. The viral polymerase inhibitor 7-deaza-2′-C-methyladenosine is a potent inhibitor of in vitro Zika virus replication and delays disease progression in a robust mouse infection model. PLoS Negl Trop Dis 10:e0004695. doi: 10.1371/journal.pntd.0004695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacNamara FN. 1954. Zika virus: a report on three cases of human infection during an epidemic of jaundice in Nigeria. Trans R Soc Trop Med Hyg 48:139–145. doi: 10.1016/0035-9203(54)90006-1. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y, Cui X, Wu N, Song R, Yang W, Zhang W, Fan D, Chen Z, An J. 2017. A unique case of human Zika virus infection in association with severe liver injury and coagulation disorders. Sci Rep 7:11393. doi: 10.1038/s41598-017-11568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, Mermel C, Barbie DA, Awad T, Zhou X, Nguyen T, Piqani B, Li C, Golub TR, Meyerson M, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. 2008. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A 105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu J, Smith K, Hsieh PN, Mburu YK, Chattopadhyay S, Sen GC, Sarkar SN. 2010. High-throughput screening for TLR3-IFN regulatory factor 3 signaling pathway modulators identifies several antipsychotic drugs as TLR inhibitors. J Immunol 184:5768–5776. doi: 10.4049/jimmunol.0903559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker N, Porter AC. 2004. Identification of a novel gene family that includes the interferon-inducible human genes 6-16 and ISG12. BMC Genomics 5:8. doi: 10.1186/1471-2164-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J, Li Y, Qi Y, Zhang Y, Zhang L, Wang Z, Zhang X, Gui L. 2014. Coordinated regulation of autophagy and apoptosis determines endothelial cell fate during dengue virus type 2 infection. Mol Cell Biochem 397:157–165. doi: 10.1007/s11010-014-2183-3. [DOI] [PubMed] [Google Scholar]
- 32.Qi Y, Li Y, Zhang Y, Zhang L, Wang Z, Zhang X, Gui L, Huang J. 2015. IFI6 inhibits apoptosis via mitochondrial-dependent pathway in dengue virus 2 infected vascular endothelial cells. PLoS One 10:e0132743. doi: 10.1371/journal.pone.0132743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eletto D, Dersh D, Argon Y. 2010. GRP94 in ER quality control and stress responses. Semin Cell Dev Biol 21:479–485. doi: 10.1016/j.semcdb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolen CR, Ding S, Robek MD, Kleinstein SH. 2014. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology 59:1262–1272. doi: 10.1002/hep.26657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgil D, Polacek C, Harris E. 2006. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J Virol 80:2976–2986. doi: 10.1128/JVI.80.6.2976-2986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue B, Yang D, Wang J, Xu Y, Wang X, Qin Y, Tian R, Chen S, Xie Q, Liu N, Zhu H. 2016. ISG12a restricts hepatitis C virus infection through the ubiquitination-dependent degradation pathway. J Virol 90:6832–6845. doi: 10.1128/JVI.00352-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng X, Zou W, Xiong M, Wang Z, Engelhardt JF, Ye SQ, Yan Z, Qiu J. 2017. Human parvovirus infection of human airway epithelia induces pyroptotic cell death by inhibiting apoptosis. J Virol 91:e01533-17. doi: 10.1128/JVI.01533-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tahara E Jr, Tahara H, Kanno M, Naka K, Takeda Y, Matsuzaki T, Yamazaki R, Ishihara H, Yasui W, Barrett JC, Ide T, Tahara E. 2005. G1P3, an interferon inducible gene 6-16, is expressed in gastric cancers and inhibits mitochondrial-mediated apoptosis in gastric cancer cell line TMK-1 cell. Cancer Immunol Immunother 54:729–740. doi: 10.1007/s00262-004-0645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta R, Forloni M, Bisserier M, Dogra SK, Yang Q, Wajapeyee N. 2016. Interferon alpha-inducible protein 6 regulates NRASQ61K-induced melanomagenesis and growth. Elife 55:e16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheriyath V, Glaser KB, Waring JF, Baz R, Hussein MA, Borden EC. 2007. G1P3, an IFN-induced survival factor, antagonizes TRAIL-induced apoptosis in human myeloma cells. J Clin Invest 117:3107–3117. doi: 10.1172/JCI31122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gytz H, Hansen MF, Skovbjerg S, Kristensen ACM, Hørlyck S, Jensen MB, Fredborg M, Markert LD, McMillan NA, Christensen EI, Martensen PM. 2017. Apoptotic properties of the type 1 interferon induced family of human mitochondrial membrane ISG12 proteins. Biol Cell 109:94–112. doi: 10.1111/boc.201600034. [DOI] [PubMed] [Google Scholar]
- 42.Smidt KCJ, Hansen LL, Søgaard TMM, Petersen LK, Knudsen UB, Martensen PM. 2003. A nine-nucleotide deletion and splice variation in the coding region of the interferon induced ISG12 gene. Biochim Biophys Acta Mol Basis Dis 1638:227–234. doi: 10.1016/S0925-4439(03)00087-5. [DOI] [PubMed] [Google Scholar]
- 43.Chen J, Liang Y, Yi P, Xu L, Hawkins HK, Rossi SL, Soong L, Cai J, Menon R, Sun J. 2017. Outcomes of congenital Zika disease depend on timing of infection and maternal-fetal interferon action. Cell Rep 21:1588–1599. doi: 10.1016/j.celrep.2017.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heaton BE, Kennedy EM, Dumm RE, Harding AT, Sacco MT, Sachs D, Heaton NS. 2017. A CRISPR activation screen identifies a pan-avian influenza virus inhibitory host factor. Cell Rep 20:1503–1512. doi: 10.1016/j.celrep.2017.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orchard RC, Sullender ME, Dunlap BF, Balce DR, Doench JG, Virgin HW. 2019. Identification of antinorovirus genes in human cells using genome-wide CRISPR activation screening. J Virol 93:e01324-18. doi: 10.1128/JVI.e01324-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarkar SN, Peters KL, Elco CP, Sakamoto S, Pal S, Sen GC. 2004. Novel roles of TLR3 tyrosine phosphorylation and PI3 kinase in double-stranded RNA signaling. Nat Struct Mol Biol 11:1060. doi: 10.1038/nsmb847. [DOI] [PubMed] [Google Scholar]
- 49.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gack MU, Nistal-Villán E, Inn K-S, García-Sastre A, Jung JU. 2010. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J Virol 84:3220–3229. doi: 10.1128/JVI.02241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holden KL, Harris E. 2004. Enhancement of dengue virus translation: role of the 3′ untranslated region and the terminal 3′ stem-loop domain. Virology 329:119–133. doi: 10.1016/j.virol.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 53.Clyde K, Barrera J, Harris E. 2008. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology 379:314–323. doi: 10.1016/j.virol.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stern O, Hung YF, Valdau O, Yaffe Y, Harris E, Hoffmann S, Willbold D, Sklan EH. 2013. An N-terminal amphipathic helix in dengue virus nonstructural protein 4A mediates oligomerization and is essential for replication. J Virol 87:4080–4085. doi: 10.1128/JVI.01900-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sklan EH, Serrano RL, Einav S, Pfeffer SR, Lambright DG, Glenn JS. 2007. TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. J Biol Chem 282:36354–36361. doi: 10.1074/jbc.M705221200. [DOI] [PubMed] [Google Scholar]
- 56.Sobarzo A, Eskira Y, Herbert AS, Kuehne AI, Stonier SW, Ochayon DE, Fedida-Metula S, Balinandi S, Kislev Y, Tali N, Lewis EC, Lutwama JJ, Dye JM, Yavelsky V, Lobel L. 2015. Immune memory to Sudan virus: comparison between two separate disease outbreaks. Viruses 7:37–51. doi: 10.3390/v7010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.