This study shows that only a single introduction of SINV into a new geographical area is required for spread and establishment, provided that the requisite vector(s) and reservoir(s) of epizootological and epidemiological importance are present. Furthermore, we present the first report of recombination between two strains of SINV in nature. Our study increases the knowledge on new introductions and dispersal of arboviruses in general and of SINV in particular.
KEYWORDS: evolution, phylogeny, Sindbis virus
ABSTRACT
Bird-hosted viruses have the potential to be transported over large areas of the world and to be transmitted in distant geographical regions. Sindbis virus (SINV) is a mosquito-borne alphavirus that is locally amplified in a bird-mosquito enzootic cycle and distributed all over the Old World and Australia/Oceania. Sindbis virus genotype I (SINV-I) is the cause of disease outbreaks in humans in South Africa as well as in northern Europe. To trace the evolutionary history and potential strain-disease association of SINV-I, we sequenced 36 complete genomes isolated from field material in Europe, as well as in Africa and the Middle East, collected over 58 years. These were analyzed together with 30 additional published whole SINV-I genomes using Bayesian analysis. Our results suggested that SINV-I was introduced only once to northern Europe from central Africa, in the 1920s. After its first introduction to Sweden, it spread east and southward on two separate occasions in the 1960s and 1970s. Another introduction from central Africa to southern/central Europe seems to have occurred, and where these two introductions meet, one recombination event was detected in central Europe. In addition, another recombinant strain was found in central Africa, where the most divergent SINV-I strains also originated.
IMPORTANCE This study shows that only a single introduction of SINV into a new geographical area is required for spread and establishment, provided that the requisite vector(s) and reservoir(s) of epizootological and epidemiological importance are present. Furthermore, we present the first report of recombination between two strains of SINV in nature. Our study increases the knowledge on new introductions and dispersal of arboviruses in general and of SINV in particular.
INTRODUCTION
Viruses hosted by birds have the potential to be transported over large areas of the world, connecting countries and regions that are well separated in climate and habitat. The occurrence of human disease from vector-borne bird viruses is dependent on the availability and abundance of competent vectors in proximity to the bird hosts and humans in different regions. Thus, these viruses need to be able to replicate in various vector and host species, as the geographic distribution often spans tropical as well as subtropical or temperate regions, with unique sets of mosquito and bird species.
Mosquito-borne bird viruses include a number of alphaviruses (Togaviridae), e.g., Sindbis virus (SINV) and Western equine encephalitis virus (WEEV), and flaviviruses (Flaviviridae), e.g., West Nile virus (WNV), Japanese encephalitis virus (JEV), Murray Valley encephalitis virus (MVEV), St. Louis encephalitis virus (SLEV), Rocio virus (ROCV), and Usutu virus (USUV) (1–5). In addition to their dependence on avian hosts, all these viruses also utilize vectors from the same genus of mosquitoes, Culex. This genus has species representatives throughout the world; thus, it has the potential to provide suitable vector species in both tropical and temperate regions (2, 6–8). Human cases emerge from an infectious mosquito bite and are considered an effect of spillover from a viremic bird population. The viremia produced in humans is not enough to infect a mosquito, and consequently, humans are dead-end hosts of the virus (2).
SINV is a positive single-stranded RNA virus that has a wide distribution throughout the Old World. It has a genome size of about 11.7 kb and has two open reading frames (ORFs) that encode four nonstructural (nsP1 to -4) and five structural (C, E3, E2, 6K/TF, and E1) proteins, respectively (9). Based on phylogenetic analyses of the partial E2 gene, a total of six genotypes (genotypes I to VI) have been identified (10). SINV genotype I (SINV-I) has been isolated from Europe, Africa, and the Middle East; SINV-II and SINV-VI have been isolated from Australia; SINV-III has been isolated from Southeast Asia; SINV-IV has been isolated from Asia and the Middle East; and SINV-V (also referred to as Whataroa virus) has been isolated from New Zealand (10).
SINV-I is the only genotype that has been associated with outbreaks of human disease, and it was first isolated from mosquitoes in Cairo, Egypt, in 1952 (11). Outbreaks have been reported from South Africa and northern Europe, and the disease goes under several names: Pogosta, Ockelbo disease, Karelian fever, and Sindbis fever. Signs and symptoms include fever, exanthema, arthralgia, myalgia, and cases of arthritis that can remain for years (12, 13).
The first cases of Sindbis fever in Fennoscandia were reported from Sweden in the 1960s, and later, this disease was shown to be associated with a strain of SINV-I isolated from mosquitoes in Edsbyn, Sweden (14). Cases of Sindbis fever are sporadically diagnosed in central Sweden (15), and in recent years, there has also been an outbreak of Sindbis fever in northern Sweden (16). Several larger outbreaks have been observed in Finland and in South Africa (17, 18). However, as mentioned above, SINV-I has also been isolated from other parts of Africa, Europe, and the Middle East, without any reports of disease outbreaks. Previous studies indicated that SINV-I has been transported with northward-migrating birds, connecting South Africa and Fennoscandia (10). The driving forces behind SINV-I outbreaks and the details of SINV-I movements between continents and regions are still unknown.
To further elucidate the evolutionary history of SINV-I and its dispersal patterns, we have sequenced and analyzed the complete genomes of 36 new strains, in addition to 30 previously sequenced publicly available strains, from various vector and host species, spanning 10 countries and 58 years.
RESULTS
Evolutionary history of SINV-I.
The phylogenetic tree of the complete coding sequence of SINV-I contained 64 strains (2 strains were excluded due to recombination [see below]) and showed that the strains from central Africa were basal in each clade. According to the parsimony principle (i.e., that the best hypothesis is the one that requires the fewest evolutionary changes), this indicates that SINV-I originated in central Africa and diverged into two main clades with well-supported values (posterior probability, 1.0) (Fig. 1). Clade A contained all the strains from Sweden, Finland, Norway, Germany, and Russia and shared a common ancestor with strains from clade B, which included strains from central African countries (i.e., Central African Republic, Cameroon, and Kenya). Clades A and B shared a common ancestor with the strains from South Africa (clade C). The other main clade (clade D) contained the prototype SINV-I strain from Egypt (1952), which clustered with sequences from other countries in the Middle East and adjacent regions (i.e., Egypt, Israel, Saudi Arabia, and Azerbaijan), southern/central Europe (Slovak Republic and Italy), and east Africa (Kenya). Clade D shared a most recent common ancestor with clade E, which included strains from central Africa (Uganda) and South Africa. However, these two clades (clades D and E) contained only 15 sequences, including some from unconventional hosts, and therefore, a more detailed analysis of this group could not be made.
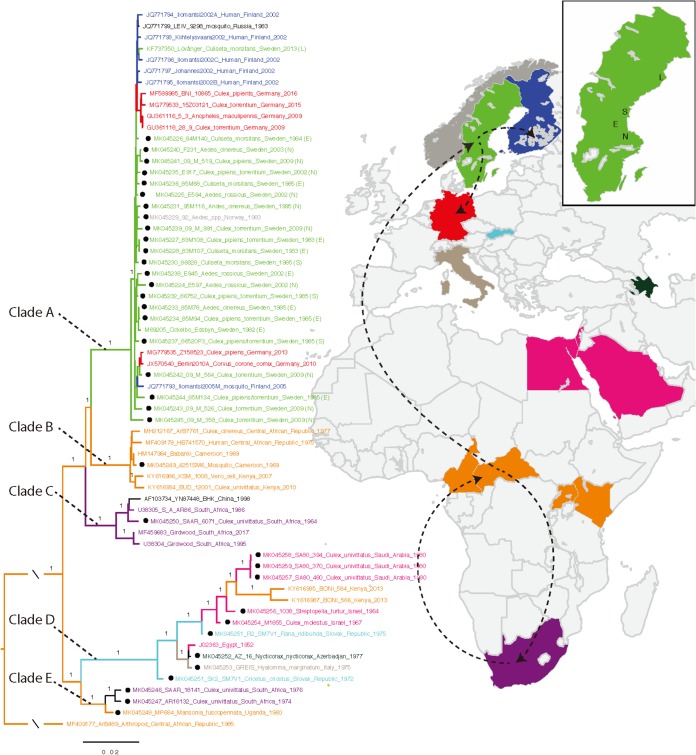
FIG 1.
Spread of SINV-I from Africa to Europe. Shown is a Bayesian phylogenetic tree of SINV-I based on concatenated nucleotide ORF sequences (MrBayes). The colors indicate Sweden (green); Finland (blue); Germany (red); Egypt, Israel, and Saudi Arabia (pink); Kenya and the Central African Republic (orange); Slovak Republic (light blue); Italy (gray); and South Africa (purple). The roots in orange indicate that SINV-I originated in central Africa. Main SINV-I dispersal routes from Africa to and within northern/central Europe are indicated with dashed lines. L, N, E, and S represent locations in Sweden (Lövånger, Nedre Dalälven, Edsbyn, and Sundsvall, respectively). Posterior probability values above 0.95 are shown for the branches. Dots indicate the sequences generated from this study.
The most parsimonious interpretation is that SINV-I strains circulating in central and northern Europe were introduced from central Africa rather than from South Africa, as previously hypothesized (10). Analysis of clade A, which contains most European strains, shows that the basal position was held by a strain from Nedre Dalälven, Sweden (09_M_358; GenBank accession number MK045245), suggesting that SINV-I was first introduced from central Africa to Sweden, with subsequent circulation there, and then further dispersed from Sweden to other parts of northern, eastern, and central Europe (Fig. 1). This was further confirmed by the phylogenetic tree based on amino acid sequences of SINV-I, which had the same topology (Fig. 2).
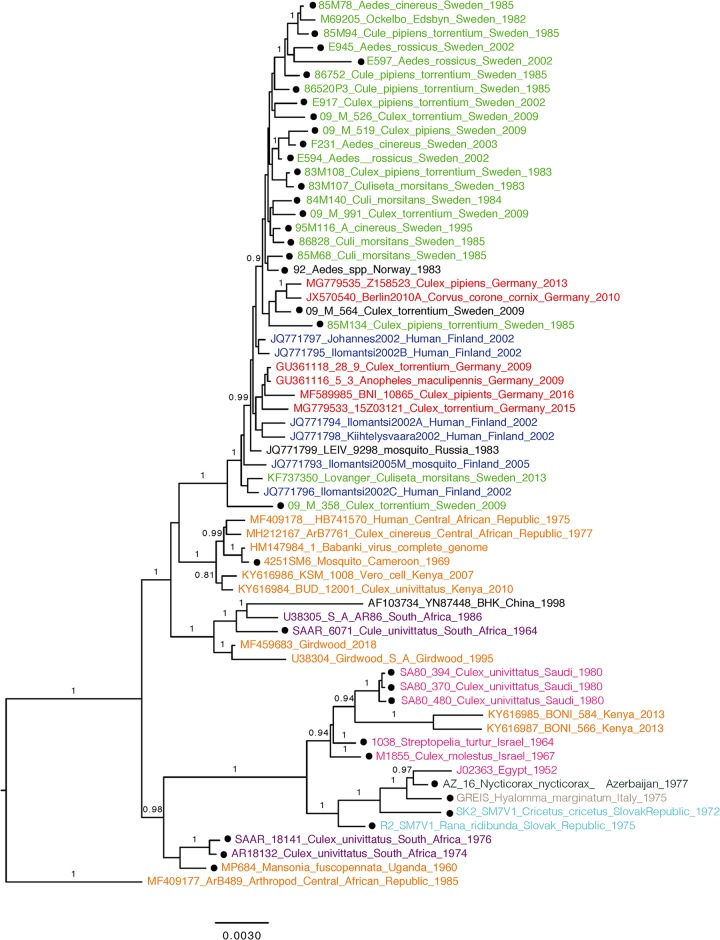
FIG 2.
Bayesian phylogenetic tree of SINV-I based on the complete open reading frame (ORF) amino acid sequences resulting from MrBayes. Dots indicate the sequences generated from this study. Posterior probability values above 0.8 are shown for the branches.
Evolutionary rates.
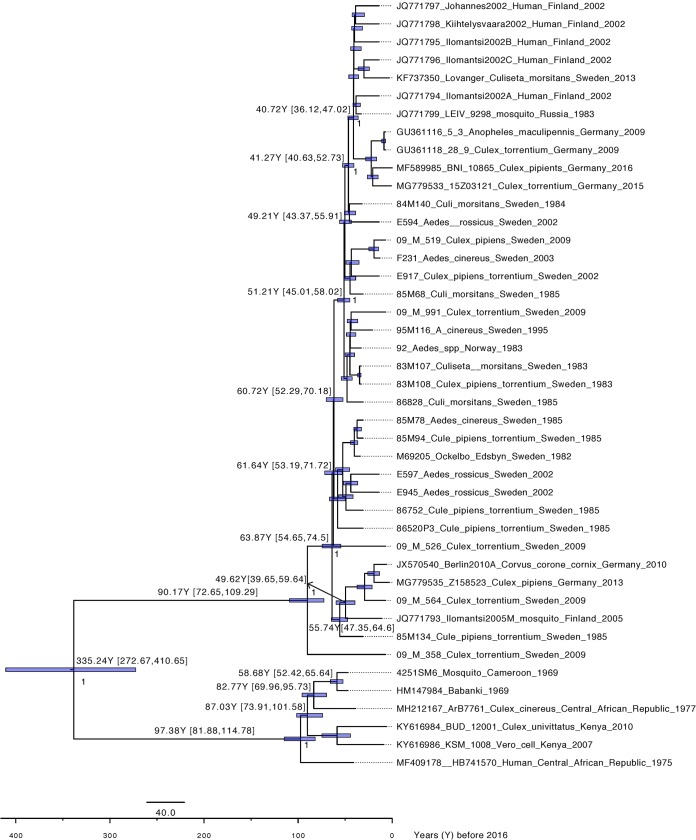
A TempEst analysis showed that the whole data set (64 sequences) did not have any temporal structure (correlation coefficient, −0.0904). However, the data set including sequences from clades A and B showed a temporal signal (43 sequences; correlation coefficient, 0.5128) and was subsequently used to estimate the evolutionary rate of clades A and B. Following model testing, the evolutionary history of clades A and B of SINV-I was reconstructed with a strict molecular clock mode and a coalescent exponential population demographic model. The evolutionary rate for this data set is 5.45 × 10−5 substitutions (95% highest posterior density [HPD], 4.40 × 10−5 to 6.56 × 10−5) (Table 1). The time to the most recent common ancestor (tMRCA) estimated that clade A dated back to 93 years ago (95% HPD, 76 to 112 years), indicating that SINV-I was introduced into Sweden in the 1920s. This was then followed by two separate introductions of SINV-I from Sweden into Finland and Germany around the 1960s and the 1970s, respectively, which corresponds to epidemiological data on SINV infection in northern Europe (Fig. 3). Notably, we also estimate that clade A and clade B diverged from their African ancestors more than 300 years ago (95% HPD, 1605 to 1743) and perhaps longer if the data have been impacted by strong purifying selection (19). Hence, prior to the emergence of SINV-I in northern Europe, there were more than 300 years of viral diversity and spread that are still unaccounted for.
TABLE 1.
Identities of nucleotide and amino acid sequences of SINV-I strains within different clades
| Identity categorya | % identity |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CDS | nsP1 | nsP2 | nsP3 | nsP4 | CP | E3 | E2 | 6K | E1 | |
| Total (nt) | 87.9–100 | 91.8–100 | 85.8–100 | 84.4–100 | 88–100 | 88.7–100 | 86.6–100 | 87.6–99.9 | 88.1–100 | 89.1–100 |
| Total (aa) | 96.3–100 | 97.1–100 | 96.9–100 | 91.1–100 | 97.5–100 | 92.2–100 | 94.5–100 | 95.3–100 | 94.2–100 | 96.2–100 |
| Within clade A (nt) | 99.1–100 | 99.2–100 | 98.9–100 | 99–100 | 98.9–100 | 96.8–100 | 98.8–100 | 99–99.9 | 97.5–100 | 99–100 |
| Within clade A (aa) | 99.1–100 | 98.9–100 | 99.3–100 | 89.3–100 | 99.3–100 | 94.4–100 | 98.2–100 | 99–100 | 96.2–98.1 | 99.3–100 |
| Within clade B (nt) | 99.1–99.9 | 99.1–99.8 | 99.1–99.9 | 99.3–99.9 | 99.1–100 | 98.8–100 | 99.4–100 | 99–99.8 | 98.8–100 | 99.1–99.9 |
| Within clade B (aa) | 99.7–100 | 99.3–99.8 | 99.8–100 | 99.6–100 | 99.7–100 | 99.2–100 | 100 | 99.8–100 | 100 | 99.1–100 |
| Within clade C (nt) | 98.3–99.8 | 98.6–99.9 | 98.1–99.6 | 97.6–99.6 | 98.7–99.7 | 98.7–100 | 97.6–100 | 98.3–99.8 | 95.4–100 | 98.4–100 |
| Within clade C (aa) | 98.9–99.6 | 98.5–100 | 98.5–99.6 | 97.6–99.5 | 98.9–100 | 99.2–100 | 96.4–100 | 98.8–100 | 96.2–100 | 98.6–100 |
| Within clade D (nt) | 87.9–100 | 96.1–100 | 94.3–100 | 95.7–100 | 96.1–100 | 95.7–100 | 97–100 | 94.9–100 | 93.2–100 | 97–100 |
| Within clade D (aa) | 96.3–100 | 97.9–100 | 97.7–100 | 96.6–100 | 98–100 | 97.7–100 | 100 | 97.3–100 | 98.1–100 | 98.6–100 |
| Within clade E (nt) | 99.2–99.6 | 99.4–99.7 | 98.9–99.8 | 98.5–99.3 | 99.3–99.4 | 99.1–99.7 | 98.8–99.4 | 99.3–99.8 | 93.1–98.8 | 99.3–100 |
| Within clade E (aa) | 99.6–99.9 | 99.8–100 | 99.8–100 | 99–99.6 | 99.8–100 | 99.2–100 | 100 | 99.8–100 | 94.2–98.1 | 94.8–100 |
nt, nucleotides; aa, amino acids.
FIG 3.
Dated phylogenetic tree of clades A and B. Posterior probability values above 0.95 are shown in the nodes. The values on the branches indicate years before 2016 and 95% highest posterior density (HPD) values.
Phylogeny of the partial E2 gene.
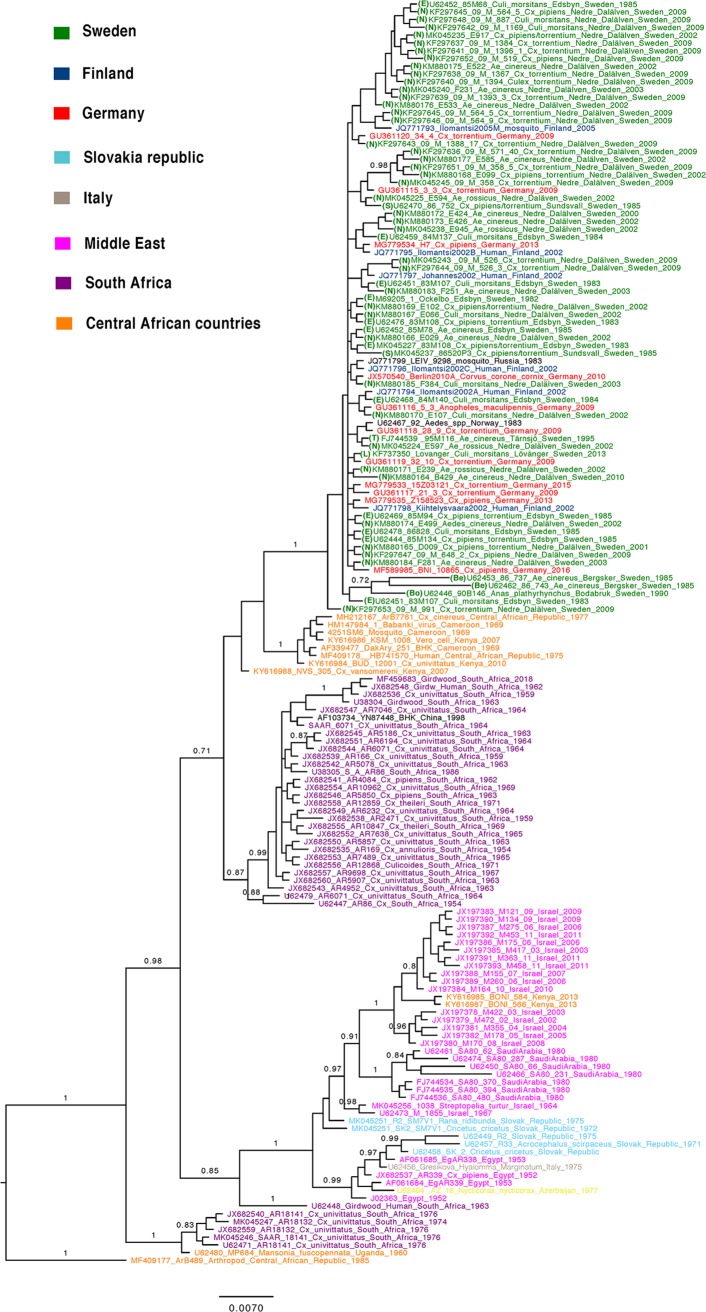
We also performed the same analysis on 163 sequences of the partial E2 alignment (Fig. 4), and the results showed that it could not discriminate any additional within-clade geographical structure compared to the analysis of the complete ORFs. Northern/eastern SINV strains also clustered together in the partial E2 phylogeny. However, it failed to detect the origin of this clade, as both the central African and South African clades were mixed together, and the overall posterior probability support was low.
FIG 4.
Bayesian phylogenetic tree of SINV-I based on partial E2 sequences. Posterior probability values above 0.8 are shown for the branches.
Genetic diversity of SINV-I.
The SINV-I genome was highly conserved, with an average nucleotide similarity of 96.5% (range, 87.9% to 100%). The similarity of the nonstructural protein genes nsP1 (97.4%; range, 91.8% to 100%), nsP2 (96.3%; range, 85.8% to 100%), nsP3 (95.4%; range, 84.4% to 100%), and nsP4 (96.4%; range, 88% to 100%) showed similar levels compared to the structural protein genes capsid (96.7%; range, 88.7% to 100%), E3 (96.7%; range, 86.6% to 100%), E2 (96.2%; range, 87.6% to 99.9%), 6K/TF (96.8%; range, 88.1% to 100%), and E1 (97.5%; range, 89.1% to 100%), with an average nucleotide similarity ranging from 95.4% (nsP3) to 97.5% (E1).
A comparison of nucleotide and amino acid differences showed that the highest degree of observed divergence was found in clade D (Table 1). Within clades A, B, and C, where most strains from regions with reported outbreaks were located, no specific differences could be observed that clearly separated strains isolated from human patients from other strains (Fig. 5). Thus, direct disease association could not be accounted for by any unique amino acid substitution.
FIG 5.
Comparison of amino acid variations across the SINV-I genome among the strains in clades A, B, and C. The colors of the strains indicate Sweden (green), Finland (blue), Germany (red), Kenya and the Central African Republic (orange), and South Africa (purple).
Recombination within SINV-I.
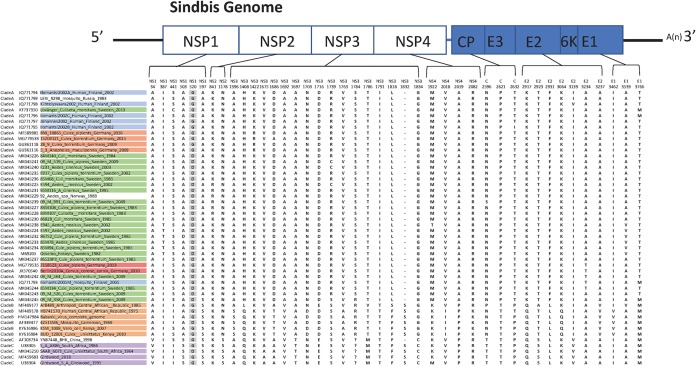
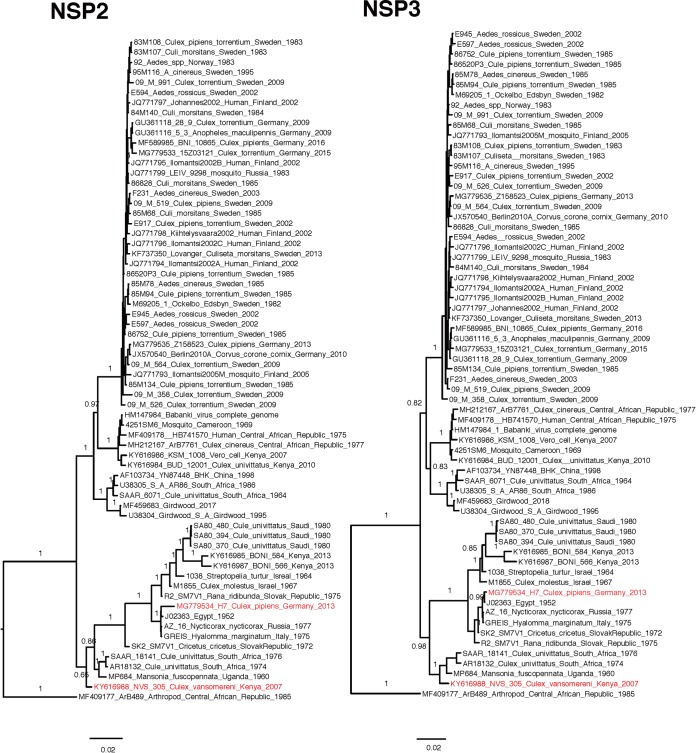
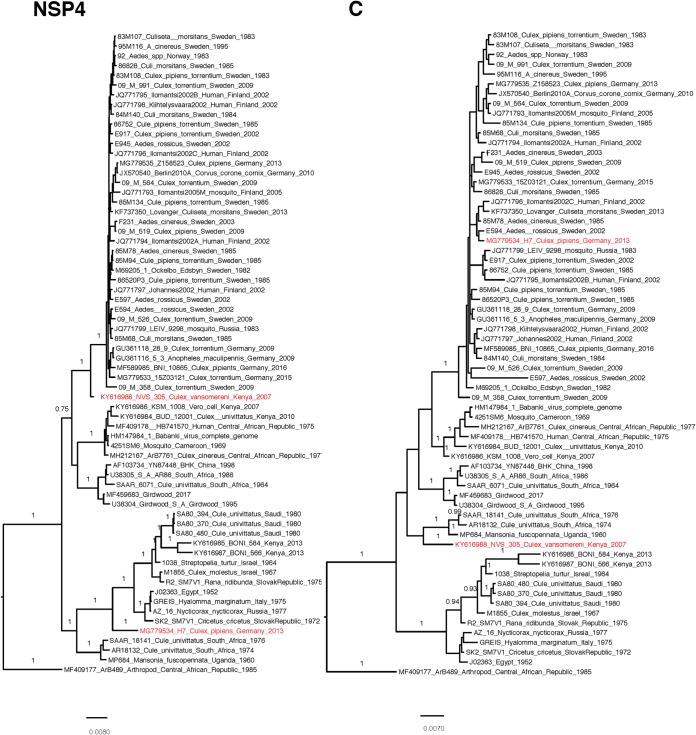
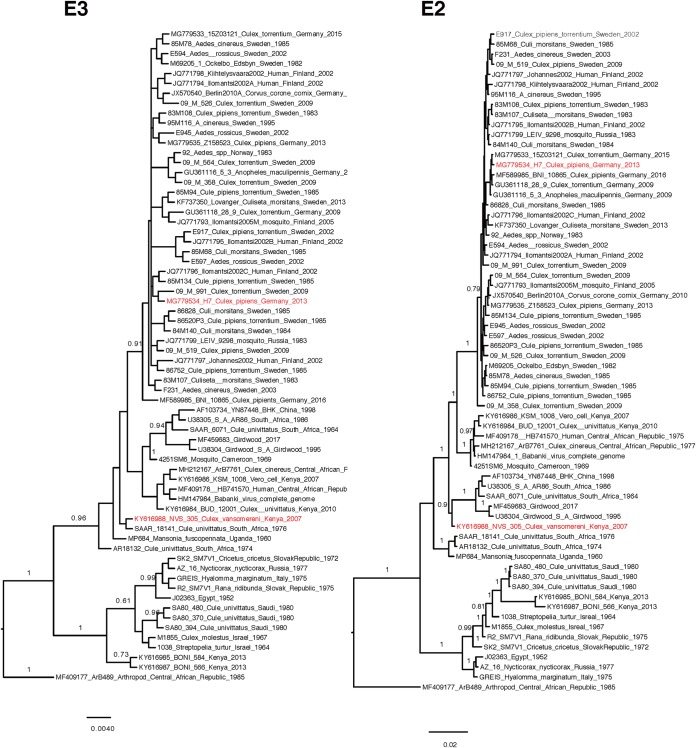
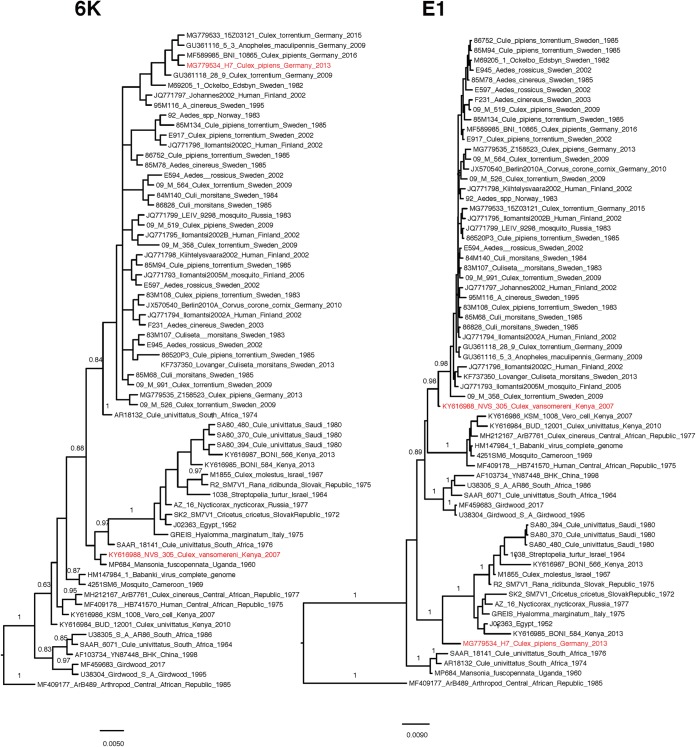
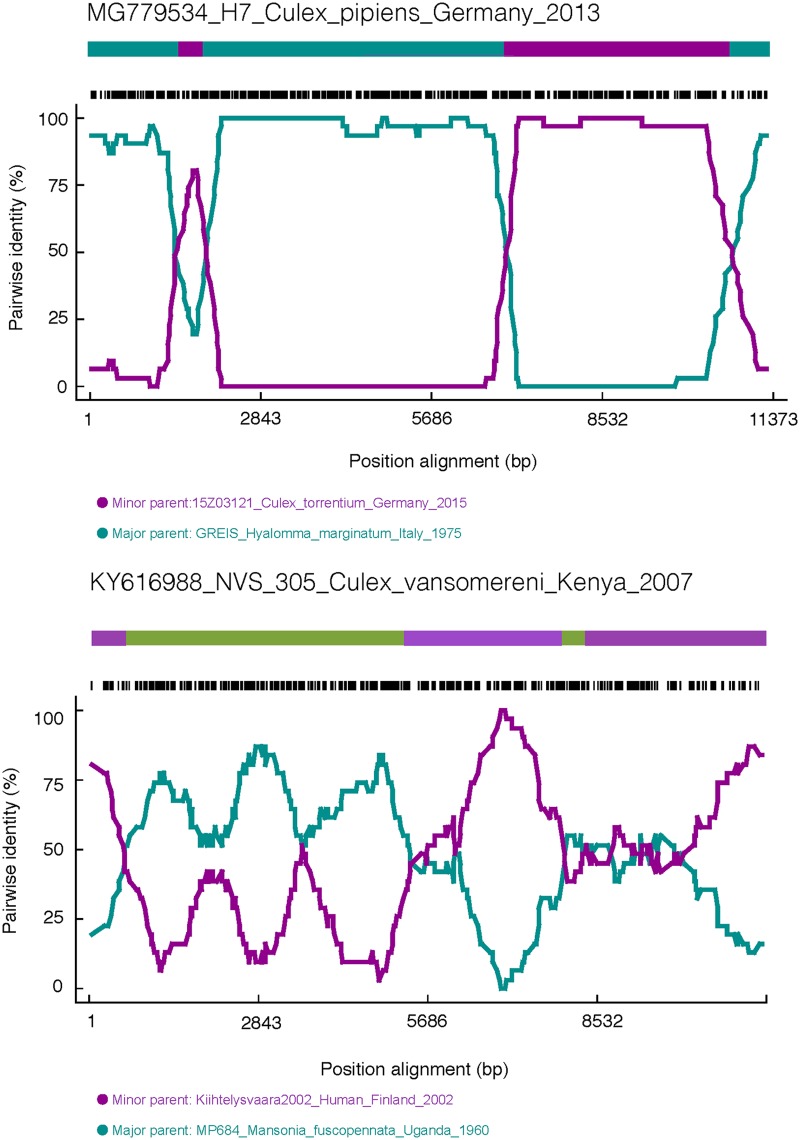
Two recombination events in the alignment of the complete ORFs were found, showing that the strains NVS_305_Culex_vansomereni_Kenya_2007 (GenBank accession number KY616988) and H7_Culex_pipiens_Germany_2013 (GenBank accession number MG779534) have undergone recombination (Fig. 6). Interestingly, the phylogenies of nsP4 and E1 for NVS_305_Culex_vansomereni_Kenya_2007 were incongruous with those of other genes. Based on nsP4 and E1, this strain clustered in clade A together with the strains from northern Europe, whereas based on the other genes, NVS_305_Culex_vansomereni_Kenya_2007 is a member of clade E. For H7_Culex_pipiens_Germany_2013, incongruences of phylogenies were largely based on structural versus nonstructural protein-coding genes. This strain clustered in clade D on the basis of nonstructural genes and the (structural) E1 gene, whereas the other structural genes (C, E3, E2, and 6K/TF) of this strain were found to be more related to the strains from clade A. This recombinant strain indicates a second introduction of SINV-I to Germany, arriving from the south, which recombined with the strain introduced from the north. The potential breakpoints for these two strains were also confirmed by several methods in RDP4 (Table 2 and Fig. 7).
FIG 6.
Bayesian phylogenetic trees based on the complete ORFs and the separated genes (nsP1 to -4, C, E3, E2, 6K/TF, and E1) of fully sequenced SINV-I genomes. Sequences that have undergone recombination are highlighted in red. CDS, coding sequence.
TABLE 2.
Recombination analysis for strains H7_Culex_pipiens_Germany_2013 and NVS_305_Culex_vansomereni_Kenya_2007a
| Recombination strain | Event | Minor parental sequence (breakpoint positions [bp]) | Major parental sequence |
P value determined by detection method |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RDP | GENECONV | Bootscan | Maxchi | Chimera | SiSscan | Phylpro | LARD | 3Seq | ||||
| H7_Culex_pipiens_Germany_2013 | 1 | 15Z03121_Culex_torrentium_Germany_2015 (6897–10655) | GREIS_Hyalomma_marginatum_Italy_1975 | 1.07E−78 | 2.76E−70 | 2.46E−76 | 1.35E−33 | 8.76E−34 | 4.41E−39 | NS | NS | 3.43E−12 |
| E594_Aedes__rossicus_Sweden_2002 | SK2_SM7V1_Cricetus_cricetus_SlovakRepublic_1972 | |||||||||||
| 84M140_Culi_morsitans_Sweden_1984 | J02363_Egypt_1952 | |||||||||||
| 83M108_Culex_pipiens_torrentium_Sweden_1983 | AZ_16_Nycticorax_nycticorax_Russia_1977 | |||||||||||
| 2 | 15Z03121_Culex_torrentium_Germany_2015 (1512–1930) | AZ_16_Nycticorax_nycticorax_Russia_1977 | 1.35E−23 | 2.90E−22 | 6.01E−20 | 1.86E−05 | 1.83E−05 | 4.61E−05 | NS | NS | 3.43E−12 | |
| E594_Aedes_rossicus_Sweden_2002 | SK2_SM7V1_Cricetus_cricetus_SlovakRepublic_a_1972 | |||||||||||
| 84M140_Culi_morsitans_Sweden_1984 | GREIS_Hyalomma_marginatum_Italy_1975 | |||||||||||
| NVS_305_Culex_vansomereni_Kenya_2007 | 1 | Kiihtelysvaara2002_Human_Finland_2002 (5258–7890) | MP684_Mansonia_fuscopennata_Uganda_1960 | 7.77E−06 | 2.56E−06 | 0.001478066 | 1.63E−07 | 0.034855155 | 1.21E−10 | NS | NS | 8.71E−09 |
| E594_Aedes__rossicus_Sweden_2002 | SAAR_18141_Cule_univittatus_South_Afria_1976 | |||||||||||
| 84M140_Culi_morsitans_Sweden_1984 | Culex_univittatus_South_Africa_1974 | |||||||||||
| 2 | JQ771793_Ilomantsi2005M_mosquito_Finland_2005 (8274–596) | SAAR_18141_Cule_univittatus_South_Afria_1976 | 4.45E−06 | 5.18E−05 | 0.000194763 | 2.12E−06 | 2.27E−06 | 3.71E−06 | NS | NS | 0.010431756 | |
| E594_Aedes_rossicus_Sweden_2002 | Culex_univittatus_South_Africa_1974 | |||||||||||
| 84M140_Culi_morsitans_Sweden_1984 | MP684_Mansonia_fuscopennata_Uganda_1960 | |||||||||||
The minor parent is the parent contributing the smaller fraction of sequence. The major parent is the parent contributing the larger fraction of sequence. Breakpoint positions (in base pairs) are relative to E594_Aedes_rossicus_Sweden_2002. NS indicates that no significant P value was recorded for this recombination event using this method.
FIG 7.
Recombination scheme for strains H7_Culex_pipiens_Germany_2013 and NVS_305_Culex_vansomereni_Kenya_2007. The recombination events were checked by using RDP in RDP3, and the window size (the number of polymorphic sites per window) was set to 30.
DISCUSSION
In this study, we have investigated the evolutionary history and dispersal pattern of SINV-I, by phylogenetic analyses of 66 full-genome sequences isolated from regions all throughout its geographical range. The current long-standing hypothesis has been that SINV-I was introduced to northern Europe from South Africa by migratory birds (20–22). This has also been supported by reports of a similar disease occurring in these two regions (12, 18). However, our analyses suggest that the most likely origin of all strains isolated in northern Europe is a single introduction of SINV-I into Sweden from central Africa rather than from South Africa. The specific region from which the virus was exported is, however, uncertain, due to the few and geographically limited isolates of SINV available from Africa. In addition, our results indicate that SINV-I strains were further exported from Sweden to Finland, Russia, and Germany, presenting an eastward and southward dispersal, in contrast to the more commonly proposed northward dispersal of pathogens (23).
The intercontinental dispersals of several bird-hosted viruses, such as WNV and USUV, have been linked to the northward movement of migratory birds (1, 24). In the present study, there is no indication of multiple introductions of SINV-I to northern Europe; thus, northward transport with migratory birds is likely to be a very rare event. This supports conclusions from previous studies on SINV-I phylogeny and the low SINV-I antibody prevalence in northward-migrating birds (20, 25). However, the composition of clade D, containing Middle Eastern and southern/central European strains, implied that southern/central Europe might have had three introductions of SINV-I, two from northern Europe and one from central Africa.
The southward dispersal of SINV-I could be explained by the autumn migration of the main amplifying host species. The enzootic circulation of SINV-I occurs mainly in August, with thrushes of the genus Turdus as the main amplifying host of SINV-I in Sweden (15, 21). These Turdus species breed in Sweden and migrate southward in August to October to spend the winter in central and southwestern Europe (26). The SINV-neutralizing antibody prevalence in redwing (Turdus iliacus), songthrush (Turdus philomelos), and fieldfare (Turdus pilaris) sometimes exceeds 70% in Sweden (27), which indicates that a substantial number of migrating thrushes are likely to include viremic individuals dispersing SINV-I to local mosquito populations on their way south.
The first report of Sindbis fever in humans in Fennoscandia was from Sweden in 1967 (28). However, prior to this report, there were already signs indicating the existence of SINV in Europe: specific anti-SINV antibodies were found in human sera in northern Italy and Finland already in 1965 (29, 30). Antibody screenings of 5,000 human serum samples in Finland in 1958 to 1964 and in Austria before 1963 (31), however, reported that no antibodies were present. Our dating analysis showed that SINV-I was introduced into Sweden in the 1920s and spread east and southward on two separate occasions to Finland and Germany in the 1960s and 1970s. This is well in agreement with epidemiological data from Finland but also indicates that SINV-I was circulating undetected in Sweden for many decades before it was reported to cause disease. After the confirmation of the first SINV human case in Sweden, sporadic cases as well as recurrent outbreaks have been reported annually in Sweden (32) and Finland (33, 34), as have occasional outbreaks of hundreds or even thousands of human cases. In Finland, SINV-I has been a notifiable disease since 1995, and numbers of reported cases are 10-fold higher than in Sweden, where the awareness of Sindbis fever is considerably lower. Further investigations are needed to clarify if this difference is due to underreporting in Sweden or if it reflects real differences in Sindbis fever occurrence. In both countries, however, the introduction of SINV-I was a rare one-time event, and the virus successfully managed to become established in an enzootic cycle locally, with occasional cases of spillover to humans.
Previous studies have used the E2 gene as a proxy for genotyping SINV-I strains (10). This study confirms that it is a good marker for genotyping; however, it has a limited ability to resolve detailed dispersal patterns. Reconstructing the phylogenies based on separate genes also did not improve the resolution of the phylogeny, despite many more sequences being used in the partial E2 phylogeny. Thus, the best-resolved phylogeny is by using concatenated ORF sequences.
We detected two strains (NVS_305_Culex_vansomereni_Kenya_2007 and H7_Culex_pipiens_Germany_2013) that most likely underwent a recombination event. This is, to our knowledge, the first time that a recombination event has been detected for SINV-I in nature. A previous recombination event between a Sindbis-like virus and Eastern equine encephalomyelitis virus (EEEV) in South America has had a large impact on the alphavirus genus, giving rise to WEEV, Highlands J virus, and Fort Morgan virus (35, 36). The ancestral recombinant obtained E1 and E2 proteins from the Sindbis-like virus and the remaining genes from EEEV (35). The three recombinants, and their variants (e.g., Buggy Creek virus, a variant of Fort Morgan virus), are all vector-borne viruses that infect birds in the New World, but only WEEV has a clear association with human disease.
SINV-I, as for all alphaviruses, evolves relatively slowly (5), which is indicated by the comparatively low genetic diversity observed within the clades and by the finding that model selection favored the constant clock model in this study. Our results showed few genetic differences within SINV-I, especially within the clade of northern European strains (99.1 to 100% overall similarity). The E2 gene encodes a glycoprotein that is associated with the pathogenicity of SINV infections (37, 38). Single substitutions can cause considerable differences in arbovirus vector specificity and pathogenicity, as shown for chikungunya virus (CHIKV) and Zika virus (39, 40). However, we could not detect any clear nucleotide, or any amino acid, differences between SINV-I strains from regions with disease and those without disease (Fig. 5). The reasons for the lack of reported disease cases can be many. In central Africa, several severe diseases are endemic, and underdetection and underreporting would be highly likely. An alternative explanation could be that various ecological factors have an impact on the efficiency of disease transmission, such as a lack of competent and abundant vectors. When considering the lack of human cases in central European countries, this might be a plausible explanation, as studies on human antibodies against SINV-I showed a very low prevalence (41). Studies on antibody prevalence in regions of endemicity present 3% in Sweden and 9% in Finland (25, 32, 42). Thus, humans in Germany do not seem to become infected with SINV-I as often as humans do in areas of endemicity of Fennoscandia. Ecological studies have shown that the abundance of the most competent vector of SINV-I, Culex torrentium, is decreasing southward in Europe (43, 44). One could speculate that there is a critical abundance of Cx. torrentium that needs to be reached for an efficient-enough transmission of SINV-I in the bird population to allow spillover infections of humans. Likewise, the abundance of the bridge vector Aedes cinereus is likely of importance for the transmission of virus from birds to humans (3). Other ecological factors, such as variations in host choice by the vector and mosquito longevity, could also contribute to explain the differences in transmission. In summary, the geographical limitation of SINV fever outbreaks needs to be further investigated in studies on the pathogenicity of the different strains as well as in ecological studies on the vector and host populations in different regions.
In conclusion, our results suggest that SINV-I was successfully introduced only once into northern Europe from central Africa, probably via migratory birds. This introduction led to the establishment of endemic SINV circulation in Sweden, and from there, SINV-I spread to other parts of northern, eastern, and central Europe. The emergence of reported human cases is potentially due to synergistic effects, including awareness of the disease in the community, ecological circumstances, and undiscovered host and viral genetic factors.
MATERIALS AND METHODS
Virus strains.
SINV strains for the present study are listed in Table 3 and were originally isolated from a number of sources, including mosquitoes, birds, and humans (e.g., see references 10, 20, and 45; J. O. Lundström, J. Hesson, M. Schafer, O. Ostman, T. Semmler, M. Weidmann, and M. Pfeffer, unpublished data). All strains sequenced in this study were propagated in Vero cells as described previously by Hesson et al. (45) or in suckling mice as described previously (46) and stored at −80°C.
TABLE 3.
Sindbis virus genotype I strains included in this study
| Isolate | Yr of isolation | Location(s) (region)a | Isolation sourceb | E2 GenBank accession no. | GenBank accession no. | Reference(s) |
|---|---|---|---|---|---|---|
| E597 | 2002 | Sweden (N) | Aedes rossicus | MK045224 | This study | |
| E594 | 2002 | Sweden (N) | A. rossicus | MK045225 | This study | |
| 84M140 | 1984 | Sweden (E) | Culiseta morsitans | MK045226 | This study | |
| 83M108 | 1983 | Sweden (E) | Culex pipiens/Cx. torrentium | MK045227 | This study | |
| 83M107 | 1983 | Sweden (E) | Cs. morsitans | MK045228 | This study | |
| Vmork1/92 | 1983 | Norway | Aedes spp. | U62467 | MK045229 | This study, 22 |
| 86828 | 1985 | Sweden (E) | Cs. morsitans | MK045230 | This study | |
| 95M116 | 1995 | Sweden (N) | A. cinereus | FJ744539 | MK045231 | This study, 10 |
| 86752 | 1985 | Sweden (S) | Cx. pipiens/Cx. torrentium | MK045232 | This study | |
| 85M78 | 1985 | Sweden (E) | A. cinereus | U62460 | MK045233 | This study, 22 |
| 85M94 | 1985 | Sweden (E) | Cx. pipiens/Cx. torrentium | U62469 | MK045234 | This study, 22 |
| E917 | 2002 | Sweden (N) | Cx. pipiens/Cx. torrentium | MK045235 | This study | |
| 85M68 | 1985 | Sweden (E) | Cs. morsitans | U62452 | MK045236 | This study, 22 |
| 86520P3 | 1985 | Sweden (S) | Cx. pipiens/Cx. torrentium | MK045237 | This study | |
| E945 | 2002 | Sweden (N) | A. rossicus | MK045238 | This study | |
| 09_M_991 | 2009 | Sweden (N) | Cx. torrentium | KF297653 | MK045239 | This study, 45 |
| F231 | 2003 | Sweden (N) | A. cinereus | MK045240 | This study | |
| 09_M_519 | 2009 | Sweden (N) | Cx. pipiens | KF297652 | MK045241 | This study, 45 |
| 09_M_564 | 2009 | Sweden (N) | Cx. torrentium | KF297646 | MK045242 | This study, 45 |
| 09_M_526 | 2009 | Sweden (N) | Cx. torrentium | KF297644 | MK045243 | This study, 45 |
| 85M134 | 1985 | Sweden (E) | Cx. pipiens/Cx. torrentium | U62444 | MK045244 | This study |
| 09_M_358 | 2009 | Sweden (N) | Cx. torrentium | KF297651 | MK045245 | This study, 45 |
| SAAR_18141 | 1976 | South Africa | Culex univittatus | U62471 | MK045246 | This study, 22 |
| AR18132 | 1974 | South Africa | Cx. univittatus | U62463 | MK045247 | This study, 22 |
| MP684 | 1960 | Uganda | Mansonia fuscopennata | U62480 | MK045248 | This study, 22 |
| 4251SM6 | 1969 | Cameroon | Mosquito | AF339477 | MK045249 | This study; R. M. Kinney and M. Pfeffer, unpublished data |
| SAAR_6071 | 1964 | South Africa | Cx. univittatus | U62479 | MK045250 | This study, 22 |
| SK2_SM7V1 | 1972 | Slovakia | Cricetus cricetus (M) | U62458 | MK045251 | This study, 22 |
| AZ_16 | 1977 | Russia, Azerbaijan | Nycticorax nycticorax (B) | U62464 | MK045252 | This study, 22 |
| GREIS | 1975 | Italy | Hyalomma marginatum (T) | U62456 | MK045253 | This study, 22 |
| M1855 | 1967 | Israel | Cx. pipiens molestus | U62473 | MK045254 | This study, 22 |
| R2_SM7V1 | 1975 | Slovakia | Rana ridibunda (A) | U62459 | MK045255 | This study, 22 |
| 1038 | 1964 | Israel | Streptopelia turtur (B) | U62443 | MK045256 | This study, 22 |
| SA80_480 | 1980 | Saudi Arabia | Cx. univittatus | FJ744536 | MK045257 | This study, 10 |
| SA80_394 | 1980 | Saudi Arabia | Cx. univittatus | FJ744535 | MK045258 | This study, 10 |
| SA80_370 | 1980 | Saudi Arabia | Cx. univittatus | FJ744534 | MK045259 | This study, 10 |
| BNI_10865 | 2016 | Germany | Cx. pipiens | MF589985 | 6 | |
| 15Z03121 | 2015 | Germany | Cx. torrentium | MG779533 | 6 | |
| LEIV_9298 | 1983 | Russia | Mosquito | JQ771799 | 17 | |
| Kiihtelysvaara2002 | 2002 | Finland | Homo sapiens | JQ771798 | 17 | |
| Johannes2002 | 2002 | Finland | Homo sapiens | JQ771797 | 17 | |
| Edsbyn | 1982 | Sweden | Mosquito | M69205 | 58 | |
| Ilomantsi2002B | 2002 | Finland | Homo sapiens | JQ771795 | 17 | |
| Ilomantsi2002A | 2002 | Finland | Homo sapiens | JQ771794 | 17 | |
| 28_9 | 2009 | Germany | Cx. torrentium | GU361118 | 41 | |
| 5_3 | 2009 | Germany | Anopheles maculipennis | GU361116 | 41 | |
| Ilomantsi2002C | 2002 | Finland | Homo sapiens | JQ771796 | 17 | |
| Lovanger | 2013 | Sweden (L) | Cs. morsitans | KF737350 | 16 | |
| Z158523 | 2013 | Germany | Cx. pipiens | MG779535 | 6 | |
| Berlin2010A | 2010 | Germany | Corvus corone cornix (B) | JX570540 | 59 | |
| Ilomantsi2005M | 2005 | Finland | Mosquito | JQ771793 | 17 | |
| NVS_305 | 2007 | Kenya | Culex vansomereni | KY616988 | 60 | |
| ArB7761 | 1977 | Central African Republic | Cx. cinereus | MH212167 | 46 | |
| HB741570 | 1975 | Central African Republic | Homo sapiens | MF409178 | V. Tricou, E. Nakoune, B. Selekon, M. Kazanji, and N. Berthet, unpublished data | |
| Babanki | 1969 | Cameroon | Mosquito | HM147984 | 47 | |
| KSM_1008 | 2007 | Kenya | Culex spp. | KY616986 | 60 | |
| BUD_12001 | 2010 | Kenya | Cx. univittatus | KY616984 | 60 | |
| Girdwood_2017 | 1962 | South Africa | Homo sapiensc | MF459683 | 61 | |
| Girdwood_SA | 1962 | South Africa | Homo sapiensc | U38304 | 62 | |
| YN87448 | 1998 | China | Unknown | AF103734 | G. L. Zhou, G. D. Liang, L. Li, S. H. Fu, Q. Jin, H. L. Zhang, W. L. Huang, and Y. D. Hou, unpublished data | |
| Prototype AR399 | 1952 | Egypt | Cx. pipiens/Cx. univittatus | J02363 | 63 | |
| S_A_AR86 | 1954 | South Africa | Culex spp. | U38305 | 62 | |
| H7 | 2013 | Germany | Cx. pipiens | MG779534 | 6 | |
| BONI_584 | 2013 | Kenya | Aedes tricholabis | KY616985 | 60 | |
| BONI_566 | 2013 | Kenya | A. ochraceus | KY616987 | 60 | |
| ArB489 | 1985 | Central African Republic | Arthropod | MF409177 |
Letters in parentheses indicate regions within Sweden shown in Fig. 1 (N, Nedre Dalälven; E, Edsbyn; S, Sundsvall; L, Lövånger).
Letters in parentheses indicate the isolation source M, mammal; B, bird; T, tick; A, amphibian.
This strain was isolated from a human in 1962 and sequenced twice.
RNA extraction and sequencing.
Total RNA of the SINV-infected cell culture supernatant was extracted using the QIAamp viral RNA minikit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. cDNA was synthesized using the RevertAid H minus first-strand cDNA synthesis kit (Thermo Scientific, Vilnius, Lithuania) in a 20-μl mixture containing 2 μl of random hexamers (10 pmol/μl), 2 μl 10 M deoxynucleoside triphosphate (dNTP), and 5 μl RNA. The presence of SINV RNA was confirmed by quantitative PCR (qPCR) as described previously (41).
PCRs, using modified oligonucleotide primers that cover the complete genome of SINV (17) (Table 4), were performed using Phusion Flash (Thermo Scientific, Lithuania). PCR amplicons from the same strain were pooled and purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Illumina sequencing libraries were constructed using multiplex PCR products (1 ng of input DNA) and the Nextera XT DNA library prep kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Sequencing was performed on an Illumina MiSeq instrument using MiSeq reagent kit version 2 (Illumina, San Diego, CA, USA). Assembly of the sequence data was done using the CLC genome workbench, using the SINV Babanki strain (GenBank accession number HM147984) as the reference sequence (47). Low-coverage regions were closed by conventional PCR, with primers designed according to known sequences in the flanking regions.
TABLE 4.
Primers used in this study
| Oligonucleotide | Sequence | Positionsa | Length of the product (bp) |
|---|---|---|---|
| SINV F1 | GAATCRAACAGCCGACCAAT | 25–44 | 641 |
| SINV R1 | GTCGGCCCAGTTAGTGTTGT | 646–665 | |
| SINV F2 | ACTGGATTGGCTTTGACACC | 583–602 | 485 |
| SINV R2 | CGGGATATACGTGCACACAG | 1048–1067 | |
| SINV F3 | CAAACAATAGCGAGGGCTTC | 979–998 | 793 |
| SINV R3 | GAGGTTGGTGAAACGACGAT | 1752–1771 | |
| SINV F4 | TGGCAGACAAAGACATCGAG | 1606–1625 | 681 |
| SINV R4 | GTGCCGTGACAGTTGACTTG | 2267–2286 | |
| SINV F5 | GCGATGCGTTAAGAAGGAAG | 2096–2115 | 1,088 |
| SINV R5 | ACTGGCAACCGGTAAGTACG | 3164–3183 | |
| SINV F6 | GAGGACTGGGAAGCTGAACA | 3030–3049 | 1,062 |
| SINV R6 | TCCRTCTCTTGTACCCTCRT | 4072–4091 | |
| SINV F7 | GRCCAGAKTGYGTCTCAAGCA | 3959–3978 | 600 |
| SINV R7 | TCGATTCGTTCCTTCCACTT | 4539–4558 | |
| SINV F8 | ATCAAGTCTGTCGCCATTCC | 4401–4420 | 1,086 |
| SINV R8 | CATGGATACCCCACCAAAAG | 5467–5486 | |
| SINV F9 | TTTAGCGGATCGGACAATTC | 5238–5257 | 1,298 |
| SINV R9 | GCGGTGACGAACTCAGTAG | 6517–6535 | |
| SINV F10 | CTGGAYTCAGCGACATTCAA | 6423–6442 | 1,127 |
| SINV R10 | TTGCTCTGGGCAAAAGTTCT | 7530–7549 | |
| SINV F11 | GCCCTGCTAGATGAAACGAA | 7416–7435 | 1,171 |
| SINV R11 | TATCGTAGGCCTCGTGGTTC | 8567–8586 | |
| SINV F12 | GGATAACTCAGGTCGGGTTG | 8312–8331 | 768 |
| SINV R12 | TTACCGTGAACGGGAGGTAG | 9060–9079 | |
| SINV F13 | AGCGTGACGGTTAGCATAGC | 8964–8983 | 909 |
| SINV R13 | CAGCGAAGTTGGAATTACGG | 9853–9872 | |
| SINV F14 | TACCATCGCCATCCTGTGTA | 9714–9733 | 949 |
| SINV R14 | CTGGTTTCATCGCTCCGTAT | 10 643–10 662 | |
| SINV F15 | ACGGAGTTACACCAGGAACG | 10 516–10 535 | 1,121 |
| SINV R15 | TATGCACCAYGCTTCCTCAG | 11 617–11 636 |
The reference genome was from reference 17.
Phylogenetic analysis.
Publicly available full-genome sequences of SINV were retrieved from the National Center for Biotechnology Information (NCBI) website (https://www.ncbi.nlm.nih.gov/). All sequences, 66 in total, were then aligned using MAFFT with default settings, followed by manual refinement using AliView (48, 49).
Several alignments were constructed based on (i) the complete open reading frames (ORFs), (ii) the single genes (nsP1 to -4, C, E1, E2, E3, and 6K/TF), (iii) complete and partial E2 sequences, and (iv) the complete ORFs excluding recombinant sequences. The best-fit evolutionary nucleotide substitution model following jModelTest analysis, GTR+F+I+G4 (general time-reversible model with empirical base frequencies, allowing for a proportion of invariable sites, and a discrete gamma model with default 4 rate categories), was used in all phylogenetic analyses (50). Potential recombination events were investigated using the Phi test in SPLITS TREE 4.0 (51), Simplot version 3.5.1 (52), and RDP3 (53). Recombination events were determined using RDP, GENECONV, Bootscan, Maxchi, Chimera, SiSscan, Phylpro, LARD, and 3Seq. In addition, alignment 4 was translated into amino acids, and amino acid positions were compared using AliView.
To reconstruct the evolutionary history of the SINV-I complex, Bayesian phylogenetic trees of the complete ORFs and separated genes (nsP1 to -4, C, E3, E2, 6K/TF, and E1) of SINV-I were constructed by employing MrBayes v.3.2.6 (54). Bayesian analysis consisted at least 5 million Bayesian Monte Carlo Markov chain (MCMC) generations sampling every 1,000 generations. The run was continued until convergence was obtained (average deviation, <0.01) and with a 25% burn-in. To further infer the evolutionary rates and divergence time of SINV-I, we first performed a regression of root-to-tip genetic distances against date of sampling by using TempEst (55). The whole data set (64 sequences) showed no clocklike structure (correlation coefficient, −0.0904). Thus, only the sequences from clades A and B (43 sequences; correlation coefficient, 0.5128) were used for the evolutionary rate estimation. We employed six different combinations of demographic and molecular clock models (S2) and ran 50 million Bayesian MCMC generations sampling every 1,000 generations, implemented in BEAST version 2.3.1 (56) (Table 5). Model comparison performed by using a marginal-likelihood estimator in two approaches, path sampling (PS) and stepping-stone sampling (SS), selected strict clock and exponential population as a better model for data analysis, with the log Bayesian factor (BF) value over at least 25. In all analyses, strain ArB489 from central Africa, isolated in 1985 (GenBank accession number MF409177), was used to root the tree. All computations were run using the CIPRES computational cluster (http://www.phylo.org/index.php/). Finally, trees were viewed and edited using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
TABLE 5.
Model comparison by BEAST analysisa
| Molecular clock model | Demographic model | Mutation rate | 95% HPD | Log marginal likelihood (SS) | Log marginal likelihood (PS) |
|---|---|---|---|---|---|
| Relaxed clock, exponential | Coalescent exponential population | 1.045 × 10−4 | 5.6508 × 10−5, 1.6026 × 10−4 | −23,632.61 | −23,632.33 |
| Relaxed clock, log normal | Coalescent exponential population | 1.5723 × 10−4 | 6.6297 × 10−5, 2.6341 × 10−4 | −23,632.24 | −23,631.94 |
| Relaxed clock, exponential | Coalescent Bayesian skyline | 1.225 × 10−4 | 6.8056 × 10−5, 1.8561 × 10−4 | −23,622.27 | −23,621.7 |
| Relaxed clock, normal | Coalescent Bayesian skyline | 1.257 × 10−4 | 7.4641 × 10−5, 1.8208 × 10−4 | −23,623.27 | −23,623.38 |
| Strict | Coalescent exponential population | 5.4535 × 10−5 | 4.3983 × 10−5, 6.5605 × 10−5 | −23,757.07 | −23,756.55 |
| Strict | Coalescent Bayesian skyline | 5.476 × 10−5 | 4.2365 × 10−5, 6.7008 × 10−5 | −23,746.58 | −23,746.95 |
95% HPD, 95% highest posterior density; SS, stepping-stone sampling; PS, path sampling.
A number of studies have previously used partial E2 sequences for creating SINV phylogenies (10, 22, 57). To investigate whether phylogenies based on partial E2 sequences give the same results as the phylogenies based on the complete ORF, and to be able to compare more strains, we also constructed phylogenies using all strains sequenced in this study and all available partial E2 sequences from GenBank (170 in total [minimum, 313 nucleotides {nt}; maximum, 2,200 nt]) (10).
Data availability.
All newly sequenced strains have been deposited in GenBank under accession numbers MK045224 to MK045259.
ACKNOWLEDGMENTS
We thank Ge Mengyun for technical assistance.
This study was supported by the Swedish Research Council (2017-05807). J.C.H. was supported by the Swedish Society for Medical Research and E. and R. Börjeson’s Foundation. J.H.-O.P. was supported by FORMAS grant 2015-710.
REFERENCES
- 1.Engel D, Jöst H, Wink M, Borstler J, Bosch S, Garigliany MM, Jost A, Czajka C, Luhken R, Ziegler U, Groschup MH, Pfeffer M, Becker N, Cadar D, Schmidt-Chanasit J. 2016. Reconstruction of the evolutionary history and dispersal of Usutu virus, a neglected emerging arbovirus in Europe and Africa. mBio 7:e01938-15. doi: 10.1128/mBio.01938-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Go YY, Balasuriya UB, Lee CK. 2014. Zoonotic encephalitides caused by arboviruses: transmission and epidemiology of alphaviruses and flaviviruses. Clin Exp Vaccine Res 3:58–77. doi: 10.7774/cevr.2014.3.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lundström JO. 1999. Mosquito-borne viruses in western Europe: a review. J Vector Ecol 24:1–39. [PubMed] [Google Scholar]
- 4.Petersen LR, Brault AC, Nasci RS. 2013. West Nile virus: review of the literature. JAMA 310:308–315. doi: 10.1001/jama.2013.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan Y, Lam TT-Y, Heberlein-Larson LA, Smole SC, Auguste AJ, Hennigan S, Halpin RA, Fedorova N, Puri V, Stockwell TB, Shilts MH, Andreadis T, Armstrong PM, Tesh RB, Weaver SC, Unnasch TR, Ciota AT, Kramer LD, Das SR. 2018. Large-scale complete-genome sequencing and phylodynamic analysis of Eastern equine encephalitis virus reveals source-sink transmission dynamics in the United States. J Virol 92:e00074-18. doi: 10.1128/JVI.00074-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheuch DE, Schäfer M, Eiden M, Heym EC, Ziegler U, Walther D, Schmidt-Chanasit J, Keller M, Groschup MH, Kampen H. 2018. Detection of Usutu, Sindbis, and Batai viruses in mosquitoes (Diptera: Culicidae) collected in Germany, 2011–2016. Viruses 10:E389. doi: 10.3390/v10070389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogels CB, Göertz GP, Pijlman GP, Koenraadt CJ. 2017. Vector competence of European mosquitoes for West Nile virus. Emerg Microbes Infect 6:e96. doi: 10.1038/emi.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeller H. 2012. Is Japanese encephalitis emerging in Europe? Euro Surveill 17(32):pii=20242 https://www.eurosurveillance.org/content/10.2807/ese.17.32.20242-en. [DOI] [PubMed] [Google Scholar]
- 9.Saikku P. 1984. Alphaviruses in Europe. Academy of Medical Sciences, Petrozavodsk, former Soviet Union. [Google Scholar]
- 10.Lundström JO, Pfeffer M. 2010. Phylogeographic structure and evolutionary history of Sindbis virus. Vector Borne Zoonotic Dis 10:889–907. doi: 10.1089/vbz.2009.0069. [DOI] [PubMed] [Google Scholar]
- 11.Taylor RM, Hurlbut HS, Work TH, Kingston JR, Frothingham TE. 1955. Sindbis virus: a newly recognized arthropod transmitted virus. Am J Trop Med Hyg 4:844–862. doi: 10.4269/ajtmh.1955.4.844. [DOI] [PubMed] [Google Scholar]
- 12.Gylfe Å, Ribers Å, Forsman O, Bucht G, Alenius GM, Wallberg-Jonsson S, Ahlm C, Evander M. 2018. Mosquitoborne Sindbis virus infection and long-term illness. Emerg Infect Dis 24:1141–1142. doi: 10.3201/eid2406.170892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skogh M, Espmark A. 1982. Ockelbo disease: epidemic arthritis-exanthema syndrome in Sweden caused by Sindbis-virus like agent. Lancet i:795–796. [DOI] [PubMed] [Google Scholar]
- 14.Espmark A, Niklasson B. 1984. Ockelbo disease in Sweden: epidemiological, clinical, and virological data from the 1982 outbreak. Am J Trop Med Hyg 33:1203–1211. doi: 10.4269/ajtmh.1984.33.1203. [DOI] [PubMed] [Google Scholar]
- 15.Lundström JO, Vene S, Saluzzo JF, Niklasson B. 1993. Antigenic comparison of Ockelbo virus isolates from Sweden and Russia with Sindbis virus isolates from Europe, Africa, and Australia: further evidence for variation among alphaviruses. Am J Trop Med Hyg 49:531–537. doi: 10.4269/ajtmh.1993.49.531. [DOI] [PubMed] [Google Scholar]
- 16.Bergqvist J, Forsman O, Lärsson P, Naslund J, Lilja T, Engdahl C, Lindström A, Gylfe Å, Ahlm C, Evander M, Bucht G. 2015. Detection and isolation of Sindbis virus from mosquitoes captured during an outbreak in Sweden, 2013. Vector Borne Zoonotic Dis 15:133–140. doi: 10.1089/vbz.2014.1717. [DOI] [PubMed] [Google Scholar]
- 17.Sane J, Kurkela S, Putkuri N, Huhtamo E, Vaheri A, Vapalahti O. 2012. Complete coding sequence and molecular epidemiological analysis of Sindbis virus isolates from mosquitoes and humans, Finland. J Gen Virol 93:1984–1990. doi: 10.1099/vir.0.042853-0. [DOI] [PubMed] [Google Scholar]
- 18.Storm N, Weyer J, Markotter W, Kemp A, Leman PA, Dermaux-Msimang V, Nel LH, Paweska JT. 2014. Human cases of Sindbis fever in South Africa, 2006–2010. Epidemiol Infect 142:234–238. doi: 10.1017/S0950268813000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forrester NL, Wertheim JO, Dugan VG, Auguste AJ, Lin D, Adams AP, Chen R, Gorchakov R, Leal G, Estrada-Franco JG, Pandya J, Halpin RA, Hari K, Jain R, Stockwell TB, Das SR, Wentworth DE, Smith MD, Kosakovsky Pond SL, Weaver SC. 2017. Evolution and spread of Venezuelan equine encephalitis complex alphavirus in the Americas. PLoS Negl Trop Dis 11:e0005693. doi: 10.1371/journal.pntd.0005693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francy DB, Jaenson TG, Lundström JO, Schildt EB, Espmark A, Henriksson B, Niklasson B. 1989. Ecologic studies of mosquitoes and birds as hosts of Ockelbo virus in Sweden and isolation of Inkoo and Batai viruses from mosquitoes. Am J Trop Med Hyg 41:355–363. doi: 10.4269/ajtmh.1989.41.355. [DOI] [PubMed] [Google Scholar]
- 21.Lundström JO, Lindström KM, Olsen B, Dufva R, Krakower DS. 2001. Prevalence of Sindbis virus neutralizing antibodies among Swedish passerines indicates that thrushes are the main amplifying hosts. J Med Entomol 38:289–297. doi: 10.1603/0022-2585-38.2.289. [DOI] [PubMed] [Google Scholar]
- 22.Norder H, Lundström JO, Kozuch O, Magnius LO. 1996. Genetic relatedness of Sindbis virus strains from Europe, Middle East, and Africa. Virology 222:440–445. doi: 10.1006/viro.1996.0441. [DOI] [PubMed] [Google Scholar]
- 23.Randolph SE, Rogers DJ. 2010. The arrival, establishment and spread of exotic diseases: patterns and predictions. Nat Rev Microbiol 8:361–371. doi: 10.1038/nrmicro2336. [DOI] [PubMed] [Google Scholar]
- 24.Charrel RN, Brault AC, Gallian P, Lemasson JJ, Murgue B, Murri S, Pastorino B, Zeller H, de Chesse R, de Micco P, de Lamballerie X. 2003. Evolutionary relationship between Old World West Nile virus strains. Evidence for viral gene flow between Africa, the Middle East, and Europe. Virology 315:381–388. doi: 10.1016/S0042-6822(03)00536-1. [DOI] [PubMed] [Google Scholar]
- 25.Kurkela S, Ratti O, Huhtamo E, Uzcátegui NY, Nuorti JP, Laakkonen J, Manni T, Helle P, Vaheri A, Vapalahti O. 2008. Sindbis virus infection in resident birds, migratory birds, and humans, Finland. Emerg Infect Dis 14:41–47. doi: 10.3201/eid1401.070510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snow DW, Perrins C (ed). 1999. The birds of the western Palearctic, the concise edition, vol 2 Passerines. Oxford University Press, New York, NY. [Google Scholar]
- 27.Hesson JC, Lundström JO, Tok A, Östman Ö, Lundkvist Å. 2016. Temporal variation in Sindbis virus antibody prevalence in bird hosts in an endemic area in Sweden. PLoS One 11:e0162005. doi: 10.1371/journal.pone.0162005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skogh M, Westermark SE. 1973. August exanthema with arthralgia. Abstr Annu Meet Swedish Dermatol Soc, Linköping, Sweden. [Google Scholar]
- 29.Balducci M, Verani P, Lopes MC, Gregorig B. 1967. Survey for antibodies against arthropod-borne viruses in man and animals in Italy. II. Serologic status of human beings in a northern Italian region (Gorizia province). Am J Trop Med Hyg 16:211–215. doi: 10.4269/ajtmh.1967.16.211. [DOI] [PubMed] [Google Scholar]
- 30.Saikku P. 1969. Occurrence of group A hemagglutination inhibiting activity in Finnish serum material, 375–378. In Arboviruses of the California Complex and the Bunyamwera Group: Proceedings of the Symposium Held at Smolenice Near Bratislava October 18–21, 1966 Slovak Academy of Sciences, Bratislava, Czechoslovakia. [Google Scholar]
- 31.Kunz C. 1963. Demonstration of hemagglutination-inhibiting arborvirus antibodies in the population of Austria. Zentralbl Bakteriol Orig 190:174–182. (In German.) [PubMed] [Google Scholar]
- 32.Lundström JO, Vene S, Espmark A, Engvall M, Niklasson B. 1991. Geographical and temporal distribution of Ockelbo disease in Sweden. Epidemiol Infect 106:567–574. doi: 10.1017/s0950268800067637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brummer-Korvenkontio M, Vapalahti O, Kuusisto P, Saikku P, Manni T, Koskela P, Nygren T, Brummer-Korvenkontio H, Vaheri A. 2002. Epidemiology of Sindbis virus infections in Finland 1981-96: possible factors explaining a peculiar disease pattern. Epidemiol Infect 129:335–345. doi: 10.1017/S0950268802007409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sane J, Guedes S, Kurkela S, Lyytikäinen O, Vapalahti O. 2010. Epidemiological analysis of mosquito-borne Pogosta disease in Finland, 2009. Euro Surveill 15(2):pii=19462 https://www.eurosurveillance.org/content/10.2807/ese.15.02.19462-en. [DOI] [PubMed] [Google Scholar]
- 35.Hahn CS, Lustig S, Strauss EG, Strauss JH. 1988. Western equine encephalitis virus is a recombinant virus. Proc Natl Acad Sci U S A 85:5997–6001. doi: 10.1073/pnas.85.16.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allison AB, Stallknecht DE, Holmes EC. 2015. Evolutionary genetics and vector adaptation of recombinant viruses of the Western equine encephalitis antigenic complex provides new insights into alphavirus diversity and host switching. Virology 474:154–162. doi: 10.1016/j.virol.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis NL, Fuller FJ, Dougherty WG, Olmsted RA, Johnston RE. 1986. A single nucleotide change in the E2 glycoprotein gene of Sindbis virus affects penetration rate in cell culture and virulence in neonatal mice. Proc Natl Acad Sci U S A 83:6771–6775. doi: 10.1073/pnas.83.18.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu W, Deng L, Wei Y, Wang H, Wang J, Liang G. 2015. A substitution in nsP1 combined with a double substitution in E2 glycoprotein renders Sindbis-like virus XJ-160 fully neurovirulent for adult mice. Virus Res 196:1–4. doi: 10.1016/j.virusres.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 39.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. 2007. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan L, Huang XY, Liu ZY, Zhang F, Zhu XL, Yu JY, Ji X, Xu YP, Li G, Li C, Wang HJ, Deng YQ, Wu M, Cheng ML, Ye Q, Xie DY, Li XF, Wang X, Shi W, Hu B, Shi PY, Xu Z, Qin CF. 2017. A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science 358:933–936. doi: 10.1126/science.aam7120. [DOI] [PubMed] [Google Scholar]
- 41.Jöst H, Bialonski A, Storch V, Günther S, Becker N, Schmidt-Chanasit J. 2010. Isolation and phylogenetic analysis of Sindbis viruses from mosquitoes in Germany. J Clin Microbiol 48:1900–1903. doi: 10.1128/JCM.00037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahlm C, Eliasson M, Vapalahti O, Evander M. 2014. Seroprevalence of Sindbis virus and associated risk factors in northern Sweden. Epidemiol Infect 142:1559–1565. doi: 10.1017/S0950268813002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hesson JC, Rettich F, Merdić E, Vignjević G, Ostman O, Schäfer M, Schaffner F, Foussadier R, Besnard G, Medlock J, Scholte EJ, Lundström JO. 2014. The arbovirus vector Culex torrentium is more prevalent than Culex pipiens in northern and central Europe. Med Vet Entomol 28:179–186. doi: 10.1111/mve.12024. [DOI] [PubMed] [Google Scholar]
- 44.Lundström JO, Turell MJ, Niklasson B. 1990. Effect of environmental temperature on the vector competence of Culex pipiens and Cx. torrentium for Ockelbo virus. Am J Trop Med Hyg 43:534–542. doi: 10.4269/ajtmh.1990.43.534. [DOI] [PubMed] [Google Scholar]
- 45.Hesson JC, Verner-Carlsson J, Larsson A, Ahmed R, Lundkvist Å, Lundström JO. 2015. Culex torrentium mosquito role as major enzootic vector defined by rate of Sindbis virus infection, Sweden, 2009. Emerg Infect Dis 21:875–878. doi: 10.3201/eid2105.141577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sem Ouilibona RC, Tchetgna Simo HD, Vickos U, Berthet N, Nakouné E. 2018. Full-length genome sequence of a Sindbis virus strain isolated from Culex cinereus in 1977 in Bozo, Central African Republic. Genome Announc 6(26):e00455-18. doi: 10.1128/genomeA.00455-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forrester NL, Palacios G, Tesh RB, Savji N, Guzman H, Sherman M, Weaver SC, Lipkin WI. 2012. Genome-scale phylogeny of the alphavirus genus suggests a marine origin. J Virol 86:2729–2738. doi: 10.1128/JVI.05591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 51.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 52.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 55.Rambaut A, Lam TT, Carvalho LM, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adouchief S, Smura T, Sane J, Vapalahti O, Kurkela S. 2016. Sindbis virus as a human pathogen—epidemiology, clinical picture and pathogenesis. Rev Med Virol 26:221–241. doi: 10.1002/rmv.1876. [DOI] [PubMed] [Google Scholar]
- 58.Shirako Y, Niklasson B, Dalrymple JM, Strauss EG, Strauss JH. 1991. Structure of the Ockelbo virus genome and its relationship to other Sindbis viruses. Virology 182:753–764. doi: 10.1016/0042-6822(91)90616-J. [DOI] [PubMed] [Google Scholar]
- 59.Eiden M, Ziegler U, Keller M, Müller K, Granzow H, Jöst H, Schmidt-Chanasit J, Groschup MH. 2014. Isolation of Sindbis virus from a hooded crow in Germany. Vector Borne Zoonotic Dis 14:220–222. doi: 10.1089/vbz.2013.1354. [DOI] [PubMed] [Google Scholar]
- 60.Sigei F, Nindo F, Mukunzi S, Ng’ang’a Z, Sang R. 2018. Evolutionary analyses of Sindbis virus strains isolated from mosquitoes in Kenya. Arch Virol 163:2465–2469. doi: 10.1007/s00705-018-3869-8. [DOI] [PubMed] [Google Scholar]
- 61.Kutchko KM, Madden EA, Morrison C, Plante KS, Sanders W, Vincent HA, Cruz Cisneros MC, Long KM, Moorman NJ, Heise MT, Laederach A. 2018. Structural divergence creates new functional features in alphavirus genomes. Nucleic Acids Res 46:3657–3670. doi: 10.1093/nar/gky012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simpson DA, Davis NL, Lin SC, Russell D, Johnston RE. 1996. Complete nucleotide sequence and full-length cDNA clone of S.A.AR86 a South African alphavirus related to Sindbis. Virology 222:464–469. doi: 10.1006/viro.1996.0445. [DOI] [PubMed] [Google Scholar]
- 63.Strauss EG, Rice CM, Strauss JH. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All newly sequenced strains have been deposited in GenBank under accession numbers MK045224 to MK045259.