Abstract
The Achilles heel of anticancer treatments is intrinsic or acquired resistance. Among many targeted therapies, the DNA repair inhibitors show limited efficacy due to rapid emergence of resistance. We examined evolution of cancer cells and tumors treated with AsiDNA, a new DNA repair inhibitor targeting all DNA break repair pathways. Effects of AsiDNA or Olaparib were analyzed in various cell lines. Frequency of AsiDNA- and olaparib-resistant clones was measured after 2 weeks of continuous treatment in KBM7 haploid cells. Cell survivals were also measured after one to six cycles of 1-week treatment and 1-week recovery in MDA-MB-231 and NCI-H446. Transcriptomes of cell populations recovering from cyclic treatments or mock treatment were compared. MDA-MB-231 xenografted models were treated with three cycles of AsiDNA to monitor the effects of treatment on tumor growth and transcriptional modifications. No resistant clones were selected after AsiDNA treatment (frequency < 3x10−8) in treatment conditions that generate resistance to olaparib at a frequency of 7.2x10−7 resistant clones per treated cell. Cyclic treatments promote cumulative sensitivity characterized by a higher mortality of cells having undergone previous treatment cycles. This sensitization was stable, and transcriptome analysis revealed a major gene downregulation with a specific overrepresentation of genes coding for targets of DNA-PK. Such changes were also detected in tumor models which showed impaired growth after cycles of AsiDNA treatment.
Introduction
Conventional anticancer treatments and, more recently, targeted therapies have improved the control of tumors. Nonetheless, the rate of therapy failure is high, primarily due to side effects that limit dose escalation and the onset of resistance during treatment. Targeting tumors based on their genetic defects has revolutionized the era of cancer treatment and precision medicine. Targeted therapies yield high rates of initial response, although most responding tumors fail to achieve a complete response. Furthermore, the development of acquired resistance is nearly universal in patients who respond initially to therapy. Targeted drugs often produce remission for only a limited period because intrinsic or acquired resistance to treatment by the malignant cells leads to relapse, progression, and eventually death [1], [2]. Although the ability of cancer to evolve has been traditionally perceived as a major problem in curing it, it has been increasingly suggested that it could also inspire novel therapies [3].
Poly-ADP-ribose-polymerase (PARP) inhibitors, which mainly target the base excision and single-strand break repair pathways, have been tested for many years and have become a potential supplement to conventional chemotherapy, show increasing evidence of the appearance of resistance during treatment [4]. Molecular mechanisms that give rise to resistance to PARP inhibitor therapy usually involve mutations of the target that prevent interaction with the drug, overexpression of the target, or activation of alternative pathways [5]. Genetic instability, a characteristic of almost all cancers [6], facilitates the emergence of resistance and could be particularly enhanced by treatments inhibiting DNA repair. We have recently developed a new inhibitor of the DNA repair, AsiDNA, based on the Dbait concept [7], [8]. AsiDNA molecules are cell-permeant small double-stranded DNA molecules that mimic double-strand breaks. Inside the cells, they bind to and activate both PARP and DNA-dependent protein kinase (DNA-PK), thus triggering an inappropriate DNA repair signal, which in turn prevents the recruitment and the activity of enzymes required for homologous recombination and nonhomologous end joining at endogenous genomic damage site [7], [9] (Supplementary Figure S1). The unique mechanism of action of AsiDNA, which activates two independent target enzymes and inhibits several repair pathways, differs from other DNA repair inhibitors. Indeed, intrinsic resistance of tumor cells to a single exposure of AsiDNA has been shown to be associated with multiple parameters such as high mRNA expression level; copy number gains; and mutations in processes involved in cell proliferation, cell survival, epithelial-mesenchymal transition, and cell motility [10]. In this study, the potential of AsiDNA, in comparison with other anticancer treatments such as PARP inhibitors, to promote acquired resistance was evaluated by performing repeated cyclic treatments. Unexpectedly, no resistant clones could be obtained after AsiDNA treatment. We explored possible mechanisms explaining the low occurrence of acquired resistance to AsiDNA.
Materials and Methods
Cell Lines and Treatment
The MDA-MB-231, MDA-MB-468, HCC1143, THP-1, U-937, NCI-H446, MCF-10A, MCF-12A, and TK6 cell lines were purchased from the ATCC. The BC227 cell line was obtained from the Institut Curie (30) and the KBM7 cell line from Dr. Brummelkamp (NKI, the Netherlands). Cells were grown according to the supplier's instructions and maintained at 37 °C in a humidified atmosphere at 5% CO2, except MDA-MB-231 cells (0% CO2). Cell lines were verified by short tandem repeat profiling (Geneprint 10, Promega) at 10 different loci (TH01, D21S11, D5S818, D13S317, D7S820, D16S539, CSF1PO, AMEL, vWA, TPOX) and tested negative for Mycoplasma contamination with the VenorGeM Avance Kit (Biovalley). Haploid KBM7 cells were purified by flow cytometry before each experiment, and mutagenized haploid KBM7 cells (KBM7mut) were obtained by incubating the cells for 24 hours with 0.01 μg/ml 4NQO. PARP inhibitors, AZD-2281 (olaparib) and BMN673 (talazoparib), were purchased from Medchem Express (Princeton, USA) and diluted in DMSO to a stock concentration of 10 mM. Imatinib and 6-thioguanine (6-TG) were purchased from Sigma Aldrich (Saint Louis, USA).
For continuous (acute) treatment, KBM7mut cells were seeded in 96-well plates at a density of 2x104 cells/well in the presence of AsiDNA (15 plates: 1440 independent populations), olaparib (10 plates: 960 independent populations), imatinib (15 plates: 960 independent populations), or 6-TG (15 plates: 960 independent populations). The doses of AsiDNA (5 μM), olaparib (5 μM), imatinib (1 μM), and 6-TG (0.5 μg/ml) were chosen to result in 5% survival (relative to nontreated cells). Resistance frequency was estimated by the ratio of growing populations on the total number of treated cells.
Cells were treated by repeated cycles of 2 weeks. Each cycle started on day 0 and ended on day 14. On day 0, cells were seeded in six-well culture plates with 2x104 cells per well and incubated for 24 hours at 37°C before addition of the drug (5 μM AsiDNA, 10 μM olaparib, or 0.1 μM talazoparib) or kept growing with no drug addiction. Cells were harvested on day 7, washed, and counted after staining with 0.4% trypan blue (Sigma Aldrich, St Louis, USA). Survival to each cycle is calculated at day 7 by the ratio of living cells with drug treatment on the number of living cells without drug addition at the last cycle. After counting at day 7, cells were seeded again in six-well culture plates with 2x104 cells per well, medium was changed 24 hours after incubation to remove dead cells, and the cells were allowed to recover for 1 week. Another cycle of treatment/recovery was then started for up to five cycles, depending on the cell line.
ELISA Measurement of PARylation
A sandwich ELISA was used to detect Poly(ADP-Ribose) (PAR) polymers. Cells were boiled in PathScan Sandwich ELISA Lysis Buffer (Cell Signaling Technology) supplemented with 1 mM phenylmethanesulfonyl fluoride (Sigma). Cell extracts were then diluted in Superblock buffer (Thermo Scientific) prior to the ELISA. A 96-well polystyrene plate (Thermo Scientific Pierce White Opaque) was coated with 100 μl per well carbonate buffer (1.5 g/l sodium carbonate Na2CO3, 3 g/l NaHCO3) containing the capture antibody (mouse anti-PAR at 4 μg/ml, Trevigen 4335) overnight at 4°C, after which it was washed with PBST solution. The wells were then blocked with Superblock at 37°C for 1 hour. Then, 10 μl of cell extract was added to 65 μl of Superblock, applied to each well in triplicate, and incubated overnight at 4°C, after which it was washed with PBST solution. Then the detection antibody (Rabbit anti-PAR, Trevigen 4336, diluted 1/1000 in PBS/2% milk/1% mouse serum) was added and incubated for 1 hour at room temperature. After washing, secondary antibody HRP-conjugated anti-rabbit (Abcam, ab97085, diluted 1/5000 in PBS/2% milk/1% mouse serum) was applied to each well for 1 hour. To read out, 75 μl of substrate for the enzyme (Supersignal Pico, Pierce) was added to each well. The optical absorbance (OD λ = 425 nm) was recorded at various time points (1, 5, and 15 minutes).
γ-H2AX Quantification by Flow Cytometry
Cells were treated with different doses of AsiDNA for 24 hours and then fixed and permeabilized with cold (−20°C) 70% ethanol for 2 hours. After washing with PBS, the cells were further permeabilized with 0.2% Triton in PBS for 30 minutes at RT, washed in PBS, and incubated with anti–γ-H2AX antibody (05-636 Millipore) in 2% BSA in PBS. After washing with PBS, and cells were incubated with an Alexa Fluor 488–conjugated secondary antibody. Fluorescence intensities were determined with a FACSCanto (BD- Biosciences). Data were analyzed using FlowJo software (Tree Star, CA).
Expression Array Data Analysis
Affymetrix Human Gene 2.1 Array datasets were controlled using Expression console (Affymetrix), and further analyses and visualization were made using EASANA (GenoSplice, www.genosplice.com), which is based on the FAST DB release 2016_1 annotations of GenoSplice. For more details, refer to Supplementary Materials and Methods. Genes coding for proteins interacting with DNA-PK (136 genes) were defined using uniting KEGG pathways hsa03450 and hsa04110. Genes coding for proteins interacting with PARP (238 genes) were defined using uniting KEGG pathways hsa03410, hsa04064, and hsa04210.
In Vivo Experiments
MDA-MB-231 cell-derived xenografts (CDXs) were obtained by injecting 5.106 cells into the mammary fat pad of 6- to 8-week-old adult female nude NMRI-nu Rj:NMRI-Foxn1nu/Foxn1nu mice (Janvier). The animals were housed at least 1 week before tumor engraftment under controlled conditions of light and dark (12 hours/12 hours), relative humidity (55%), and temperature (21°C). Mice were randomized into different treatment groups of 10-15 animals when engrafted tumors reached 80-250 mm3. AsiDNA was injected systemically (intraperitoneal administration) according to each treatment protocol. Tumor growth was evaluated three times a week using a caliper, and tumor volume was calculated using the following formula: (length × width × width)/2. Mice were followed for up to 3 months and ethically sacrificed when the tumor volume reached 1500 mm3. The Local Animal Experimentation Ethics Committee approved all experiments. The authorization to perform animal studies (#01593.02) was delivered by the French Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche.
Statistical Analysis
All statistical analyses were performed using the two-tailed Student t test, except for data in Supplementary Figure S3 analyzed with the Mann-Whitney test.
Results
AsiDNA activity in Haploid KBM7 Cells
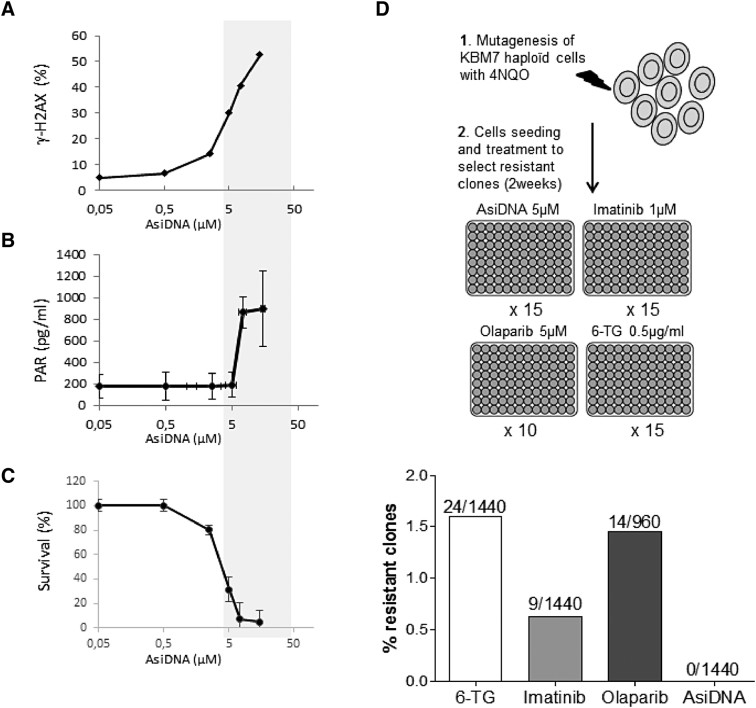
We first screened for genetic mutants that develop resistance to AsiDNA during 2 weeks of treatment in human KBM7 leukemia cells. Most of the genome of KBM7 cells is haploid, except chromosome 8, which might help to rapidly reveal recessive mutations that could provide resistance to treatment [11]. We analyzed AsiDNA activity in KBM7 cells by monitoring the activation of DNA-PK and PARP. Both enzymes were activated to modify their targets after interacting with the AsiDNA DNA moiety which mimics a double-strand break. KBM7 cells treated with AsiDNA showed dose-dependent phosphorylation of the histone H2AX (γH2AX), a target of DNA-PK, and Poly(ADP-Ribose) (PAR) polymer accumulation (Figure 1, A and B). Cell death was detected at doses corresponding to DNA-PK and PARP activation (Figure 1C), validating the use of this cell line for further analysis of resistance to AsiDNA.
Figure 1.
Effect of AsiDNA and occurrence of resistance in KBM7 cells. Cells were treated with increasing doses of AsiDNA and assessed for (A) H2AX phosphorylation (γH2AX) and (B) PARP hyperactivation by measuring cellular PARylation 24 hours after treatment (n = 3), and (C) survival in the presence of AsiDNA 6 days after treatment (n = 3). The gray rectangle indicates the response dose range to AsiDNA, showing a correlation between cytotoxic effects and targets activation. Data are represented as mean ± SD. (D) Independent mutagenized KBM7 cell cultures (N) were treated for 2 weeks with AsiDNA (5 μM), olaparib (5 μM), imatinib (1 μM), or 6-TG (0.05 μg/ml). Doses were chosen to result in 5% survival (relative to nontreated cells); resistant cultures (N) were identified as those with high density (>5 105 cells/well) relative to the rest of the populations (frequencies of resistant clones are indicated as N/n).
Continuous Treatment with AsiDNA Does Not Promote Resistance
We increased the mutation rate of KBM7 cells to facilitate resistance by pretreating them with the chemical agent 4-NQO, which induces substitutions in both guanine and adenine residues distributed randomly throughout most of the genome [12]. Mutagenized (KBM7mut) and nonmutagenized (KBM7) cells showed no significant differences in sensitivity to AsiDNA (data not shown). For resistance selection, we continuously treated the cells during 2 weeks at doses inducing more than 90% death. A total of 1440 independent cultures, corresponding to a total of 2.9 107 seeded KBM7mut cells, were treated with AsiDNA, the tyrosine kinase inhibitor imatinib, or 6-TG, and their growth was analyzed. No resistant clones were selected in cultures treated by AsiDNA (resistance frequency <0.34 10−7). In contrast, we recovered 24 independent resistant clones from samples treated with 6-TG (resistance frequency ~8.3 10−7) and 9 resistant clones from imatinib-treated samples (resistance frequency ~3.1 10−7) (Figure 1D). As AsiDNA acts as an inhibitor of DNA repair, we investigated whether such behavior is typical of DNA-repair inhibitors by analyzing the emergence of resistant clones to the PARP inhibitor olaparib under similar conditions. Olaparib resistance occurred at a high frequency, similar to imatinib and 6-TG resistance, with 14 clones growing among the 960 independently treated populations (resistance frequency ~7.2 10−7) (Figure 1D).
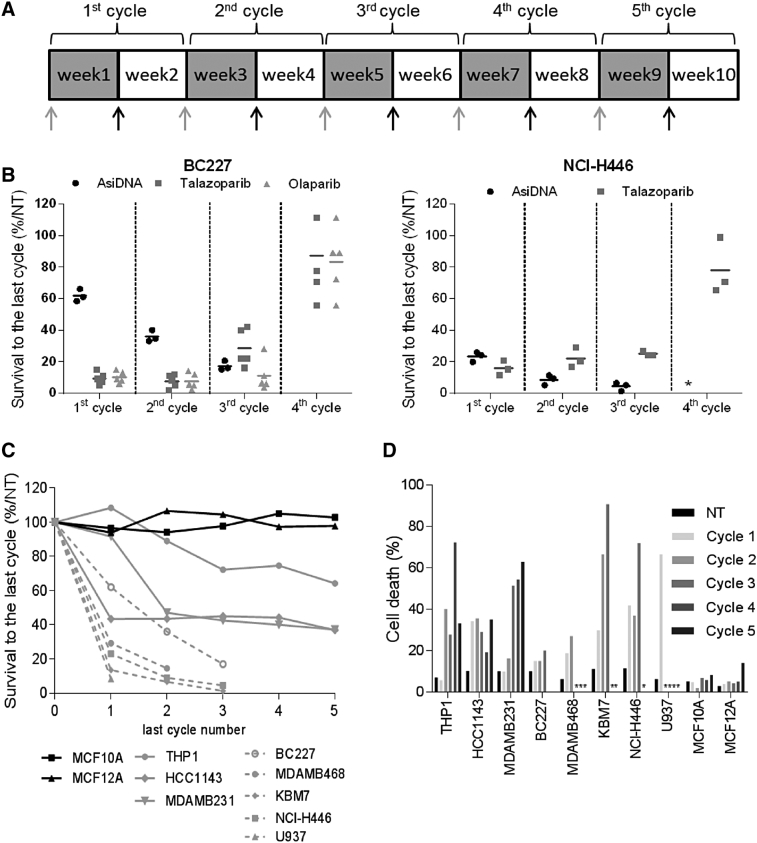
Cyclic Treatment with AsiDNA Does Not Promote Resistance
Clinically relevant models are developed with the aim of mimicking the conditions that cancer patients experience during chemotherapy. A pulsed treatment strategy is often used, in which the cells recover in drug-free media after a period of treatment. Such protocols appear to facilitate the emergence of resistance [13]. Therefore, we subjected KBM7mut cells to repeated cycles of treatment to increase the possibility of generating AsiDNA-resistant clones (Figure 2A). Treatment cycles consisted of 1 week of treatment and 1 week of recovery. We did not obtain any resistant clones from AsiDNA-treated populations under three treatment cycles of such conditions, after which the cells did not recover (Supplementary Figure S2).
Figure 2.
Effect of repeated treatment with AsiDNA. Cells were treated for 1 week and grown without treatment for an additional week for one to five cycles. Survival was monitored at every cycle by counting the cells at the end of the treatment with AsiDNA as described in Material and Methods. (A) Scheme of treatment. Gray arrows indicate change of growth medium with AsiDNA (gray) and without (black). Gray square: week of AsiDNA treatment; white square: no treatment. (B, C, D) Effect of previous repeated cycles of treatment on survival of different cell lines to 1 week of treatment. Survival was estimated by the ratio of treated cells to nontreated cells after 1 week of treatment at each cycle. Numbers of cycles in abscissa indicate the number of the last treatment analyzed for the survival. (B) Survival of BC227 or NCI-H446 independent cell populations (n = 3-5) during repeated cycles of treatment with AsiDNA (black circles), olaparib (gray triangle), or talazoparib (gray square). (C) Cell lines: nontumor (black line): MCF-10A (square), MCF-12A (triangle); tumor with intermediate sensitization (gray line): THP1 (circle), MDA-MB-231 (triangle), and HCC1143 (diamond); tumor with rapid sensitization (gray dotted line): MDA-MB-468 (circle), KBM7 (diamond), U937 (triangle), NCI-H446 (square), and BC227 (empty circle). (D) The percentage of dead cells was calculated as the number of dead cells of the total number of counted cells at NT, and after cycle 1, 2, 3, 4, and 5. *No recovery after the previous treatment cycle.
The KBM7 cell line was chosen for resistant mutant selection to increase the chances of detecting recessive mutations that give a resistant phenotype. It is possible that the lack of resistant clone selection could have been an artifact of this specific cell line. Therefore, we performed the same resistance screen on two other tumor cell lines—BC227, a BRCA2 mutant breast cancer cell line, and NCI-H446, a small cell lung cancer cell line—that are both intrinsically sensitive to PARP inhibitors and AsiDNA (Figure 2A). The efficacy of the screening method was confirmed by the isolation of resistant populations to the PARP inhibitors olaparib or talazoparib between cycle 3 and cycle 4 of treatment (Figure 2B). Indeed, all the BC227 and NCI-H446 populations submitted to PARP inhibitor selection pressure became resistant, raising the question of the clinical benefit of long-term maintenance monotherapy with PARP inhibitors. No resistance appeared during AsiDNA treatment under the same treatment conditions. Surprisingly, the sensitivity of the entire populations treated with one cycle of AsiDNA increased with number of previous treatment cycles, with no recovery of any of the independently AsiDNA-treated populations after the fourth cycle of treatment (Figure 2B). To understand if this increase of sensitivity is a general effect of AsiDNA treatment independent of the genetic background of the tumors, we further studied the evolution of AsiDNA sensitivity during repeated cycles of treatment in eight representative tumor cell lines with different sensitivity to AsiDNA (Supplementary Table 1) that included models of different tumor types derived from breast cancer (MDA-MB-231, MDA-MB-468, and HCC1143, BC227), myeloid leukemia (THP1, U937, KBM7), and lung cancer (NCI-H446). No resistance to AsiDNA was observed, and cancer cells became increasingly sensitive to AsiDNA treatment, independently of their initial sensitivity or origin (Figure 2C). This effect was associated with high mortality in the population, indicating an increase in the toxicity of AsiDNA with each cycle of treatment (Figure 2D). The increase in sensitivity mainly occurred during the first three cycles in all cell lines. In contrast, the nontumor mammary epithelial cell lines MCF-10A and MCF-12A did not show sensitivity to AsiDNA even after five cycles of treatment (Figures 2, C and D). Overall, these resistance selection studies demonstrate that acquired resistance is much less likely to occur after AsiDNA treatment than after any other anticancer treatments, such as inhibitor imatinib, or 6-TG and PARP inhibitors.
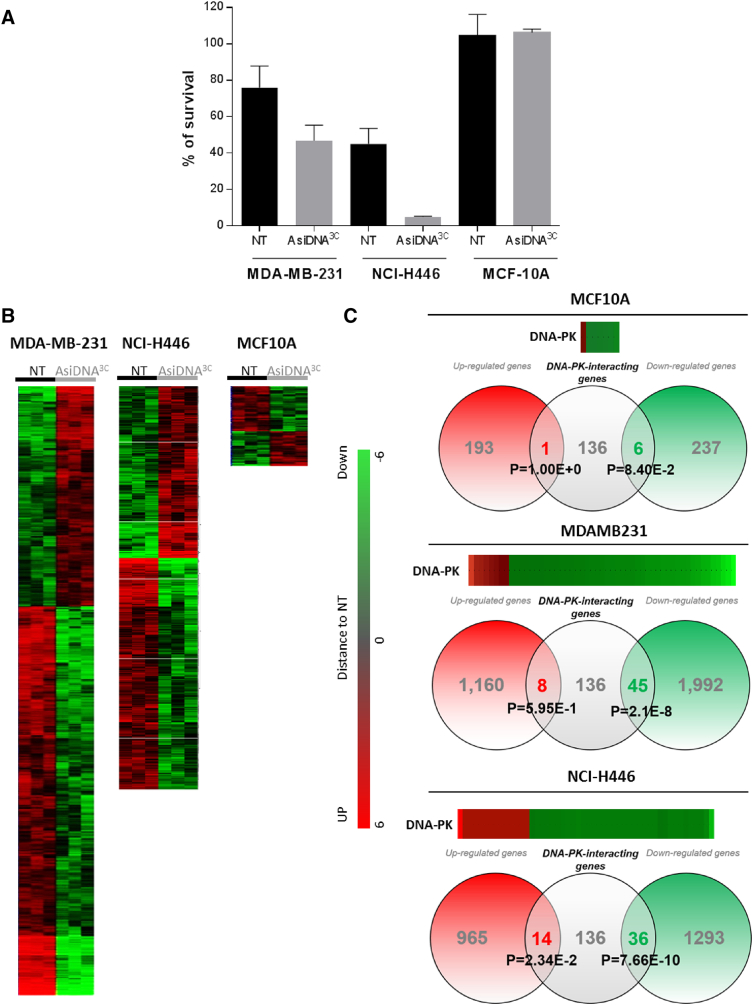
AsiDNA Treatment Induces Long-Lasting Transcriptional Modifications
The increase in sensitivity to AsiDNA observed in several tumor cell lines suggests that the population evolved during the treatment cycles. The increase in sensitivity to AsiDNA observed after three cycles of treatment (labeled/marked AsiDNA3C) was stable and lasted for at least 3 months of continuous growth without treatment (Supplementary Figure S3). Interestingly, all independent populations from a given cell line end up with similar sensitivity after cyclic treatment. To further understand the evolution of the treated populations, we compared the transcriptome of three independent populations of two AsiDNA-treated tumor cell lines (MDA-MB-231AsiDNA3C and NCI-H446AsiDNA3C) and one AsiDNA-treated nontumor cell line (MCF-10AAsiDNA3C) to their mock-treated counterparts (MDA-MB-231NT, NCI-H446NT, and MCF-10ANT, respectively) (Figure 3A). Transcriptional analysis showed that AsiDNA-treated independent populations from a same cell line display a similar extensive transcriptional change after three cycles of AsiDNA. Hierarchical clustering of regulated genes revealed high global gene expression differences between nontreated and treated populations of MDA-MB-231 and NCI-H446 tumor cells and very few differences between the treated and nontreated nontumor MCF-10A cells (Figure 3B). Interestingly, there was a large excess of downregulated genes in AsiDNA-treated populations, with approximately two-fold more genes downregulated than upregulated ones when compared to nontreated populations. Detailed analysis of the cellular pathways involving the main AsiDNA targets, DNA-PK and PARP1, revealed significant enrichment in the number of downregulated genes coding for proteins modified by or interacting with activated DNA-PK (MDA-MB-231, P = 2.110−8; NCI-H446, P = 7.66 10−10) (Figure 3C).
Figure 3.
Gene expression profile after AsiDNA repeated treatments. MDA-MB-231 and NCI-H446 tumor cells or MCF-10A nontumor independent cell populations were treated with three cycles of AsiDNA (according to Figure 2A), tested for AsiDNA sensitivity before transcriptome analysis: (A) survival to AsiDNA of MDA-MB-231 (n = 8), NCI-H446 (n = 3), MCF-10A (n = 2). Data are mean ± SD. (B) transcriptional change 2 weeks after the third cycle of treatment. Hierarchical clustering of regulated genes. (C) Heatmap analysis showing the distance of AsiDNA three- cycle–treated (AsiDNA3C) to mock-treated (NT) cell populations and Venn diagram representation with genes interacting with DNA-PK. Number of genes upregulated (red circle) or downregulated (green circle) in AsiDNA-treated populations is indicated. Distribution of genes interacting with DNA-PK according to KEGG classification in each group is indicated in the gray circle. Overrepresentation of DNA-PK interacting genes was calculated using a Student t test.
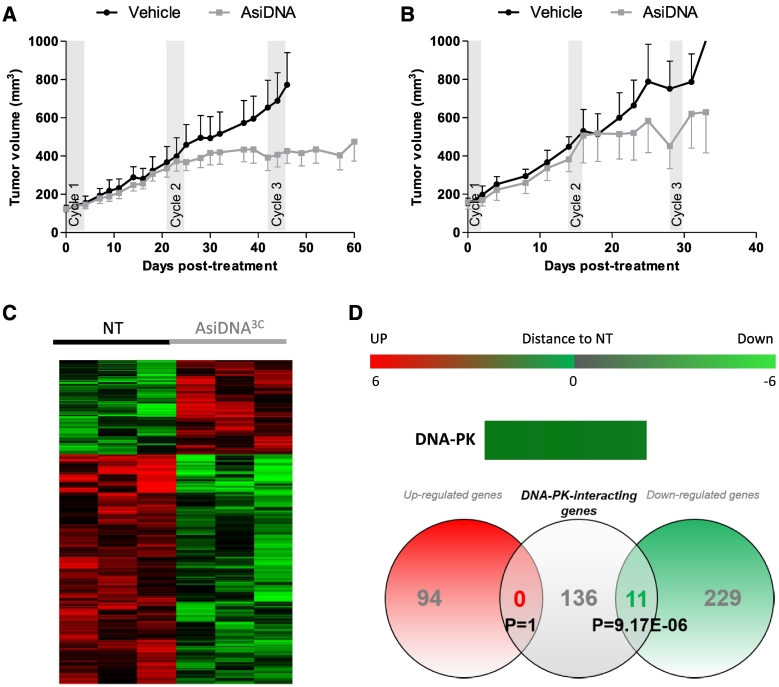
Evolution of AsiDNA-Treated Tumors
We validated in vivo the increase of sensitivity to AsiDNA upon cyclic treatments in MDA-MB-231 cell-derived xenografts. As tumor cells do not grow at the same speed in tumors and culture medium, we tested two different cyclic treatment protocols with 1 or 2 weeks of recovery between the weeks of treatment in two independent experiments (Figure 4, A and B). In both protocols, the tumors did not respond to the first cycle of treatment but stopped growing at the second treatment cycle, suggesting that tumors might also develop sensitivity with repeated treatment. Such sensitization did not depend upon the rest time between cycles: either 17 (Figure 4A) or 12 days (Figure 4B). These results are consistent with in vitro data, which showed a large increase in sensitivity to AsiDNA starting from the second cycle of AsiDNA treatment (Figure 2C). Moreover, transcriptomic analysis of tumors having received three cycles of treatment (with a schedule of treatment sessions every 3 weeks) and harvested 1 week after the last treatment revealed transcriptome change as compared to untreated tumors of the same size (Figure 4C). Gene expression was preferentially downregulated after AsiDNA treatment (230 genes downregulated and 93 upregulated), and DNA-PK targets were significantly overrepresented in the downregulated genes (P = 9.17 E-06) (Supplementary Table S2). These results are totally consistent with our previous in vitro data.
Figure 4.
Effect of repeated AsiDNA treatment in MDA-MB-231 cell-derived xenografts. Growth of MDA-MD-231 cell-derived xenografted tumors mock treated (black lines) or treated (gray lines) with three cycles of AsiDNA (gray rectangles) with different schedules: (A) 5 consecutive days of injections (5 mg/injection) followed by 17 days of rest (vehicle, n = 6; AsiDNA, n = 8) or (B) 3 consecutive days (15 mg/injection) followed by 12 days of rest (vehicle, n = 5; AsiDNA, n = 10). Data are represented as mean ± SEM. (C, D) Transcriptomic analysis of three tumors mock-treated (NT) or treated with AsiDNA (AsiDNA3C). Three mice from each group treated with protocol B were sacrificed at day 38 and analyzed: (C) hierarchical clustering of regulated genes and heatmap analysis. (D) Venn diagram representation of genes differentially expressed and genes interacting with DNA-PK. Overrepresentation of DNA-PK was calculated by Student t test.
Discussion
AsiDNA is a first-in-class DNA repair inhibitor that prevents the recruitment of repair enzymes to sites of DNA damage, either by acting as bait for DNA repair proteins or by inducing false DNA damage signaling, obscuring the detection of DNA breaks. It acts on enzymes involved in the different DNA repair pathways, such as homologous recombination, nonhomologous end-joining, base excision repair, and single-strand break repair, providing a broad DNA repair inhibitor activity rather than a specific protein targeting [8].
In this study, we exposed tumor cells to continuous or cyclic treatments of AsiDNA and analyzed after each treatment their sensitivity to this novel class of DNA repair inhibitor. No increase of survival that could reveal evolution toward resistance was observed. In contrast, in many different cell lines, independently of their initial (or intrinsic) sensitivity to AsiDNA, each cycle of AsiDNA treatment potentiates the sensitivity of the cells to AsiDNA. In total, cell lines derived from breast adenocarcinoma triple-negative B or ductal A, monocytic acute leukemia, chronic myelogenous leukemia, monocytic lymphoma, and lung cancer adenocarcinoma showed similar behavior during cyclic treatment with AsiDNA. Transcriptomic analysis revealed that independent populations evolved in the same way under AsiDNA treatment. The treated cells contained an excess of downregulated genes as compared to untreated cells. This unique behavior was specific to tumor cells since nontumor cells showed only moderate transcriptomic changes and no increase in sensitivity to AsiDNA upon repeated cycles of treatment. Two classes of tumor cell evolution with cyclic AsiDNA treatment were observed: the “sensitive” one that reached a stage where they did not recover from a treatment and another class more “resistant” that showed a sensitization in the first cycles and then reached a plateau value with almost no more variations in sensitivity with time. The belonging to one or the other class did not depend upon the origin of the tumor cells, the initial sensitivity to AsiDNA, or the growth speed. So far, we have no clear explanation for it. However, we are exploring the possibility that “resistant” tumor populations could be heterogeneous at the beginning of treatment containing subpopulations of cells in a “sensitive” state that would progressively disappear and other cells in a “nonresponsive” state close to the nontumor cells state. Actually, transcriptome analysis shows that several oncogenes are downregulated in three-cycle–treated cells.
The observation that MDA-MB-231 tumor cells were in vitro not sensitive to a first cycle of treatment but became sensitive after the second cycle was reproduced in vivo in two independent animal experiments, suggesting that such peculiar evolution of the tumor cells under treatment is conserved in tumors. We observed that, in AsiDNA-treated tumor cells in vitro and in vivo, the downregulation of many genes including DNA-PK pathway enzymes was always correlated with the increased sensitivity to AsiDNA. The causal link between both properties remains to be demonstrated.
Increasing the mutation rate by genotoxic therapies places surviving cells under intense evolutionary selective pressure, favoring Darwinian dynamics [14]. Indeed, resistant cells initially present in the tumors or generated during treatment (by the drug's mutagenic effect) are then positively selected under the pressure of treatment. Here, we show that repeated treatments with AsiDNA do not favor the emergence of resistance. This result indicates that preexisting resistant clones are rare in the population and/or that events that could lead to AsiDNA-resistant phenotype also lead to other associated defects that preclude population invasion as, for example, proliferation defect or cell cycle perturbations. However, the unusual evolution of the tumor cells under AsiDNA treatment toward increased sensitivity suggests the evolution of all the treated cells in a new state in absence of any selection process. Indeed, one cannot exclude that tumor cells accumulate damage or genetic instability at each cycle of AsiDNA treatment due to unrepaired or misrepaired chromosomes that will require a constitutive repair activity and make them more sensitive to new AsiDNA treatments. Moreover, the transcriptional change induced by the treatment could reduce the ability of the cells to recover from new AsiDNA treatments.
In conclusion, our finding that acquired resistance to AsiDNA is much less likely to occur than resistance to other anticancer treatments, such as PARP inhibitors, allows the consideration of long-term maintenance monotherapy with AsiDNA in the clinic, with a very low risk of the emergence of resistance.
Acknowledgments
Acknowledgements
We thank Laura Bellassen, Nathalie Berthault, and Marie-Christine Lienafa for their participation in this project. We thank Pierre de la Grange and Ariane Jolly from Genosplice for their help in the gene expression analysis, and the Genomics platform of the Curie Institute for the SNP array analysis.
Data Availability
All data generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Footnotes
Conflict of interest: W. J., P. L., and F. B. are employees of Onxeo. M. D. is a consultant for Onxeo.
Funding: This work was supported by the SIRIC-Curie, the Institut Curie, the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, and the IRS “NanoTheRad” of University Paris-Sud (Paris-Saclay). Funding for open access charge was from Institut Curie. W. J. was supported by a CIFFRE-ANRT fellowship (2013/0907). M. K. was supported by a Marie Sklodowska-Curie European fellowship ITN-RADIATE No. 642623. S. T. was supported by the Institut National du Cancer (TRANSLA13-081).
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.06.006.
Appendix A. Supplementary data
Supplementary Figure S1. Representation and formula of AsiDNA. AsiDNA, a 64-nucleotide (nt) oligodeoxyribonucleotide consisting of two 32-nt strands of complementary sequence connected through a 1,19-bis (phospho)-8-hydraza-2-hydroxy-4-oxa-9-oxo-nonadecane linker, with a cholesterol at the 5′-end and three phosphorothioate internucleotide linkages at each of the 5′ and the 3′ ends (Agilent, USA). The sequence is: 5′-X GsCsTs GTG CCC ACA ACC CAG CAA ACA AGC CTA GA L - CLTCT AGG CTT GTT TGC TGG GTT GTG GGC AC sAsGsC-3′.
Supplementary Figure S2. Repeated cycles of AsiDNA treatment did not recover resistance. Ninety-six different KBM7mut populations were treated with several cycles of AsiDNA (5 μM). Survival is percentage of living cells relative to nontreated condition (NT). NA, nonapplicable because no cells recovered after the previous cycle of AsiDNA treatment. Mean values (black lines).
Supplementary Figure S3. Duration of AsiDNA-induced changes in sensitivity to treatment. Independent populations pretreated with three cycles of AsiDNA (gray) or mock treated (black) were cultured without AsiDNA. They were assayed at different times after the end of the pretreatment for their sensitivity to AsiDNA (cell survival was estimated by the ratio of treated cells on nontreated (NT) cells). Dotted lines indicate the mean value of the survival of both populations. Mann-Whitney test (P<10-5).
Supplementary tables
References
- 1.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13(10):714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 2.Sirisena ND, Deen K, Mandawala DEN, Herath P, Dissanayake VHW. The pattern of KRAS mutations in metastatic colorectal cancer: a retrospective audit from Sri Lanka. BMC Res Notes. 2017;10(1):392. doi: 10.1186/s13104-017-2731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6(12):924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 4.Montoni A, Robu M, Pouliot E, Shah GM. Resistance to PARP-inhibitors in cancer therapy. Front Pharmacol. 2013;4:18. doi: 10.3389/fphar.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neel DS, Bivona TG. Resistance is futile: overcoming resistance to targeted therapies in lung adenocarcinoma. NPJ Precis Oncol. 2017;1 doi: 10.1038/s41698-017-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11(3):220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 7.Quanz M, Berthault N, Roulin C, Roy M, Herbette A, Agrario C. Small-molecule drugs mimicking DNA damage: a new strategy for sensitizing tumors to radiotherapy. Clin Cancer Res. 2009;15(4):1308–1316. doi: 10.1158/1078-0432.CCR-08-2108. [DOI] [PubMed] [Google Scholar]
- 8.Dutreix M, Devun F, Herath N, Noguiez-Hellin P. Dbait: a new concept of DNA repair pathways inhibitor from bench to bedside. In: Pollard J, Curtin N, editors. Targeting the DNA damage response for anti-cancer therapy. Springer International Publishing; Cham: 2018. pp. 359–373. [Google Scholar]
- 9.Croset A, Cordelieres FP, Berthault N, Buhler C, Sun JS, Quanz M. Inhibition of DNA damage repair by artificial activation of PARP with siDNA. Nucleic Acids Res. 2013;41(15):7344–7355. doi: 10.1093/nar/gkt522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jdey W, Thierry S, Popova T, Stern MH, Dutreix M. Micronuclei frequency in tumors is a predictive biomarker for genetic instability and sensitivity to the DNA repair inhibitor AsiDNA. Cancer Res. 2017;77(16):4207–4216. doi: 10.1158/0008-5472.CAN-16-2693. [DOI] [PubMed] [Google Scholar]
- 11.Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326(5957):1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 12.Downes DJ, Chonofsky M, Tan K, Pfannenstiel BT, Reck-Peterson SL, Todd RB. Characterization of the mutagenic spectrum of 4-nitroquinoline 1-oxide (4-NQO) in Aspergillus nidulans by whole genome sequencing. G3 (Bethesda) 2014;4(12):2483–92 doi 10.1534/g3.114.014712. [DOI] [PMC free article] [PubMed]
- 13.McDermott M, Eustace AJ, Busschots S, Breen L, Crown J, Clynes M. In vitro development of chemotherapy and targeted therapy drug-resistant cancer cell lines: a practical guide with case studies. Front Oncol. 2014;4:40. doi: 10.3389/fonc.2014.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillies RJ, Verduzco D, Gatenby RA. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer. 2012;12(7):487–493. doi: 10.1038/nrc3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Representation and formula of AsiDNA. AsiDNA, a 64-nucleotide (nt) oligodeoxyribonucleotide consisting of two 32-nt strands of complementary sequence connected through a 1,19-bis (phospho)-8-hydraza-2-hydroxy-4-oxa-9-oxo-nonadecane linker, with a cholesterol at the 5′-end and three phosphorothioate internucleotide linkages at each of the 5′ and the 3′ ends (Agilent, USA). The sequence is: 5′-X GsCsTs GTG CCC ACA ACC CAG CAA ACA AGC CTA GA L - CLTCT AGG CTT GTT TGC TGG GTT GTG GGC AC sAsGsC-3′.
Supplementary Figure S2. Repeated cycles of AsiDNA treatment did not recover resistance. Ninety-six different KBM7mut populations were treated with several cycles of AsiDNA (5 μM). Survival is percentage of living cells relative to nontreated condition (NT). NA, nonapplicable because no cells recovered after the previous cycle of AsiDNA treatment. Mean values (black lines).
Supplementary Figure S3. Duration of AsiDNA-induced changes in sensitivity to treatment. Independent populations pretreated with three cycles of AsiDNA (gray) or mock treated (black) were cultured without AsiDNA. They were assayed at different times after the end of the pretreatment for their sensitivity to AsiDNA (cell survival was estimated by the ratio of treated cells on nontreated (NT) cells). Dotted lines indicate the mean value of the survival of both populations. Mann-Whitney test (P<10-5).
Supplementary tables
Data Availability Statement
All data generated and/or analyzed during the current study are available from the corresponding author on reasonable request.