Rummeliibacillus sp. strain TYF005 is a thermophilic bacterium with high ethanol (8% vol/vol) and salt (13% wt/vol) tolerance that was isolated from spoilage vinegar. Here, we report the draft genome sequence of this strain, which has 117 scaffolds with a total genome size of 3.7 Mb and a 34.4% GC content.
ABSTRACT
Rummeliibacillus sp. strain TYF005 is a thermophilic bacterium with high ethanol (8% vol/vol) and salt (13% wt/vol) tolerance that was isolated from spoilage vinegar. Here, we report the draft genome sequence of this strain, which has 117 scaffolds with a total genome size of 3.7 Mb and a 34.4% GC content.
ANNOUNCEMENT
The genus Rummeliibacillus was first described in the United States in 2009. R. stabekisii, a physiologically recalcitrant microorganism that came from the surface of a spacecraft, was the first species to be described (1). The genus comprises three species, namely, R. stabekisii, R. pycnus (1), and R. suwonensis (2). Although there have been a few studies on the genus, its application potential has been highlighted in biotechnology. For instance, it can convert palm oil mill effluent into terpolymer polyhydroxyalkanoate and biodiesel (3), and it has potential for biomineralization (4). Furthermore, a thermally stable arginase from R. pycnus is used in the production of l-ornithine (5, 6). In this study, we report the draft genome sequence of Rummeliibacillus sp. strain TYF005, which was isolated from spoilage vinegar in Shanxi, China, using de Man, Rogosa, and Sharpe (MRS) agar medium (7) with the dilution spread plate method (8).
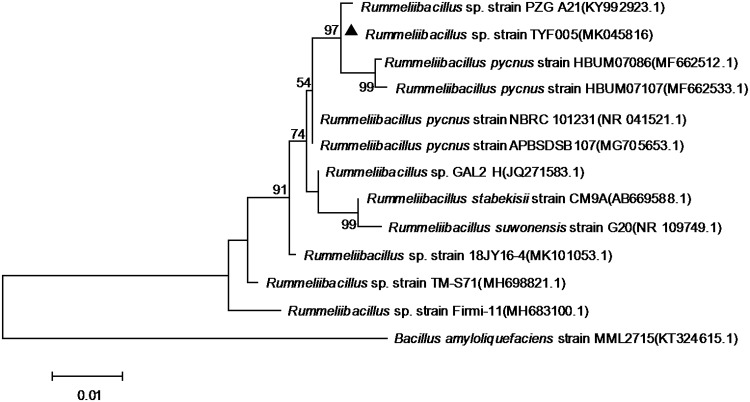
TFY005 was cultured in minimal medium (MM) broth (9) and MRS broth medium for 5 days with shaking at 200 rpm in an Erlenmeyer flask to investigate the ability to degrade corn straw and the tolerance to pH, temperature, alcohol, and NaCl (2). In all cases, the optical density at 600 nm (OD600) was measured to determine the cell growth. The strain TYF005 shows the ability to grow in MM culture medium containing 5% (wt/vol) natural corn straw powder as the sole carbon source. The optimal growth conditions in MRS broth include a pH of 5.0 to 8.5, alcohol and NaCl concentrations of 1 to 8% (vol/vol) and 1 to 13% (wt/vol), respectively, and a temperature of 30 to 55°C. The 16S rRNA gene was amplified by PCR with the universal primers 27F and 1492R as previously described (10). Based on analysis of a phylogenetic tree of the 16S rRNA for the most closely related species (Fig. 1), the strain was identified and designated Rummeliibacillus sp. strain TYF005.
FIG 1.
16S rRNA gene phylogeny of Rummeliibacillus sp. strain TYF005. The 16S rRNA gene sequences of related taxa were obtained from GenBank, and multiple alignments were performed with the CLUSTAL W program (22). Phylogenetic analysis based on the 16S rRNA sequence was conducted using the maximum likelihood method based on the Kimura 2-parameter model (23) in MEGA7 (24) with 1,000 replications in a bootstrap test. The initial tree for the heuristic search was obtained automatically by applying the neighbor-join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with the superior log likelihood value. Bootstrap values of >50% are shown. Bacillus amyloliquefaciens strain MML2715, which comes from the next closest genus, was used as an outgroup.
A single colony of TYF005 was inoculated in MRS broth at 45°C for 24 hours, and 1.5 ml of the liquid culture was aliquoted for the DNA extraction and purification using a NucleoSpin tissue kit (TaKaRa Bio, Japan) following the manufacturer’s instructions. The extracted DNA was used with the TruSeq DNA sample prep kit (Illumina, CA, USA) to generate Illumina shotgun paired-end (400-bp) sequence libraries, which were sequenced on an Illumina HiSeq 2000 platform. A total of 7,627,629 raw paired-end reads with 2,303,543,958 bp were generated. Low-quality reads (quality score, 15), short reads (length, <25), and adaptors were removed, producing high-quality reads totaling 2,124,220,316 bp containing 6,884,863 paired-end reads and 714,099 single reads. The draft genome was assembled using SOAPdenovo v2.04 (11) (assembly parameter k = 31), and local hole filling and base correction for assembly results were acquired using GapCloser v1.12 (12), both with default settings. The protein sequences of the genes which were predicted using GLIMMER v3.02 (13) with default settings were searched against the nonredundant (NR) (14), Clusters of Orthologous Groups (COG) (15), STRING (16), gene ontology (GO) (17), and KEGG (18) databases using BLAST v2.2.28+ to obtain annotation information. rRNA genes and tRNA genes were predicted using Barrnap v0.4.2 and tRNAscan-SE v1.3.1 (19), both with default parameters. The sequencing protocol generated 170× coverage of the genome.
The genome of Rummeliibacillus sp. strain TYF005 has a size of 3.7 Mb with a GC content of 34.4%. The assembly resulted in 107 scaffolds (>10,00 bp), with the largest scaffold being 376,591 bp. The scaffold N50 and N90 values were 68,863 bp and 17,202 bp, respectively. The genome contains 3,610 genes, including 4 rRNA genes and 33 tRNA genes. The strain TYF005 has the ability to simultaneously utilize pentose and hexose. The genome contains genes for deferrochelatase (dye-decolorizing peroxidase) and laccase, which are involved in lignin degradation (20, 21). Furthermore, 253 genes are involved in biosynthesis of secondary metabolites. These results suggest that this strain has value in the conversion of straw-based biomass and various biotechnological processes, especially for some industrial processes requiring high temperature and high alcohol and salt concentrations.
Data availability.
The whole-genome sequence (WGS) of Rummeliibacillus sp. strain TYF005 has been deposited under the BioProject number PRJNA421055 at DDBJ/ENA/GenBank and under the accession number QGPZ00000000. The version described in this paper is the first version. The raw sequencing reads are available as SRA data with the number SRS2763992.
ACKNOWLEDGMENT
This work was supported by the Foundation of Shanxi Province (grant numbers 20180008, 201703D121044, and 201603D2110805).
REFERENCES
- 1.Vaishampayan P, Miyashita M, Ohnishi A, Satomi M, Rooney A, Duc MTL, Venkateswaran K. 2009. Description of Rummeliibacillus stabekisii gen. nov., sp. nov. and reclassification of Bacillus pycnus Nakamura et al. 2002 as Rummeliibacillus pycnus comb. nov. Int J Syst Evol Micr 59:1094–1099. doi: 10.1099/ijs.0.006098-0. [DOI] [PubMed] [Google Scholar]
- 2.Her J, Kim J. 2013. Rummeliibacillus suwonensis sp. nov., isolated from soil collected in a mountain area of South Korea. J Microbiol 51:268–272. doi: 10.1007/s12275-013-3126-5. [DOI] [PubMed] [Google Scholar]
- 3.Junpadit P, Suksaroj TT, Boonsawang P. 2017. Transformation of palm oil mill effluent to terpolymer polyhydroxyalkanoate and biodiesel using Rummeliibacillus pycnus strain TS 8. Waste Biomass Valor 8:1247–1256. doi: 10.1007/s12649-016-9711-1. [DOI] [Google Scholar]
- 4.Mudgil D, Baskar S, Baskar R, Paul D, Shouche YS. 2018. Biomineralization potential of Bacillus subtilis, Rummeliibacillus stabekisii and Staphylococcus epidermidis strains in vitro isolated from speleothems, Khasi Hill Caves, Meghalaya, India. Geomicrobiol J 35:675–694. doi: 10.1080/01490451.2018.1450461. [DOI] [Google Scholar]
- 5.Kai H, Tao Z, Bo J, Mu W, Ming M. 2016. Characterization of a thermostable arginase from Rummeliibacillus pycnus SK31.001. J Mol Catal B Enzym 133:S68–S75. doi: 10.1016/j.molcatb.2016.11.020. [DOI] [Google Scholar]
- 6.Huang K, Zhang T, Jiang B, Yan X, Mu W, Miao M. 2017. Overproduction of Ummeliibacillus pycnus arginase with multi-copy insertion of the arg R.pyc cassette into the Bacillus subtilis chromosome. Appl Microbiol Biotechnol 101:6039–6048. doi: 10.1007/s00253-017-8355-9. [DOI] [PubMed] [Google Scholar]
- 7.Yu Z, Ng IS, Yao C, Lu Y. 2014. Orthogonal array deciphering MRS medium requirements for isolated Lactobacillus rhamnosus ZY with cell properties characterization. J Biosci Bioeng 118:298–304. doi: 10.1016/j.jbiosc.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 8.Wu J, Ma Y, Zhang F, Chen F. 2012. Biodiversity of yeasts, lactic acid bacteria and acetic acid bacteria in the fermentation of “Shanxi aged vinegar”, a traditional Chinese vinegar. Food Microbiol 30:289–297. doi: 10.1016/j.fm.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Ilmén M, Saloheimo A, Onnela ML, Penttil ME. 1997. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl Environ Microbiol 63:1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. 2008. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol 74:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li R, Li Y, Kristiansen K, Wang J. 2008. SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 12.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a Web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35:52–57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christian VM, Jensen LJ, Michael K, Samuel C, Tobias D, Beate K, Berend S, Peer B. 2007. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res 35:358–362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, Bateman A. 2009. Rfam: updates to the RNA families database. Nucleic Acids Res 37:136–140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nawrocki EP, Kolbe DL, Eddy SR. 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25:1335. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahmanpour R, Rea D, Jamshidi S, Fülöp V, Bugg TD. 2016. Structure of Thermobifida fusca DyP-type peroxidase and activity towards Kraft lignin and lignin model compounds. Arch Biochem Biophys 594:54–60. doi: 10.1016/j.abb.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 21.Roth S, Spiess AC. 2015. Laccases for biorefinery applications: a critical review on challenges and perspectives. Bioprocess Biosyst Eng 38:2285–2313. doi: 10.1007/s00449-015-1475-7. [DOI] [PubMed] [Google Scholar]
- 22.Aiyar A. 2000. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol Biol 132:221–241. [DOI] [PubMed] [Google Scholar]
- 23.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The whole-genome sequence (WGS) of Rummeliibacillus sp. strain TYF005 has been deposited under the BioProject number PRJNA421055 at DDBJ/ENA/GenBank and under the accession number QGPZ00000000. The version described in this paper is the first version. The raw sequencing reads are available as SRA data with the number SRS2763992.