Abstract
Whilst the potential of neural stem cell (NSC)-based treatment is recognized worldwide and seems to offer a promising therapeutic option for stroke treatments, there is currently no full understanding regarding the effects of hypoxic and baicalein-enriched fraction (BEF) preconditioning approaches on the therapeutic potential of these cells for stroke. The potential of preconditioned NSC can be determined based on the expression of several key neuroprotective genes using qRT-PCR technique. However, prior to that, it is imperative and extremely important to carefully select reference gene(s) for accurate qRT-PCR data normalization to avoid error in data interpretation. This study aimed to evaluate the stability of ten candidate reference genes via comprehensive analysis using three algorithms software: geNorm, NormFinder and BestKeeper. Our results revealed that HPRT1 and RPL13A were the most reliable reference genes for BEF-preconditioned NSCs, but ironically, HPRT1 was ranked as the least stable reference gene for hypoxic-preconditioned NSCs. On the other hand, RPLP1 and RPL13A were selected as the most stably expressed pair of reference genes for hypoxic-preconditioned NSCs. In conclusion, this study has pointed out the importance of identifying valid reference genes and has presented the first significant validation on best reference genes recommended for qRT-PCR study involves NSC preconditioned with hypoxia or with BEF extracted from Oroxylum indicum medicinal plant.
Keywords: Cell biology, Cell culture, Stem cells research, Gene expression, Molecular biology, Baicalein-enriched fraction (BEF) preconditioning, Hypoxic preconditioning, Neural stem cell (NSC), Quantitative real time-polymerase chain reaction (qRT-PCR), Reference gene selection
1. Introduction
Ischemic stroke is caused by physical blockage of blood flow to brain, typically by blood clot or cholesterol plaque, resulting in an area of ischemia where the brain tissue becomes irreversibly damaged due to lack of oxygen and nutrients. Neuronal cell apoptosis that occurs following ischemic stroke can be reduced if the occlusion is removed quickly and tissue reperfusion is restored early (Tachibana et al., 2017). Although current pharmacological drugs such as tissue plasminogen activator, thrombolytic agents and statin allow rapid restoration of blood supply (Carandang, 2016; Xian et al., 2016), these treatments are unable to trigger regeneration of new viable neuronal cells to replace the damaged brain tissue and thus cannot completely restore the brain function. As a consequence, poor prognosis and high rate of recurrent events are observed among the stroke patients (Cheah et al., 2016). This is indicated by previous report which showed that about 13% of stroke patients who had suffered a first stroke developed a second stroke within a year and the observed mortality rate was 7-fold higher among the patients who experienced a recurrent stroke (Aarnio et al., 2014). The constraint of these therapies has led to the demand for an alternative therapy. Recently, cell grafting has emerged as a potential therapy for the stroke (Locatelli et al., 2009).
Neural stem cells (NSCs) were first identified within the central nervous system (CNS) in 1992 (Reynolds and Weiss, 1992). These cells are multipotent progenitors that are able to grow and self-renew in culture for extensive periods, yet retaining a stable differentiation capacity to give rise to matured, functional brain cells (Shi et al., 2008). In animal studies, NSC transplantation successfully improved brain function by regenerating the damaged brain tissue (Stroemer et al., 2009; Zhao et al., 2008). However, the major limitation of stem cell-based therapy is its inability to retain most of the donor stem cells after grafting into hypoxic sites with excessive free radicals, glutamate excitotoxicity and inflammation (Fischer et al., 2009; Roybon et al., 2006). A study showed that as little as 0.09% retention rate of grafted stem cells was observed in mice with transient middle cerebral artery occlusion (MCAO) (Bacigaluppi et al., 2009). Low cell survival rate limits the capacity of stem cell to repair the injured area and this poses a much more difficult challenge to the stem cell-based therapy for stroke.
Several strategies have been applied to circumvent this limitation. In our lab, hypoxic and baicalein-enriched fraction (BEF) preconditioning of NSCs were employed to enhance the stem cell-based therapy. Hypoxic preconditioning involves brief periods of hypoxia that triggers various protective signaling pathways and enhances resilience to ischemia (Sharp et al., 2004; Wei et al., 2017). Transcriptional responses to hypoxia are mediated by the hypoxia-inducible factor (HIF) (Sharp et al., 2004). Hypoxic preconditioning has been shown to promote NSC survival after transplantation into ischemic rat brain (Wakai et al., 2015). On the other hand, BEF preconditioning involves brief periods of exposure to baicalein compounds extracted from Oroxylum indicum - a medicinal herb belongs to family of Bignoniaceae that is commonly found in many regions of South-east Asia (Harminder et al., 2011). In Malaysia, O. indicum is commonly known as “Beko” and the young leaves as well as the fruits are consumed raw as salad by the local community. The baicalein active compound is postulated to trigger intrinsic neuroprotective mechanisms and enhance resilience to subsequent lethal insults (Harminder et al., 2011). It also has been documented as HIF stabilizer under normoxic conditions (Cho et al., 2008). Hence, BEF preconditioning could exert neuroprotective effects mimicking the hypoxic preconditioning under ambient conditions. We are determined to investigate the fundamental gene pathway involved in both preconditioning strategies in enhancing NSC survival rate within an ishemic environment using qRT-PCR based analysis.
qRT-PCR is the most well-known technique for absolute and relative quantification of mRNA transcription level. This technique offers greater sensitivity and specificity than Northen blot and semi-quantitative PCR (Rumlow et al., 2016). The expression of target gene is often normalized to reference gene to eliminate sample-to-sample biases. Reference genes are genes that are expressed constantly in all cells to maintain essential cellular functions (Scrideli et al., 2008). However, there is no universal reference gene for qRT-PCR as the expression of reference gene could vary under different experimental conditions, tissue types or species. As such, normalization using non-validated reference genes may introduce errors in gene studies (Gutierrez et al., 2008). Since the validity and accuracy of qRT-PCR are highly dependent on the reference gene, hence, it is imperative to identify the ideal candidate reference genes prior to qRT-PCR in this study.
Here, we evaluated the suitability of ten reference genes primarily used for gene expression study in human cells, namely β-actin (ACTB), eukaryotic initiation factor 4A (eIF4A), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), hypoxanthine phosphoribosyl transferase 1 (HPRT1), importin 8 (IPO8), peptidyl-prolyl isomerase A (PPIA), ribosomal protein L13A (RPL13A), 60S ribosomal protein large P1 (RPLP1), TATA box binding protein (TBP) and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ) for qRT-PCR normalization of hypoxic and BEF-preconditioned human NSCs, respectively. These candidate genes were carefully selected based on literature review of relevant studies which examined over 7,000 human genes in major tissue types to identify and designate housekeeping genes (Hsiao et al., 2001; Lee et al., 2007). The information of the candidate reference genes are summarized in Table 1. In this study, three algorithms software: geNorm, Normfinder and BestKeeper were used to analyse the stability of these candidate reference genes. This is because a single statistical approach could not possibly cover all variables associated with gene expression studies. Therefore, interpretation based on only one calculation algorithms can be associated with false positive results and misleading conclusions.
Table 1.
Main physiological function and accession number of candidate reference genes used in this study.
| Gene | Main physiological function | Accession no. |
|---|---|---|
| ACTB | Formation of cellular cytoskeleton | NM_001101.3 |
| eIF4A | Translation initiation | NM_014740.3 |
| GAPDH | Carbohydrate metabolism | NM_001289745.1 |
| HPRT1 | Nucleotide synthesis | NM_000194.2 |
| IPO8 | Nuclear protein import | NM_006390.3 |
| PPIA | Protein biosynthesis | NM_021130.4 |
| RPLP1 | Protein synthesis | NM_001003.2 |
| RPL13A | Protein biosynthesis | NM_012423.3 |
| TBP | Transcription factor | NM_003194.4 |
| YWHAZ | Signal transduction | NM_001135699.1 |
2. Materials and methods
2.1. Neural stem cell culture
Human neural stem cell line derived from the National Institutes of Health (NIH) approved H9 human embryonic stem cells (H9-hNSCs) were purchased from GIBCO, USA. The H9-hNSCs were seeded at a density of 1.0 × 105 cells/cm2 in a CELLstartTM CTSTM (GIBCO, USA) coated T25 flask added with 3 mL of complete StemPro NSC SFM culture medium comprised of 1X KnockoutTM D-MEM/F-12 (GIBCO, USA), 2 mM GlutaMAXTM–I Supplement (GIBCO, USA), 20 ng/mL basic Fibroblast Growth Factor (GIBCO, USA), 20 ng/mL Epidermal Growth Factor (GIBCO, USA) and 2% StemPro Neural Supplement (GIBCO, USA). The cells were allowed to adhere for 24 h in a CO2 incubator at 37 °C in the presence of 5% CO2. The medium was replaced with equal volume of pre-warmed complete StemPro NSC SFM every two days and cultured until confluence prior to subsequent preconditioning treatments.
2.2. Study design
In present study, H9-hNSCs were cultured and subjected to two groups. Group 1: H9-hNSCs preconditioned under hypoxic cell culture condition (hypoxic-preconditioned H9-hNSCs) vs. H9-hNSCs without hypoxic preconditioning (non-hypoxic H9-hNSCs; control); and Group 2: H9-hNSCs preconditioned with BEF treatment (BEF-preconditioned H9-hNSCs) vs. H9-hNSCs without the BEF treatment (non-BEF H9-hNSCs; control). Reference genes were carefully selected from each of these groups, respectively.
2.3. Hypoxic preconditioning of H9-hNSCs
Confluent H9-hNSCs were subjected to hypoxic cell culture condition set at 2% O2 for 24 h at 37 °C using a tri-gas hypoxic incubator flushed with 5% CO2, 2% O2 and 93% N2. On the other hand, the non-hypoxic H9-hNSCs (control) were subjected to normoxic cell culture condition maintained at 21% O2 and 5% CO2 for 24 h using a standard CO2 incubator.
2.4. BEF preconditioning of H9-hNSCs
Fresh leaves of O. indicum were collected from Kampung Pasir Parit, Pasir Mas (LGPS coordinate: latitude 5.905471, longitude 102.1884469). The plant material was verified by Dr. Rahmad Zakaria and voucher specimens (USM. Herbarium 11751) were deposited in the Universiti Sains Malaysia herbarium by Mr. V. Shunmugam (Fig. 1). The leaves were oven-dried at 50 °C and then crumbled into powder. 25 g of the O. indicum powder was transferred into a thimble added with 300 mL of petroleum ether solvent. The temperature of the extraction was set at the boiling point of petroleum ether (42–62 °C) and the extraction was carried out until the color of solvent turned clear. Then, the solvent was discarded and replaced with 500 mL of methanol. The temperature of the extraction was the adjusted to the boiling point of methanol (62–65°C) and the extraction was carried out again until the color of solvent turned clear. The clear solvent was collected and dried using rotary evaporator at 40 °C for 12 h. The dried crude extract was subjected to column chromatography packed with 70 g of Diaion HP20 resin and flushed with mixtures of distilled water and methanol at gradient concentrations to obtain baicalein-enriched fraction (BEF). Confluent H9-hNSCs were subjected to BEF preconditioning by culturing the cells with complete StemPro NSC SFM medium added with 1.56 μg/mL of BEF for 24 h at 37 °C in a standard humidified chamber with 21% O2 and 5% CO2. The concentration was selected based on a series of optimization procedure done in our laboratory (data not shown). On the other hand, the non-BEF H9-hNSCs were cultured with complete StemPro NSC SFM medium added with 0.1% DMSO for 24 h at 37 °C in a standard humidified chamber with 21% O2 and 5% CO2.
Fig. 1.
Voucher specimen of Oroxylum indicum plant (USM Herbarium 11751) verified by Dr. Rahmad Zakaria and deposited in the Universiti Sains Malaysia (USM) herbarium by Mr. V. Shunmugam. The leaves of O. indicum plant were used to extract baicalein-enriched fraction (BEF) for H9-hNSCs preconditioning treatment in this study.
2.5. Primer design and optimization
Primers were designed using Primer3Plus program (Table 2). Each primer pair flanking the exon-exon boundary were designed to avoid genomic DNA amplification that possibly present in the sample. The designated primer sequences were BLAST against human genome database to avoid any possibility of homologous sequences.
Table 2.
List of primer sequences used in this study.
| Gene | Forward (5′ to 3′) | Reverse (5′ to 3′) | Product size |
|---|---|---|---|
| ACTB | ATTGGCAATGAGCGGTTC | TGAAGGTAGTTTCGTGGATGC | 91 |
| eIF4A | GAAGCTGGATTACGGACAGC | GCACGTGTCCTTAGGCTTCT | 87 |
| GAPDH | CCAAGGTCATCCATGACAAC | ACAGTCTTCTGGGTGGCAGT | 82 |
| HPRT1 | TGAGGATTTGGAAAGGGTGT | CCTCCCATCTCCTTCATCAC | 90 |
| IPO8 | AGCTGGCTTGGCAGTTTATT | GCTGAATACGAGGCAGGAAT | 118 |
| PPIA | AGGGTTCCTGCTTTCACAGA | CATAGATGGACTTGCCACCA | 93 |
| RPLP1 | AGCCGGTGTAAATGTTGAGC | CAGATGAGGCTCCCAATGTT | 81 |
| RPL13A | GGATGAACACCAACCCTTCC | AACACCTTGAGACGGTCCAG | 133 |
| TBP | TTCGGAGAGTTCTGGGATTG | GGATTATATTCGGCGTTTCG | 117 |
| YWHAZ | CCGTTACTTGGCTGAGGTTG | TTGCATTTCCTTTTTGCTGA | 106 |
2.6. Quantitative real-time PCR (qRT-PCR)
Briefly, total RNA was isolated from H9-hNSCs using Trizol reagent (Thermo Scientific, USA). Isolated RNA samples underwent DNase treatment using Ambion TURBO DNA-free™ DNase Treatment and Removal Reagents (Thermo Scientific, USA). cDNA were synthesized from 1 μg of total RNA samples RevertAidTM H minus first strand cDNA Synthesis kit (Thermo Scientific, USA). The qRT-PCR was carried out in 20 μL total volume reactions contained 1X Power SYBR Green Master Mix (Thermo Scientific, USA), 2 μL of 10X diluted cDNA template and 0.1 μM of forward and reverse primers using Applied Biosystems 7500 Real-Time PCR system. The thermal cycling program was set up with an initial heat activation step at 95 °C for 10 min followed by 40 cycles of denaturation step at 95 °C for 15 s, an annealing step at 57 °C for 30 s and an extension step at 72 °C for 45 s. Fluorescence was measured at the end of extension step (72 °C for 45 s). At the end of the PCR reactions, dissociation or melt curves were run by heating PCR products at 95 °C for 15 s, cooling to 60 °C for 1 min and gradually re-heating to 95 °C for 15 s. Dissociation curve analysis was performed to confirm specificity of amplification. Moreover, in order to confirm the primer specificity and accuracy, agarose gel electrophoresis analysis was done to verify the PCR product size. PCR efficiency (E) was calculated based on the data obtained from the exponential phase of each individual amplification plot via equation E = [10(−1/slope)-1] × 100%.
2.7. Data analysis
The stability and ranking of all candidate reference genes were evaluated using geNorm, NormFinder and BestKeeper. To compare the transcription level of the reference genes, the average cycle threshold (CT) value of duplicate reactions was used as input for subsequent analysis using the geNorm, NormFinder and BestKeeper programs.
2.8. geNorm analysis
geNorm software was downloaded from http://allserv.urgent.be/∼jvdesomp/genorm/index.html. The CT value was transformed into linear scale expression quantity, Q using the standard formula Q = EΔCT where where E is the exponential amplification and ΔCT is the difference between the lowest CT value and sample CT value (Vandesompele et al., 2002). Reference gene with the highest expression has the highest Q, which was set to 1. Q values were then used as the input of Microsoft Excel for geNorm analysis. The software generated an average expression stability measure, M as the mean of the stability values of all other reference genes. The most stably expressed reference gene deemed the lowest M value. geNorm also was used to determine the gene expression stability by calculating the average pairwise variation, V(n/n+1) of a pair particular reference genes with all other candidate genes of the study. The cutoff value for pairwise variation was set at 0.15 (Vandesompele et al., 2002).
2.9. NormFinder analysis
NormFinder was obtained from http://www.mdl.dk/publicationsnormfinder.html. Similar to geNorm analysis, the CT value was converted to linear scale expression quantity (Q) by standard formula Q = EΔCT (Andersen et al., 2004). The highest relative quantity for each reference gene was set to 1. The transformed Q values were then used as input data for NormFinder program. NormFinder estimated the overall expression variation of all reference genes and the variation between sample subgroups. Stability value (M) for each gene which represented expression variability within samples and between groups were calculated. Lower M indicated lower intra- and inter-group variations, hence implying a higher stability (Andersen et al., 2004).
2.10. BestKeeper analysis
The BestKeeper application was obtained from http://www.genequantification.com/bestkeeper.html. The average raw CT values were used directly as the input data. The geometric mean of the CT values of all reference genes were grouped together to obtain the BestKeeper Index (Pfaffl et al., 2004). BestKeeper index was determined based on three criteria: The pairwise correlation coefficient (r) between each reference gene, the standard deviation (SD) of the CT values between the entire data set and the coefficient of variance (CV) of the CT values between the entire data set. Stable reference gene had r value closer to 1, while lower SD and CV value indicated lower variability and high stability of genes across samples. Besides, BestKeeper also excluded any reference gene with SD value more than 1 which corresponding to inconsistent input amount. In this study, the stability of all reference genes were ranked according to the three criteria (r, SD and CV) individually and subsequently calculated the mean of the rankings to generate the final rank of each reference genes (Olias et al., 2014).
2.11. Comprehensive analysis
The three software applied in this study utilized different algorithms in ranking the stability of reference genes. Therefore a geometric mean of the stability values obtained from all three software was calculated as comprehensive ranking to ensure a comprehensive analysis of 10 references genes based on the stability at different aspects to select the most reliable reference gene for qRT-PCR analysis in this study (Ma et al., 2016; Xie et al., 2012).
3. Results
3.1. PCR amplification specificity and efficiency for candidate reference genes
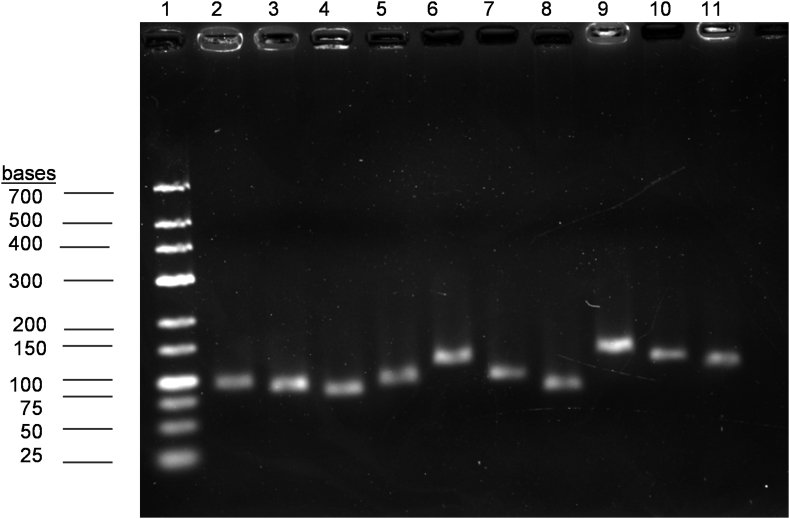
Amplification specificity for each primer pair was successfully confirmed by a single peak in the melting curve analysis (Fig. 2). The amplification specificity was further validated by a 3% agarose gel with a specific PCR product of expected size yielded for each primer pair (Fig. 3). Amplification efficiency of each primer pair was also successfully determined using serial dilution curves (Fig. 4). The amplification efficiency of all the reference genes ranged from 91.6% to 113.9% with a positive correlation coefficient (R2) ranged from 0.9986 to 0.9990 (Table 3). Efficiency between 90% and 110% can be considered as good. The amplification efficiency test showed that all primer pairs consistently performed throughout the amplification.
Fig. 2.
Determination of qRT-PCR primer amplification specificity for (A) ACTB, (B) eIF4A, (C) GAPDH, (D) HPRT1, (E) IPO8, (F) PPIA, (G) RPLP1, (H) RPL13A, (I) TBP and (J) YWHAZ based on melt curve analysis. Single melt curve indicated that only the desired product was specifically amplified in each reaction. d(RFU)/dT = Relative fluorescence units (RFU) with time (T).
Fig. 3.
Representative image of agarose gel (3%) electrophoresis showing amplification of a specific qRT-PCR product of the expected size for each reference gene (n = 3). No primer dimers and other unspecific amplification were found in the gel. Lane 1: GeneRulerTM low range DNA ladder; Lane 2: qRT-PCR product amplifying ACTB (91 bp); Lane 3: eIF4A (87 bp); Lane 4: GAPDH (82 bp); Lane 5: HPRT1 (90 bp); Lane 6: IPO8 (118 bp); Lane 7: PPIA (93 bp); Lane 8: RPLP1 (81 bp); Lane 9: RPL13A (133 bp); Lane 10: TBP (117 bp) and Lane 11: YWHAZ (106 bp).
Fig. 4.
Determination of qRT-PCR primer amplification efficiency for (A) ACTB, (B) eIF4A, (C) GAPDH, (D) HPRT1, (E) IPO8, (F) PPIA, (G) RPLP1, (H) RPL13A, (I) TBP and (J) YWHAZ based on standard curves calculation. The standard curve was generated by plotting the mean CT values against the 2-fold serial dilutions of cDNA.
Table 3.
The amplification efficiency and R2 obtained from standard curves for each candidate reference gene used in the study.
| Gene | Efficiency (E = [10(-1/slope)-1] × 100%) | Correlation coefficient (R2) |
|---|---|---|
| ACTB | 105.6 | 0.9998 |
| eIF4A | 98.0 | 0.9996 |
| GAPDH | 107.5 | 0.9992 |
| HPRT1 | 104.9 | 0.9986 |
| IPO8 | 108.9 | 0.9990 |
| PPIA | 91.7 | 0.9999 |
| RPLP1 | 91.6 | 0.9990 |
| RPL13A | 94.4 | 0.9998 |
| TBP | 113.9 | 0.9991 |
| YWHAZ | 106.1 | 0.9997 |
3.2. Expression level of candidate reference genes
The expression levels of all these 10 candidate reference genes were relatively similar across hypoxic-preconditioned and non-hypoxic H9-hNSCs except eIF4A and HPRT1 which showed significant downregulation under hypoxic condition (Fig. 5A). On the other hand, the expression levels of candidate reference genes across BEF-preconditioned and non-BEF H9-hNSCs were relatively similar except TBP which showed significant increase after BEF preconditioning (Fig. 5B). Due to their unstable transcription expressions across samples, eIF4A, HPRT1 and TBP would not be the reliable reference genes for normalization in their respective group. However, selection of suitable reference genes based on solely the raw CT value may mask genomic context, hence, more appropriate measures using statistical approach were carried out in subsequent steps to determine the reliability of each reference gene for qRT-PCR normalization.
Fig. 5.
The expression levels (CT value) of candidate reference genes in (A) hypoxic-preconditioned vs. non-hypoxic H9-hNSCs and (B) BEF-preconditioned vs. non-BEF H9-hNSCs. The average CT values were relatively similar across both cells in two respective groups, except the eIF4A, HPRT1 and TBP. The values were expressed as mean CT ± SEM from three independent experiments. Statistical analysis was performed using the two-tailed Student's t-test (*p < 0.05).
3.3. Expression stability of candidate reference genes by geNorm
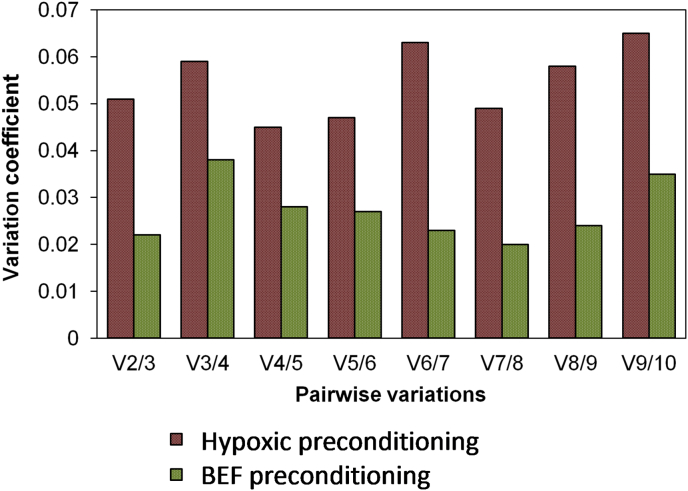
geNorm ranked the candidate reference genes differently across hypoxic- and BEF-preconditioned H9-hNSCs (Table 4). RPL13A and RPLP1 were ranked the most stable pair of reference genes whilst HPRT1 ranked the least stable reference gene across hypoxic- and non-hypoxic H9-hNSCs. To our surprise, the least stable reference gene (HPRT1) across hypoxic- and non-hypoxic H9-hNSCs was ranked the most stable reference genes across BEF- and non-BEF H9-hNSCs together with RPL13A. On the other hand, TBP was ranked the least stable in the BEF group. The V2/3 score which was lower than the cutoff value of 0.15 supported the use of two reference genes for qRT-PCR normalization of both hypoxic- and BEF-preconditioned H9-hNSCs (Fig. 6).
Table 4.
geNorm analysis of the candidate reference genes across hypoxic- and BEF-preconditioned H9-hNSCs. The most stably expressed reference genes had lowest M value.
| Ranking | Hypoxic |
BEF |
||
|---|---|---|---|---|
| M | Gene | M | Gene | |
| 1 | 0.0817 | RPL13A/RPLP1 | 0.0513 | RPL13A/HPRT1 |
| 3 | 0.1313 | ACTB | 0.0942 | PPIA |
| 4 | 0.1889 | TBP | 0.0985 | ACTB |
| 5 | 0.2063 | GAPDH | 0.1211 | RPLP1 |
| 6 | 0.2343 | PPIA | 0.1390 | YWHAZ |
| 7 | 0.2957 | IPO8 | 0.1520 | IPO8 |
| 8 | 0.3213 | YWHAZ | 0.1609 | eIF4A |
| 9 | 0.3663 | eIF4A | 0.1787 | GAPDH |
| 10 | 0.4237 | HPRT1 | 0.2157 | TBP |
Fig. 6.
Determination of the optimal number of reference genes based on geNorm analysis for hypoxic- and BEF-preconditioned H9-hNSCs. Cutoff value of 0.15 indicates the minimum number of reference genes for valid normalization. This result suggested a minimum two reference genes are necessary for qRT-PCR normalization.
3.4. Expression stability of candidate reference genes by NormFinder
The reference genes for hypoxic- and BEF-preconditioned groups ranked by NormFinder was found consistent to geNorm analysis. RPLP1 was selected as the most stably expressed gene whilst HPRT1 deemed the least stable for hypoxic-preconditioned H9-hNSCs. On the other hand, HPRT1 was the most reliable gene whilst TBP ranked the least reliable for BEF-preconditioned H9-hNSCs (Table 5).
Table 5.
NormFinder analysis for best candidate reference genes between hypoxic- and BEF-preconditioned H9-hNSCs. The most stably expressed reference genes had lowest M value.
| Ranking | Hypoxic |
BEF |
||
|---|---|---|---|---|
| M | Gene | M | Gene | |
| 1 | 0.094 | RPLP1 | 0.053 | HPRT1 |
| 2 | 0.14 | GAPDH | 0.056 | PPIA |
| 3 | 0.159 | TBP | 0.064 | RPL13A |
| 4 | 0.172 | IPO8 | 0.076 | ACTB |
| 5 | 0.176 | RPL13A | 0.090 | RPLP1 |
| 6 | 0.177 | YWHAZ | 0.093 | IPO8 |
| 7 | 0.216 | PPIA | 0.099 | YWHAZ |
| 8 | 0.288 | eIF4A | 0.118 | eIF4A |
| 9 | 0.365 | ACTB | 0.156 | GAPDH |
| 10 | 0.388 | HPRT1 | 0.218 | TBP |
3.5. Expression stability of candidate reference genes by BestKeeper
The ranking of reference gene based on BestKeeper SD, CV and r values exhibited differences across hypoxic- and BEF-preconditioned group (Table 6). Based on the geometric mean, RPLP1 was the most stably expressed reference gene whilst HPRT1 ranked the least reliable for qRT-PCR normalization of hypoxic-preconditioned and non-hypoxic H9-hNSCs. On the other hand, geometric mean revealed that GAPDH was the most stably expressed reference gene, while RPLP1 was the least reliable reference gene across BEF-preconditioned and non-BEF H9-hNSCs (Table 6).
Table 6.
Ranking of candidate reference genes across hypoxic- and BEF-preconditioned H9-hNSC based on BestKeeper analysis. Geometric mean was calculated for the final ranking.
| Hypoxic |
Final rank | BEF |
Final rank | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Ranking based on |
Geometric mean | Gene | Ranking based on |
Geometric mean | ||||||
| SD | CV | r | SD | CV | r | ||||||
| RPLP1 | 1 | 1 | 9 | 2.08 | 1 | GAPDH | 1 | 1 | 10 | 2.15 | 1 |
| ACTB | 2 | 2 | 10 | 3.42 | 2 | HPRT1 | 4 | 3 | 3 | 3.3 | 2 |
| RPL13A | 3 | 3 | 5 | 3.56 | 3 | YWHAZ | 2 | 4 | 5 | 3.42 | 3 |
| IPO8 | 7 | 7 | 1 | 3.66 | 4 | RPL13A | 3 | 5 | 4 | 3.91 | 4 |
| TBP | 4 | 4 | 6 | 4.58 | 5 | IPO8 | 4 | 2 | 8 | 4.00 | 5 |
| YWHAZ | 8 | 8 | 2 | 5.04 | 6 | TBP | 10 | 10 | 1 | 4.64 | 6 |
| GAPDH | 5 | 5 | 7 | 5.59 | 7 | PPIA | 8 | 9 | 2 | 5.24 | 7 |
| eIF4A | 9 | 9 | 3 | 6.24 | 8 | ACTB | 7 | 7 | 5 | 6.25 | 8 |
| PPIA | 6 | 6 | 8 | 6.6 | 9 | eIF4A | 6 | 6 | 9 | 6.87 | 9 |
| HPRT1 | 10 | 10 | 3 | 6.69 | 10 | RPLP1 | 8 | 8 | 7 | 7.65 | 10 |
3.6. Overall expression stability of candidate reference genes
The order of stability ranking obtained from all three programs was similar but not identical. This discrepancy in ranking might attribute to distinct underlying theoretical algorithms of each program. Overall, RPLP1 was ranked as the most reliable reference gene for qRT-PCR normalization of hypoxic preconditioning group (Table 7). HPRT1 was overall ranked as the most unstable reference gene for hypoxic-preconditioned and non-hypoxic H9-hNSCs (Table 7). To summarize, the comprehensive ranking from the most to the least stable reference gene for hypoxic preconditioning group was RPLP1, RPL13A, ACTB, TBP, GAPDH, IPO8, YWHAZ, eIF4A, PPIA and HPRT1. To be congruent with recommendation by pairwise variation analysis, the two best performing reference genes, RPLP1 and RPL13A were selected for qRT-PCR normalization of hypoxic preconditioning group.
Table 7.
Comprehensive ranking of the 10 candidate reference genes based on geometric mean of geNorm, NormFinder and BestKeeper ranking across hypoxic- and BEF-preconditioned H9-hNSCs.
| Hypoxic |
BEF |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | geNorm | Norm-Finder | Best-Keeper | Overall ranking | No. | geNorm | Norm-Finder | Best-Keeper | Overall ranking |
| 1 | RPL13A/RPLP1 | RPLP1 | RPLP1 | RPLP1 | 1 | RPL13A/HPRT1 | HPRT1 | GAPDH | HPRT1 |
| 2 | GAPDH | ACTB | RPL13A | 2 | PPIA | HPRT1 | RPL13A | ||
| 3 | ACTB | TBP | RPL13A | ACTB | 3 | PPIA | RPL13A | YWHAZ | PPIA |
| 4 | TBP | IPO8 | IPO8 | TBP | 4 | ACTB | ACTB | RPL13A | GAPDH |
| 5 | GAPDH | RPL13A | TBP | GAPDH | 5 | RPLP1 | RPLP1 | IPO8 | YWHAZ |
| 6 | PPIA | YWHAZ | YWHAZ | IPO8 | 6 | YWHAZ | IPO8 | TBP | ACTB |
| 7 | IPO8 | PPIA | GAPDH | YWHAZ | 7 | IPO8 | YWHAZ | PPIA | IPO8 |
| 8 | YWHAZ | eIF4A | eIF4A | eIF4A | 8 | eIF4A | eIF4A | ACTB | RPLP1 |
| 9 | eIF4A | ACTB | PPIA | PPIA | 9 | GAPDH | GAPDH | eIF4A | eIF4A |
| 10 | HPRT1 | HPRT1 | HPRT1 | HPRT1 | 10 | TBP | TBP | RPLP1 | TBP |
On the other hand, the comprehensive ranking for BEF preconditioning group in descending stability was HPRT1, RPL13A, PPIA, GAPDH, YWHAZ, ACTB, IPO8, RPLP1, eIF4A and TBP. Overall, HPRT1 was ranked as the most reliable reference gene for qRT-PCR normalization of BEF-preconditioned and non-BEF H9-hNSCs. TBP was overall ranked as the most unstable reference gene. The two most reliable reference genes, HPRT1 and RPL13A were selected to normalize gene expression data of BEF preconditioning group (Table 7).
4. Discussion
This study provided the first validation on reliability of ten frequently used candidate reference genes for hypoxic-preconditioned and BEF-precondiitoned H9-hNSCs, respectively. We found that both experimental conditions required different reference genes even they are from the same cell type, demonstrating that reference genes can be regulated depending on experimental conditions.
The cell types under investigation in this study were H9-hNSCs preconditioned with either hypoxia or BEF. The expression levels of all ten candidate reference genes were similarly regulated in both groups, except the eIF4A, HPRT1 and TBP. Downregulation of eIF4A in H9-hNSCs after hypoxic preconditioning was concomitant with previous studies which also reported significant decrease of eIF4A in human cells cultured under low oxygen condition (Ho et al., 2016). eIF4A encodes for protein functions as a subunit of the eukaryotic initiation factor (eIF) 4F (eIF4F) complex comprised on several initiation factors including the RNA helicase eIF4A, the cap-binding eIF4B and the scaffold eIF4G which altogether mediates the binding of mRNA to the ribosome (Jackson et al., 2010). In normoxia, the eIF4F complex typically initiates protein synthesis routinely. However, under hypoxia, low O2 concentration inhibits binding of eIF4E to eIF4G, thereby inhibiting eIF4F activity, as well as its counterpart, the eIF4A (Connolly et al., 2006). This explained our finding which showed significant decreased eIF4A expression level in hypoxic-preconditioned H9-hNSCs compared to the non-hypoxic cells. Further, this also support our finding that eIF4A was among the least stable reference genes for hypoxia group. On the other hand, HPRT1 has a central role in the generation of purine nucleotides through the purine salvage pathway. HPRT1 encodes hypoxanthine phosphoribosyltransferase which catalyzes conversion of hypoxanthine to inosine monophosphate and guanine to guanosine monophosphate via transfer of the 5-phosphoribosyl group from 5-phosphoribosyl 1-pyrophosphate, and this biochemistry process consumes energy (Keebaugh et al., 2007). Under hypoxia, as adenosine triphosphate (ATP) generation by oxidative phosphorylation begins to fall off due to O2 limitation, the cellular ATP demands of most mammalian cells and tissues tend to remain constant, trying to avoid unnecessary consumption of ATP. This could be the reason of the significant reduction of HPRT1 expression in hypoxic-preconditioned cells. Thus, we are not surprised when all three algorithms software: geNorm, NormFinder and BestKeeper revealed that HPRT1 as the least stable reference genes for hypoxic preconditioning group. Nonetheless, the least stable reference gene for BEF preconditioning was not elf4A or HPRT1. For this group of cells, TBP scored the lowest rank in stability as compared to other nine candidate reference genes. Furthermore, TBP also was the only reference gene that showed significant different in expression level among the BEF-preconditioned vs. non BEF-preconditioned H9-hNSCs. Hence, it was not recommended for qRT-PCR normalization of BEF-preconditioned H9-hNSCs.
Out of the 10 reference genes evaluated, RPLP1 was consistently ranked by geNorm, NormFinder and BestKeeper as the most suitable reference gene across hypoxic preconditioning group. The RPLP1 gene was documented as crucial ribosomal protein for progenitor cell proliferation, embryonic and brain development (Perucho et al., 2014). However, to our surprise, the least stable reference gene (HPRT1) for hypoxic preconditioning group was found to be the most stable reference gene in BEF preconditioning group. In a previous study done by Amable et al. (2013), HPRT1 was identified as one of the best reference gene in human bone marrow-derived mesenchymal stromal cells and dermal fibroblasts (Amable et al., 2013). Therefore, we suggest that the discrepancies of HPRT1 expression between cells indicated that it was regulated by experimental condition. This contradictory finding, again, highlighted the importance of identifying valid reference genes for each experimental set-up prior to gene expression analysis for accurate normalization of qRT-PCR data.
Normalization with multiple reference genes is becoming more prevalent. It was reported in previous studies that normalization based on multiple candidate genes provided the advantage of more accurate normalization calculation than a single gene (Tan et al., 2011; Tricarico et al., 2002). Based on geNorm analysis, we found that an optimum of two housekeeping genes were needed to perform a valid normalization for both hypoxic and BEF preconditioning group. In this analysis, RPLP1 and RPL13A were selected as the best combination for normalization of gene expression between hypoxic and non-hypoxic H9-hNSCs while HPRT1 and RPL13A were selected as the best two genes for normalization of gene expression between BEF- and non-BEF H9-hNSCs.
5. Conclusions
Ideally, reference genes should exhibit constant expression level across all tissue types and experimental settings. However, this study has pointed out that such reference genes do not exist. Not only that, we also found that the best reference gene for one experimental condition could be the worst for another experimental condition even using the same cell types. Therefore, this novel study clearly elucidated the importance of identifying valid reference genes for each experimental set-up prior to gene expression analysis. In conclusion, comprehensive analysis using geNorm, NormFinder and BestKeeper analysis revealed that RPLP1 and RPL13A were the best reference genes for hypoxic-preconditioned human NSCs whilst HPRT1 and RPL13A were the most reliable for BEF-preconditioned human NSCs.
Declarations
Author contribution statement
In Nee Kang: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Chong Yew Lee: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Suat Cheng Tan: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by Universiti Sains Malaysia (USM) Research University Individual (RUI) Grant (Grant no: 1001/PPSK/812140).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Aarnio K., Haapaniemi E., Melkas S., Kaste M., Tatlisumak T., Putaala J. Long-term mortality after first-ever and recurrent stroke in young adults. Stroke. 2014;45:2670–2676. doi: 10.1161/STROKEAHA.114.005648. [DOI] [PubMed] [Google Scholar]
- Amable P.R., Teixeira M.V., Carias R.B., Granjeiro J.M., Borojevic R. Identification of appropriate reference genes for human mesenchymal cells during expansion and differentiation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen C.L., Jensen J.L., Ørntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Bacigaluppi M., Pluchino S., Peruzzotti-Jametti L., Kilic E., Kilic U., Salani G., Brambilla E., West M.J., Comi G., Martino G., Hermann D.M. Delayed post-ischaemic neuroprotection following systemic neural stem cell transplantation involves multiple mechanisms. Brain. 2009;132:2239–2251. doi: 10.1093/brain/awp174. [DOI] [PubMed] [Google Scholar]
- Carandang R.A. Thirty years after the national institute of neurological disorders and stroke recombinant tissue plasminogen activator trial: a new era for stroke therapy. JAMA Neurol. 2016;73:265–267. doi: 10.1001/jamaneurol.2015.4453. [DOI] [PubMed] [Google Scholar]
- Cheah W.K., Hor C.P., Zariah A.A., Looi I. A review of stroke Research in Malaysia from 2000 - 2014. Med. J. Malays. 2016;71:58–69. [PubMed] [Google Scholar]
- Cho H., Lee H.Y., Ahn D.R., Kim S.Y., Kim S., Lee K.B., Lee Y.M., Park H., Yang E.G. Baicalein induces functional hypoxia-inducible factor-1alpha and angiogenesis. Mol. Pharmacol. 2008;74:70–81. doi: 10.1124/mol.107.040162. [DOI] [PubMed] [Google Scholar]
- Connolly E., Braunstein S., Formenti S., Schneider R.J. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol. Cell. Biol. 2006;26:3955–3965. doi: 10.1128/MCB.26.10.3955-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U.M., Harting M.T., Jimenez F., Monzon-Posadas W.O., Xue H., Savitz S.I., Laine G.A., Cox C.S., Jr. Pulmonary passage is a major obstacle for intravenous stem cell delivery: the pulmonary first-pass effect. Stem Cells Dev. 2009;18:683–692. doi: 10.1089/scd.2008.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L., Mauriat M., Guenin S., Pelloux J., Lefebvre J.F., Louvet R., Rusterucci C., Moritz T., Guerineau F., Bellini C., Van Wuytswinkel O. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 2008;6:609–618. doi: 10.1111/j.1467-7652.2008.00346.x. [DOI] [PubMed] [Google Scholar]
- Harminder, Singh V., Chaudhary A.K. A review on the taxonomy, ethnobotany, chemistry and pharmacology of Oroxylum indicum Vent. Indian J. Pharm. Sci. 2011;73:483–490. doi: 10.4103/0250-474X.98981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.J.D., Wang M., Audas T.E., Kwon D., Carlsson S.K., Timpano S., Evagelou S.L., Brothers S., Gonzalgo M.L., Krieger J.R., Chen S., Uniacke J., Lee S. Systemic reprogramming of translation efficiencies on oxygen stimulus. Cell Rep. 2016;14:1293–1300. doi: 10.1016/j.celrep.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao L.L., Dangond F., Yoshida T., Hong R., Jensen R.V., Misra J., Dillon W., Lee K.F., Clark K.E., Haverty P., Weng Z., Mutter G.L., Frosch M.P., MacDonald M.E., Milford E.L., Crum C.P., Bueno R., Pratt R.E., Mahadevappa M., Warrington J.A., Stephanopoulos G., Gullans S.R. A compendium of gene expression in normal human tissues. Physiol. Genom. 2001;7:97–104. doi: 10.1152/physiolgenomics.00040.2001. [DOI] [PubMed] [Google Scholar]
- Jackson R.J., Hellen C.U., Pestova T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keebaugh A.C., Sullivan R.T., Thomas J.W. Gene duplication and inactivation in the HPRT gene family. Genomics. 2007;89:134–142. doi: 10.1016/j.ygeno.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Lee S., Jo M., Lee J., Koh S.S., Kim S. Identification of novel universal housekeeping genes by statistical analysis of microarray data. J. Biochem. Mol. Biol. 2007;40:226–231. doi: 10.5483/bmbrep.2007.40.2.226. [DOI] [PubMed] [Google Scholar]
- Locatelli F., Bersano A., Ballabio E., Lanfranconi S., Papadimitriou D., Strazzer S., Bresolin N., Comi G.P., Corti S. Stem cell therapy in stroke. Cell. Mol. Life Sci. 2009;66:757–772. doi: 10.1007/s00018-008-8346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K.S., Li F., Liang P.Z., Chen X.W., Liu Y., Gao X.W. Identification and validation of reference genes for the normalization of gene expression data in qRT-PCR analysis in Aphis gossypii (Hemiptera: Aphididae) J. Insect Sci. 2016;16 doi: 10.1093/jisesa/iew003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olias P., Adam I., Meyer A., Scharff C., Gruber A.D. Reference genes for quantitative gene expression studies in multiple avian species. PLoS One. 2014;9 doi: 10.1371/journal.pone.0099678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perucho L., Artero-Castro A., Guerrero S., Ramón y Cajal S., Lleonart M.E., Wang Z.-Q. RPLP1, a crucial ribosomal protein for embryonic development of the nervous system. PLoS One. 2014;9 doi: 10.1371/journal.pone.0099956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W., Tichopad A., Prgomet C., Neuvians T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Reynolds B.A., Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Roybon L., Ma Z., Asztely F., Fosum A., Jacobsen S.E., Brundin P., Li J.Y. Failure of transdifferentiation of adult hematopoietic stem cells into neurons. Stem Cells. 2006;24:1594–1604. doi: 10.1634/stemcells.2005-0548. [DOI] [PubMed] [Google Scholar]
- Rumlow A., Keunen E., Klein J., Pallmann P., Riemenschneider A., Cuypers A., Papenbrock J. Quantitative expression analysis in Brassica napus by northern blot analysis and reverse transcription-quantitative PCR in a complex experimental setting. PLoS One. 2016;11 doi: 10.1371/journal.pone.0163679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrideli C.A., Carlotti C.G.J., Okamoto O.K., Andrade V.S., Cortez M.A., Motta F.J., Lucio-Eterovic A.K., Neder L., Rosemberg S., Oba-Shinjo S.M., Marie S.K., Tone L.G. Gene expression profile analysis of primary glioblastomas and non-neoplastic brain tissue: identification of potential target genes by oligonucleotide microarray and real-time quantitative PCR. Neuro Oncol. 2008;88:281–291. doi: 10.1007/s11060-008-9579-4. [DOI] [PubMed] [Google Scholar]
- Sharp F.R., Ran R., Lu A., Tang Y., Strauss K.I., Glass T., Ardizzone T., Bernaudin M. Hypoxic preconditioning protects against ischemic brain injury. Neurotherapeutics. 2004;1:26–35. doi: 10.1602/neurorx.1.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Sun G., Zhao C., Stewart R. Neural stem cell self-renewal. Crit. Rev. Oncol. Hematol. 2008;65:43–53. doi: 10.1016/j.critrevonc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroemer P., Patel S., Hope A., Oliveira C., Pollock K., Sinden J. The neural stem cell line CTX0E03 promotes behavioral recovery and endogenous neurogenesis after experimental stroke in a dose-dependent fashion. Neurorehabilitation Neural Repair. 2009;23:895–909. doi: 10.1177/1545968309335978. [DOI] [PubMed] [Google Scholar]
- Tachibana M., Ago T., Wakisaka Y., Kuroda J., Shijo M., Yoshikawa Y., Komori M., Nishimura A., Makihara N., Nakamura K., Kitazono T. Early reperfusion after brain ischemia has beneficial effects beyond rescuing neurons. Stroke. 2017;48:2222–2230. doi: 10.1161/STROKEAHA.117.016689. [DOI] [PubMed] [Google Scholar]
- Tan S.C., Carr C.A., Yeoh K.K., Schofield C.J., Davies K.E., Clarke K. Identification of valid housekeeping genes for quantitative RT-PCR analysis of cardiosphere-derived cells preconditioned under hypoxia or with prolyl-4-hydroxylase inhibitors. Mol. Biol. Rep. 2011;39:4857–4867. doi: 10.1007/s11033-011-1281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricarico C., Pinzani P., Bianchi S., Paglierani M., Distante V., Pazzagli M., Bustin S.A., Orlando C. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal. Biochem. 2002;309:293–300. doi: 10.1016/s0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. research0034.0031-0034.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakai T., Narasimhan P., Sakata H., Wang E., Yoshioka H., Kinouchi H., Chan P.H. Hypoxic preconditioning enhances neural stem cell transplantation therapy after intracerebral hemorrhage in mice. J. Cereb. Blood Flow Metab. 2015;36(12) doi: 10.1177/0271678X15613798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z.Z., Zhu Y.B., Zhang J.Y., McCrary M.R., Wang S., Zhang Y.B., Yu S.P., Wei L. Priming of the cells: hypoxic preconditioning for stem cell therapy. Chin. Med. J. 2017:2361–2374. doi: 10.4103/0366-6999.215324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian Y., Federspiel J.J., Grau-Sepulveda M., Hernandez A.F., Schwamm L.H., Bhatt D.L., Smith E.E., Reeves M.J., Thomas L., Webb L., Bettger J.P., Laskowitz D.T., Fonarow G.C., Peterson E.D. Risks and benefits associated with prestroke Antiplatelet therapy among patients with Acute ischemic stroke treated with intravenous tissue plasminogen activator. JAMA Neurol. 2016;73:50–59. doi: 10.1001/jamaneurol.2015.3106. [DOI] [PubMed] [Google Scholar]
- Xie F., Xiao P., Chen D., Xu L., Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Xie P., Zhu X.F., Cai Z.Y. Neural stem cell transplantation and nerve growth factor promote neurological recovery in rats with ischemic stroke. Nan Fang Yi Ke Da Xue Xue Bao. 2008;28:1123–1126. [PubMed] [Google Scholar]