Abstract
The lungs are one of the most common target organs of diabetic injury in patients with diabetes. Long non-coding RNA (lncRNA) smoke and cancer-associated lncRNA 1 (SCAL1), also known as lung cancer associated transcript 1 (LUCAT1), is known to have a pivotal role in lung cancer. The aim of the current study was to investigate the potential involvement of SCAL1 in diabetic lung disease. The expression levels of SCAL1 were determined by reverse transcription-quantitative PCR in serum samples from healthy controls (n=40), diabetic patients without lung disease (n=56) and diabetic patients with diabetic lung disease (n=44). Receiver operating characteristic analysis was used to evaluate the diagnostic value of serum SCAL1 in discriminating diabetic patients with diabetic lung disease from diabetic patients without lung disease and healthy controls. Pearson's correlation analysis was performed to examine the correlation between SCAL1 and inducible nitric oxide synthase (iNOS) mRNA expression levels in blood and lung tissue samples. Expression levels of iNOS and NO production following treatment with high (30 mM) glucose were examined by western blot analysis and NO assay, respectively. The expression levels of SCAL1 were significantly downregulated in diabetic patients with diabetic lung disease, and downregulated serum expression levels of SCAL1 effectively distinguished diabetic patients with diabetic lung disease from diabetic patients without lung disease and healthy controls. Treatment with high glucose significantly upregulated SCAL1 expression in normal lung cells. Furthermore, the overexpression of SCAL1 inhibited iNOS protein expression and reduced NO production in cells treated with high glucose. In conclusion, the current study demonstrated that lncRNA SCAL1 inhibits iNOS protein expression in lung cells under high-glucose conditions, which suggests that SCAL1 may have potential in the treatment of patients with diabetic lung disease.
Keywords: diabetic, lung, long non-coding RNA, smoke and cancer-associated lncRNA, lung cancer associated transcript 1, inducible nitric oxide synthase
Introduction
Diabetes mellitus is a group of common metabolic disorders that has become a major public health worldwide (1). Diabetes currently affects 6.4% of adults (285 million) globally and its incidence is expected to increase to 7.7% by 2030, affecting 439 million adults (2). In developing countries, the incidence of diabetes is predicted to increase by 60% in the next 20 years (2). Although the incidence of diabetes in China is relatively low, this may change in the future due to changes in lifestyle and increased psychological and physiological stress (3). In 2016, the World Health Organization revealed that diabetes causes ~1.5 million (2.7%) deaths every year, while the major cause of death in diabetic patients is glucotoxicity-induced complications in major organs (4). Although the lung is one of the least studied organs in diabetes, a number of studies have revealed that the lung is a common target of glucotoxicity-induced diabetic injury (5).
The pathogenesis of diabetic lung disease is complex and remains largely unknown (6). Excessive nitric oxide (NO) in lung tissue can cause platelet activation and induce chronic inflammation, which in turn leads to damaged lung capillary endothelium and microangiopathy (7). Nitric oxide synthases (NOSs) are a family of enzymes, which catalyze the production of NO from L-arginine (8). The inhibition of NOSs production under high-glucose conditions may prevent the occurrence of diabetic lung disease by protecting lung cells via reducing the inflammatory response.
Smoke and cancer-associated lncRNA 1 (SCAL1), also known as lung cancer associated transcript 1 (LUCAT1), is a long non-coding RNA (lncRNA) that was initially identified to play a role in lung cancer (9). In lung cancer, SCAL1 interact with downstream NRF2 in airway epithelial cells to mediate oxidative stress protection and regulate gene expression. In view of the roles of NOSs in the oxidative metabolism (8), the present study hypothesized that SCAL1 may also interact with NOSs. The current study aimed to investigate the role of SCAL1 in diabetic lung disease and its possible interactions with nitric oxide synthase (iNOS) to provide potential novel treatment targets.
Materials and methods
Patients and specimens
A total of 56 patients with type 2 diabetes without lung disease (diabetic group) and 44 patients with type 2 diabetes with pneumonia (diabetic lung group) were included in the current study. All patients with type 2 diabetes were diagnosed according to the criteria established by Chinese Medical Association (2014) and enrolled at Hebei General Hospital between March 2015 and January 2017. Inclusion criteria were as follows: i) patients received treatment for the first time; ii) patients with normal major organ function; and iii) patients willing to participate in the study. Exclusion criteria were as follows: i) detection of other severe diseases or other respiratory diseases; ii) patients aged ≥70 years (chronic disease prevalence rates are increased in patients aged ≥70 years); and iii) patients with lung inflammation caused by other factors. In addition, 40 healthy controls who received physical examinations were included in the current study as the control group. No significant differences in age and gender were observed among the three groups (Table I). Blood samples were collected from all participants on the first day of admission. Lung biopsies were obtained from 25/44 (57%) patients in the diabetic lung group. This study was approved by the Ethics Committee of Hebei General Hospital (Shijiazhuang, China). All participants and/or their families provided written informed consent.
Table I.
Basic information of participants.
| Group | n | Male (n) | Female (n) | Age range, years | Mean age, years |
|---|---|---|---|---|---|
| Control | 40 | 26 | 14 | 24–69 | 46.0±6.4 |
| Diabetes | 56 | 30 | 26 | 26–69 | 44.8±5.2 |
| Diabetic lung | 44 | 24 | 20 | 23–67 | 45.1±5.1 |
Cell culture and transfection
Normal human lung cell line BEAS-2B (ATCC® CRL-9609™) was purchased from the American Type Culture Collection (ATCC). Cells were cultured in ATCC-formulated Eagle's Minimum Essential Medium (cat. no. 30-2003; ATCC) supplemented with 10% FBS (Sangon Biotech Co., Ltd.) and maintained at 37°C in a 5% CO2-humidified incubator. The full-length SACL1 cDNA fragment was obtained by PCR amplification and cloned into the linearised pIRSE2 vector (Clontech Laboratories, Inc.) to generate the SACL1 expression vector. Lipofectamine® 2000 reagent (Thermo Fisher Scientific, lnc.) was initially mixed with expression vectors (10 nm) to form transfection reagent-vector complexes, prior to transfection. Cells were transfected with transfection reagent-vector complexes at 37°C for 5 h at 37°C. Cells were subsequently washed with fresh ATCC-formulated Eagle's Minimum Essential Medium to avoid cytotoxicity. Cells transfected with empty pIRSE2 vector were used as the negative control (NC), while untransfected cells were used as the control (C). SACL1 overexpression was confirmed by RT-qPCR at 24 h post-transfection and cells were collected at this time pointfor subsequent experimentation.
NO assay
Total NO in the cell culture supernatant was examined using the Nitric Oxide Detection kit (cat. no. ADI-917-010; Enzo Life Sciences, Inc.), according to the manufacturer's protocol.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from blood, tissue and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. For in vitro experiments, BEAS-2B cells were treated with D-glucose (5, 10, 20, 30 and 40 mM) for 5, 10 and 15 h at 37°C. Total RNA was reverse transcribed into cDNA using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific, Inc.). The following conditions were used for RT: 50°C for 15 min and 80°C for 10 min. qPCR was subsequently performed using SYBR® Green Real-Time PCR Master mix (Thermo Fisher Scientific, Inc.). The following primer pairs were used for qPCR: lncRNA SACL1 forward, 5′-GTGTCAAGCTCGGATTGCCT-3′ and reverse, 5′-GAGCCCACACACTCAGGTTC-3′; iNOS forward, 5′-CCCTTCCGAAGTTTCTGGCAGCAG-3′ and reverse, 5′-GGCTGTCAGAGCCTCGTGGCTTTGG-3′; and β-actin forward, 5′-GACCTCTATGCCAACACAGT-3′ and reverse, 5′-AGTACTTGCGCTCAGGAGGA-3′. The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 40 sec, 40 cycles of 95°C for 20 sec and 57°C for 30 sec. All data were quantified using the 2−ΔΔCq method (10) and normalized to the internal control β-actin.
Western blot analysis
Total protein was extracted from cells using RIPA buffer (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. For in vitro experiments, cells were treated with D-glucose (30 mM) for 15 h at 37°C. Total protein was quantified using a bicinchoninic acid assay and 20 µg protein/lane was separated via SDS-PAGE on a 10% gel. The separated proteins were transferred onto PVDF membranes, and then blocked for 1 h at room temperature with 5% skimmed milk. Membranes were washed and incubated with primary antibodies against iNOS (1:1,200; ab3523) and GAPDH (1:1,000; ab9485; both Abcam) overnight at 4°C. Membranes were washed and then incubated with horseradish peroxidase-labeled anti-rabbit IgG secondary antibody (1:1,000; MBS435036; MyBioSource) for 1 h at room temperature. Protein bands were visualized using Pierce™ ECL Western Blotting Substrate (Pierce; Thermo Fisher Scientific, Inc.). Protein expression was quantified using ImageJ v1.48 software (National Institutes of Health) and normalized to the loading control GAPDH.
Statistical analysis
Data were presented as the mean ± standard deviation. All statistical analyses were performed using GraphPad Prism software (version 6.0; GraphPad Software). One-way analysis of variance followed by the least significant difference test was used to analyze differences among multiple groups. Correlation analyses were performed using Pearson's correlation analysis. Receiver operating characteristic (ROC) curve analysis was performed to evaluate the diagnostic value of serum lncRNA SCAL1 in diabetic patients with diabetic lung disease compared with healthy controls. P<0.05 was considered to indicate a statistically significant difference.
Results
Serum levels of lncRNA SCAL1 were significantly downregulated in diabetic patients with diabetic lung disease
Differentially expressed genes between healthy controls and patients with a disease can often indicate the involvement of certain genes in a disease. The relative expression level of lncRNA SACL1 was determined by RT-qPCR in serum samples from healthy controls, diabetic patients without lung disease and diabetic patients with diabetic lung disease. The serum expression level of lncRNA SACL1 was significantly decreased in diabetic patients with diabetic lung disease compared with diabetic patients without lung disease and healthy controls (P<0.05; Fig. 1). Serum levels of SACL1 were decreased in diabetic patients without lung disease compared with healthy controls; however, the difference was not statistically significant.
Figure 1.
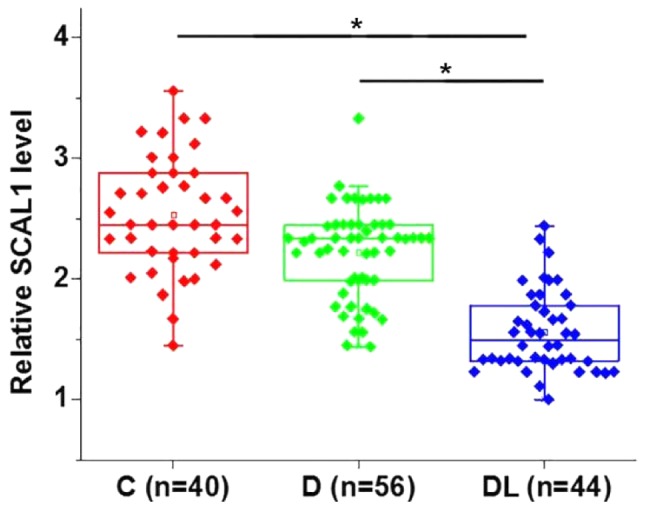
Serum levels of lncRNA SCAL1. Serum levels of lncRNA SCAL1 were significantly downregulated in diabetic patients with diabetic lung disease. *P<0.05 as indicated by lines. lncRNA, long non-coding RNA; SCAL1, smoke and cancer-associated lncRNA 1; C, control group; D, diabetic group; DL, diabetic lung group.
Low serum lncRNA SCAL1 distinguishes diabetic patients with diabetic lung disease from diabetic patients without diabetic lung disease and healthy controls
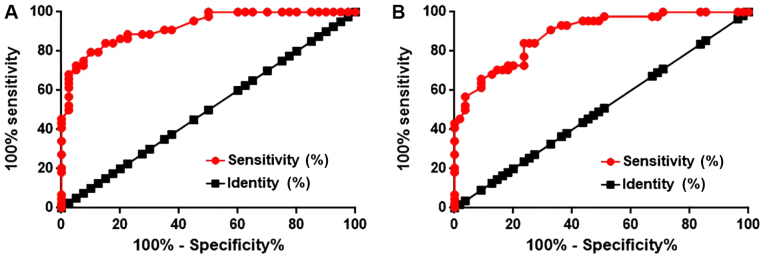
ROC curve analysis determined that the area under the curve (AUC) was 0.9233 (95% confidence interval: 0.8679–0.9769) with a standard error of 0.02735 (Fig. 2A). In addition, ROC analysis was used to evaluate the diagnostic value of serum lncRNA SCAL1 in discriminating diabetic patients with diabetic lung disease from diabetic patients without diabetic lung disease. The AUC was 0.8876 (95% confidence interval: 0.8254–0.9498) with a standard error of 0.03174 (Fig. 2B). An AUC>0.65 indicates the potential diagnostic value of a certain indicator for a disease (11). Therefore, serum levels of lncRNA SCAL1 can be used to effectively distinguish diabetic lung patients from healthy controls and diabetic patients without lung disease.
Figure 2.
ROC analysis of serum lncRNA SCAL1. ROC analysis was used to examine the diagnostic value of serum lncRNA SCAL1 to distinguish diabetic patients with diabetic lung disease from (A) healthy controls and (B) diabetic patients without lung disease. ROC, receiver operating curve; lncRNA, long non-coding RNA; SCAL1, smoke and cancer-associated lncRNA 1.
Expression levels of lncRNA SCAL1 are significantly upregulated in normal human lung cells following treatment with high glucose
D-glucose (5, 10, 20, 30 and 40 mM) was used to treat the BEAS-2B normal human lung cell line for 5, 10 and 15 h, respectively. The relative SCAL1 expression levels were significantly increased in lung cells treated with high glucose (10, 20, 30 and 40 mM) concentrations in a time- and dose-dependent manner compared with the control group (P<0.05; Fig. 3).
Figure 3.
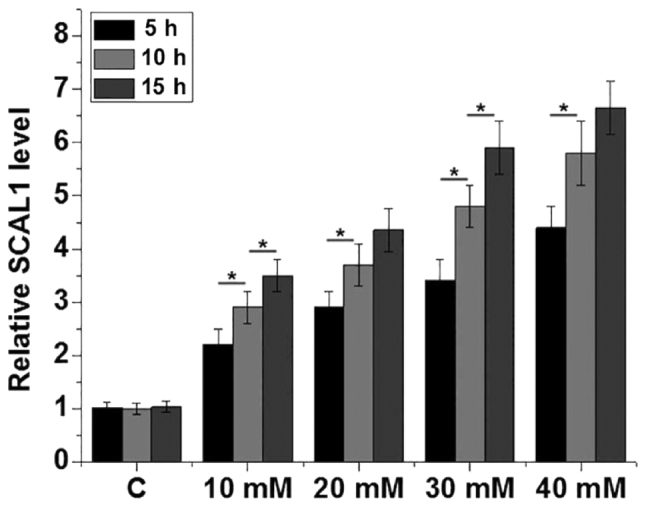
Relative expression levels of lncRNA SCAL1 were upregulated following treatment with high glucose. The relative expression levels of lncRNA SCAL1 were determined by reverse transcription-quantitative PCR in the BEAS-2B normal human lung cell line following treatment with various concentrations (5, 10, 20, 30 and 40 mM) of glucose. *P<0.05 as indicated by lines. LncRNA, long non-coding RNA; SCAL1, smoke and cancer-associated lncRNA 1; C, control.
SCAL1 and iNOS mRNA expression in blood and lung tissue samples are negatively correlated in diabetic patients with diabetic lung disease
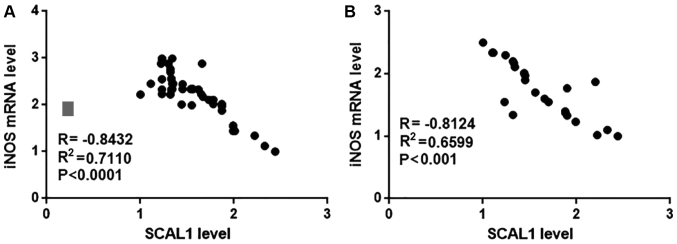
Pearson correlation analysis was used to examine the correlation between SCAL1 and iNOS mRNA expression in blood and lung tissue samples from diabetic patients with diabetic lung disease. Pearson's correlation analysis indicated a significant negative correlation between SCAL1 and iNOS mRNA expression levels in blood (R=−0.8432, P<0.0001; Fig. 4A) and lung tissue samples (R2=−0.8124, P<0.001; Fig. 4B) from diabetic patients with diabetic lung disease. No significant correlation was observed between SCAL1 and iNOS expression levels in blood and lung tissue samples from diabetic patients without diabetic lung disease or healthy controls (data not shown).
Figure 4.
Correlation of SCAL1 and iNOS mRNA expression in blood and lung tissue samples from diabetic patients with diabetic lung disease. The relative lncRNA SCAL1 expression level was inversely correlated with iNOS mRNA expression in (A) blood and (B) lung tissue samples from patients with diabetic lung disease. SCAL1, smoke and cancer-associated lncRNA 1; iNOS, inducible nitric oxide synthase; lncRNA, long non-coding RNA.
SCAL1 overexpression inhibits iNOS expression and reduces NO production in normal human lung cells
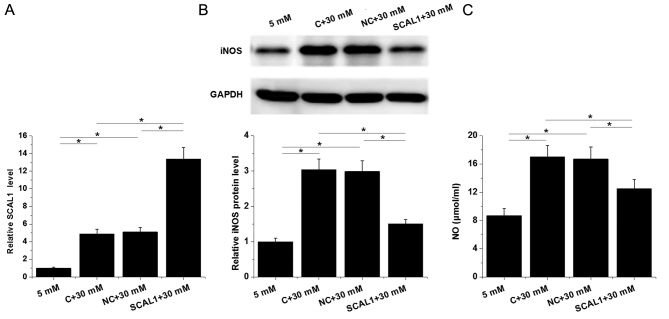
The expression of lncRNA SACL1 was determined by RT-qPCR in normal human lung cells following transfection with SCAL1 expression vector and treatment with D-glucose (30 mM). Untransfected cells and cells transfected with empty circular pIRSE2 vector were used as control and negative control groups, respectively (Fig. 5). The relative expression level of lncRNA SACL1 was significantly increased in the control group following treatment with high (30 mM D-glucose) glucose compared with the group treated with low (5 mM D-glucose) glucose (Fig. 5A). In addition, the relative expression level of lncRNA SACL1 was significantly increased in cells following transfection with SCAL1 expression vector compared with the negative control, indicating that the transfection was successful (P<0.05; Fig. 5A).
Figure 5.
SCAL1 overexpression inhibited iNOS expression and reduced NO production in lung cells under high-glucose conditions. (A) The relative SCAL1 expression level was determined by reverse transcription-quantitative PCR in the BEAS-2B normal human lung cell line following transfection with SCAL1 expression vector and treatment with high glucose. (B) The relative iNOS protein expression level was determined by western blotting in BEAS-2B cells following transfection with SCAL1 expression vector and treatment with high glucose. (C) NO production was examined in the supernatant of BEAS-2B cells following transfection with SCAL1 expression vector and treatment with high glucose. *P<0.05 as indicated by lines. SCAL1, smoke and cancer-associated lncRNA 1; iNOS, inducible nitric oxide synthase; NO, nitric oxide; C, control; NC, negative control.
The relative protein expression level of iNOS, and secretion of NO into the cell supernatant were significantly upregulated in cells treated with high glucose compared with those treated with low glucose (P<0.05; Fig. 5B and C). The overexpression of SCAL1 inhibited iNOS protein expression and reduced NO production following treatment with high glucose compared with the high glucose-treated controls (P<0.05; Fig. 5B and C). Furthermore, no significant differences were observed in the protein expression level of iNOS and NO production in cells following the overexpression of SCAL1 and treatment with low glucose compared with control cells following treatment with low glucose (data not shown).
Discussion
Excessive NO production plays a pivotal role in the development of diabetic lung disease (7). The current study demonstrated that SCAL1, a lncRNA with a role in lung cancer, may be involved in the pathogenesis of diabetic lung disease. It also indicated that underlying mechanism of SCAL1 in diabetic lung may be via the downregulation of iNOS expression and reduced NO production.
Glucotoxicity, which is caused by a high-glucose environment in patients with diabetes, affects the expression levels of several genes as well as lncRNAs (12,13). Altered expression of lncRNAs may participate in the pathogenesis of diabetes-associated complications by controlling blood glucose concentrations (14,15). SCAL1 is upregulated in human lung cancer cell lines in response to cigarette smoke (9). In the current study, serum expression levels of SCAL1 were decreased in diabetic patients without lung disease compared with healthy controls, although the difference was not observed to be significant. However, in vitro experiments performed in the present study suggest that the expression level of lncRNA SCAL1 was upregulated under high-glucose conditions. Therefore, it is suggested that the downregulation of SCAL1 expression in patients with diabetic lung disease may be induced by the formation of lung lesions.
Diabetic lung disease is one of the least studied diabetes-associated complications and its pathogenesis is largely unknown, leading to difficulties in disease diagnosis. A previous study reported that caveolin-1 overexpression is associated with structural modifications of endothelial cells in patients with diabetic lung disease, indicating the potential application of caveolin-1 as a diagnostic biomarker for diabetic lung disease (16). The development of human disease is usually associated with changes in certain substances in the blood, and the detection of those substances may provide guidance for disease diagnosis (17). In the current study, the relative expression levels of SCAL1 were significantly downregulated in patients with diabetic lung disease, and downregulated serum expression levels of SCAL1 effectively distinguished diabetic patients with diabetic lung disease from diabetic patients without lung disease and healthy controls. Therefore, serum SCAL1 may serve as a potential diagnostic biomarker for diabetic lung disease in diabetic patients.
Excessive production of NO is thought to be involved in the pathogenesis of different human diseases, including diabetic lung disease (7,18,19) and, therefore, inhibition of NO production may suppress the development of those diseases. In the current study, the lncRNA SCAL1 expression level was inversely correlated with the iNOS mRNA expression level in blood and lung tissue samples from patients with diabetic lung disease. Furthermore, SCAL1 overexpression inhibited iNOS protein expression and NO production in normal lung cells. Therefore, SCAL1 may be used as a therapeutic target for the prevention and treatment of diabetic lung disease. The current study demonstrated a potential interaction between SCAL1 and iNOS in lung tissue under high-glucose conditions; however, whether this interaction is direct or indirect remains unknown and further experimentation is required to further examine this interaction.
The current study is limited by the small sample size; therefore, future studies with a larger sample size are required to further confirm the conclusions made. In addition, the underlying molecular mechanism by which SCAL1 regulates iNOS remains unclear and requires further investigation.
In conclusion, the relative expression levels of SCAL1 were significantly downregulated in diabetic patients with diabetic lung disease. SCAL1 expression may suppress diabetic lung disease by inhibiting iNOS protein expression, and thereby reducing the production of NO in lung tissues.
Acknowledgements
Not applicable.
Funding
No funding received.
Availability of data and materials
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
PL designed experiments. PL and NZ performed experiments. PL, FP, YG and LC assisted experiments and analyzed data. PL drafted the manuscript and all authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Hebei General Hospital (Shijiazhuang, China). All participants and/or their families provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Piero MN, Nzaro GM, Njagi JM. Diabetes mellitus-a devastating metabolic disorder. Asian J Biomed Pharm. 2015;5:1–7. doi: 10.15272/ajbps.v4i40.645. [DOI] [Google Scholar]
- 2.Shaw JE, Sicree RA, Zimmet PZ. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pract. 2010;87:4–14. doi: 10.1016/j.diabres.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Li MZ, Su L, Liang BY, Tan JJ, Chen Q, Long JX, Xie JJ, Wu GL, Yan Y, Guo XJ, Gu L. Trends in prevalence, awareness, treatment, and control of diabetes mellitus in mainland China from 1979 to 2012. Int J Endocrinol. 2013 doi: 10.1155/2013/753150. 753150, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roglic G. WHO Global report on diabetes: A summary. Int J Noncommun Dis. 2016;1:3–8. doi: 10.4103/2468-8827.184853. [DOI] [Google Scholar]
- 5.Pitocco D, Fuso L, Conte EG, Zaccardi F, Condoluci C, Scavone G, Incalzi RA, Ghirlanda G. The diabetic lung-a new target organ. Rev Diabet Stud. 2012;9:23–25. doi: 10.1900/RDS.2012.9.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsia CC, Raskin P. The diabetic lung: Relevance of alveolar microangiopathy for the use of inhaled insulin. Am J Med. 2005;118:205–211. doi: 10.1016/j.amjmed.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal A. Diabetic Lung: A Sweet Kiss of Impending Death. EC Pulmonology and Respiratory Medicine. 2018;7:341–342. [Google Scholar]
- 8.Leferink NG, Hay S, Rigby SE, Scrutton NS. Towards the free energy landscape for catalysis in mammalian nitric oxide synthases. FEBS J. 2015;282:3016–3029. doi: 10.1111/febs.13171. [DOI] [PubMed] [Google Scholar]
- 9.Thai P, Statt S, Chen CH, Liang E, Campbell C, Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol. 2013;49:204–211. doi: 10.1165/rcmb.2013-0159RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 12.Kaizer EC, Glaser CL, Chaussabel D, Banchereau J, Pascual V, White PC. Gene expression in peripheral blood mononuclear cells from children with diabetes. J Clin Endocrinol Metab. 2007;92:3705–3711. doi: 10.1210/jc.2007-0979. [DOI] [PubMed] [Google Scholar]
- 13.Reynier F, Pachot A, Paye M, Xu Q, Turrel-Davin F, Petit F, Hot A, Auffray C, Bendelac N, Nicolino M, et al. Specific gene expression signature associated with development of autoimmune type-I diabetes using whole-blood microarray analysis. Genes Immun. 2010;11:269–278. doi: 10.1038/gene.2009.112. [DOI] [PubMed] [Google Scholar]
- 14.Alvarez ML, DiStefano JK. The role of non-coding RNAs in diabetic nephropathy: potential applications as biomarkers for disease development and progression. Diabetes Res Clin Pract. 2013;99:1–11. doi: 10.1016/j.diabres.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 15.He X, Ou C, Xiao Y, Han Q, Li H, Zhou S. LncRNAs: Key players and novel insights into diabetes mellitus. Oncotarget. 2017;8:71325–71341. doi: 10.18632/oncotarget.19921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uyy E, Antohe F, Ivan L, Haraba R, Radu DL, Simionescu M. Upregulation of caveolin-1 expression is associated with structural modifications of endothelial cells in diabetic lung. Microvasc Res. 2010;79:154–159. doi: 10.1016/j.mvr.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Tobolowsky FA, Wada N, Martinez-Maza O, Magpantay L, Koletar SL, Palella FJ, Jr, Brown TT, Lake JE. Brief report: Circulating markers of fibrosis are associated with immune reconstitution status in HIV-infected men. PLoS One. 2018;13:e0191606. doi: 10.1371/journal.pone.0191606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuste JE, Tarragon E, Campuzano CM, Ros-Bernal F. Implications of glial nitric oxide in neurodegenerative diseases. Front Cell Neurosci. 2015;9:322. doi: 10.3389/fncel.2015.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura T, Lipton SA. Protein S-nitrosylation as a therapeutic target for neurodegenerative diseases. Trends Pharmacol Sci. 2016;37:73–84. doi: 10.1016/j.tips.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.