Abstract
Immune checkpoint blockade (ICB) therapy has achieved remarkable clinical benefit in non‐small‐cell lung cancer (NSCLC), but our understanding of biomarkers that predict the response to ICB remain obscure. Here we integrated somatic mutational profile and clinicopathologic information from 113 NSCLC patients treated by ICB (CTLA‐4/PD‐1). High tumor mutation burden (TMB) and neoantigen burden were identified significantly associated with improved efficacy in NSCLC immunotherapy. Furthermore, we identified apolipoprotein B mRNA editing enzyme, catalytic polypeptide‐like (APOBEC) mutational signature was markedly associated with responding of ICB therapy (log‐rank test, P = .001; odds ratio (OR), 0.18 [95% CI, 0.06‐0.50], P < .001). The association with progression‐free survival remained statistically significant after controlling for age, sex, histological type, smoking, PD‐L1 expression, hypermutation, smoking signature and mismatch repair (MMR) (HR, 0.30 [95% CI, 0.12‐0.75], P = .010). Combined high TMB with APOBEC signature preferably predict immunotherapy responders in NSCLC cohort. The CIBERSORT algorithm revealed that high APOBEC mutational activity samples were associated with increased infiltration of CD4 memory activated T cells, CD8+ T cells and natural killer (NK) cells, but reduced infiltration of regulatory T cells. Besides, individual genes mutation of IFNGR1 or VTCN1 were only found in responders; however, the PTEN mutation was only found in non‐responders (Fisher's exact test, all P < .05). These findings may be applicable for guiding immunotherapy for patients with NSCLC.
Keywords: APOBEC signature, immune checkpoint blockade, neoantigen burden, non‐small‐cell lung cancer, tumor mutation burden
Abbreviations
- APOBEC
apolipoprotein B mRNA editing catalytic polypeptide‐like family of enzymes
- B2M
β2 microglobulin
- CTLA4,
also known as CD152, cytotoxic T‐lymphocyte protein 4
- EGFR
epidermal growth factor receptor
- HR
hazards ratio
- ICB
immune checkpoint blockade
- IFNGR1
interferon gamma receptor 1
- KRAS KRAS proto‐oncogene
GTPase
- LAG3
lymphocyte activation gene 3 protein
- MMR
mismatch repair
- NA
not available
- NB
neoantigen burden
- NMF
non‐negative matrix factorization,
- NSCLC
non‐small‐cell lung cancer
- OR
odds ratio
- PD‐1
programmed cell death protein 1
- PD‐L1,
also known as CD274, B7H1, programmed cell death 1 ligand 1
- PTPN2
tyrosine‐protein phosphatase non‐receptor type 2
- SNV
single nucleotide variants
- TCGA
the Cancer Genome Atlas
- TMB
non‐synonymous tumor mutation burden
- TML
total tumor mutation load
- TP53
tumor protein p53
- VTCN1
V‐set domain containing T cell activation inhibitor 1
1. INTRODUCTION
Immune checkpoint blockades (ICB) therapy have demonstrated durable antitumor effects in the treatment of non‐small‐cell lung cancer (NSCLC) and other tumors.1, 2, 3, 4 Recent genomic mutation studies have elucidated potential driver genetic aberrations underlying NSCLC immune responses.5, 6, 7 Cancer genomic and transcriptome features can contribute to the response to ICB such as higher tumor mutation burden (TMB), PD‐L1 expression and interferon‐γ (IFNγ) signaling were correlated with survival benefits from anti‐CTLA‐4 and/or anti‐PD‐1 therapy.5, 8, 9 Somatic mutations can cause the presence of tumor‐specific neoantigen (neopeptide fragments), some of which with unique qualities serve as T‐cell targets10 and can be exploited to identify responders to immune checkpoint inhibitors.11, 12
The mutational signatures are fingerprints of endogenous and exogenous factors that have acted over the course of tumorigenesis and heterogeneity. Smoking signatures and MMR signatures have been reported in immune responses in lung cancer patients.6, 13 The APOBEC signature, characterized by C>T mutations at TpCpW (where W = A or T) trinucleotide sequences, has been found in bladder, breast, cervical, and non‐small‐cell lung cancers (NSCLCs),14, 15 and attributed to the activity of the AID/APOBEC family of cytidine deaminases.16 The APOBEC gene family have been found to be associated with interferon‐associated anti‐virus immunity activity.17 Nevertheless, any possible association between the APOBEC mutational signature and the NSCLC immune response remains unknown.
Individual gene mutation may play a vital role in guiding immunotherapy. Recent studies have found that IFNGR1, a subunit of the IFN‐γ receptor, acted in IFN‐γ pathways and regulated the immune response.18 Loss of function of PTEN was been previously reported to increase resistance to immune checkpoint blockade (ICB) in melanoma19 and uterine leiomyosarcoma.20
Features of the tumor microenvironment (TME) have been also associated with response to ICB therapy. Baseline levels of tumor‐infiltrating CD8+ T cells and CD4+ T cells are correlated with the response to immune therapy.21 Enriched genes involved in the pathways for IL2‐STAT5, TNF‐α signaling via NF‐κB were also reported to have a connection with immunotherapy.22
The purpose of this study was to characterize mutational signatures and prognosticators in patients diagnosed with NSCLC who were treated using ICB. The jointly interrogating NSCLC genomic data and clinical information were collected from published immune therapy studies. We consider that these findings may be applicable for guiding immunotherapy treatment for NSCLC patients.
2. MATERIALS AND METHODS
2.1. Genomic data and clinical information
Somatic mutations data (70 965 coding somatic mutations) for the aggregated immunotherapy cohort were acquired from recent publications5, 6, 7 (Table S1). All previously called somatic mutations were re‐annotated by oncotator23 against hg19 reference genomics database. Both peptides resulting from wild‐type and mutated sequences for predicted binding affinities scores below 500 nM to patient human leukocyte antigen (HLA) are defined as neoantigens (Table S2).6, 10 Patients with confirmed complete or partial response were considered to be responders; patients with stable disease, progressive disease, or not evaluable were considered to be non‐responders. All patients had stage IV NSCLC and were treated by PD‐1 or PD‐1 plus CTLA‐4 blockades. Detailed clinical information including age, gender, histological type, PD‐L1 expression status (antibody and cutoff level), smoking status, ICB types, progression‐free time, and status were also collected from these studies and are illustrated in Table S3. The overall clinical characteristics were summarized and are listed in Table S4. Somatic mutation, gene expression data and clinical information from 998 NSCLC samples (lung adenocarcinoma, n = 509; lung squamous‐cell carcinoma, n = 489) in the Cancer Genome Atlas (TCGA) were downloaded from the Genome Data Commons site (https://portal.gdc.cancer.gov).
2.2. Deciphering mutational signature operative in the genome
The framework advised by Kim et al24, 25 was used to extract mutational signatures of aggregated samples (n = 113) and TCGA NSCLC cohorts. We used the framework based on Bayesian variant nonnegative matrix factorization that can automatically determine the optimal number of mutational signatures. The mutation portrait matrix A was factorized into 2 nonnegative matrices W and H, where W represent mutational processes and H represent the corresponding mutational activities. The rows of matrix A are the 96 base substitutions in trinucleotide sequence contexts, and its columns are the NSCLC samples of both cohorts. The extracted mutational portrait of NSCLC was compared by cosine similarity against the Catalogue of Somatic Mutations in Cancer (COSMIC, v85, released 08‐May‐18).26, 27
2.3. Extracted signature vs immune response
The extracted mutational signatures were stratified as binary variables (i.e No and Yes) in the multivariate model. The classified method is according to a previous study, in which a signature was considered significant if it contributed to more than 100 substitutions or more than 25% of total mutation activities.26 We used Generalized Linear Models and Fit Proportional Hazards Regression Model to perform multivariate regression analyses.
2.4. GSEA and network analysis
The gene expression profile was collected from the NSCLC cohort of TCGA. Patients of APOBEC mutational activity above the top quartile of this cohort were defined as the APOBEC high activity group (n = 250) and below the bottom quartile were defined as the APOBEC low activity group (n = 250). The R package limma28 was used to evaluate differential expression of each gene in TCGA NSCLC samples with different APOBEC signature activities. Specifically, gene expression data were normalized and then fed to lmFit and eBayes functions in the R limma package. The differential expression statistics obtained from the eBayes function were used as input to perform gene set enrichment analysis29 on the HallMarker gene set (download from MSigDB database v6.2, updated: July 2018). The fast gene set enrichment analysis algorithm implemented in Bioconductor R package fgsea was used.
2.5. Tumor‐infiltrating lymphocyte cells analysis
CIBERSORT (http://cibersort.stanford.edu/) was used to conduct the estimation of the abundances of tumor‐infiltrating lymphocyte (TIL) cell types30 with the gene expression profile of each patient. Gene expression data of 250 low APOBEC signature activity samples and 250 high activity samples were also collected from TCGA.
2.6. Statistical analyses
Statistical analyses in this study were generated by R‐3.2.3. Quantitative data are presented as the median. Continuous variables between groups were compared using the Mann‐Whitney U test. The Spearman Correlation Coefficient was used to analyze the correlation between two quantitative variables. The association between mutational signatures and immunotherapy response status was tested by logistic regression analysis and adjusted for age, sex, smoking, PD‐L1, histological type, and hypermutation, OR was used for evaluate response capability. Kaplan‐Meier survival analysis and Cox proportional hazards model were used to analyze the association between mutational signatures and prognosis with the R survival package (Survminer 2.40‐1). We used stan_lm from the R package rstanarm (version 2.13.1) to perform multivariate Bayesian logistic regression analyses. All comparisons were two‐sided with an alpha level of .05, and Benjamini‐Hochberg correction was applied to control for the false discovery rate (FDR) for multiple hypothesis testing.31
3. RESULTS
3.1. Tumor mutation characteristics associated with immunotherapy benefit
Somatic mutational profiles of 113 NSCLC patients from previous genomic immune therapy studies were analyzed. A median of 468 mutations per sample (range from 11 to 2921) in 70 964 coding somatic mutations were collected from 3 previously published research studies. Overall, we found that elevated total tumor mutation load (TML, including silent variants) and non‐synonymous tumor mutation burden (TMB) were all significantly associated with an improved immune response, but TMB had a more strong association than TML (Wilcoxon rank sum test, TMB: P = 3.1e‐5, TML: P = 8.7e‐3; AUC, TMB: 0.74, TML: 0.65, P = 0.039; Figure S1). Subsequently, we then sought to determine the association between aggregate neoantigen properties and clinical benefit. The high neoantigen burden (NB, median served as cutoff) was significantly associated with improved ICB treatment progression‐free survival and immune response (log‐rank test, P = 0.047, Wilcoxon rank sum test, P = 0.002) (Figure S2A,B). The association between NB with immune response remained significant, even after controlling for age, gender, smoke history and PD‐L1 expression (HR, 0.48 [95% CI: 0.28‐0.8], P = 0.005) (Figure S2C). We next assessed mutations in individual genes (such as common oncogenic driver mutations, TP53, EGFR, KRAS, and PTEN; and immune response associated genes, B2M, PD‐L1, IFNGR1 and PTPN2) that may influence response or resistance to ICB treatment. Mutations in PTEN were only found in non‐responders (5 in 74, 6.8%), whereas mutations in IFNGR1 or VTCN1 were only found in responders (IFNGR1, 4 in 39, 10.2%; VTCN1, 3 in 39, 7.7%). TP53 and EGFR findings were not statistically significance possibly due to small numbers (responder vs non‐responder: TP53, 64.1% vs 48.6%; EGFR, 7.7% vs 14.9%, P > .05). In the current cohort, mutations in CD274 (PD‐L1), B2M, and PTPN2 were all rare, occurring in only one patient with non‐synonymous mutation (Figure 1).
Figure 1.
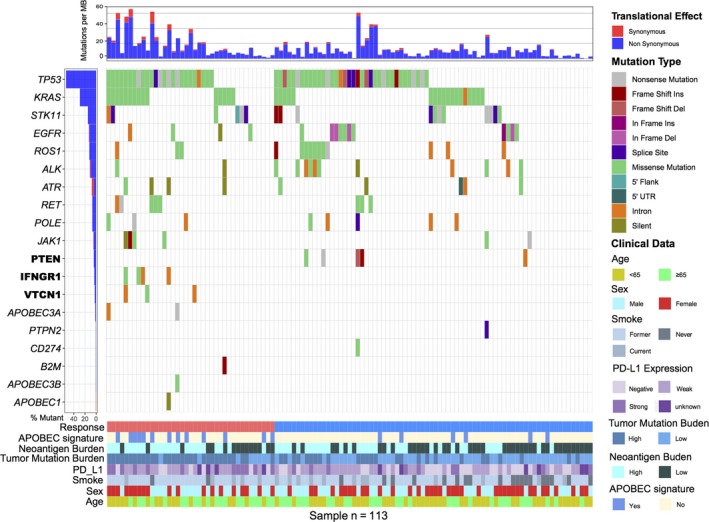
Mutational landscape of immune‐related genes in NSCLC patients treated with immune checkpoint blockade therapy. The left panel indicates gene mutation frequency, the upper panel shows mutational prevalence with respect to synonymous and non‐synonymous mutations, the middle panel depicts the gene mutation landscape across analyzed cases with different mutation types color coded differently, and the bottom panel displays clinical features such as immune response status, sex, histological type, smoking status, PD‐L1 expression, and APOBEC signature. Differentially responsive genes are highlighted in upper left bold
3.2. Mutational signatures operative in aggregated NSCLC cohort
To gain further insights into the mutational processes operative in responder vs non‐responder, we delineated the mutation signatures from the somatic mutation data. The overall mutational pattern was mainly dominated by C > T and C > A mutations and the responding vs non‐responding group had no obvious difference in the single‐nucleotide variant (SNV) mutation pattern (Figures 2A and S3A). Subsequently, we extracted 6 mutational signatures (i.e signatures 2, 4, 6, 7, 16 and an unknown signature) from the NSCLC with varying mutational activities (Figure 2B). A heat map depicting these 6 mutational signatures and COSMIC signatures is shown in Figure S3B. We observed that signature 2 (mutation activity, 4033/39 966, 10.1%), characterized by C>T mutations at TpCpW (where W = A or T) trinucleotide sequences, has been attributed to activity of the AID/APOBEC family of cytidine deaminases (APOBEC).14 Signature 4, the most prevalent signature, accounted for 24 178 in 39 966 mutations (60.5%) and was found to be associated with smoking in lung cancer. Signature 6 (5289/39 966, 13.2%), characterized by C>T mutations, is thought to be associated with defective DNA mismatch repair (MMR) and has been found in microsatellite unstable tumors. Signature 7 (3006/39 966, 7.5%) is associated with large numbers of CC>TT dinucleotide mutations at dipyrimidines and found predominantly in melanoma, possibly due to ultraviolet light exposure. Signature 16 (1942/39 966, 4.8%) contains an extremely strong transcriptional strand bias for T>C mutations in the ApTpN context and may be associated with alcohol consumption32 (Figure 2C).
Figure 2.
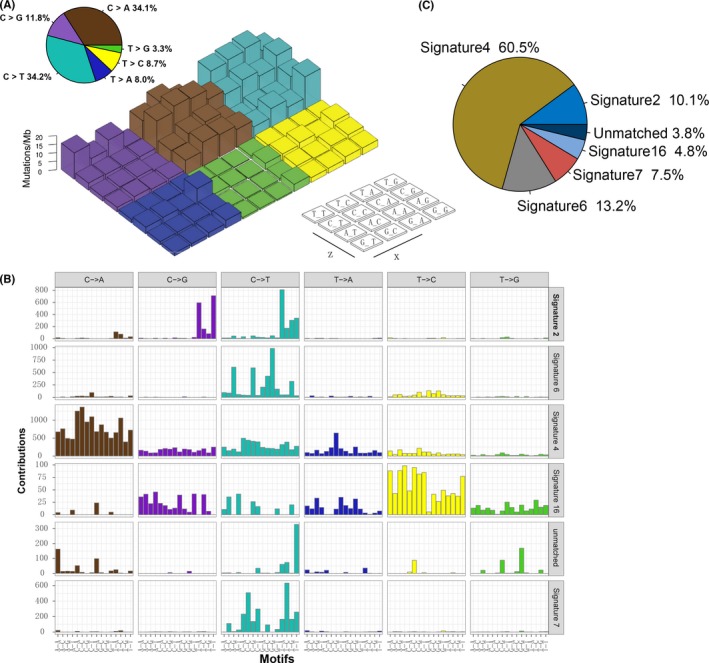
Mutational signatures extracted from the aggregated NSCLC dataset. A, Lego plot representation of mutation patterns in 113 NSCLC samples. Single‐nucleotide substitutions are divided into 6 categories with 16 surrounding flanking bases. The pie chart in the upper left shows the proportion of 6 categories of mutation patterns. B, The mutational activities of corresponding extracted mutational signatures (signatures 2, 4, 6, 7, 16, 26, and unmatched, named COSMIC signature). C, The mutational activities of corresponding mutational signatures displayed as a pie chart
3.3. Mutational signatures correlated with the immune response
To identify mutagenic factors that are responsible for the immune response, logistic regression and survival analyses were performed to determine the relationship between mutation signatures and immune response. Of interest, we observed that patients with signature 2 (associated with APOBEC family of cytidine deaminases) were markedly associated with better immune responses. The objective response rate for NSCLC patients was 68.4% with the APOBEC signature vs 27.6% without the APOBEC signature, and the median progression‐free survival was not reached (95% CI, 17.1 to NA) vs 5.9 mo (95% CI, 3.8‐7.9), (HR, 0.27 [95% CI: 0.12‐0.63], P = 0.002; log‐rank test, P = .001, Figure 3A). To rule out the possibility that associations between APOBEC mutational signature and immune response were affected by confounding factors, we included age, sex, PD‐L1 expression, smoking history, histological type, MMR signature, smoking signature, and hypermutation in the multivariate model. Associations between APOBEC mutations with immunotherapy survival (HR, 0.31 [95% CI: 0.12‐0.78], P = .010; Figure 3B) and immune response status (OR, 0.10 [95%CI: 0.02‐0.44], P = 0.004; Figure 3C) remained statistically significant. APOBEC mutational activities were strongly correlated with tumor mutation burden and neoantigen burden (TMB, Spearman's R = 0.59, P < .001, Figure S4 bottom; NB, Spearman's R = 0.54, P < .001; Figure S4 top). Furthermore, TMB and NB were greater in patients with the APOBEC signature than those without (TMB, median, 272 vs 160, P = .016, Figure 3D left; NB, median, 576 vs 215, P = 0.005; Figure 3D right). To identify the most suitable biomarkers for immune response and rule out the influence of confounding factors, we included mutations in IFNGR1, VTCN1 and PTEN, APOBEC signature, TMB and NB status, PD‐L1 expression, age, and sex in the Bayesian logistic regression model. Association between APOBEC signature and immunotherapy response remained statistically significant after controlling for such factors (OR, 0.23 [95% CI, 0.07‐0.82], P = .017; Figure S5).
Figure 3.
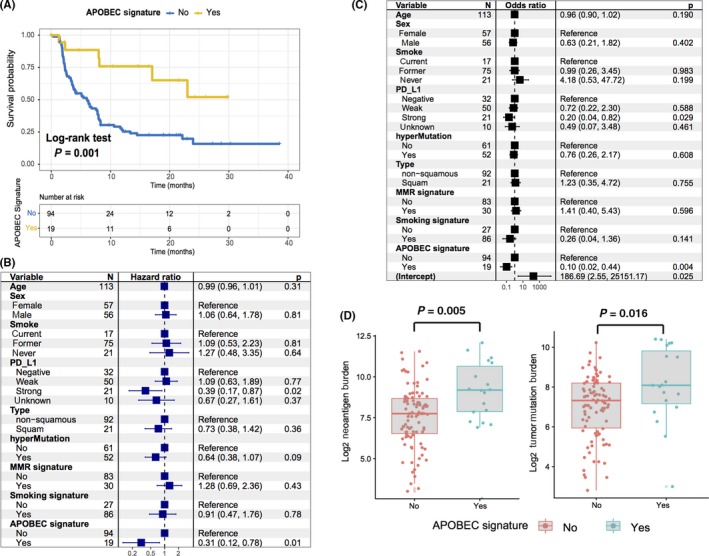
APOBEC mutational signature associated with immunotherapy benefit. A, Kaplan‐Meier survival analysis stratified by APOBEC mutational signatures status. The association between mutational activity of the APOBEC signature with progression‐free survival (B) and immune response status (C) were displayed in a forest plot. Age, sex, smoking status, histological type, PD‐L1, hypermutation, MMR, and smoking signature were likewise taken into account. Square data markers indicate estimated hazard ratios (B) and odds ratios (C). Error bars represent 95% CIs. APOBEC signature is associated with increased tumor mutation burden and neoantigen burden (D)
3.4. Further validation of APOBEC mutational signature in the TCGA cohort
Of the 998 patients with matched somatic mutation and gene expression data in the TCGA Project NSCLC cohort, 509 (51.0%) were lung adenocarcinoma (LUAD) patients and 489(49.0%) were lung squamous cell carcinoma (LUSC) patients. We extracted 5 mutation signatures (i.e signatures 2, 4, 5, 6, and 7 according to COSMIC; Figure S6A) from TCGA with varying mutational activities (Figure S6B). The extracted most prevalent mutational signatures included smoking signature 4, which accounted for 221 831 of the 349 106 total mutations (63.5%), and signature 5, which accounted for 58 449/349 106 (16.7%). APOBEC‐associated signature 2 contributed to 45 773 of the 349 106 total mutations (13.1%) and ultraviolet (UV) light exposure signature 7 (14 146/349 106, 4.0%), MMR signature 6 (8905/319 106, 2.8%) (Figure S6C). As in the ICB treatment cohort, NSCLC samples with an APOBEC signature had a significantly higher TMB than those without (Wilcoxon rank sum test, P = 0.024). Interestingly, patients with an APOBEC signature in TCGA not only had no benefit in survival but also exhibited a tendency for a worse prognosis, suggesting that the mutations are specific predictors of response to immunotherapy and not simply chemotherapy (Figure S6D).
We also investigated the potential mechanism behind APOBEC mutagenesis signature and immunotherapy response. Gene set enrichment analysis (GSEA) on Hallmarker sets revealed that enrichment of genes involved in IL2‐STAT5 signaling was significantly altered in samples with high mutational APOBEC activities, whereas MYC targets, PI3K–AKT–mTOR associated pathways were enriched in the low activities group (q < .001, Figures 4A and S7A). Furthermore, we found that immune checkpoint associated genes CD274 (PD‐L1), LAG3, VTCN1 (Figure S7B) and antigen processing presentation associated genes TAP1, TAP2, PTPN2 (Figure S7C) were also highly expressed in the high APOBEC mutation group.
Figure 4.
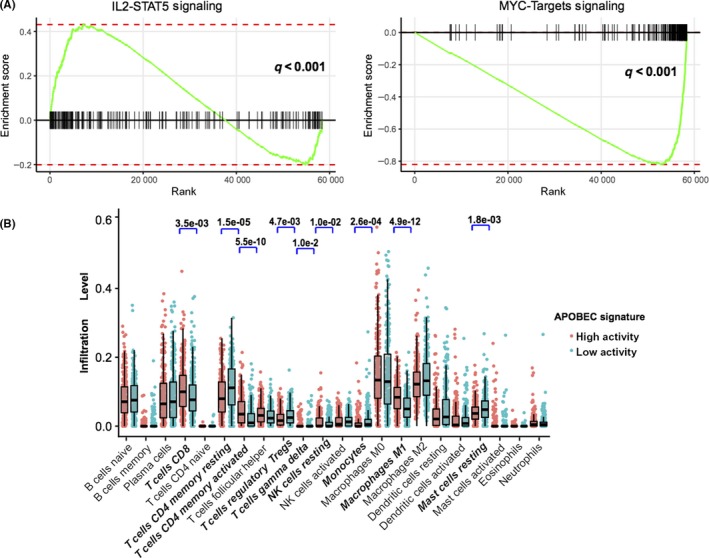
Significantly enriched pathways and tumor‐infiltrating leukocytes in high APOBEC mutational activities samples. A, GSEA was performed on RNA‐seq data from pretreatment tumors in TCGA NSCLC cohorts (high APOBEC mutational activity samples vs low activity, n = 250) using the hallmark gene sets. Enrichment plots show increased expression of the IL‐2‐STAT5 gene set and decreased expression of the MYC targets gene set in the high mutational activity samples. B, Tumor‐infiltrating leukocyte (TIL) level in high APOBEC mutational activity vs low activity samples were estimated using the CIBERSORT algorithm. Bold fonts represent the significantly different infiltrated immunocytes
Moreover, we evaluated (with use of the CIBER algorithm) the abundance of TIL cells in the NSCLC microenvironment using TCGA gene expression data. We found that CD8+ T cells, CD4 memory activated T cells, NK cells, and M1 macrophages were enriched in the high mutational APOBEC activity group, in contrast CD4 memory rested T cells, monocytes, resting mast cells and regulatory T cells were enriched in the low mutational activity group (Figure 4B).
4. DISCUSSION
The development of anti‐PD‐1 and anti‐CTLA‐4 agents has revolutionized therapeutic strategies for NSCLC and multiple cancers. However, the relationship between the genomic landscape and the benefit from immune therapy remains obscure.5, 13, 22, 33 Therefore, we carried out a meta‐analysis of the data from 113 NSCLC patients treated by immunotherapy and identified that mutational APOBEC signature and individual gene mutations in IFNGR1, VTCN1, and PTEN were significantly associated with e immune responde. We further identified a signature in data from TCGA dataset of 998 samples from patients with NSCLC and found that interferon gamma‐associated IL2‐STAT5 pathways were significantly enriched in the APOBEC high mutation samples.
Mutations patterns in APOBEC signatures are commonly found to be due to the local hypermutation found in some cancers, and known as kataegis, potentially also implicating AID/APOBEC enzymes in this process. Recent studies have reported that interferon induces APOBEC gene families that are involved in antiviral infection, tissue inflammation, and enhanced host immune responses.17, 34 Furthermore, there is early evidence pointing out that APOBEC correlates with overexpression of the immune checkpoint molecule PD‐1 ligands, potentially leading to the development of immune exhaustion.35 Therefore, reversing immune exhaustion with an anti‐PD‐1/L1 antibody may sensitize APOBEC‐associated tumor cells to the immune response.
Recent evidence has indicated that a series of molecular features influence immunotherapy efficacy, including expression of immune checkpoint molecules,36, 37, 38 infiltration of CD8+ T cells,39, 40 oncogenic pathways,19 defects in antigen processing and presentation,41 TMB5, 8 and tumor‐specific neoantigen.33, 42 We found that APOBEC signature mutational activity upregulation is positively associated with PD‐L1, LAG‐3 immune checkpoint associated gene markers, and CD8+ T‐cell and CD4+ T‐cell immune cell infiltration makers. Furthermore, APOBEC signature mutational activity was also identified as correlated with TMB and NB. Furthermore, given that the APOBEC gene family is correlated with interferon‐mediated immune reactions,34, 43 and the IL2‐STAT5 pathway gene involvement in the interferon gamma (IFN‐γ) signaling pathway,44 we speculated that the APOBEC mutational signature may affect differential expression of IFN‐associated genes and influence the response to ICB treatment. These findings indicated that an APOBEC mutational signature may sensitizes tumors to immunotherapy and predict the immune response.
Although TP53, EGFR, and STK11 mutations were not found to be significantly associated with the immune response, possibly due to the sample size, we identified that IFNGR1, VTCN1 were exclusively associated with the response and PTEN was associated with resistance in our series. It has been reported that mutations in IFNGR1, VTCN1, and PTEN are connected to the immune response in immunotherapy,18, 20, 45 IFNGR1 was also found to be mainly mutated in patients with the APOBEC signature (3 of 4 in APOBEC signature carriers, χ2 test, P = .015).
In patients with advanced NSCLC who were treated with ICB monotherapy, multiple reports have identified an association between increased TMB and NB with a likelihood of clinical benefit.5, 6, 8 Combined TMB and APOBEC signatures may preferably predict responders in immunotherapy than using either signature alone (12 responders in 13 predict responding samples, positive predictive value, 92.3%). The mechanisms underlying the association between TMB and the APOBEC signature with immunotherapy is not entirely clear. A leading hypothesis detailed that the mutation pattern of APOBEC drives a kataegic‐like mutation signature and produces replication stress‐associated chromosomal instability in cancers,46, 47 suggesting that the APOBEC signature may concur with high somatic mutation.
Although we utilized multiple datasets for analysis, the only dataset that contained both whole‐exome sequencing and RNA expression data was TCGA. As a result, association data between mutational patterns and gene expression, including analysis in immune cell infiltration and oncogenic pathways need further validation. In addition, the mutational activities of each signatures in TCGA and our datasets were differential distribution, for example the MMR signature 6 is significantly higher than TCGA cohort (signature 6:13.2% vs 2.8%, χ2 test, P < .001), signature 16 (16.5%) was observed exclusively in our aggregate datasets, and signature 5 (16.7%) exclusively in TCGA dataset. This difference may be because there was a higher proportion of squamous‐cell carcinoma samples in TCGA than in the aggregated cohort (46% vs 19%; χ2 test, P < .001).
In this study, we assembled and characterized genomic data and clinical information from 113 patients treated with ICB to determine whether tumor genetic landscape affects clinical benefit. These studies have identified putative a genomic mutation signature and molecular biomarkers in response to ICB, demonstrating the complex interplay of host and tumor in the treatment response. However, the mechanisms underlying the association between APOBEC signature with improved immunotherapy is still unclear and further studies in other cancer types are warranted.
DISCLOSURE
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting information
ACKNOWLEDGMENTS
The author thanks Professors XC L. and KX C. for unselfish help and valuable suggestions, and thanks MY for protocols and helpful discussions. HC thanks the inimitable care and support of WC over the past years, will you spend the rest of your life with me?
Chen H, Chong W, Teng C, Yao Y, Wang X, Li X. The immune response‐related mutational signatures and driver genes in non‐small‐cell lung cancer. Cancer Sci. 2019;110:2348‐2356. 10.1111/cas.14113
Funding information
Science and technology planning project of Binzhou Medical college, (Grant/Award Number: ‘BY2017KJ02’) Top talent training program of the first affiliated hospital of PLA Army Medical University, (Grant/Award Number: ‘SWH2018BJKJ‐12’) National Natural Science Foundation of China, (Grant/Award Number: ‘81803215’).
Chen and Chong contributed equally to this work.
Contributor Information
Xin Wang, Email: wangxinmarine@126.com.
Xue Li, Email: yolandadalee@yahoo.com.
REFERENCES
- 1. Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous‐cell non‐small‐cell lung cancer. N Engl J Med. 2015;373:123‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Escudier B, Motzer RJ, Sharma P, et al. Treatment beyond progression in patients with advanced renal cell carcinoma treated with nivolumab in CheckMate 025. Eur Urol. 2017;72:368‐376. [DOI] [PubMed] [Google Scholar]
- 3. Wolchok JD, Chiarion‐Sileni V, Gonzalez R, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377:1345‐1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ferris RL, Blumenschein G, Fayette J, et al. Nivolumab for recurrent squamous‐cell carcinoma of the head and neck. N Engl J Med. 2016;375:1856‐1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hellmann MD, Nathanson T, Rizvi H, et al. Genomic features of response to combination immunotherapy in patients with advanced non‐small‐cell lung cancer. Cancer Cell. 2018;33(843–852):e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD‐1 blockade in non‐small cell lung cancer. Science. 2015;348:124‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anagnostou V, Smith KN, Forde PM, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non‐small cell lung cancer. Cancer Discov. 2017;7:264‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rizvi H, Sanchez‐Vega F, La K, et al. Molecular determinants of response to anti‐programmed cell death (PD)‐1 and anti‐programmed death‐ligand 1 (PD‐L1) blockade in patients with non‐small‐cell lung cancer profiled with targeted next‐generation sequencing. J Clin Oncol. 2018;36:633‐641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cristescu R, Mogg R, Ayers M, et al. Pan‐tumor genomic biomarkers for PD‐1 checkpoint blockade‐based immunotherapy. Science. 2018;362:eaar3593 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Balachandran VP, Łuksza M, Zhao JN, et al. Identification of unique neoantigen qualities in long‐term survivors of pancreatic cancer. Nature. 2017;551:512‐516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown SD, Warren RL, Gibb EA, et al. Neo‐antigens predicted by tumor genome meta‐analysis correlate with increased patient survival. Genome Res. 2014;24:743‐750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim S, Kim HS, Kim E, et al. Neopepsee: accurate genome‐level prediction of neoantigens by harnessing sequence and amino acid immunogenicity information. Ann Oncol. 2018;29:1030‐1036. [DOI] [PubMed] [Google Scholar]
- 13. Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD‐1 blockade. Science. 2017;357:409‐413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roberts SA, Lawrence MS, Klimczak LJ, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970‐976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Middlebrooks CD, Banday AR, Matsuda K, et al. Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC‐signature mutations in tumors. Nat Genet. 2016;48:1330‐1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peng G, Lei KJ, Jin W, Greenwell‐Wild T, Wahl SM. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon‐induced anti‐HIV‐1 activity. J Exp Med. 2006;203:41‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao J, Shi LZ, Zhao H, et al. Loss of IFN‐gamma pathway genes in tumor cells as a mechanism of resistance to anti‐CTLA‐4 therapy. Cell. 2016;167(397–404):e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peng W, Chen JQ, Liu C, et al. Loss of PTEN promotes resistance to T cell‐mediated immunotherapy. Cancer Discov. 2016;6:202‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. George S, Miao D, Demetri GD, et al. Loss of PTEN is associated with resistance to anti‐PD‐1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity. 2017;46:197‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism‐driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359:801‐806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramos AH, Lichtenstein L, Gupta M, et al. Oncotator: cancer variant annotation tool. Hum Mutat. 2015;36:E2423‐E2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim J, Mouw KW, Polak P, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet. 2016;48:600‐606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li X, Wu WKK, Xing R, et al. Distinct subtypes of gastric cancer defined by molecular characterization include novel mutational signatures with prognostic capability. Cancer Res. 2016;76:1724‐1732. [DOI] [PubMed] [Google Scholar]
- 26. Alexandrov LB, Nik‐Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415‐421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333‐339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ritchie ME, Phipson B, Wu DI, et al. limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li XC, Wang MY, Yang M, et al. A mutational signature associated with alcohol consumption and prognostically significantly mutated driver genes in esophageal squamous cell carcinoma. Ann Oncol. 2018;29:938‐944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Riaz N, Havel JJ, Makarov V, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171(934–949):e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Almeida RR, Raposo RAS, Coirada FC, et al. Modulating APOBEC expression enhances DNA vaccine immunogenicity. Immunol Cell Biol. 2015;93:868‐876. [DOI] [PubMed] [Google Scholar]
- 35. Boichard A, Tsigelny IF, Kurzrock R. High expression of PD‐1 ligands is associated with kataegis mutational signature and APOBEC3 alterations. Oncoimmunology. 2017;6:e1284719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matsuzaki J, Gnjatic S, Mhawech‐Fauceglia P, et al. Tumor‐infiltrating NY‐ESO‐1‐specific CD8+ T cells are negatively regulated by LAG‐3 and PD‐1 in human ovarian cancer. Proc Natl Acad Sci USA. 2010;107:7875‐7880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schumacher TN, Kesmir C, van Buuren MM. Biomarkers in cancer immunotherapy. Cancer Cell. 2015;27:12‐14. [DOI] [PubMed] [Google Scholar]
- 38. Shukuya T, Carbone DP. Predictive markers for the efficacy of anti‐PD‐1/PD‐L1 antibodies in lung cancer. J Thorac Oncol. 2016;11:976‐988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chauvin J‐M, Pagliano O, Fourcade J, et al. TIGIT and PD‐1 impair tumor antigen‐specific CD8(+) T cells in melanoma patients. J Clin Invest. 2015;125:2046‐2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tang H, Wang Y, Chlewicki L, et al. Facilitating T cell infiltration in tumor microenvironment overcomes resistance to PD‐L1 blockade. Cancer Cell. 2016;30:500. [DOI] [PubMed] [Google Scholar]
- 41. DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour‐specific antigens underlies cancer immunoediting. Nature. 2012;482:405‐409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA‐4 blockade in metastatic melanoma. Science. 2015;350:207‐211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Y, Xia Y, Han M, et al. IFN‐alpha‐mediated base excision repair pathway correlates with antiviral response against hepatitis B virus infection. Sci Rep. 2017;7:12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sa Q, Woodward J, Suzuki Y. IL‐2 produced by CD8+ immune T cells can augment their IFN‐gamma production independently from their proliferation in the secondary response to an intracellular pathogen. J Immunol. 2013;190:2199‐2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Podojil JR, Miller SD. Potential targeting of B7–H4 for the treatment of cancer. Immunol Rev. 2017;276:40‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015;5:704‐712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nikkilä J, Kumar R, Campbell J, et al. Elevated APOBEC3B expression drives a kataegic‐like mutation signature and replication stress‐related therapeutic vulnerabilities in p53‐defective cells. Br J Cancer. 2017;117:113‐123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


