Abstract
Precision medicine places significant emphasis on techniques for the identification of DNA mutations and gene expression by deep sequencing of gene panels to obtain medical data. However, other diverse information that is not easily readable using bioinformatics, including RNA modifications, has emerged as a novel diagnostic and innovative therapy owing to its multifunctional aspects. It is suggested that this breakthrough innovation might open new avenues for the elucidation of uncharacterized cancer cellular functions to develop more precise medical applications. The functional characteristics and regulatory mechanisms of RNA modifications, ie, the epitranscriptome (ETR), which reflects RNA metabolism, remains unclear, mainly due to detection methods being limited. Recent studies have revealed that N6‐methyl adenosine, the most common modification in mRNA in eukaryotes, is affected in various types of cancer and, in some cases, cancer stem cells, but also affects cellular responses to viral infections. The ETR can control cancer cell fate through mRNA splicing, stability, nuclear export, and translation. Here we report on the recent progress of ETR detection methods, and biological findings regarding the significance of ETR in cancer precision medicine.
Keywords: diagnosis, modification, MODOMICS, RNA, therapy
1. INTRODUCTION
Numerous previous studies have extensively investigated cellular biology in cancer, in terms of gene controls, which are the result of genetic information on DNA, acquired modifications in histones, dynamic regulations by proteins, and the fate determining flow of metabolites. These findings have led to the acceptance of conceptual conclusions as to whether cancer is fundamentally a genetic disease, although various factors beyond simple alterations in DNA sequences and expression orchestrate the biologically malignant phenotypes of tumors.1
It was reported that, although cellular reprogramming by induced pluripotent stem cell technology2 is fundamentally epigenetic manipulation mainly using transcription factors, subsequent studies have indicated that epigenetic regulation associated with induced pluripotent stem cell derivation could drive the development of particular types of cancer in mouse experiments, suggesting the significance of epigenetics in cancer.3, 4 In addition, in the process of cancer metastasis, the biological mechanism of epithelial‐mesenchymal transition (EMT) is controlled by epigenetic regulations rather than genetic events, suggesting a role of the transcriptional and epigenetic landscapes, and the gene regulatory network in controlling EMT states.5 In addition, it has been shown that focus on phenotypic drug development might not target the appropriate molecular variant in rare driver mutations, as variants of the same protein can express more than one phenotype.6 This indicates that a multifaceted understanding beyond genetics is required in order to ensure further precision in cancer research and medicine.
Recently, other diverse information, including RNA modifications, has emerged in cell physiology,7 technology,8 and structure chemistry.9 Although RNA modifications are not readily readable using bioinformatics, their significance has been emphasized for the diagnosis and therapy of diseases (Figure 1). Whether RNA modifications are associated with tumor heterogeneity remains to be fully elucidated.
Figure 1.
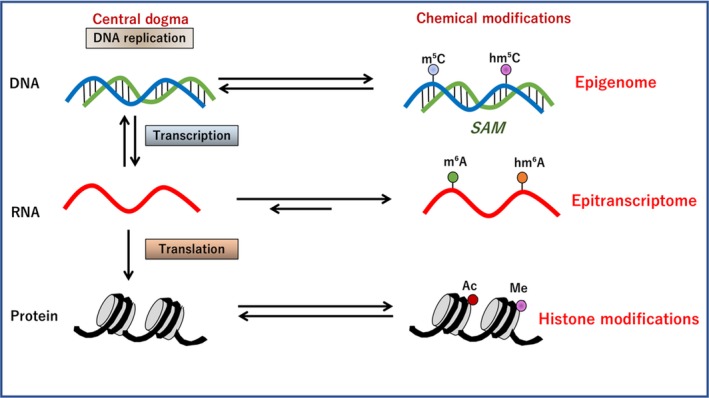
Biochemistry of the epitranscriptome (ETR) in the central dogma. The modification of DNA has been examined extensively in the term of epigenetics. According to the genetic codes on DNA, ETR denotes the modification of RNA, which is involved in the splicing and translation on ribosomes, leading to the production of proteins. The acquired histone modification is involved in the regulation of transcription.87 SAM, S‐adenosylmethionine
This systemic review considers the recently emerged modifications of ribonucleotides and epitranscriptomes (ETRs) in cancer, and discussed the significance of tumor heterogeneity. A published database of references (https://www.ncbi.nlm.nih.gov/pubmed/) between February 1959 and 2019 was reviewed using the following search terms: “RNA methylation”, 21 521 papers; “epitranscriptome”, 164 papers; and “RNA MODOMICS”, 12 papers. Information of ETRs in cancer was then updated.
2. HISTORICAL REVIEW OF THE DETECTION OF RNA MODOMICS
Since the 1960s, the important role of post‐transcriptional modification of RNAs in the synthesis of eukaryotic RNA has be traced; it was reported that one of the most notable features of rRNA synthesis is the specific methylation that occurs on the 45S precursor RNA molecule in a study of HeLa cells.10 In 1974, methylated nucleotides were identified in a study of Novikoff hepatoma cells by methyl labeling of RNA with L‐[methyl‐3H] methionine, which was detected by liquid chromatography.11 This study found that, although rRNA and tRNA possess complex patterns in base‐methylation, the distribution in mRNA was relatively simple, consisting predominantly of N6‐methyladenosine.12
Although previous studies have revealed the diversity of chemical modifications of RNA, two‐thirds of all known RNA modifications were reported before 1980, and the observed variations have remained relatively unchanged over several decades.13 A historical review of the techniques to detect RNA modifications, namely RNA MODOMICS, is summarized in Figure 2.
Figure 2.
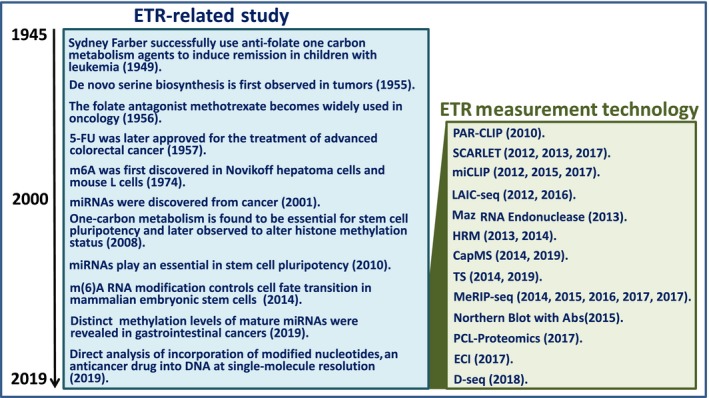
Historical view of the study of the epitranscriptome (ETR). Although ETR‐related studies began in the 1950s, ETR measurement technologies were in practice in the 21st century. 5‐FU, 5‐fluorouracil; CapMS, captured mass spectrometry; D‐seq, direct m6A sequencing; ECI, electrochemical immunosensor with silver SiO2; HRM, high‐resolution melting; LAIC‐seq, m6A levels and isoform‐characterization sequencing; MeRIP‐seq, methylated RNA immunoprecipitation sequencing; miCLIP, m6A individual nucleotide resolution cross‐linking and immunoprecipitation; miRNA, microRNA; PCL‐Proteomics, photo‐crosslinkers and quantitative proteomics; SCARLET, site‐specific cleavage and radioactive‐labeling followed by ligation‐assisted extraction and thin‐layer chromatography; TS, tunnel current sequencing
3. DATABASE FOCUSING ON RNA MODOMICS
In 1994, the RNA Modification Database was introduced as a focal source for information of naturally occurring RNA modifications.13 The database incorporated a searchable interface for detailed data of 109 currently known RNA modifications.11
In 1999, the RNA Modification Database provided a comprehensive list of 95 different natural modifications of RNA, including information on chemical structure, phylogenetic distribution, structure characterization, and chemical synthesis.14
In 2008, MODOMICS (http://modomics.genesilico.pl) was made available as a database of 119 different modifications of tRNAs, rRNAs, snRNAs, and small nucleolar RNA, regarding comprehensive information of the chemical structures of modified ribonucleosides, their synthetic pathways, the location of modified residues in RNA sequences, and RNA modifying enzymes.15 The modifications in RNAs were shown using a liquid chromatography‐mass spectrometry (LC‐MS) technique for tRNAs,16 by 15N or 13C stable isotope labeling and LC‐MS/MS in Escherichia coli and yeast,17 and by LC‐MS and LC‐MS/MS analysis in archaeal tRNA(Ile).18
The biochemical and physiological roles of modifications have been found in the decoding process of tRNAs, as examined and reviewed by Gu et al,19 and the methylation process of rRNA was investigated.20, 21, 22, 23
4. CONSENSUS SEQUENCE OF RNA MODOMICS
In molecular biology, the complementarity of DNA or RNA sequences defines the association between 2 structures, each following a lock‐and‐key association. Complementarity includes the principles of DNA replication, RNA transcription, and translation of peptides, in addition to various functions modulating the central dogma, such as RNA interference24 and competing endogenous RNAs25 and genome editing.26
It has been shown that the structural modifications of nucleotides, mainly as ribose analogs, are aimed at the application of siRNAs to limit their sensitivity to ribonucleases, poor cellular uptake, and rapid size‐mediated renal clearance. The correction of thermal asymmetry is achieved by the incorporation of nucleotide substitutions, or blocking sense strand incorporation in the RNA‐induced silencing complex. Locked nucleic acids also increase the sensitivity and specificity of short RNA and DNA targets.27 These technologies are used for the validation of potent drug targets for anticancer therapy and the silencing of thermodynamically unfavorable targets, ie, mutations of oncogenes and tumor suppressor genes and chimeric fusion genes.27 The modification of bases in DNA or RNA allows the 4 ribonucleotide residues to gain diverse functions similar to those of the side chains of amino acid residues.
Most mammalian m6A sites are found within the consensus sequence, typically “GGACU”; more precisely, Rm6ACH (R denotes G or A; H includes A, C, or U), or Pu(G>A)m6AC(A/C/U) (where Pu represents purine).28 Previous studies have revealed that the enriched binding motif, GGACC, is observed in methyltransferase‐like protein 14 (METTL14), an RNA‐binding scaffold protein that recognizes the substrate in a component of m6A methyltransferase complex, whereas the motif NHGGAC is found in METTL3, a catalytic core of m6A methyltransferase, which formed a complex with METTL14 (N indicates G, C, A, or U).28 The overlay of METTL3 and METTL14 is GUCGGAC.29 Wilms tumor 1‐associating protein (WTAP) can interact with the complex of METTL3 and METTL14 to affect methylation of the GACU site.28
5. WRITING, ERASING, AND READING IN RNA MODOMICS
Among the various conserved internal chemical modifications of RNAs, the N6‐methyladenosine (m6A) is the most prevalent modification, which is found in almost all eukaryotic RNAs,30 and the importance of these modifications has been reported.31 Generally, the m6A modification is post‐transcriptionally installed by a multicomponent N6‐adenosine methyltransferase (MT) complex.28 It was reported that the ~200‐kDa (MT) complex, which was isolated from mammalian cell nuclear extract, revealed only a 70‐kDa protein, which is involved in the inhibition of methyltransferase activity and was termed MT‐A70 or METTL3.32 A recent study showed that human METTL3 and METTL14 form a stable heterodimer core complex that functions in the deposition of cellular m6A on mammalian nuclear RNAs and catalyzes m6A RNA methylation.28 As a mammalian splicing factor, WTAP can interact with this complex and affect this methylation.28 Therefore, the core complex of METTL3‐14 with WTAP exerts a function of methylation at N6‐adenosines, as writers for ETR codes33 (Figure 3).
Figure 3.
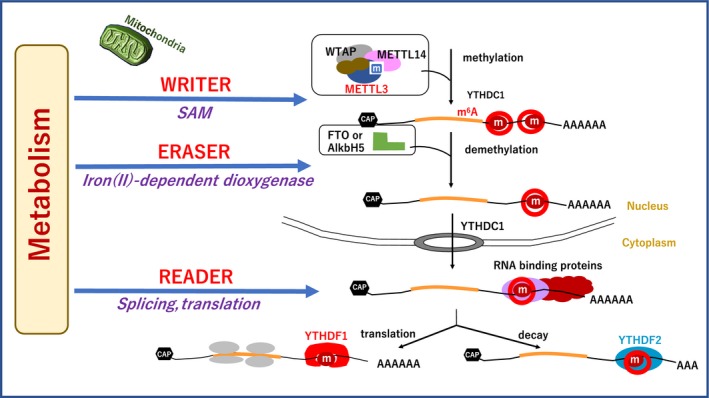
Writers, erasers, and readers of the epitranscriptome. In response to the extracellular stimuli and cellular metabolism, the methyltransferase‐like protein 3 (METTL3)/MELLT14/Wilms tumor 1‐associating protein (WTAP) writer complex is involved in the methylation of RNAs at the specific residue. Fat mass and obesity‐associated protein (FTO) or α‐ketoglutarate‐dependent dioxygenase homolog 5 (ALKBH5) is involved in demethylation, recognized by reader groups, including YT521‐B homology N6‐methyladenosine RNA binding protein 1 9YTHDF1), YTHDF2, and heterogeneous nuclear ribonucleoproteins (HNRNPA238). CAP, RNA 5'‐CAP; m, methylated base; SAM, S‐adenosylmethionine
By contrast, functionally significant demethylases that reverse this methylation, such as fat mass and obesity‐associated protein (FTO)34 and α‐ketoglutarate‐dependent dioxygenase (ALKB) homolog 5 (ALKBH5)29 have been identified. A recent study also revealed distributions of m6A in the mammalian transcriptomes35 and indicated diverse regulatory functions of this dynamic modification.36 Therefore, these factors function as erasers to ETR codes33 (Figure 3).
Readers, including heterogeneous nuclear ribonucleoproteins and YT521‐B homology (YTH) N6‐methyladenosine RNA binding protein 1 (YTHDF1) have been reported. A recent study revealed that oncogene c‐myc promotes the expression of ERT m6A reader YTHDF1 in colorectal cancer.37 Therefore, reader, writer, and eraser proteins for m6A were discovered.33 Furthermore, it is reported that YTHDF2, another YTH domain family member, is involved in the localization of bound mRNA from the translatable pool to mRNA decay sites, such as processing bodies, through a carboxy‐terminal domain of YTHDF2 selectively binding to m6A‐containing mRNA.38 Taken together, studies have indicated that the dynamic m6A modification is recognized by different binding proteins to affect the translation status and lifetime of RNA33, 37, 39 (Figure 3).
6. RNA MODOMICS IN EMBRYONAL STEM CELLS
The study of murine embryonal stem cells (ESCs) has identified METTL3 as a regulator for terminating murine naïve pluripotency.40 The preimplantation epiblasts and naïve ESCs with METTL3 knockout were depleted of the m6A modification and were viable40; however, the adequate termination of their naïve pluripotency failed and they underwent aberrant and restricted lineage priming at the postimplantation stage, which led to early embryonic lethality.40 This suggests that m6A serves a role in determining the fate of murine ESCs, which functionally influence naïve and primed pluripotency in an opposing manner. m6A predominantly and directly reduces the stability of mRNA.40 It is suggested that METTL3 is involved in the balance between pluripotency and lineage‐committed differentiation and that the depletion of METTL3 results in the stabilization of target genes; ie, METTL3‐deficient naïve pluripotent cells upregulate the already high naïve pluripotent genes to create a “hyper”‐naïve pluripotent state, and METTL3 depletion occurring in primed cells could induce the determination of lineage commitment and tip the balance toward differentiation.41
In addition to METTL3, a recent study indicated a synergistic role of METTL14. The knockdown of METTL3 and METTL14 in mouse ESCs resulted in a lack of m6A modification and loss of self‐renewal capability.42 The study indicated that m6A modification was inversely correlated with mRNA stability and gene expression, and that the human antigen R and microRNA pathways were linked to the effects. This supports the hypothesis that m6A modification is required to maintain mouse ESCs in a naïve state and might be relevant to thousands of mRNAs, long non‐coding RNAs (lncRNAs) and microRNAs in various cell types.42 In addition, a recent study indicated that the depletion of mouse and human METTL3 resulted in the impairment of ECSs exiting from a naïve state to a state of differentiation, suggesting a role of the m6A modification in marking transcriptome flexibility in cells.43
7. RNA MODOMICS IN NON‐CODING RNAS AND VIRAL INFECTION
m6A is the most prevalent and reversible modification in mRNAs and non‐coding RNAs (ncRNAs), including lncRNAs.28 The cellular ETR finely sculpts a dynamic proteome in response to diverse cues during cellular processes, including differentiation, immune tolerance, and neuronal signaling.44 The ETR of cancer emerges and chemical modifications of RNA serve a central role in the control of mRNA and ncRNA activity, which could assist in the development of anticancer drugs.45
The involvement of viral epitranscriptomics has been reported in viral RNAs that replicate in the nuclei of host cells, as shown in the study of the Rous sarcoma virus in the 1970s,46 Flaviviridae virus,47 Zika virus,48 Kaposi sarcoma‐associated herpesvirus,38 HIV,49 influenza A virus,50 and tobacco mosaic virus.51
8. RNA MODOMICS IN DIFFERENT TYPES OF CANCER
8.1. Glioma
A study on glioma indicated that the knockdown of METTL3 or METTL14 resulted in the changes in mRNA m6A enrichment and alterations in the mRNA expression of target genes, including ADAM19, which serves a critical role in glioblastoma stem cells.52 In addition, the inhibition of FTO, an m6A demethylase, reportedly suppressed tumor progression and prolonged the lifespan of glioblastoma stem cell‐grafted mice in vivo.52 This study indicated that m6A is critical for self‐renewal and tumorigenesis in glioblastoma stem cells, presumably through the regulation of target RNAs, and that m6A modification is a promising therapeutic target for glioblastoma.52
8.2. Leukemia
It has been shown that FTO is expressed at high levels in the leukemic cells of patients with acute myeloid leukemia (AML) with t(11q23)/myeloid/lymphoid or mixed‐lineage leukemia rearrangements, t(15;17)/promyelocytic leukemia ‐ retinoic acid receptor alpha (RARA), Fms‐related tyrosine kinase 3 ‐ internal tandem duplication, and/or nucleophosmin 1 mutations, suggesting that FTO serves a critical oncogenic role in AML.53 This study suggested that FTO decreases the expression of targets, including ankyrin repeat and SOCS box protein 2 and RARA, through removal of the m6A modification in these mRNAs, and thus inhibits all‐trans‐retinoic acid‐induced AML cell differentiation, which leads to leukemic transformation, providing profound insight into the disease mechanism and for therapeutic drug development.53 Additionally, METTL3 plays a critical role in cell growth of myeloid leukemia, in which METTL3 controls the function of target genes.54 METTL3 is involved in remodeling chromatin, which is necessary for the maintenance of myeloid leukaemia.54
8.3. Lung cancer
A study on METTL3‐mediated translation indicated that METTL3 enhances mRNA translation through an interaction with the translation initiation machinery, which promotes the translation of certain mRNAs, including epidermal growth factor receptor and the Hippo pathway effector transcriptional coactivator with PDZ‐binding motif (TAZ) in human cancer cells.55 This study indicated that METTL3 promotes the growth, survival, and invasion of human lung cancer cells,55 suggesting that METTL3 could be a possible target of therapy.
8.4. Breast cancer
A study on human breast cancer cells indicated that the exposure to hypoxia resulted in the stimulated expression of AlkB homolog 5 (AlkBH5), an m6A demethylase, which decreased the methylation level of NANOG mRNA, increased NANOG levels, and increased breast cancer stem cells.56 AlkB homolog 5 was involved in the demethylation of m6A in sequence AAACU in the 3′‐UTR of NANOG mRNA.56 Exposure to hypoxia stimulated expression in a hypoxia‐inducible factor (HIF)‐1α‐ and HIF‐2α‐dependent manner.56 In addition, hypoxia induced an zinc finger protein 217 (ZNF217)‐dependent inhibition of the m6A modification of mRNAs encoding NANOG and Kruppel‐like factor 4 in breast cancer.57 Zinc finger protein 217 or AlkBH5 are involved in the control of pluripotency factor expression in breast cancer in hypoxia.57
8.5. Colorectal cancer
A recent study indicated that m6A modification serves a role in the malignant behaviors of colorectal cancer.37 Epigenetic data obtained by ChIP indicated that the oncogene c‐Myc is involved in the transcription of m6A reader YTHDF1 in colorectal cancer.37 Immunohistochemical analysis of YTHDF1 showed that its expression was associated with various malignant tumor behaviors, and the expression of YTHDF1 was identified as an independent prognostic factor in patients.37
8.6. Pancreatic cancer
A previous hospital‐based case‐control study of genetic variations indicated an association between variations in the FTO gene and pancreatic cancer risk in Japan.58 The significant association of the FTO rs9939609 variant polymorphism was shown in endometrial cancer and pancreatic cancer, particularly in Asian populations, and could be a potential biomarker in the early diagnosis of these types of cancer.59 An association between the FTO rs9939609 variant and malignant pleural mesothelioma risk was also reported.60
Another study on pancreatic cancer found that METTL3‐depleted cells showed higher sensitivity to anticancer reagents, including gemcitabine, 5‐fluorouracil, cisplatin, and irradiation, suggesting that METTL3 is a potent target for enhancing therapeutic efficacy in patients with pancreatic cancer.61
8.7. Measurement and detection technology of ETR
Several technologies have been reported for the measurement and detection of ETRs. For measurement purposes, several methods have been reported, including the dot blot,53 northern blot with Ab,62 Maz RNA endonuclease (MazF is an E. coli toxin),63 high‐resolution melting (HRM),64 RNA photo‐crosslinkers and quantitative proteomics (PCL‐Proteomics),65 and electrochemical immunosensor (ECI) with silver SiO2.66
Several methods have been reported for location detection purposes, including the detection of m6A levels and isoform‐characterization sequencing (LAIC‐seq),67 direct m6A sequencing (D‐seq),68 m6A individual‐nucleotide‐resolution cross‐linking and immunoprecipitation (miCLIP),69 methylated RNA immunoprecipitation sequencing (MeRIP‐seq),35 photo activatable ribonucleoside‐enhanced cross‐linking and immunoprecipitation (PAR‐CLIP),70 site‐specific cleavage and radioactive‐labeling followed by ligation‐assisted extraction and thin‐layer chromatography (SCARLET),71 tunnel current sequencing (TS),72 and captured mass spectrometry (CapMS) (M. Konno and H. Ishii, unpubl. data, 2019) (Figures 4 and 5).
Figure 4.
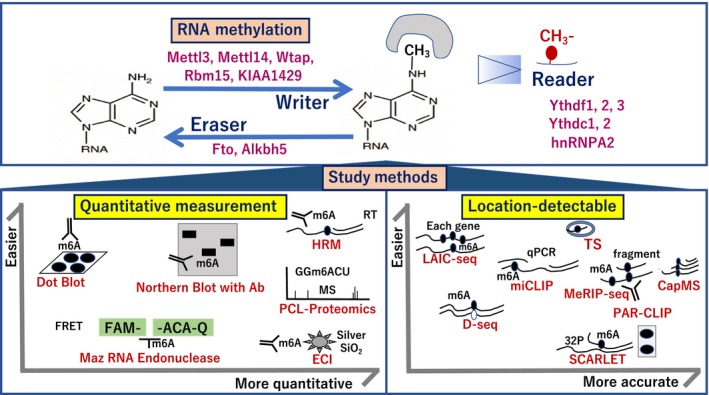
Study methods of RNA MODOMICS. For measurement purposes, several methods have been reported, including dot blot, northern blot with Ab, Maz RNA endonuclease, high‐resolution melting (HRM), photo‐crosslinkers and quantitative proteomics (PCL‐Proteomics), and electrochemical immunosensor (ECI) with silver SiO2. For location detection, several methods have been reported, including m6A levels and isoform‐characterization sequencing (LAIC‐seq), direct m6A sequencing (D‐seq), m6A individual nucleotide resolution cross‐linking and immunoprecipitation (miCLIP), methylated RNA immunoprecipitation sequencing (MeRIP‐seq), photo activatable ribonucleoside‐enhanced cross‐linking and immunoprecipitation (PAR‐CLIP), site‐specific cleavage and radioactive‐labeling followed by ligation‐assisted extraction and thin‐layer chromatography (SCARLET), tunnel current sequencing (TS), and captured mass spectrometry (CapMS). The figure indicates each technology according to ease of quantitative assessment (left) and accuracy (right). qPCR, quantitative PCR
Figure 5.
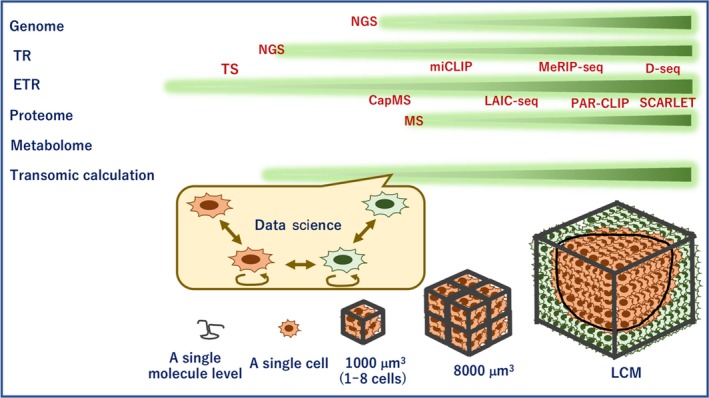
Tissue study by epitranscriptome (ETR) and other technologies. Information on genome, transcriptome (TR), ETR, proteome, and metabolome levels is assessed by various techniques, as indicated. The data obtained through each technique are analyzed by transomic approaches. Although bulk tissues are examined by laser‐capture microdissection (LCM), high‐speed next‐generation sequencing (NGS) can evaluate data of a single cell, and tunnel current sequencing and captured mass spectrometry (MS) can get obtain data for each molecule at the subcellular level of ETR. CapMS, captured mass spectrometry; D‐seq, direct m6A sequencing; LAIC‐seq, m6A levels and isoform‐characterization sequencing; MeRIP‐seq, methylated RNA immunoprecipitation sequencing; miCLIP, m6A individual nucleotide resolution cross‐linking and immunoprecipitation; PAR‐CLIP, photo activatable ribonucleoside‐enhanced cross‐linking and immunoprecipitation; SCARLET, site‐specific cleavage and radioactive‐labeling followed by ligation‐assisted extraction and thin‐layer chromatography; TS, tunnel current sequencing
Eventually TS can be applied to DNA,73 methylcytosines in DNA,74 RNA,75 post‐translational signaling,76 and DNA‐incorporated anticancer reagent trifluorothymidine (trifluridine)77 (Figure 5).
8.8. Tumor heterogeneity affected by ETRs
Epitranscriptome modifications regulate all stages of the RNA life cycle, from transcription to decay.78 As recent studies have indicated a mechanism in which RNA methylation is involved, information regarding aberrant modification is suggested to be involved in cancer initiation and progression, and serve as a biomarker for early stage diagnosis in several types of cancer.78 In response to extracellular stimuli, signals including epidermal growth factor elicit METTL3 function in collaboration with TAZ.55 Serine metabolism in one‐carbon metabolism supports the methionine cycle and DNA and RNA methylation in cancer cells.79 In terms of one‐carbon metabolism, a previous study indicated that polyamine flow and S‐adenosylmethionine metabolism exert a critical role in the maintenance of therapy‐resistant cancer stem cells.80 One‐carbon metabolism is involved in the function of signaling organelle mitochondria through mitochondrial products, including α‐ketoglutarate/2‐oxoglutarate, and succinate and fumarate interfering mechanisms to 2‐oxoglutarate and iron (II)‐dependent dioxygenases, including demethylases of DNA and RNA81 (Figure 6). Computational analysis has indicated the imbalanced mechanism of isocitrate dehydrogenase in colorectal cancer.82
Figure 6.
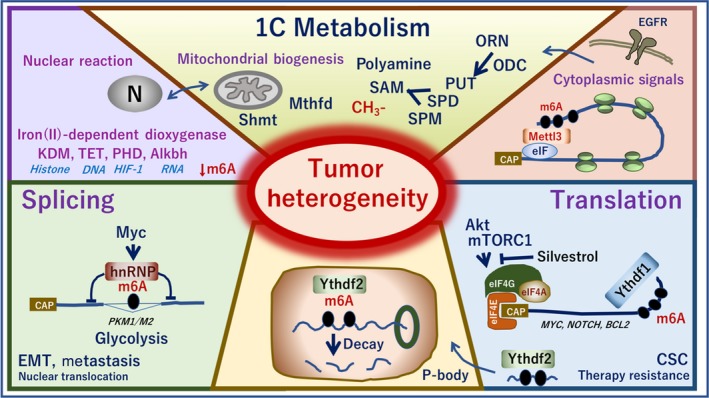
RNA MODOMICS and heterogeneity of tumors. In response to the upstream signals, including epidermal growth factors, one‐carbon metabolism serves a critical role in mitochondrial biogenesis through methylenetetrahydrofolate dehydrogenase (Mthfd) and serine hydroxymethyltransferase 2 (Shmt). Polyamine flows including metabolites, ornithine, spermidine, and spermine regulate the methylation reaction. The mitochondrial metabolites control α‐ketoglutarate/2‐oxoglutarate dioxygenase (Alkbh) and iron (II)‐dependent dioxygenase, histone lysine demethylase (KDM), and ten‐eleven translocation methylcytosine dioxygenase 1 (TET), which catalyzes the conversion of the modified DNA base 5‐methylcytosine to 5‐hydroxymethylcytosine, prolyl hydroxylase domain enzymes, an enzyme involved in the hypoxia‐inducible factor (HIF) signaling pathways. ALKBH5 demethylates specifically by oxidative demethylation m6A RNA, the most prevalent internal modification of mRNA in higher eukaryotes. m6A is involved in the RNA splicing by heterogeneous nuclear ribonucleoproteins (hnRNP) that are known as splicing factors. YT521‐B homology 6‐methyladenosine RNA binding protein 1 (YTHDF1) recognizes m6A and binds the mRNA's 3′UTR. YHTDF1 recruits the eIF4 family and induces protein translation, which is regulated by the Akt‐mTORC1 pathway. m6A markings are also involved in the P‐body decay by YTHDF2. These mechanisms are altered in tumors to various levels and contribute to the heterogeneity of tumors. ALKBH5, α‐ketoglutarate‐dependent dioxygenase ALKB homolog 5; CAP, RNA 5'‐CAP; CSC, cancer stem cell; EGFR, epidermal growth factor receptor; EMT, epithelial‐mesenchymal transition; ODC, ornithine decarboxylase; ORN, ornithine; PHD, plant homeodomain; PKM, pyruvate kinase; PUT, putrescine; SAM, S‐adenosylmethionine; SPD, spermidine; SPM, spermine
As oncogene c‐Myc is involved in the transcriptional control of ETR reader YTHDF1,37 in addition to METTL3,61 a recent study indicated that METTL3 promotes ovarian cancer growth and invasion through EMT.83 Furthermore, the RNA editing process, which irreversibly recodes primary RNA sequences by enzymes apolipoprotein B mRNA editing enzyme, catalytic polypeptide‐like and adenosine deaminase, RNA‐specific, are reported to be involved in dynamically altering RNA molecules by writers and demethylases in response to environmental conditions.84 Furthermore, it has been proposed that malignant RNA processing plays a role in the generation and maintenance of cancer stem cells through mechanisms of RNA methylation, RNA editing, and RNA splicing, the functional consequences of which contribute to aberrant regulation in human malignancies.85 Therefore, it is expected that ETR affects the heterogeneous phenotypes of tumor biology through the mechanisms of RNA splicing, nuclear‐cytoplasmic transport, translation, and regulation of gene expression by RNA interference,84 although genetic codes serve a role in defining a fundamental phenotype. Of note, ETR information is difficult to read through standard deep sequencing, and further development of measuring technologies is required.86
Whether ETR information can contribute to profiling responsiveness to anticancer reagents, and the subclass of cancer diseases, remains a challenge. To overcome this, a perceptive view for the multiomic profiling of tumor tissues, through a combination of deep sequencing data and ETR sequencing, is required to improve precision medicine for intractable cancer (Figure 7).
Figure 7.
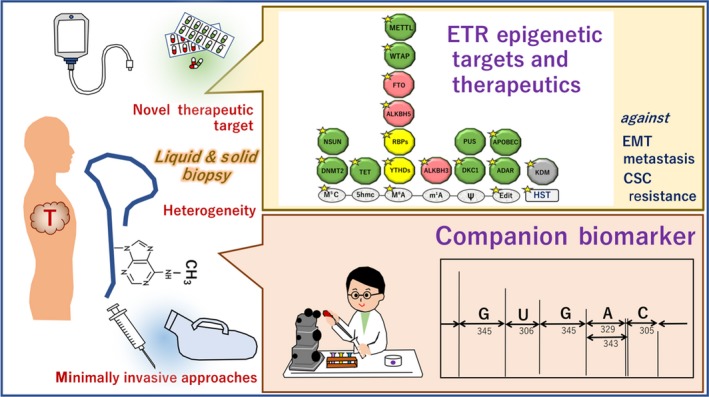
Nucleotide precision medicine by focusing on the epitranscriptome (ETR). By liquid and solid biopsy, precise information including ETR can be obtained. The ETR can be useful for companion biomarker and novel drug discovery. Writers of RNA modification (green), readers (orange), and erasers (red)55; yellow stars show targets that are reportedly involved in tumorigenesis.55 ADAR, adenosine deaminase RNA; ALKBH, α‐ketoglutarate‐dependent dioxygenase homolog; APOBEC, apolipoprotein B mRNA editing enzyme catalytic; CSC, cancer stem cell; DKC1, dyskerin pseudouridine synthase 1; DNMT2, DNA methyltransferase 2; EMT, epithelial‐mesenchymal transition; FTO, fat mass and obesity‐associated protein; HST, histones as possible therapeutics; KDM, histone demethylase; METTL3, methyltransferase‐like protein; NSUN, NOP2/Sun RNA methyltransferase; PUS, pseudouridine synthase; RBP, RNA binding protein; TET, tet methylcytosine dioxygenase; WTAP, Wilms tumor 1‐associating protein; YTHD, YTH N6‐methyladenosine RNA binding protein
9. CONCLUSION
The ETR is involved in the initiation and progression of tumors through a mechanism involving RNA splicing, nuclear‐cytoplasmic transport, translation, and the regulation of gene expression by RNA interference and editing. The present review will facilitate further development of novel technology to profile tissue RNA MODOMICS for precision medicine.
DISCLOSURE
Institutional endowments were received from Taiho Pharmaceutical, Unitech, IDEA Consultants, and Kinshu‐kai Medical Corporation (H. Ishii); Chugai, Yakult Honsha, and Merck & Co. (M. Konno). The sponsors had no role in the design or performance of the study, data collection, data management and interpretation, preparation of this article, or approval of the article.
ACKNOWLEDGMENTS
We thank every member in our laboratory for fruitful discussion, M. Ozaki and Y. Noguchi for technical assistance, and C. Otsuka for drawings. This work received financial support from grants‐in‐aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (grant nos. 17H04282, 17K19698, 16K15615, P‐DIRECT, P‐CREATE, AMED‐Japan Cancer Research Project).
Konno M, Taniguchi M, Ishii H. Significant epitranscriptomes in heterogeneous cancer. Cancer Sci. 2019;110:2318‐2327. 10.1111/cas.14095
REFERENCES
- 1. Wishart DS. Is cancer a genetic disease or a metabolic disease? EBioMedicine. 2015;2:478‐479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949‐953. [DOI] [PubMed] [Google Scholar]
- 3. Ohnishi K, Semi K, Yamamoto T, et al. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014;156:663‐677. [DOI] [PubMed] [Google Scholar]
- 4. Oshima N, Yamada Y, Nagayama S, et al. Induction of cancer stem cell properties in colon cancer cells by defined factors. PLoS ONE. 2014;9:e101735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212‐226. [DOI] [PubMed] [Google Scholar]
- 6. Nussinov R, Jang H, Tsai CJ, Cheng F. Precision medicine review: rare driver mutations and their biophysical classification. Biophys Rev. 2019;11:5‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bi Z, Liu Y, Zhao Y, et al. A dynamic reversible RNA N6 ‐methyladenosine modification: current status and perspectives. J Cell Physiol. 2019;234:7948‐7956. [DOI] [PubMed] [Google Scholar]
- 8. Krogh N, Nielsen H. Sequencing‐based methods for detection and quantitation of ribose methylations in RNA. Methods. 2019;156:5‐15. [DOI] [PubMed] [Google Scholar]
- 9. Tanzer A, Hofacker IL, Lorenz R. RNA modifications in structure prediction ‐ Status quo and future challenges. Methods. 2019;156:32‐39. [DOI] [PubMed] [Google Scholar]
- 10. Greenberg H, Penman S. Methylation and processing of ribosomal RNA in HeLa cells. J Mol Biol. 1966;21:527‐535. [DOI] [PubMed] [Google Scholar]
- 11. Czerwoniec A, Dunin‐Horkawicz S, Purta E, et al. MODOMICS: a database of RNA modification pathways update. Nucl Acids Res. 2008;2009(37):D118‐D121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71:3971‐3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cantara WA, Crain PF, Rozenski J, et al. The RNA modification database, RNAMDB: 2011 update. Nucl Acids Res. 2011;39:D195‐D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rozenski J, Crain PF, McCloskey JA. The RNA modification database: 1999 update. Nucl Acid Res. 1999;27:196‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boccaletto P, Machnicka MA, Purta E, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucl Acid Res. 2017;2018(46):D303‐D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Su D, Chan CT, Gu C, et al. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC‐coupled mass spectrometry. Nat Protoc. 2014;9:828‐841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kellner S, Ochel A, Thüring K, et al. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucl Acid Res. 2014;42:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mandal D, Köhrer C, Su D, et al. Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine. Proc Natl Acad Sci USA. 2010;107:2872‐2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gu C, Begley TJ, Dedon PC. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014;588:4287‐4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sergiev PV, Bogdanov AA, Dontsova OA. Ribosomal RNA guanine‐(N2)‐methyltransferases and their targets. Nucl Acid Res. 2007;35:2295‐2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Douthwaite S. YbeA is the m3Psi methyltransferase RlmH that targets nucleotide 1915 in 23S rRNA. RNA. 2008;14:2234‐2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Purta E, O'Connor M, Bujnicki JM, Douthwaite S. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J Mol Biol. 2008;383:641‐651. [DOI] [PubMed] [Google Scholar]
- 23. McCloskey JA, Rozenski J. The small subunit rRNA modification database. Nucl Acid Res. 2005;33:D135‐D138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fellmann C, Lowe SW. Stable RNA interference rules for silencing. Nat Cell Biol. 2014;16:10‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344‐352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim JS. Genome editing comes of age. Nat Protoc. 2016;11:1573‐1578. [DOI] [PubMed] [Google Scholar]
- 27. Lundin KE, Højland T, Hansen BR, et al. Biological activity and biotechnological aspects of locked nucleic acids. Adv Genet. 2013;82:47‐107. [DOI] [PubMed] [Google Scholar]
- 28. Liu J, Yue Y, Han D, et al. A METTL3‐METTL14 complex mediates mammalian nuclear RNA N6‐adenosine methylation. Nat Chem Biol. 2014;10:93‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011;2:611‐631. [DOI] [PubMed] [Google Scholar]
- 31. Kennedy EM, Courtney DG, Tsai K, Cullen BR. Viral epitranscriptomics. J Virol. 2017;91:e02263‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet‐binding subunit of the human mRNA (N6‐adenosine)‐methyltransferase. RNA. 1997;3:1233‐1247. [PMC free article] [PubMed] [Google Scholar]
- 33. Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6‐methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15:313‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jia G, Fu Y, Zhao X, et al. N6‐methyladenosine in nuclear RNA is a major substrate of the obesity‐associated FTO. Nat Chem Biol. 2011;7:885‐887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635‐1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dominissini D, Moshitch‐Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature. 2012;485:201‐206. [DOI] [PubMed] [Google Scholar]
- 37. Nishizawa Y, Konno M, Asai A, et al. Oncogene c‐Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2017;9:7476‐7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hesser CR, Karijolich J, Dominissini D, He C, Glaunsinger BA. N6‐methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi's sarcoma‐associated herpesvirus infection. PLoS Pathog. 2018;14:e1006995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang X, Lu Z, Gomez A, et al. N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature. 2014;505:117‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Geula S, Moshitch‐Moshkovitz S, Dominissini D, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002‐1006. [DOI] [PubMed] [Google Scholar]
- 41. Aguilo F, Walsh MJ. The N6‐Methyladenosine RNA modification in pluripotency and reprogramming. Curr Opin Genet Dev. 2017;46:77‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6‐methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Batista PJ, Molinie B, Wang J, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707‐719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maity A, Das B. N6‐methyladenosine modification in mRNA: machinery, function and implications for health and diseases. FEBS J. 2016;283:1607‐1630. [DOI] [PubMed] [Google Scholar]
- 45. Esteller M, Pandolfi PP. The epitranscriptome of noncoding RNAs in cancer. Cancer Discov 2017;7:359‐368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Beemon K, Keith J. Localization of N6‐methyladenosine in the Rous sarcoma virus genome. J Mol Biol. 1977;113:165‐179. [DOI] [PubMed] [Google Scholar]
- 47. Gokhale NS, McIntyre ABR, McFadden MJ, et al. N6‐methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe. 2016;20:654‐665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lichinchi G, Zhao BS, Wu Y, et al. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe. 2016;20:666‐673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lichinchi G, Gao S, Saletore Y, et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV‐1 infection of T cells. Nat Microbiol. 2016;1:16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Courtney DG, Kennedy EM, Dumm RE, et al. Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host Microbe. 2017;22(377–386):e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li Z, Shi J, Yu L, et al. N6‐methyl‐adenosine level in Nicotiana tabacum is associated with tobacco mosaic virus. Virol J. 2018;15:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cui Q, Shi H, Ye P, et al. m6A RNA methylation regulates the self‐renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622‐2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li Z, Weng H, Su R, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6‐methyladenosine RNA demethylase. Cancer Cell. 2017;31:127‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Barbieri I, Tzelepis K, Pandlfini L, et al. Promoter‐bound METTL3 maintains myeloid leukaemia by m6A‐dependent translation control. Nature. 2017;552:126‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang C, Samanta D, Lu H, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF‐dependent and ALKBH5‐mediated m⁶A‐demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113:E2047‐E2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang C, Zhi WI, Lu H, et al. Hypoxia‐inducible factors regulate pluripotency factor expression by ZNF217‐ and ALKBH5‐mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7:64527‐64542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lin Y, Ueda J, Yagyu K, et al. Association between variations in the fat mass and obesity‐associated gene and pancreatic cancer risk: a case‐control study in Japan. BMC Cancer. 2013;13:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huang X, Zhao J, Yang M, Li M, Zheng J. Association between FTO gene polymorphism (rs9939609 T/A) and cancer risk: a meta‐analysis. Eur J Cancer Care. 2017;26:e12464. [DOI] [PubMed] [Google Scholar]
- 60. Khella MS, Salem AM, Abdel‐Rahman O, Saad AS. The association between the FTO rs9939609 variant and malignant pleural mesothelioma risk: a case‐control study. Genet Test Mol Biomarkers. 2018;22:79‐84. [DOI] [PubMed] [Google Scholar]
- 61. Taketo K, Konno M, Asai A, et al. The epitranscriptome m6A writer METTL3 promotes chemo‐ and radioresistance in pancreatic cancer cells. Int J Oncol. 2018;52:621‐629. [DOI] [PubMed] [Google Scholar]
- 62. Mishima E, Jinno D, Akiyama Y, et al. Immuno‐northern blotting: detection of RNA modifications by using antibodies against modified nucleosides. PLoS ONE. 2015;10:e0143756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Imanishi M, Tsuji S, Suda A, Futaki S. Detection of N6‐methyladenosine based on the methyl‐sensitivity of MazF RNA endonuclease. Chem Commun. 2017;53:12930‐12933. [DOI] [PubMed] [Google Scholar]
- 64. Golovina AY, Dzama MM, Petriukov KS, et al. Method for site‐specific detection of m6A nucleoside presence in RNA based on high‐resolution melting (HRM) analysis. Nucleic Acids Res. 2014;42:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Arguello AE, DeLiberto AN, Kleiner RE. RNA chemical proteomics reveals the N6‐methyladenosine (m6A)‐regulated protein‐RNA interactome. J Am Chem Soc. 2017;139:17249‐17252. [DOI] [PubMed] [Google Scholar]
- 66. Yin H, Wang H, Jiang W, Zhou Y, Ai S. Electrochemical immunosensor for N6‐methyladenosine detection in human cell lines based on biotin‐streptavidin system and silver‐SiO2 signal amplification. Biosens Bioelectron. 2017;90:494‐500. [DOI] [PubMed] [Google Scholar]
- 67. Molinie B, Wang J, Lim KS, et al. m(6)A‐LAIC‐seq reveals the census and complexity of the m(6)A epitranscriptome. Nat Method. 2016;13:692‐698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aschenbrenner J, Werner S, Marchand V, et al. Engineering of a DNA polymerase for direct m6 a sequencing. Angew Chem Int Ed Engl. 2018;57:417‐421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Linder B, Grozhik AV, Olarerin‐George AO, Meydan C, Mason CE, Jaffrey SR. Single‐nucleotide‐resolution mapping of m6A and m6Am throughout the transcriptome. Nat Method. 2015;12:767‐772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hafner M, Landthaler M, Burger L, et al. Transcriptome‐wide identification of RNA‐binding protein and microRNA target sites by PAR‐CLIP. Cell. 2010;141:129‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. Probing N6‐methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19:1848‐1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Di Ventra M, Taniguchi M. Decoding DNA, RNA and peptides with quantum tunnelling. Nat Nanotechnol. 2016;11:117‐126. [DOI] [PubMed] [Google Scholar]
- 73. Ohshiro T, Tsutsui M, Yokota K, Taniguchi M. Quantitative analysis of DNA with single‐molecule sequencing. Sci Rep. 2018;8:8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tsutsui M, Matsubara K, Ohshiro T, Furuhashi M, Taniguchi M, Kawai T. Electrical detection of single‐methylcytosines in a DNA oligomer. J Am Chem Soc. 2011;133:9124‐9128. [DOI] [PubMed] [Google Scholar]
- 75. Ohshiro T, Matsubara K, Tsutsui M, Furuhashi M, Taniguchi M, Kawai T. Single‐molecule electrical random resequencing of DNA and RNA. Sci Rep. 2012;2:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ohshiro T, Tsutsui M, Yokota K, Furuhashi M, Taniguchi M, Kawai T. Detection of post‐translational modifications in single peptides using electron tunnelling currents. Nat Nanotechnol. 2014;9:835‐840. [DOI] [PubMed] [Google Scholar]
- 77. Ohshiro T, Komoto Y, Konno M, et al. Direct analysis of incorporation of an anticancer drug into DNA at single‐molecule resolution. Sci Rep. 2019;9(1):3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fitzsimmons CM, Batista PJ. It's complicated… m6A‐dependent regulation of gene expression in cancer. Biochim Biophys Acta Gene Regul Mech. 2019;1862:382‐393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Maddocks OD, Labuschagne CF, Adams PD, Vousden KH. Serine metabolism supports the methionine cycle and DNA/RNA methylation through de novo ATP synthesis in cancer cells. Mol Cell. 2016;61:210‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Koseki J, Matsui H, Konno M, et al. A trans‐omics mathematical analysis reveals novel functions of the ornithine metabolic pathway in cancer stem cells. Sci Rep. 2016;6:20726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ponnaluri VK, Maciejewski JP, Mukherji M. A mechanistic overview of TET‐mediated 5‐methylcytosine oxidation. Biochem Biophys Res Commun. 2013;436:115‐120. [DOI] [PubMed] [Google Scholar]
- 82. Koseki J, Colvin H, Fukusumi T, et al. Mathematical analysis predicts imbalanced IDH1/2 expression associates with 2‐HG‐inactivating β‐oxygenation pathway in colorectal cancer. Int J Oncol. 2015;46:1181‐1191. [DOI] [PubMed] [Google Scholar]
- 83. Hua W, Zhao Y, Jin X, et al. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol Oncol. 2018;151:356‐365. [DOI] [PubMed] [Google Scholar]
- 84. Witkin KL, Hanlon SE, Strasburger JA, et al. RNA editing, epitranscriptomics, and processing in cancer progression. Cancer Biol Ther. 2015;16:21‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jiang Q, Crews LA, Holm F, Jamieson CHM. RNA editing‐dependent epitranscriptome diversity in cancer stem cells. Nat Rev Cancer. 2017;17:381‐392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li X, Xiong X, Yi C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Method. 2016;14:23‐31. [DOI] [PubMed] [Google Scholar]
- 87. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293‐306. [DOI] [PubMed] [Google Scholar]
- 88. Licht K, Jantsch FM. Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications. J Cell Biol. 2016;213:15‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]


