Abstract
Deregulation of microRNAs (miRs) has been demonstrated to be involved in both the initiation and the development of non-small-cell lung cancer (NSCLC). miR-448 has been identified as a tumor suppressor in several cancer types. The aim of the present study was to explore the role of miR-448 in NSCLC. Tumor tissues and paired normal tissues were obtained from patients with NSCLC. The viability and migration of A549 cells were determined by the Cell Counting kit-8 and wound-healing assays, respectively. Gene and protein levels were detected by reverse transcription-quantitative polymerase chain reaction analysis and western blotting, respectively. The interaction between the 3′ untranslated region of sirtuin1 (SIRT1) and miR-448 was predicted by TargetScan and verified by dual luciferase reporter assay. miR-448 levels were revealed to be decreased whereas SIRT1 levels were increased in NSCLC tissues compared with normal tissues. Pearson's correlation analysis demonstrated that there was a negative correlation between miR-448 and SIRT1 mRNA levels. Overexpression of miR-448 led to reduced growth and migration ability of A549 cells. In addition, overexpression of miR-448 decreased SIRT1 mRNA and protein levels, thereby inhibiting epithelial-mesenchymal transition (EMT) and affecting EMT-associated molecules, including vimentin and E-cadherin. Dual luciferase reporter assay confirmed that SIRT1 was a direct target of miR-448. Notably, activation of SIRT1 by resveratrol treatment partially reversed the cell growth inhibition induced by miR-448 mimics. These findings suggested that the progression of NSCLC may be controlled by miR-448, which appears to hold promise as a therapeutic target for patients with NSCLC.
Keywords: non-small-cell lung cancer, microRNA-448, sirtuin1, cell proliferation, cell migration
Introduction
Lung cancer is one of the most lethal types of cancer, accounting for ~18% of cancer-associated mortalities worldwide (1). Lung cancer is divided into two major subtypes, namely small-cell lung cancer and non-small-cell lung cancer (NSCLC) (2). NSCLC accounts for >80% of all lung cancer cases and its prognosis is relatively poor due to its highly aggressive nature (3,4). It is crucial to elucidate the precise molecular mechanisms underlying NSCLC progression in order to identify novel biomarkers and therapeutic targets for the treatment of patients with NSCLC.
microRNAs (miRNAs or miRs) are small, non-coding, single-stranded RNA molecules that are involved in the regulation of gene expression in eukaryotes (5). miRNA deregulation is frequently observed in human diseases, including different types of cancer (6,7). During cancer initiation and progression, miRNAs serve an important role through downregulation of oncogenes and/or tumor suppressors, thereby affecting tumor cell proliferation, metastasis and apoptosis (8–10). In NSCLC, several miRNAs have been found to be deregulated and demonstrated to promote cancer progression or drug resistance via repression of target genes (11,12). The expression of several of those miRNAs in tumor tissues, plasma and exosomes has been considered a promising diagnostic and therapeutic biomarker for patients with NSCLC (13). miR-448 has been identified as a tumor suppressor in several cancer types, including breast, hepatocellular and colorectal cancer (14–16). A recent study reported that low expression of miR-448 predicted poor prognosis of lung squamous cell carcinoma (SCC), a subtype of NSCLC, and it suppressed SCC cell proliferation and metastasis via targeting DCLK1 (17).
Silent mating-type information regulation 2 homologue 1 (sirtuin1; SIRT1) is a member of the sirtuin family that acts as an NAD-dependent protein deacetylase (18). Through regulating the acetylation level of target proteins, such as p53, SIRT1 participates in a number of cellular events that are implicated in cancer progression (19,20). Accumulating evidence indicates that overexpression of SIRT1 promotes cancer progression, including hepatocellular carcinoma and breast cancer (21,22). In NSCLC, overexpression of SIRT1 has been associated with poor prognosis (23). However, the regulatory mechanism of SIRT1 in NSCLC remains unknown.
In the present study, a significant reduction of miR-448 levels and elevation of SIRT1 mRNA levels were observed in tumor samples from patients with NSCLC. In vitro experiments demonstrated that enhanced expression of miR-448 inhibited the proliferation and migration of A549 cells. Additionally, overexpression of miR-448 decreased SIRT1 mRNA and protein levels, thus inhibiting epithelial-mesenchymal transition (EMT). Activation of SIRT1 partially rescued miR-448-induced cell growth arrest. The findings of the present study suggest a pivotal role for miR-448 in the inhibition of NSCLC progression via targeting SIRT1.
Materials and methods
Clinical specimens
Tumor tissues and adjacent normal tissues were collected from 25 patients (17 males and 8 females; aged 35–65 years) with NSCLC undergoing surgery at Changyi People's Hospital (Weifang, China) between September 2, 2014 and May 4, 2016. None of the patients received chemotherapy or radiotherapy prior to surgery. Written informed consent was obtained from all participants and the clinical experiments were approved by the Ethics Committee of Changyi People's Hospital. All specimens were immediately stored at −80°C until use in subsequent experiments.
Cell lines and reagents
293T cells and the human NSCLC cell line A549 were purchased from American Type Culture Collection (Manassas, VA, USA) and used within 6 months. A549 cells and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific) in a 37°C incubator with 5% CO2.
miR-negative control (NC) mimics and miR-448 mimics were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences were as follows: miR-NC mimics, 5′-UCGCUUGGUGCAGGUCGGGAA-3′; miR-448 mimics, 5′-UUGCAUAUGUAGGAUGUCCCAU-3′. Transfection of 50 nM miR-NC mimics or miR-448 mimics was performed with Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After 48 h, cells were subjected to experiments.
Resveratrol was obtained from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany) and dissolved in dimethyl sulfoxide to a concentration of 50 mM for storage. For the resveratrol + miR-448 mimics group, cells were firstly transfected with 50 nM miR-448 mimics using Lipofectamine 2000, then resveratrol was diluted and added into the culture medium at a final concentration of 20 µM for SIRT1 activation for 48 h at 37°C. The DMSO + miR-448 mimics and DMSO + miR-NC mimics group was transfected with 50 nM miR-448 mimics miR-NC mimics using Lipofectamine 2000 followed by treatment with 20 µM DMSO for 48 h at 37°C.
Western blotting
Whole proteins were extracted from A549 cells using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China). Pierce BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.) was used to determine the concentration of lysates according to the manufacturer's protocol. In brief, 20 µg protein lysates were loaded into each lane of 8% gels, separated by SDS-PAGE for 30 min and transferred onto polyvinylidene fluoride (PVDF) membranes. The PVDF membranes were blocked with 5% non-fat milk for 1 h at room temperature and incubated with the indicated primary antibodies overnight at 4°C. On the next day, the PVDF membranes were incubated with the secondary antibodies for 1 h at room temperature and developed with enhanced chemiluminescence substrate (EMD Millipore, Billerica, MA, USA). Antibodies against SIRT1 (cat. no. 8469), vimentin (cat. no. 5741) and E-cadherin (cat. no. 14472; all 1:1,000) were supplied by Cell Signaling Technology, Inc. (Danvers, MA, USA). Antibodies against GAPDH (cat. no. G8795; 1:5,000) were purchased from Sigma-Aldrich; Merck KGaA. Horseradish peroxidase-conjugated anti-mouse (cat. no. SA00001-1) and anti-rabbit (cat. no. SA00001-2; both 1:10,000) secondary antibodies were purchased from ProteinTech Group, Inc. (Chicago, IL, USA). Images were captured with ImageQuant300 (GE Healthcare, Chicago, IL) and analyzed using ImageJ software (version 1.8.0; National Institutes of Health, Bethesda, MD, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from A549 cells, as well as NSCLC tissues and adjacent normal tissues using TRIzol® Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The concentration of RNA was measured with NanoDrop™ 2000 (Thermo Fisher Scientific, Inc.).
To measure miR-448 levels, RNA was reverse-transcribed into cDNA using a TaqMan MicroRNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.). RT-PCR was performed using a TaqMan MicroRNA Assay kit (Thermo Fisher Scientific, Inc.) on the CFX-96 module (Bio-Rad Laboratories, Inc., Hercules, CA, USA). U6 served as an internal control for miR-448 expression. For the detection of SIRT1, vimentin and E-cadherin mRNA levels, RNA was reverse-transcribed into cDNA using PrimeScript RT Master Mix (Takara Bio, Inc., Otsu, Japan). GAPDH served as an internal control. The thermocycling conditions for mRNA and miRNA analyses were: Pre-denaturing at 95°C for 30 sec, and 35 cycles of denaturing at 95°C for 10 sec, annealing and elongation at 60°C for 10 sec. The relative expression of gene was calculated using the 2−ΔΔCq method (24). The primer sequences were: Stem loop: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGATGGGA-3′; miR-448-forward, 5′-TCGGCAGGTTGCATATGTAGGA-3′ and reverse, 5′-CTCAACTGGTGTCGTGGA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; Vimentin forward, 5′-GACGCCATCAACACCGAGTT-3′ and reverse, 5′-CTTTGTCGTTGGTTAGCTGGT-3′; E-cadherin-forward, 5′-CGAGAGCTACACGTTCACGG-3′ and reverse, 5′-GGGTGTCGAGGGAAAAATAGG-3′; SIRT1 forward, 5′-TAGCCTTGTCAGATAAGGAAGGA-3′ and SIRT1 reverse, 5′-ACAGCTTCACAGTCAACTTTGT-3′; GAPDH forward, 5′-CCTGCACCACCAACTGCTTA-3′ and reverse, 5′-GGCCATCCACAGTCTTCTGAG-3′.
Dual luciferase reporter assay
To further investigate whether miR-448 directly regulates SIRT1 expression in A549 cells, the online bioinformatics tool TargetScan (www.targetscan.org) was used to predict miR-448 potential binding sites in SIRT1. The 3′ untranslated region (UTR) of SIRT1 mRNA was amplified from cDNA of 293T cells and ligated into the pGL3 plasmid (Promega Corporation, Madison, WI, USA) to construct pGL3-SIRT1-3′UTR-wild-type (WT). pGL3-SIRT1-3′UTR-mutant (Mut) was obtained by introducing 2 site mutations into miR-448 potential binding sites using a Quicksite mutation kit (Agilent Technologies, Inc., Santa Clara, CA, USA). pGL3-SIRT1-3′UTR-WT or pGL3-SIRT1-3′UTR-Mut was co-transfected with Renilla plasmid into A549 cells using Lipofectamine 2000, followed by transfection of miR-NC mimics or miR-448 mimics. Following 48 h, a Dual Luciferase Reporter Assay system (Promega Corporation) was used to measure the relative luciferase activity of each well. In each group, firefly luciferase activity was normalized to Renilla luciferase activity. Subsequently, the relative luciferase activity of control group was normalized to 1 and was compared with the treatment group.
Cell proliferation assay
The proliferation of A549 cells was measured using a Cell Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Subsequently, 10 µl CCK-8 solution was added into each well and sustained for 2 h. Immediately following, and 24, 48, 72 and 96 h after the incubation, the medium containing CCK-8 was transferred into a clean 96-well plate and the absorbance at 450 nm of each well was detected with a microplate reader (Bio-Rad Laboratories, Inc.).
Wound-healing assay
The cell migration ability was measured using a wound-healing assay. Briefly, 2×106 cells were plated in a 6-well plate and cultured until reaching 100% confluence. A wound was made by scratching the middle area of each well using a 10-µl pipette. After washing with PBS, DMEM without FBS was added into each well and images of the wound areas were captured immediately using an inverted microscope at a magnification of ×40. After the cells were cultured for 40 h, images of wound areas were captured again. The relative migrated areas were calculated using Image Pro-Plus version 6.0 (Media Cybernetics, Inc., Rockville, MD, USA).
Statistical analysis
All data in the present study were analyzed using GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA) and the results are expressed as the mean + standard deviation. Statistical analysis between two groups was performed using Student's t-test, and the differences between normal tissues and tumor tissues were analyzed by paired Student's t-test. The differences among three different groups were analyzed using one-way analysis of variance followed by Tukey's analysis. The correlation analysis was carried out using Pearson Correlation analysis. P<0.05 was considered to indicate statistically significant differences.
Results
Expression levels of miR-448 and SIRT1 in tumor tissues and normal tissues from patients with NSCLC
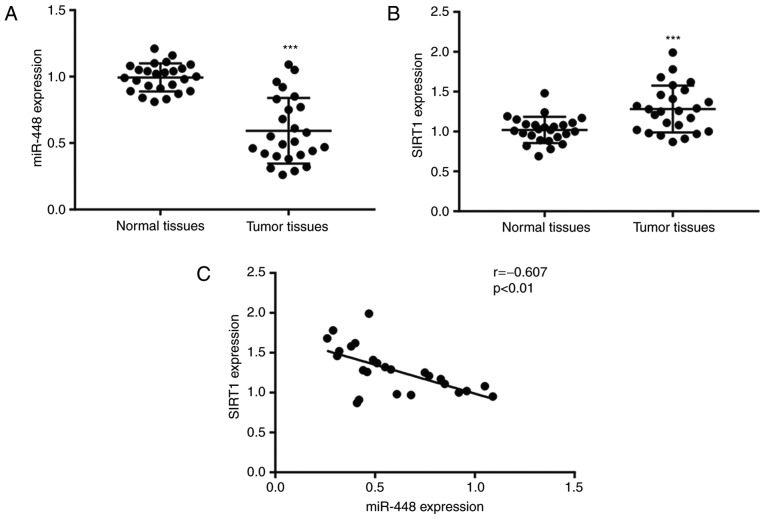
To determine the expression levels of miR-448 and SIRT1 in NSCLC, tumor tissues and matched normal tissues from 25 patients with NSCLC were used for RT-qPCR. Compared with the normal tissues, the expression of miR-448 was significantly lower, while the SIRT1 mRNA levels were significantly higher in tumor tissues (Fig. 1A and B). Their different expression pattern suggested a potential association between the two. Pearson's correlation analysis of miR-448 levels and SIRT1 mRNA levels revealed a strong negative correlation between their expressions in NSCLC tumor tissues (Fig. 1C), suggesting a potential regulatory association between them.
Figure 1.
Aberrant expression of miR-448 and SIRT1 in tumor tissues from patients with NSCLC. (A) Compared with adjacent normal tissues, miR-448 expression was decreased in tumor tissues from patients with NSCLC. (B) Compared with normal tissues, SIRT1 mRNA levels were elevated in tumor tissues from patients with NSCLC. (C) Expression of miR-448 was negatively correlated with SIRT1 expression in tumor tissues from patients with NSCLC. ***P<0.001 vs. normal tissues. miR, miroRNA; SIRT1, sirtuin1; NSCLC, non-small cell lung carcinoma.
Overexpression of miR-448 suppressed the proliferation and migration of A549 cells
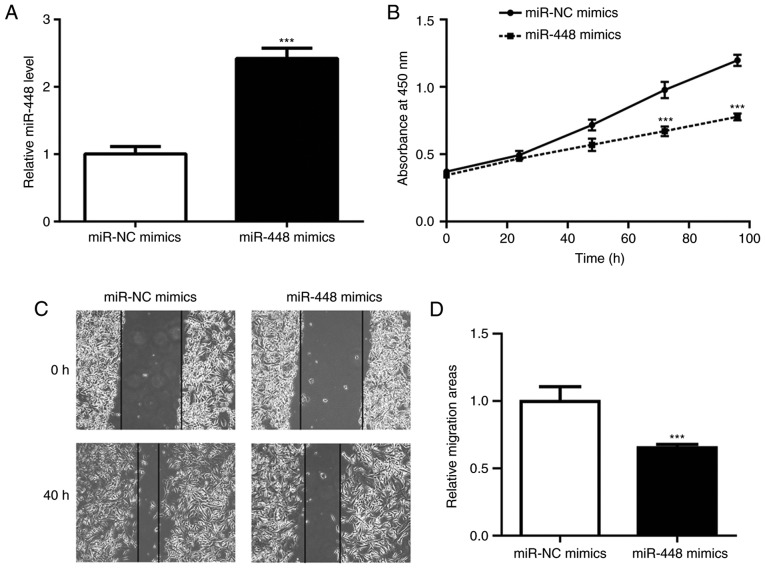
To investigate the role of miR-448 in NSCLC cells, the A549 NSCLC cell line was used to perform the associated functional studies and experiments. Transfection of miR-448 mimics markedly elevated the miR-448 level in A549 cells (Fig. 2A). Overexpression of miR-448 significantly reduced cell viability and wound healing, indicating its role in the inhibition of cell proliferation and cell migration (Fig. 2B-D).
Figure 2.
Overexpression of miR-448 inhibited the proliferation and migration of A549 cells. (A) Transfection of miR-448 mimics increased miR-448 expression in A549 cells. (B) Overexpression of miR-448 induced growth arrest in A549 cells. (C) Overexpression of miR-448 inhibited the migration of A549 cells. Representative images of migration were obtained under an inverted microscope (magnification, ×40). (D) Quantification of wound closure differences. ***P<0.001 vs. miR-NC mimics. miR, miroRNA; NC, negative control.
miR-448 repressed EMT through downregulation of SIRT1
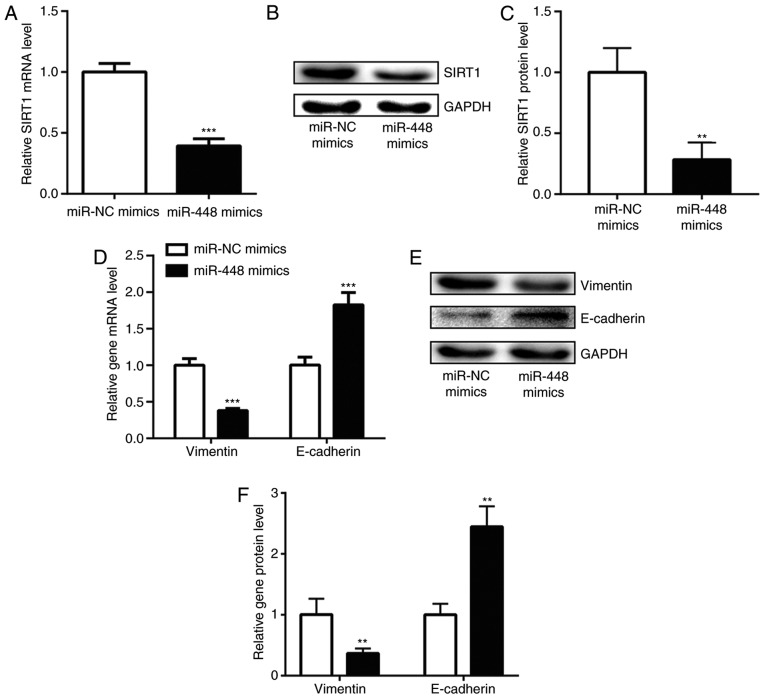
The negative correlation between miR-448 and SIRT1 mRNA levels indicated a regulatory association between miR-448 and SIRT1. Consistent with the correlation analysis, enhanced expression of miR-448 significantly decreased SIRT1 mRNA level in A549 cells (Fig. 3A). Additionally, the protein level of SIRT1 was also reduced with miR-448 overexpression (Fig. 3B and C). SIRT1 was previously reported to promote NSCLC cell proliferation and metastasis via inducing EMT (25). Increase of miR-448 levels downregulated vimentin and upregulated E-cadherin expression at both the mRNA and protein levels in A549 cells (Fig. 3D-F), suggesting that miR-448 may inhibit EMT through repression of SIRT1 expression.
Figure 3.
miR-448 overexpression repressed SIRT1 to induce epithelial-mesenchymal transition in A549 cells. (A) Overexpression of miR-448 decreased SIRT1 mRNA levels in A549 cells. (B) Overexpression of miR-448 reduced SIRT1 protein levels in A549 cells. (C) Quantification of protein expression levels. (D) Transfection of miR-448 mimics decreased vimentin and increased E-cadherin mRNA levels in A549 cells. (E) Transfection of miR-448 mimics decreased vimentin and increased E-cadherin protein levels in A549 cells. (F) Quantification of protein expression levels. **P<0.01 and ***P<0.001 vs. miR-NC mimics. miR, miroRNA; SIRT1, sirtuin1; NC, negative control.
SIRT1 is a direct target of miR-448
It is observed that there was an miR-448 binding site in the 3′UTR of SIRT1 mRNA (Fig. 4A). Subsequently, the dual luciferase reporter assay was performed to validate their association. Transfection of miR-448 mimics significantly decreased relative luciferase activity in A549 cells transfected with SIRT1-3′UTR-WT, but not SIRT1-3′UTR-Mut, suggesting that miR-448 can bind to SIRT1-3′UTR-WT (Fig. 4B). Therefore, these results confirmed that miR-448 directly binds to the 3′UTR of SIRT1 mRNA to inhibit its expression in A549 cells.
Figure 4.
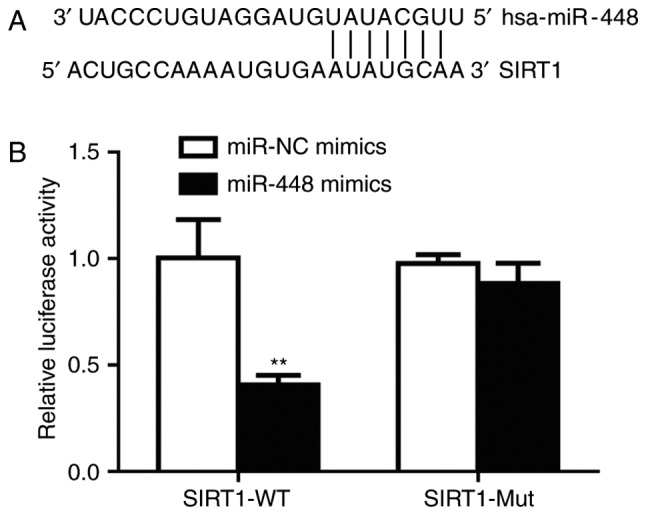
SIRT1 was revealed to be a target gene of miR-448. (A) Sequence alignment revealed that there was a complementary binding site for miR-448 on the 3′ untranslated region of SIRT1 mRNA. (B) Transfection of miR-448 mimics reduced relative luciferase activity in A549 cells transfected with SIRT1 3′UTR-WT. **P<0.01 vs. miR-NC mimics. miR, miroRNA; SIRT1, sirtuin1; NC, negative control; WT, wild type; Mut, mutant.
Elevation of SIRT1 partially rescued miR-448 mimics-induced cell growth arrest
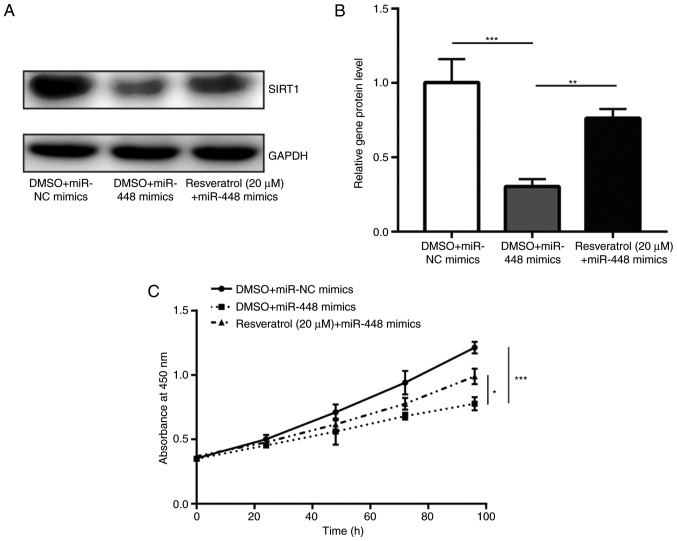
Resveratrol treatment was previously demonstrated to induce SIRT1 expression and promote cell proliferation in A549 cells (26). In the present study, it was observed that treatment with resveratrol was able to partially recover SIRT1 expression, which was downregulated by miR-448 overexpression (Fig. 5A and B). Notably, resveratrol partially reversed the cell growth inhibition induced by miR-448 mimics in A549 cells (Fig. 5C). These data indicated that miR-448 mainly relies on regulation of SIRT1 to suppress NSCLC progression.
Figure 5.
SIRT1 was pivotal for miR-448-induced cell growth arrest. (A) Transfection of miR-448 mimics reduced SIRT1 protein levels, and this effect was reversed by treatment of A549 cells with resveratrol. (B) Quantification of SIRT1 protein levels. (C) Treatment with resveratrol reversed SIRT1-induced growth arrest of A549 cells. *P<0.05, **P<0.01 and ***P<0.001. SIRT1, sirtuin1; miR, miroRNA; DMSO, dimethyl sulfoxide; NC, negative control.
Discussion
Aberrant expression of miRNA is frequent in cancer cells. The dysregulation and tumor suppressor role of miR-448 has been reported in various types of cancer (14–16). However, the role of miR-448 in NSCLC remains controversial. A previous study reported that miR-448 levels were significantly higher in plasma samples from patients with NSCLC compared with those from healthy volunteers, and that it may serve as a potential biomarker (27). Another recent study demonstrated that miR-448 was decreased in lung SCC tissues and cell lines (17). In the present study, it was observed that miR-448 was able to suppress the progression of NSCLC through targeting SIRT1.
A previous meta-analysis revealed that high expression of SIRT1 predicted poor overall survival for patients with NSCLC (23). Several miRNAs were reported to repress SIRT1 expression, thereby controlling cancer cell behavior (28,29). Particularly, miR-138 was demonstrated to suppress NSCLC cell growth, metastasis and autophagy via targeting SIRT1 (24). Although a previous study investigated miR-448 and SIRT1 in patients with diabetes mellitus (30), their role in cancer cells, particularly NSCLC cells, remains unknown. In clinical samples, the expression of SIRT1 was higher in tumor tissues and its levels were inversely correlated with miR-448 levels. In A549 cells, enhanced expression of miR-448 decreased SIRT1 mRNA and protein levels. In addition, the present dual luciferase reporter assay further supported that miR-448 could directly bind to the SIRT1 3′UTR in A549 cells. miR-448 can also bind to multiple target gene mRNAs such as EPHA7 and IGF1R in cancer cells (16,31). It was observed that activation of SIRT1 could reverse miR-448-induced cell growth inhibition. These results collectively demonstrated that miR-448 mainly relies on the regulation of SIRT1 to repress NSCLC progression.
Accumulating evidence has demonstrated that SIRT1 is an inducer of EMT and promotes cancer progression (32,33). In the present study, the expression of E-cadherin was increased while that of vimentin was decreased in A549 cells transfected with miR-448 mimics, suggesting that miR-448 reverses EMT by repressing SIRT1 expression. These findings were consistent with those of previous studies on breast cancer and hepatocellular carcinoma cells, demonstrating that miR-448 inhibits EMT (15,34). As EMT is an indicator of highly aggressive growth of cancer cells (35), miR-448 may inhibit NSCLC progression via reversing EMT. Taken together, the present findings demonstrated that miR-448 might be a promising target for the treatment of NSCLC.
Acknowledgements
Not applicable.
Funding
No funding received.
Availability of data and materials
The data and materials are available from the corresponding author.
Authors' contributions
HQ, HW and DP designed the study and acquired the data. HW collected the clinical samples. DP supervised the study, and prepared and revised the manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Changyi People's Hospital.
Patient consent for publication
Written informed consents were provided by all patients. The patients consented to the publication of data presented in the manuscript.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 2.Niklinski J, Niklinska W, Chyczewski L, Becker HD, Pluygers E. Molecular genetic abnormalities in premalignant lung lesions: Biological and clinical implications. Eur J Cancer Prev. 2001;10:213–226. doi: 10.1097/00008469-200106000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Zhou J, Song XB, He H, Zhou Y, Lu XJ, Ying BW. Prevalence and clinical profile of EGFR mutation in non-small-cell lung carcinoma patients in Southwest China. Asian Pac J Cancer Prev. 2016;17:965–971. doi: 10.7314/APJCP.2016.17.3.965. [DOI] [PubMed] [Google Scholar]
- 4.Wu S, Zhao X, Wu S, Du R, Zhu Q, Fang H, Zhang X, Zhang C, Zheng W, Yang J, Feng H. Overexpression of B7-H3 correlates with aggressive clinicopathological characteristics in non-small cell lung cancer. Oncotarget. 2016;7:81750–81756. doi: 10.18632/oncotarget.13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Mendell JT. MicroRNAs: Critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 7.Iorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu T, Wu X, Chen T, Luo Z, Hu X. Downregulation of DNMT3A by miR-708-5p inhibits lung cancer stem cell-like phenotypes through repressing wnt/β-catenin signaling. Clin Cancer Res. 2018;24:1748–1760. doi: 10.1158/1078-0432.CCR-17-1169. [DOI] [PubMed] [Google Scholar]
- 9.Pang W, Tian X, Bai F, Han R, Wang J, Shen H, Zhang X, Liu Y, Yan X, Jiang F, Xing L. Pim-1 kinase is a target of miR-486-5p and eukaryotic translation initiation factor 4E, and plays a critical role in lung cancer. Mol Cancer. 2014;13:240. doi: 10.1186/1476-4598-13-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paliouras AR, Monteverde T, Garofalo M. Oncogene-induced regulation of microRNA expression: Implications for cancer initiation, progression and therapy. Cancer Lett. 2018;421:152–160. doi: 10.1016/j.canlet.2018.02.029. [DOI] [PubMed] [Google Scholar]
- 11.Fadejeva I, Olschewski H, Hrzenjak A. MicroRNAs as regulators of cisplatin-resistance in non-small cell lung carcinomas. Oncotarget. 2017;8:115754–115773. doi: 10.18632/oncotarget.22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, Gao M, Li Z, Song L, Gao X, Han J, Wang F, Chen Y, Li W, Yang J, Han X. Role of microRNAs in metastasis of non-small cell lung cancer. Front Biosci (Landmark Ed) 2016;21:998–1005. doi: 10.2741/4436. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Q, Huang SX, Zhang F, Li SJ, Liu C, Xi YY, Wang L, Wang X, He QQ, Sun CC, Li DJ. MicroRNAs: A novel potential biomarker for diagnosis and therapy in patients with non-small cell lung cancer. Cell Prolif. 2017;50:12394. doi: 10.1111/cpr.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li QQ, Chen ZQ, Cao XX, Xu JD, Xu JW, Chen YY, Wang WJ, Chen Q, Tang F, Liu XP, Xu ZD. Involvement of NF-κB/miR-448 regulatory feedback loop in chemotherapy-induced epithelial-mesenchymal transition of breast cancer cells. Cell Death Differ. 2011;18:16–25. doi: 10.1038/cdd.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu H, Zhou X, Ma C, Chang H, Li H, Liu F, Lu J. Low expression of miR-448 induces EMT and promotes invasion by regulating ROCK2 in hepatocellular carcinoma. Cell Physiol Biochem. 2015;36:487–498. doi: 10.1159/000430114. [DOI] [PubMed] [Google Scholar]
- 16.Li B, Ge L, Li M, Wang L, Li Z. miR-448 suppresses proliferation and invasion by regulating IGF1R in colorectal cancer cells. Am J Transl Res. 2016;8:3013–3022. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Shan C, Fei F, Li F, Zhuang B, Zheng Y, Wan Y, Chen J. miR-448 is a novel prognostic factor of lung squamous cell carcinoma and regulates cells growth and metastasis by targeting DCLK1. Biomed Pharmacother. 2017;89:1227–1234. doi: 10.1016/j.biopha.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 18.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: Understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 19.Ong ALC, Ramasamy TS. Role of Sirtuin1-p53 regulatory axis in aging, cancer and cellular reprogramming. Ageing Res Rev. 2018;43:64–80. doi: 10.1016/j.arr.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009;69:1702–1705. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Xu S, Li J, Zheng L, Feng M, Wang X, Han K, Pi H, Li M, Huang X, et al. SIRT1 facilitates hepatocellular carcinoma metastasis by promoting PGC-1α-mediated mitochondrial biogenesis. Oncotarget. 2016;7:29255–29274. doi: 10.18632/oncotarget.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou Q, Tang Q, Pan Y, Wang X, Dong X, Liang Z, Huang D. MicroRNA-22 inhibits cell growth and metastasis in breast cancer via targeting of SIRT1. Exp Ther Med. 2017;14:1009–1016. doi: 10.3892/etm.2017.4590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Chen Y, Wang T, Wang W, Hu J, Li R, He S, Yang J. Prognostic and clinicopathological significance of SIRT1 expression in NSCLC: A meta-analysis. Oncotarget. 2017;8:62537–62544. doi: 10.18632/oncotarget.19244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Ye Z, Fang B, Pan J, Zhang N, Huang J, Xie C, Lou T, Cao Z. miR-138 suppresses the proliferation, metastasis and autophagy of non-small cell lung cancer by targeting Sirt1. Oncol Rep. 2017;37:3244–3252. doi: 10.3892/or.2017.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Shao K, Sun T. SIRT1 regulates the human alveolar epithelial A549 cell apoptosis induced by pseudomonas aeruginosa lipopolysaccharide. Cell Physiol Biochem. 2013;31:92–101. doi: 10.1159/000343352. [DOI] [PubMed] [Google Scholar]
- 27.Powrozek T, Krawczyk P, Kowalski DM, Kuźnar-Kamińska B, Winiarczyk K, Olszyna-Serementa M, Batura-Gabryel H, Milanowski J. Application of plasma circulating microRNA-448, 506, 4316, and 4478 analysis for non-invasive diagnosis of lung cancer. Tumour Biol. 2016;37:2049–2055. doi: 10.1007/s13277-015-3971-4. [DOI] [PubMed] [Google Scholar]
- 28.Guan Y, Rao Z, Chen C. miR-30a suppresses lung cancer progression by targeting SIRT1. Oncotarget. 2017;9:4924–4934. doi: 10.18632/oncotarget.23529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Ma J, Han X, Jia Y, Yuan H, Shui S, Guo D. MicroRNA-320 enhances radiosensitivity of glioma through down-regulation of sirtuin type 1 by directly targeting forkhead box protein M1. Transl Oncol. 2018;11:205–212. doi: 10.1016/j.tranon.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Wang DS, Cheng YS, Jia BL, Yu G, Yin XQ, Wang Y. Expression of MicroRNA-448 and SIRT1 and prognosis of obese type 2 diabetic mellitus patients after laparoscopic bariatric surgery. Cell Physiol Biochem. 2018;45:935–950. doi: 10.1159/000487287. [DOI] [PubMed] [Google Scholar]
- 31.Wu X, Yan L, Liu Y, Xian W, Wang L, Ding X. MicroRNA-448 suppresses osteosarcoma cell proliferation and invasion through targeting EPHA7. PLoS One. 2017;12:e0175553. doi: 10.1371/journal.pone.0175553. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Sun T, Jiao L, Wang Y, Yu Y, Ming L. SIRT1 induces epithelial-mesenchymal transition by promoting autophagic degradation of E-cadherin in melanoma cells. Cell Death Dis. 2018;9:136. doi: 10.1038/s41419-017-0167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray U, Roy SS, Chowdhury SR. Lysophosphatidic acid promotes epithelial to mesenchymal transition in ovarian cancer cells by repressing SIRT1. Cell Physiol Biochem. 2017;41:795–805. doi: 10.1159/000458744. [DOI] [PubMed] [Google Scholar]
- 34.Ma P, Ni K, Ke J, Zhang W, Feng Y, Mao Q. miR-448 inhibits the epithelial-mesenchymal transition in breast cancer cells by directly targeting the E-cadherin repressor ZEB1/2. Exp Biol Med (Maywood) 2018;243:473–480. doi: 10.1177/1535370218754848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Weinberg RA. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and materials are available from the corresponding author.