Abstract
Fibroblast growth factor 2 (FGF2) has been revealed to promote human periodontal ligament stem cell (PDLSC) proliferation. The abnormal proliferation of PDLSCs has also been associated with the pathogenesis of periodontitis. The long non-coding RNA, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), has been demonstrated to regulate FGF2 secretion. Therefore, MALAT1 may also be associated with periodontitis. The aim of the present study was to investigate the effect of MALAT1 overexpression on the proliferation of PDLSCs. In the current study, PDLSCs derived from healthy and periodontitis-affected teeth were collected. MALAT1 and FGF2 mRNA expression in PDLSCs was detected using reverse transcription-quantitative PCR. PDLSCs overexpressing MALAT1 were subsequently generated. PDLSC proliferation was analyzed using a Cell Counting kit-8 assay. FGF2 protein expression was detected using western blot analysis. The results revealed that MALAT1 and FGF2 mRNA were significantly upregulated in PDLSCs derived from periodontitis-affected teeth when compared with PDLSCs derived from healthy teeth. PDLSCs derived from periodontitis-affected teeth also demonstrated a significantly higher proliferation rate than PDLSCs derived from healthy teeth. MALAT1 and FGF2 mRNA expression were positively correlated in both PDLSC groups. MALAT1 overexpression promoted the proliferation of healthy and periodontitis-affected PDLSC groups and upregulated FGF2 protein expression. The present study concluded that MALAT1 overexpression promoted the proliferation of human PDLSC potentially via upregulating FGF2.
Keywords: periodontitis, long-coding RNA metastasis-associated lung adenocarcinoma transcript 1, fibroblast growth factor 2, human periodontal ligament stem cells
Introduction
Periodontitis is a chronic inflammatory disease that affects the integrity of the periodontium, which is composed of the periodontal ligament (PDL), gingiva and alveolar bone (1). Periodontitis leads to tooth loss and may also induce systemic disorders, including adverse pregnancy outcomes, atherosclerosis, rheumatoid arthritis, aspiration pneumonia or cancer (2–4). Periodontitis affects approximately 45–75% of adults worldwide (1–4). In developing countries, such as China, poor overall oral health has led to periodontitis becoming a commonly diagnosed disorder in stomatology departments (5). Periodontitis is generally treated using systemic antibiotics; however long-term use is common and more severe disorders, such as the development of resistance, are inevitable (6). Therefore, the identification of a novel therapeutic target for the treatment of periodontitis is required.
The abnormal increased proliferation of periodontal ligament stem cells (PDLSCs) has been demonstrated to be associated with the pathogenesis of periodontitis (7,8). It has been reported that fibroblast growth factor 2 (FGF2) promotes the proliferation and differentiation of human PDLSCs (8). This suggests that FGF2 may be involved in the pathogenesis of periodontitis. In addition, FGF2 protein expression and secretion has been revealed to be positively regulated by long non-coding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) (9), which is a well-characterized lncRNA in cancer biology (10,11). In the current study, the involvement of MALAT1 and FGF2 in periodontitis was assessed and the results demonstrated that MALAT1 promoted the expression of FGF2 and the proliferation of PDLSCs, which serve a role in periodontitis.
Materials and methods
Patients
All patients included in the current study were diagnosed and treated at The Stomatological Hospital of Southern Medical University (Guangzhou, China) from January 2017 to January 2018. All patients were treated with antibiotics in combination with pocket reduction surgery. The diagnostic criteria for periodontitis and the exclusion and inclusion criteria for the present study were essentially the same as described by Zheng et al (7). The only difference between the studies is that fish patients in the study by Zheng et al did not consume a diet rich in fish. Disease-free third molars and premolars were collected during orthodontic treatment from 12 patients (8 male; 4 female), and diseased teeth were collected during tooth extraction from 12 patients (7 males; 5 females) with generalized chronic periodontitis. All patients included in the current study were 30–40 years of age. The present study was approved by the Ethics Committee of The Stomatological Hospital of Southern Medical University. All participants were informed of the experimental protocol and signed informed consent.
Cell culture and transfection
PDL tissues were obtained from the central root of healthy and periodontitis-affected teeth. PDL tissues were cut into pieces (1 mm3) and cultured in a six-well plate filled with α-MEM (Sigma-Aldrich; Merck KGaA) containing 100 µM/l ascorbic acid, 0.292 mg/ml glutamine, 10% FBS (Sigma-Aldrich; Merck KGaA) and 100 U/ml penicillin/streptomycin. Cells were subsequently cultured in an incubator at 37°C and 5% CO2.
To overexpress MALAT1, full-length MALAT1 cDNA was digested using NheI restriction enzymes and amplified using PCR and inserted into a NheI-linearized pEGFPC3 vector (Clontech Laboratories, Inc.) to generate a MALAT1 expression vector. Primer sequences were as follows: Forward, 5′-CGCAGCCTGCAGCCCGAGAC-3′ and reverse, 5′-CCCAATCAAGATTTTTTTATTC-3′. PCRs were carried out using Pfu DNA Polymerase kit (Promega Corporation) with the following thermocycling conditions: 95°C for 5 min, followed by 35 cycles of 95°C for 10 sec, 55°C for 10 sec and 72°C for 90 sec. To silence MALAT1 expression, MALAT1 small interfering RNA (siRNA, 5′-CCACUUACUGCUUUAAGUUGG-3′) or negative control siRNA (5′-UUCUCCGAACGUGUCACGUdTdT-3′) were designed and synthesized by Sangon Biotech Co., Ltd. A total of 10 nM vectors or 50 nM siRNAs were transfected into 4×105 cells using Lipofectamine 2000® reagent (cat. no. 11668-019; Invitrogen; Thermo Fisher Scientific, Inc.). Control cells were untransfected and negative control cells were transfected with empty vectors or negative control siRNA. Subsequent experiments were performed at 24 h after transfection.
RNA isolation and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from PDLSCs using TRIzol reagent (Thermo Fisher Scientific, Inc.). cDNA was synthesized using a SuperScript III Reverse Transcriptase kit (Thermo Fisher Scientific, Inc.) according to the following thermocycling conditions: 25°C for 5 min, 51°C for 20 min and 75°C for 15 min. SYBR® Green Real-Time PCR Master Mixes (Thermo Fisher Scientific, Inc.) was used to prepare all PCR reactions. Reaction conditions were as follows: 95°C for 50 sec, followed by 40 cycles of 95°C for 15 sec and 58°C for 20 sec. The following primers were used: MALAT1 forward, 5′-CAGTGGGGAACTCTGACTCG-3′, and reverse, 5′-GTGCCTGGTGCTCTCTTACC-3′; FGF2 forward, 5′-GCACUGAAACGAACUGGGCAGUAU-3′, and reverse, 5′-UAUACUGCCCAGUUCGUUUCAGUGC-3′; β-actin forward, 5′-GACCTCTATGCCAACACAGT-3′, and reverse, 5′-AGTACTTGCGCTCAGGAGGA-3′. This experiment was performed in triplicate and MALAT1 expression was normalized to the β-actin endogenous control using the 2−∆∆Cq method (12).
Cell proliferation assay
Following confirmation of MALAT1 overexpression in PDLSCs compared with the controls, PDLSCs were subsequently collected and single cell suspensions were prepared with a final cell density of 3×104 cells/ml. Each well of a 96-well plate was filled with 3×103 cells in 0.1 ml cell suspension. Cells were cultured in an incubator at 37°C and 5% CO2 for 24, 48, 72 and 96 h, before 10 µl Cell Counting kit-8 (CCK-8) solution was added at the end of each time point. Cells were cultured at 37°C for an additional 4 h and a Fisherbrand™ accuSkan™ GO UV/Vis Microplate Spectrophotometer (Thermo Fisher Scientific, Inc.) was used to measure optical density (OD) values at 450 nm. Cell proliferation was normalized to the sample with the highest OD value at 96 h, which was set to 100%.
Western blot analysis
Subsequent to transfection and RT-qPCR analysis, PDLSCs were collected and mixed with RIPA buffer (Thermo Fisher Scientific, Inc.) to extract total protein, and protein concentrations were measured using a BCA kit (Thermo Fisher Scientific, Inc.). Protein samples (30 µg/lane) were denatured and separated by 10% SDS-PAGE. Following gel transfer, PVDF membranes were then blocked with 5% skimmed milk at room temperature for 2 h. Samples were then incubated with rabbit anti-human primary antibodies against FGF2 (1:1,300; cat. no. ab8880; Abcam) and GAPDH (1:1,300; cat. no. ab8245; Abcam) at 4°C overnight. Following that, membranes were further incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (1:1,300; cat. no. ab6721; Abcam) at 24°C for 2 h. Signal development was performed using ECL™ Detection Reagents GE Healthcare (Sigma-Aldrich; Merck KGaA). Signals were detected using a MYECL™ Imager (Thermo Fisher Scientific, Inc.). ImageJ software (version 1.34; National Institutes of Health) was used to normalize FGF2 expression to the GAPDH endogenous control.
Statistical analysis
GraphPad Prism 6 software (GraphPad Software, Inc.) was used for statistical analysis. Data are expressed as the mean ± standard deviation. A Student's t-test was used to perform statistical comparisons between two groups, and one-way ANOVA followed by a Fisher's least significant difference test was used to perform comparisons among multiple groups. Pearson's correlation coefficient was used for correlational analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
MALAT1 and FGF2 mRNA are upregulated in PDLSCs derived from periodontitis-affected teeth
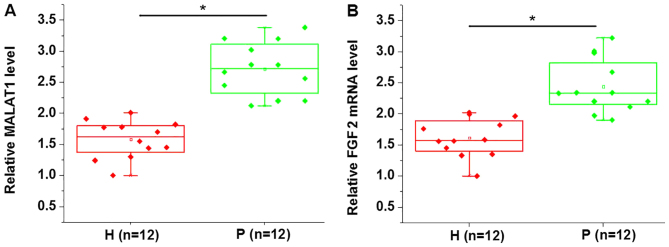
MALAT1 and FGF2 mRNA expression in PDLSCs derived from periodontitis-affected and healthy teeth was detected using RT-qPCR analysis. As presented in Fig. 1, the expression of MALAT1 (Fig. 1A) and FGF2 mRNA (Fig. 1B) was significantly upregulated in PDLSCs derived from periodontitis-affected teeth when compared with PDLSCs derived from healthy teeth.
Figure 1.
MALAT1 and FGF2 mRNA expression in PDLSCs derived from periodontitis-affected and healthy teeth. The expression of (A) MALAT1 and (B) FGF2 mRNA in PDLSCs derived from periodontitis-affected and healthy teeth. *P<0.05, as indicated. MALAT1, metastasis-associated lung adenocarcinoma transcript 1; FGF2, fibroblast growth factor 2; PDLSC, human periodontal ligament stem cells; H, healthy teeth; P, periodontitis-affected teeth.
MALAT1 and FGF2 mRNA expression are positively correlated in PDLSCs derived from healthy and periodontitis-affected patients
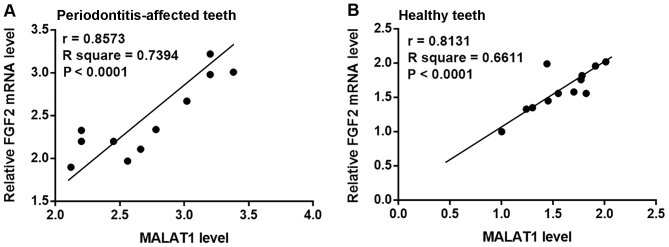
Pearson's correlation coefficient analysis of MALAT1 and FGF2 mRNA expression in PDLSCs derived from periodontitis-affected and healthy teeth was performed. As presented in Fig. 2, a significant positive correlation between the expression of MALAT1 and FGF2 mRNA in PDLSCs derived from periodontitis-affected teeth (Fig. 2A) and healthy teeth (Fig. 2B) was observed.
Figure 2.
Correlation between MALAT1 and FGF2 mRNA expression in periodontitis-affected and healthy PDLSCs. Pearson correlation coefficient analysis of MALAT1 and FGF2 mRNA expression in PDLSCs derived from (A) periodontitis-affected and (B) healthy teeth. MALAT1, metastasis-associated lung adenocarcinoma transcript 1; FGF2, fibroblast growth factor 2; PDLSC, human periodontal ligament stem cells.
PDLSCs derived from periodontitis-affected teeth reveal a higher cell proliferation rate than PDLSCs derived from healthy teeth
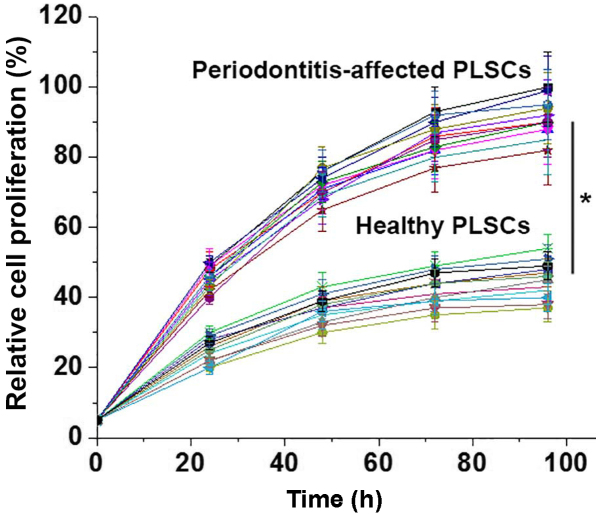
A CCK-8 kit was used to detect the proliferation of PDLSCs derived from periodontitis-affected teeth and PDLSCs derived from healthy teeth. As presented in Fig. 3, the proliferation rates of the 12 PDLSCs samples derived from periodontitis-affected teeth were significantly higher than those of the 12 cases of PDLSCs derived from healthy teeth.
Figure 3.
Relative proliferation of human periodontal ligament stem cells derived from periodontitis-affected and healthy teeth. *P<0.05, as indicated.
MALAT1 promotes the proliferation of PDLSCs derived from periodontitis-affected and healthy teeth
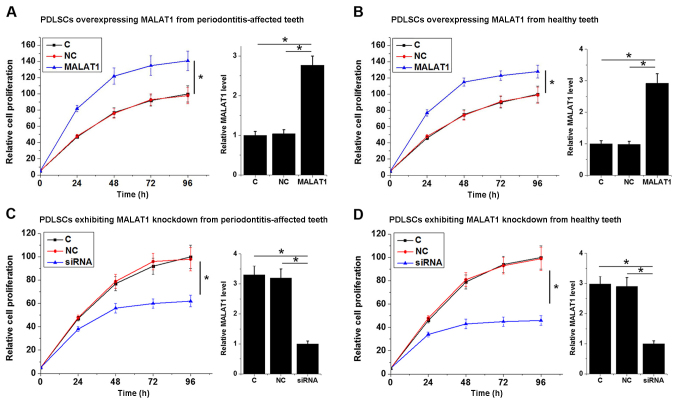
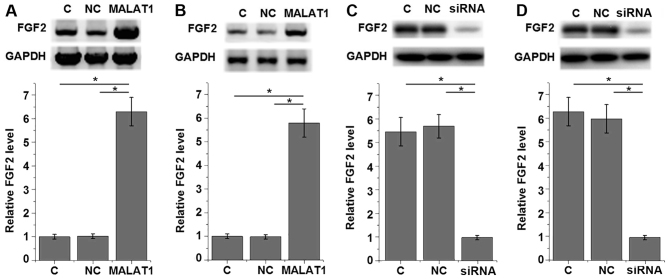
To investigate the involvement of MALAT1 in the proliferation of PDLSCs, a MALAT1 expression vector was transfected into each of the 12 PDLSC samples derived from periodontitis-affected teeth and the PDLSCs derived from 12 healthy teeth samples. The results shown in Fig. 4 are representative data. Cell proliferation was detected using a CCK-8 assay. As presented in Fig. 4, MALAT1 overexpression significantly promoted the proliferation of PDLSCs derived from periodontitis-affected teeth (P<0.05; Fig. 4A) and healthy teeth (P<0.05; Fig. 4B). By contrast, MALAT1 knockdown significantly inhibited the proliferation of PDLSCs derived from periodontitis-affected teeth (P<0.05; Fig. 4C) and healthy teeth (P<0.05; Fig. 4D). Transfections were confirmed by qPCR and data were presented on the right part of each section. The data revealed that the expression level of MALAT1 was significantly upregulated after the transfection of expression vectors and was significantly downregulated after the transfection of siRNAs (P<0.05; Fig. 4).
Figure 4.
Relative cell proliferation of PDLSCs exhibiting MALAT1 overexpression and knockdown. Relative cell proliferation and MALAT1 mRNA levels of PDLSCs overexpressing MALAT1 from (A) periodontitis-affected and (B) healthy teeth. Relative cell proliferation and MALAT1 mRNA levels of PDLSCs exhibiting MALAT1 knockdown from (C) periodontitis-affected and (D) healthy teeth. Transfections were confirmed by qPCR and data were presented on the right part of each section. *P<0.05, as indicated. PDLSC, human periodontal ligament stem cells; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; C, control cells; NC, negative control cells; siRNA, small interfering RNA.
MALAT1 upregulates FGF2 protein expression in PDLSCs derived from healthy individuals and patients with periodontitis
To further investigate the interaction between MALAT1 and FGF2 in PDLSCs, a MALAT1 expression vector was transfected into each of the 12 PDLSCs samples derived from periodontitis-affected teeth and the PDLSCs derived from 12 healthy teeth samples. The results shown in Fig. 5 are representative data. FGF2 expression was detected using western blot analysis. As presented in Fig. 5, MALAT1 overexpression significantly upregulated FGF2 expression in PDLSCs derived from periodontitis-affected teeth (P<0.05; Fig. 5A) and healthy teeth (P<0.05; Fig. 5B) when compared with the control and negative control group. By contrast, MALAT1 knockdown significantly downregulated FGF2 expression in PDLSCs derived from periodontitis-affected teeth (P<0.05; Fig. 5C) and healthy teeth (P<0.05; Fig. 5D) when compared with their respective control and negative control groups.
Figure 5.
Effect of MALAT1 overexpression on FGF2 protein expression in PDLSCs. Relative FGF2 expression in PDLSCs overexpressing MALAT1 from (A) periodontitis-affected and (B) healthy teeth. Relative FGF2 expression in PDLSCs exhibiting MALAT1 knockdown from (C) periodontitis-affected and (D) healthy teeth. *P<0.05, as indicated. MALAT1, metastasis-associated lung adenocarcinoma transcript 1; FGF2, fibroblast growth factor 2; PDLSC, human periodontal ligament stem cells; C, control cells; NC, negative control cells; siRNA, small interfering RNA.
Discussion
The present study demonstrated that lncRNAs serve pivotal roles in cancer biology and lncRNA MALAT1 was revealed to be associated with the pathogenesis of periodontitis. The results also revealed that lncRNA MALAT1 may promote the development of periodontitis by upregulating the expression of FGF2 and promoting the proliferation of PDLSCs.
The onset and development of periodontitis is accompanied by changes in the expression of a large variety of lncRNAs, indicating their association with the pathogenesis of this disease (13). However, studies assessing the expression and functionality of specific lncRNAs in periodontitis are required. In a recent study, Wang et al (14) demonstrated that lncRNA-POIR functions as a positive regulator of osteogenic differentiation, which is impaired in periodontitis, indicating a protective role of lncRNA-POIR in periodontitis (14). In an additional study, the ANRIL lncRNA was demonstrated to participate in the pathogenesis of periodontitis via interacting with multiple signaling pathways (15). MALAT1 is a well-characterized oncogenic lncRNA, which exhibits upregulated expression in a variety of human cancer types (10,11). In the present study, MALAT1 expression was revealed to be significantly upregulated in PDLSCs derived from periodontitis-affected teeth, suggesting an involvement of MALAT1 in periodontitis.
FGF2 expression stimulates periodontal regeneration, indicating its potential clinical importance as a therapeutic target in novel periodontitis treatments (16,17). The current study observed a significant upregulation of FGF2 mRNA expression in PDLSCs derived from periodontitis-affected teeth when compared with PDLSCs derived from healthy teeth. This result indicates the potential involvement of FGF2 in the pathogenesis of periodontitis. FGF2 interacts with lncRNAs through certain mediators (such as miRNAs) to achieve its biological functions, such as the regulation of vascular endothelial cell apoptosis (18). In a previous study, the MALAT1 lncRNA was demonstrated to be positively correlated with FGF2 protein expression and secretion in thyroid cancer (9). Similarly, MALAT1 and FGF2 mRNA expression was revealed to be positively correlated in PDLSCs derived from both periodontitis-affected and healthy teeth in the present study. In addition, MALAT1 overexpression was associated with upregulation of FGF2 protein in PDLSCs derived from periodontitis-affected and healthy teeth. These results suggest that MALAT1 is a positive regulator of FGF2 in periodontitis, and the regulatory effects of MALAT1 on FGF2 are unlikely to be mediated by periodontitis-specific factors.
PDLSCs possess immunomodulatory properties through the secretion of a variety of cytokines, including interleukin (IL)-6, IL-8 and monocyte chemotactic protein 1 (19). It has been previously reported that abnormally accelerated PDLSC proliferation is associated with the pathogenesis of periodontitis (8). In the current study, a significantly higher proliferation rate was observed in all 12 PDLSC samples derived from periodontitis-affected teeth when compared with the 12 PDLSC samples derived from healthy teeth. MALAT1 was also revealed to be a likely activator of PDLSCs based on the observation that MALAT1 overexpression led to the significantly increased proliferation of PDLSCs derived from both periodontitis-affected and healthy teeth. However, the molecular mechanisms underlying this effect is currently unknown and future studies should aim to elucidate this mechanism. In addition, the present study is limited by a small sample size. Therefore, future studies that include a larger sample size are required to validate the results of the current study. In conclusion, MALAT1 and FGF2 are upregulated in periodontitis. In addition, MALAT1 may serve a role in periodontitis by promoting FGF2 expression and the proliferation of PDLSCs.
Acknowledgements
Not applicable.
Funding
The current study received financial support from Natural Science Foundation of Tibet Autonomous Region (grant no. XZ2017ZR-ZYZ37) and The Scientific Research Project of Southern Medical University Stomatological Hospital (grant no. PY2018014)
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
PC and MR designed the experiments. PC, YH and YW performed the experiments. SL and HC collected and analyzed the data. MR drafted manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The Stomatological Hospital of Southern Medical University.
Patient consent for publication
Patients provide consent for the possible publication of this paper.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hajishengallis G. Periodontitis: From microbial immune subversion to systemic. Inflammation. Nat Rev Immunol. 2015;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lundberg K, Wegner N, Yucel-Lindberg T, Venables PJ. Periodontitis in RA-the citrullinated enolase connection. Nat Rev Rheumatol. 2010;6:727–730. doi: 10.1038/nrrheum.2010.139. [DOI] [PubMed] [Google Scholar]
- 3.Kebschull M, Demmer RT, Papapanou PN. ‘Gum bug, leave my heart alone!’-epidemiologic and mechanistic evidence linking periodontal infections and atherosclerosis. J Dent Res. 2010;89:879–902. doi: 10.1177/0022034510375281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madianos PN, Bobetsis YA, Offenbacher S. Adverse pregnancy outcomes (APOs) and periodontal disease: pathogenic mechanisms. J Periodontol. 2013;84(Suppl):S170–S180. doi: 10.1902/jop.2013.1340015. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Q, Li Z, Wang C, Shen T, Yang Y, Chotivichien S, Wang L. Prevalence and predictors for periodontitis among adults in China, 2010. Glob Health Action. 2014;7:24503. doi: 10.3402/gha.v7.24503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cionca N. Use and misuse of systemic antibiotics in periodontitis treatment. Oral Health Prev Dent. 2017;15:305–306. doi: 10.3290/j.ohpd.a38864. [DOI] [PubMed] [Google Scholar]
- 7.Zheng W, Wang S, Wang J, Jin F. Periodontitis promotes the proliferation and suppresses the differentiation potential of human periodontal ligament stem cells. Int J Mol Med. 2015;36:915–922. doi: 10.3892/ijmm.2015.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JH, Um S, Jang JH, Seo BM. Effects of VEGF and FGF-2 on proliferation and differentiation of human periodontal ligament stem cells. Cell Tissue Res. 2012;348:475–484. doi: 10.1007/s00441-012-1392-x. [DOI] [PubMed] [Google Scholar]
- 9.Huang JK, Ma L, Song WH, Lu BY, Huang YB, Dong HM, Ma XK, Zhu ZZ, Zhou R. LncRNA-MALAT1 promotes angiogenesis of thyroid cancer by modulating tumor-associated macrophage FGF2 protein secretion. J Cell Biochem. 2017;118:4821–4830. doi: 10.1002/jcb.26153. [DOI] [PubMed] [Google Scholar]
- 10.Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014;15:2971–2977. doi: 10.7314/APJCP.2014.15.7.2971. [DOI] [PubMed] [Google Scholar]
- 11.Tian X, Xu G. Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: Systematic review and meta-analysis. BMJ Open. 2015;5:e008653. doi: 10.1136/bmjopen-2015-008653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Zou Y, Li C, Shu F, Tian Z, Xu W, Xu H, Tian H, Shi R, Mao X. lncRNA expression signatures in periodontitis revealed by microarray: The potential role of lncRNAs in periodontitis pathogenesis. J Cell Biochem. 2015;116:640–647. doi: 10.1002/jcb.25015. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Wu F, Song Y, Li X, Wu Q, Duan Y, Jin Z. Long noncoding RNA related to periodontitis interacts with miR-182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients. Cell Tissue Res. 2016;7:e2327. doi: 10.1038/cddis.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bochenek G, Häsler R, El Mokhtari NE, König IR, Loos BG, Jepsen S, Rosenstiel P, Schreiber S, Schaefer AS. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum Mol Genet. 2013;22:4516–4527. doi: 10.1093/hmg/ddt299. [DOI] [PubMed] [Google Scholar]
- 16.Kitamura M, Akamatsu M, Machigashira M, Hara Y, Sakagami R, Hirofuji T, Hamachi T, Maeda K, Yokota M, Kido J, et al. FGF-2 stimulates periodontal regeneration: results of a multi-center randomized clinical trial. J Dent Res. 2011;90:35–40. doi: 10.1177/0022034510384616. [DOI] [PubMed] [Google Scholar]
- 17.Murakami S. Periodontal tissue regeneration by signaling molecule(s): What role does basic fibroblast growth factor (FGF-2) have in periodontal therapy? Periodontol. 2000;56:188–208. doi: 10.1111/j.1600-0757.2010.00365.x. [DOI] [PubMed] [Google Scholar]
- 18.Lu W, Huang SY, Su L, Zhao BX, Miao JY. Long noncoding RNA LOC100129973 suppresses apoptosis by targeting miR-4707-5p and miR-4767 in vascular endothelial cells. Sci Rep. 2016;6:21620. doi: 10.1038/srep21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrukhov O, Hong JS, Andrukhova O, Blufstein A, Moritz A, Rausch-Fan X. Response of human periodontal ligament stem cells to IFN-γ and TLR-agonists. Sci Rep. 2017;7:12856. doi: 10.1038/s41598-017-12480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.