Abstract
Tumor protein p53-inducible nuclear protein 1 (TP53INP1) is a tumor suppressor associated with malignant tumor metastasis. In addition, it has been reported that hsa-microRNA (miR)-3934 serves key roles in various types of lung cancer, including small-cell lung carcinomas (SCLC) and non-SCLC (NSCLC). Therefore, the present study aimed to determine the effects of miR-3934-5p on cell proliferation and apoptosis, and on sensitivity to cisplatin (DDP). Reverse transcription-quantitative polymerase chain reaction analysis and western blotting were conducted for the analysis of mRNA and protein expression, respectively. Furthermore, the target of miR-3934-5p was investigated using a luciferase reporter assay and apoptosis was analyzed by flow cytometry. The results demonstrated that miR-3934-5p was upregulated in NSCLC tissues and A549 cells. Increases in the half-maximal inhibitory concentration (IC50) and the expression of miR-3934-5p were observed in the A549/DDP group. miR-3934-5p mimic promoted the expression of miR-3934-5p and the IC50 of the A549 cells. miR-3934-5p inhibitor downregulated miR-3934-5p and reduced the IC50 of A549/DDP cells. miR-3934-5p was revealed to target the 3′-untranslated region of TP53INP1. The downregulation of miR-3934-5p significantly suppressed the proliferation and promoted the apoptosis of A549/DDP cells, which were reversed by transfection with TP53INP1 small interfering (si)RNA. The protein and mRNA expression levels of TP53INP1, B-cell lymphoma 2 (Bcl-2)-associated-X and p21 were significantly increased, whereas those of Bcl-2 were significantly decreased in the miR-3934-5p inhibitor group, which was significantly reduced by TP53INP1 siRNA transfection. miR-3934-5p, as a tumor suppressor in NSCLC, may promote the sensitivity of cells to DDP by targeting TP53INP1, associated with the suppression of cell proliferation and promotion of apoptosis.
Keywords: microRNA-3934, cisplatin sensitivity, tumor protein p53-inducible nuclear protein 1, non-small cell lung carcinoma
Introduction
Lung cancer is the most common type of cancer worldwide with a high incidence (1,800,000 new cases estimated in 2012) and mortality rate (1,600,000 cases annually) (1). In addition, lung cancer is the leading cause of cancer-associated mortality (2); ~80% of patients with lung cancer are diagnosed with non-small cell lung cancer (NSCLC) (3). The 1-year survival rate of metastatic NSCLC is only 20% (4). Genetic mutations of c-ros oncogene 1, receptor tyrosine kinase, v-Raf murine sarcoma viral oncogene homolog B1, human epidermal growth factor receptor 2 and NNG-HOS transforming have been detected in NSCLC (5). Cisplatin (DDP) is one of the most effective anticancer agents, and is widely used in the treatment of solid tumors, including those in lung, brain and ovarian cancer (6). The mechanisms of DDP or other platinum-based compounds for the treatment of cancer involve inducing the death of cancer cells via damaging DNA and inhibiting DNA synthesis and mitosis, and promoting apoptosis, production of reactive oxygen species, lipid peroxidation, p53 signaling activation and cell cycle arrest. Therefore, the intrinsic and extrinsic pathways of apoptosis are activated (6). However, treatment with DDP leads to adverse side effects, therefore, novel therapeutic strategies are urgently required.
MicroRNAs (miRNAs/miRs) are small, non-coding RNAs (~22 nucleotides) that regulate gene expression at the post-transcriptional level by suppressing the translation of corresponding mRNAs by to binding its 3′-untranslated region (3′UTR) (7). Therefore, miRNAs serve vital roles in the occurrence and progression of cancer, associated with the migration, invasive, proliferative and apoptotic potentials of cancer cells (8,9). It has been demonstrated that miRNAs are potential biomarkers and therapeutic targets in certain types of cancer (7). It was reported that miRNA (miR)-200c targets zinc finger E-box binding homeobox 1 protein or inhibits the phosphoinositide 3-kinase/protein kinase B signaling pathway to enhance gefitinib sensitivity; miR-143, miR-134 and miR-218-5p can directly bind the 3′UTR of epidermal growth factor to suppress the migration and proliferation of tumor cells in NSCLC (10). Evidence also shows that miR-3934 is upregulated in colorectal cancer cells (11). Furthermore, it was revealed that the expression of hsa-miR-3934 is correlated with survival rate and disease development in patients with NSCLC (12). However, the roles of miR-3934 and its target gene in NSCLC remain unclear.
Tumor protein 53-induced nuclear protein 1 (TP53INP1) is a known stress-induced protein located on chromosome 8q22 (13). TP53INP1, a major regulator of p53 in response to oxidative stress, regulates the activity of p73by binding to p53-responsive elements (14). Additionally, TP53INP1 has a tumor-suppressive effect by regulating metabolic homeostasis (15). Previous studies have suggested that TP53INP1 acts as a tumor suppressor in breast, prostate and colorectal cancer (14,16,17). Furthermore, TP53INP1 was identified as a critical target of miR-125b associated with the metastasis of NSCLC (18). It has been reported that TP53INP1 is associated with the sensitivity of cancer cells to DDP in hepatocellular carcinoma and lung cancer (19,20); however, the exact roles of TP53INP1 require further investigation.
In the present study, TP53INP1 was identified as a target of miR-3934-5p via TargetScan analysis. DDP-based chemotherapy regimens are standard therapeutic strategies in advanced stage NSCLC (20); however, the incidence of DDP resistance is ≤63% of NSCLC cases. Resistance remains an obstacle in chemotherapy and seriously affects the survival rate of patients with NSCLC (21). Therefore, the present study aimed to investigate whether miR-3934-5p targets TP53INP1 to affect cell sensitivity to DDP.
Materials and methods
Patients and tissues
A total of 30 pairs of tumor tissues and paracarcinoma tissues were obtained from patients with NSCLC, and fixed in 10% formalin. The present study was approved by the Ethics Committee of the People's Hospital of Yucheng (Yucheng, China). Informed consent was obtained from all patients prior to the experiments.
Cell lines
A549 (CRM-CCL-185) and BEAS-2B (CRL-9609) cells were purchased from the American Type Culture Collection (Manassas, VA, USA) and A549/DDP (GF452) cells were obtained from Gefan Biotechnology Co., Ltd. (Shanghai, China). The cells were maintained in RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.) at 37°C in an atmosphere of 5% CO2.
Plasmids and transfection
The A549 and A549/DDP cells were seeded into 24-well plates, and incubated in RPMI-1640 medium at 37°C with 5% CO2 overnight until 60–70% confluence was attained. Subsequently, 100 µM miR-3934 5p mimics and inhibitor, in addition to 1 µg TP53INP1-small interfering (si)RNA and TP53INP1 NC siRNA were obtained from Qiagen GmbH (Hilden, Germany), and were transfected into A549 and A549/DDP cells (2×105 cells) using 1 µl Lipo6000™ transfection reagent (Beyotime Institute of Biotechnology, Shanghai, China). Transfection was performed at 37°C with 5% CO2 for 6 h. Untreated cells were used as the control group. The transfected cells were incubated for a further 48 h. The cells were divided into four groups: Control (untreated), DDP + miR-negative control (NC) inhibitor group, DDP + miR-3934-5p inhibitor group, and DDP + TP53INP-siRNA + miR-3934-5p inhibitor group. The sequence of TP53INP1-1 siRNA and its NC were as followed: TP53INP1 siRNA, 5′-CACUAAUAACCCGGUUGACU-3′; TP53INP1 NC siRNA, 5′-UUCUCCGAACGUGUCACGUTT-3′. The sequences of miR-3934-5p mimics and inhibitor and the NC were as followed: miR-3934-5p mimics, 5′-CACAGCCCUUCCUGUCCCCAGUUUUCAGGUGUGGAAACUGAGGCAGGAGGCAGUGAAGUAACUUGCUCAGGUUGCACAGCUGGGAAGUGGAGCAGGGAUUUGAAUCC-3′ and miR-NC mimics: 5′-UUCUCCGAACGUGUCACGUTT-3′; miR-3934-5p inhibitor: 5′-CUACCCCAUCAGACCCAAGGG-3′ and miR-NC inhibitor: 5′-UUGUACUACACAAAAGUACUG-3′.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
RT-qPCR analysis was used for the analysis of miR-3934-5p and mRNA expression. RNA was extracted using TRIzol® reagent (Thermo Fisher Scientific, Inc.) from tissues and cells at 70% confluence. Subsequently, a NanoDrop 2000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.) was used to determine RNA concentration. In addition, DNA polymerase (APExBIO, Houston, TX, USA) and M-MLV reverse transcriptase (Promega Corporation, Madison, WI, USA) were used for cDNA synthesis and PCR. The thermocycling conditions were as follows: 98°C for 15 min, and 96°C for 5 min (initiation), 92°C for 30 sec (denaturation), 65°C for 30 sec (annealing) and 62°C for 1 min (extension) for 40 cycles. The expression of miR-3934-5p was normalized to that of U6: Forward, 5′-GCCAGCTCCTACATCTCAGC-3′ and reverse, 5′-AGCCTGACTTGCTAGTGGATTAT-3′; U6, forward: 5′-CTCGCTTCGGCAGCACA-3′, reverse: 5′-AACGCTTCACGAATTTGCGT-3′. GAPDH was used as the internal reference for the normalization of mRNA expression. The sequence of the primers used were listed as followed: TP53INP1, forward, 5′-TGAACACATTTGCCTTGTGAA-3′, and reverse, 5′-GGCAAAGTGCTGTGCTGTT-3′; Bax, forward: 5′-CACCAGCTCTGAACAGATCATGA-3′, and reverse: 5′-TCAGCCCATCTTCTTCCAGATGT-3′; Bcl-2, forward: 5′-CACCCCTGGCATCTTCTCCTT-3′, and reverse: 5′-AGCGTCTTCAGAGACAGCCAG-3′; p21, forward: 5′-CCGAAGTCAGTTCCTTGTGG-3′, and reverse, 5′-GCCATTAGCGCATCACAG-3′; GAPDH, forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′, and reverse: 5′-GGCTGTTGTCATACTTCTCATGG-3′. In addition, the abundance of RNAs were calculated using the 2−ΔΔCq method (22).
Western blotting
The protein expression levels were evaluated by western blotting; the proteins were isolated from tissues and cells using radioimmunoprecipitation assay lysis buffer (Santa Cruz Biotechnology, Inc., Hercules, CA, USA) and protein concentration was determined via a Bicinchoninic Acid kit (Thermo Fisher Scientific, Inc.). Subsequently, 15% SDS-PAGE was utilized for protein separation (2 µl protein per lane). The proteins were then transferred onto a PVDF membrane (Membrane Solutions, Kent, WA, USA), which was blocked with 5% skimmed milk for 2 h. The membrane was then incubated with primary antibodies, including anti-B-cell lymphoma 2 (Bcl-2; cat. no. ab32124, 1:1,000, Abcam, Cambridge, UK), anti-Bcl-2-associated-X (Bax; cat. no. ab32503, 1:5,000, Abcam), anti-p21 (cat. no. ab36669, 1:2,000, Abcam), anti-TP53INP1 (cat. no. 17872-1-AP, 1:2,000, ProteinTech Group, Inc., Chicago, IL, USA) and anti-β-actin (cat. no. ab8226, 1:3,000, Abcam) at room temperature for 1 h. Subsequently, incubation with a horseradish peroxidase-conjugated secondary antibody (cat. no. ab6721, 1:2,000, Abcam) was conducted at room temperature for 45 min. An enhanced chemiluminescence kit (cat. no. 35050, Thermo Fisher Scientific, Inc.) was used for visualization, and protein expression was analyzed using ImageJ software 1.46 (National Institutes of Health, Bethesda, MD, USA).
MTT assay
At 48 h post-plasmid transfection, an MTT assay was performed for the assessment of proliferation. The formazan crystals were dissolved by adding dimethyl sulfoxide. The optical density was then measured at 595 nm using a microplate reader.
Flow cytometry
Apoptosis was assessed via flow cytometric analysis. The A549 cells were transfected with mimics or inhibitors and treated with DDP, and were then seeded into 12-well plates (2×104 cells/well) and incubated at 37°C with 5% CO2. The cells then were stained with propidium iodide (10 µg/ml, Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and Annexin V-fluorescein isothiocyanate (50 µg/ml, BD Biosciences, Franklin Lakes, NJ, USA) for 15 min at room temperature in the dark. Subsequently, the cells were analyzed by flow cytometry (FACScan; BD Biosciences).
Luciferase reporter assay
TargetScan (http://www.targetscan.org/vert_71/) was used for target prediction. Analysis revealed that TP53INP1 was a potential target of miR-3934-5p due to sequence complementarity between the 3′UTR of TP53INP1 mRNA and miR-3934-5p. In addition, a luciferase reporter assay was performed to verify the findings of bioinformatics analysis. The A549 cells were plated into 96-well plates (2×103 cells/well) and incubated in RPMI-1640 medium until 60% confluence was attained. Subsequently, the co-transfection of 50 ng miR-3934-5p mimics, TP53INP1-3′UTR-wild-type (WT) or TP53INP1-3′UTR-mutant (MUT) plasmids was performed using Lipo6000™ transfection reagent (0.2 µl). Furthermore, the luciferase activity was normalized to that of Renilla luciferase. A luciferase assay kit (Thermo Fisher Scientific, Inc.) was used to evaluate luciferase activity according to the manufacturer's protocols.
Statistical analysis
Each experiment was performed in triplicate and all data are presented as the mean ± standard deviation. Comparisons between two groups were made using a Student's t-test, and one-way analysis of variance followed by a Newman-Keuls post hoc test was performed for the comparison of multiple groups using SPSS 14.0 (SPPS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-3934-5p in tissues and A549 cells
The expression of miR-3934-5p in tumor tissues and cells was assessed by RT-qPCR analysis. As shown in Fig. 1A, the expression of miR-3934-5p was significantly increased in tumor tissues compared with that in normal tissues. Furthermore, the expression of miR-3934-5p was upregulated in A549 cells compared with that in BEAS-2B cells (Fig. 1B). Therefore, miR-3934-5p may be involved in NSCLC.
Figure 1.
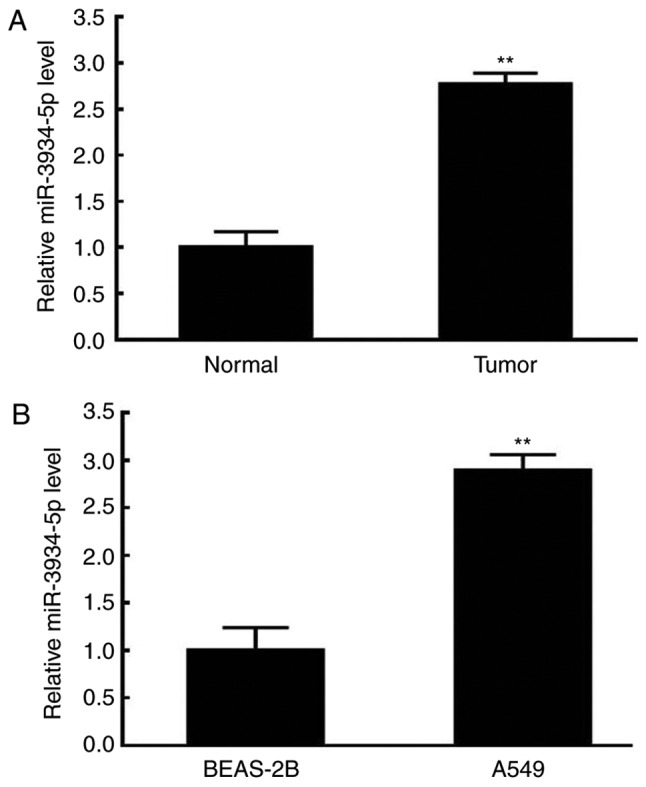
Upregulation of miR-3934-5p in tissues and A549 cells. The expression of miR-3934-5p in tissues and cells were evaluated by reverse transcription-quantitative polymerase chain reaction analysis. (A) Expression of miR-3934-5p in normal (paracarcinoma) and NSCLC tumor tissues. (B) Expression of miR-3934-5p in the BEAS-2B normal lung cell line and A549 NSCLC tumor cell line. **P<0.01 vs. Normal tissues or BEAS-2B cells. miR, microRNA; NSCLC, non-small cell lung carcinoma.
Alterations in the half-maximal inhibitory concentration (IC50) and expression of miR-3934-5p following DDP treatment
The IC50 and expression of miR-3934-5p of A549 cells were determined via an MTT assay and RT-qPCR analysis following DDP treatment. As presented in Fig. 2A, an increase in the IC50 was observed in the A549/DDP group. Additionally, a significant increase in the expression of miR-3934-5p was observed in the A549/DDP cells (Fig. 2B). The increased IC50 of A549/DDP cells suggested reduced sensitivity to DDP, which may be associated with the upregulated expression of miR-3934-5p.
Figure 2.
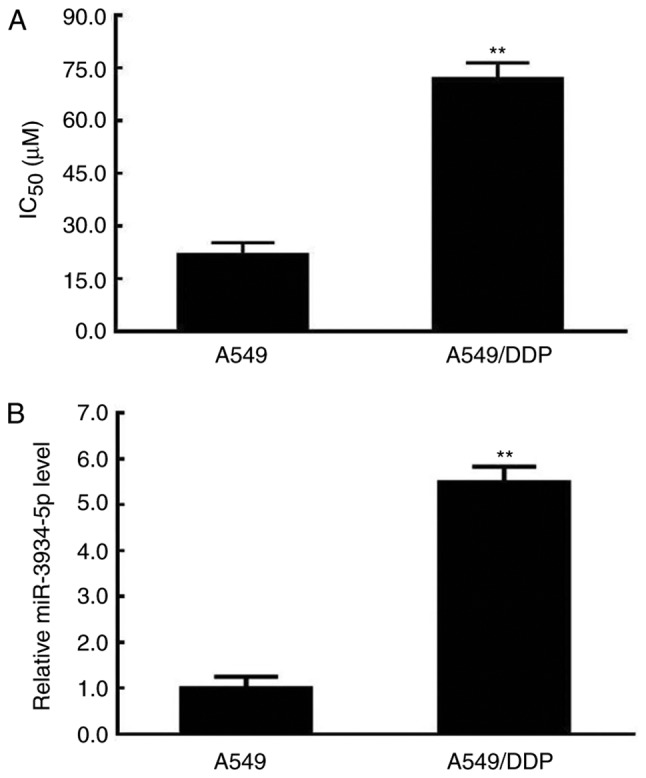
Increased IC50 and expression of miR-3934-5p following DDP treatment. The IC50 was determined by an MTT assay and the expression of miR-3934-5p was assessed by reverse transcription-quantitative polymerase chain reaction analysis. (A) IC50 value of untreated A549 cells and of A549/DDP cells. (B) Expression of miR-3934-5p in A549 cells and A549/DDP cells. A549/DDP represents A549 cells treated with DDP. **P<0.01 vs. A549 cells. miR, microRNA; IC50, half-maximal inhibitory concentration; DDP, cisplatin.
Effects of miR-3934-5p mimics on the IC50 of A549 cells
The miR-3934-5p mimics and the NC plasmids were transfected into A549 cells. The expression of miR-3934-5p and the IC50 values were determined by RT-qPCR and MTT assays, respectively. As presented in Fig. 3A and B, increases in the expression of miR-3934-5p and the IC50 were observed in the miR-3934-5p mimics-transfected A549 cells.
Figure 3.
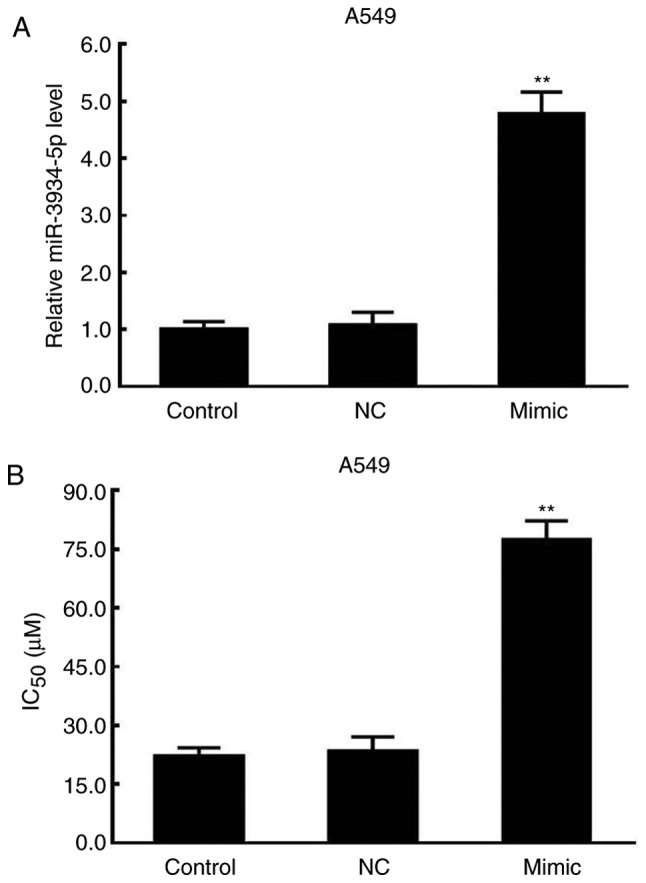
Increased miR-3934-5p and IC50 by miR-3934-5p mimic in A549 cells. A549 cells were transfected with miR-3934-5p mimic and negative control. The IC50 was determined using an MTT assay and the expression of miR-3934-5p was assessed by reverse transcription-quantitative polymerase chain reaction analysis. (A) Expression of miR-3934-5p in A549 cells. (B) IC50 of A549 cells. **P<0.01 vs. NC. miR, microRNA; Control, untreated cells; Mimic, cells transfected with miR-3934-5p mimic; NC, cells treated with miR-negative control mimics; IC50, half-maximal inhibitory concentration.
Effects of miR-3934-5p inhibitor on the IC50 of A549/DDP cells
The miR-3934-5p inhibitor and the NC plasmids were transfected into A549/DDP cells. The results revealed reductions in the expression of miR-3934-5p and the IC50 in the miR-3934-5p inhibitor group (Fig. 4A and B). Therefore, the effects of DDP on cell viability may be associated with the expression of miR-3934-5p.
Figure 4.
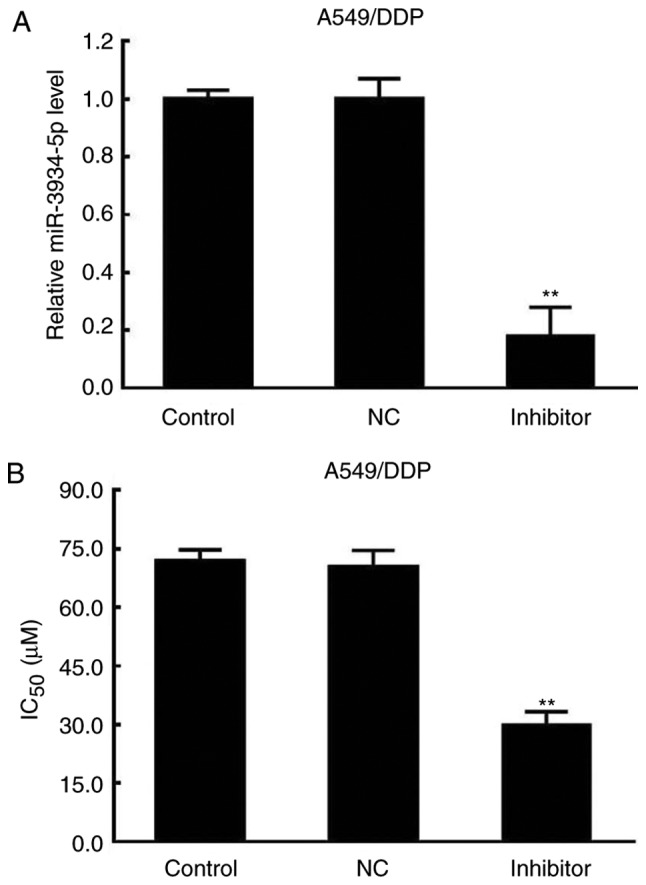
Decreased miR-3934-5p and IC50 by miR-3934-5p inhibitor in A549/DDP cells. A549/DDP cells were transfected with miR-3934-5p inhibitor and NC. The IC50 was determined using an MTT assay and the expression of miR-3934-5p was assessed by reverse transcription- quantitative polymerase chain reaction analysis in A549/DDP cells. (A) Expression of miR-3934-5p in A549/DDP cells. (B) IC50 value of A549/DDP cells. **P<0.01 vs. NC. miR, microRNA; IC50, half-maximal inhibitory concentration; NC, negative control; DDP, cisplatin.
Therefore, miR-3934-5p mimics transfection led to a significant reduction in the sensitivity of A549 cells to DDP, whereas the downregulation of miR-3934-5p significantly increased the sensitivity of A540/DDP cells to DDP.
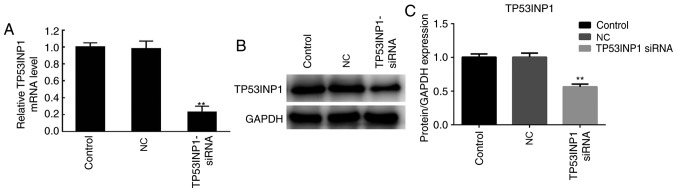
Expression of TP53INP1 following transfection with TP53INP1 siRNA
Following treatment with DDP, the A549 cells were transfected with TP53INP1 siRNA and control plasmids to induce the downregulation of TP53INP1. The expression of TP53INP was then determined by RT-qPCR analysis and western blotting. As shown in Fig. 5A, the results demonstrated that the mRNA expression levels of TP53INP were significantly decreased in the TP53INP1 siRNA group compared with those in the NC group. In addition, the protein expression levels of TP53INP1 were markedly decreased in the TP53INP1 siRNA group compared with those in the NC group (Fig. 5B and C). Therefore, TP53INP1 was downregulated at the protein and mRNA levels following TP53INP1 siRNA transfection.
Figure 5.
TP53INP1 is downregulated following TP53INP1 siRNA transfection. A549/DDP cells were treated TP53INP1 siRNA and control plasmids to establish a TP53INP1 downregulation system. (A) mRNA expression of TP53INP1 was determined by reverse transcription-quantitative polymerase chain reaction analysis. (B) Protein expression of TP53INP1 was determined by western blotting. (C) Quantitative analysis of protein expression. **P<0.01 vs. NC group. TP53INP1, tumor protein p53-inducible nuclear protein 1; siRNA, small interfering RNA; Control, untreated group; NC, cells transfected with siRNA negative control plasmids; TP53INP1 siRNA, cells transfected with TP53INP1 siRNA; DDP, cisplatin.
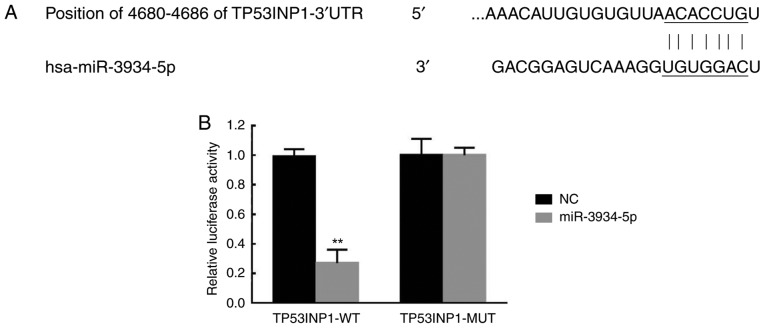
TP53INP1 is a target of miR-3934-5p
The miR-3934-5p mimics and control plasmids were transfected into the TP53INP1-3′UTR-WT-treated and TP53INP1-3′UTR-MUT-treated A549 cells. As presented in Fig. 6A, TP53INP1 was predicted as a target of miR-3934-5p due to sequence complementarity in the 3′UTR of TP53INP1 mRNA according to TargetScan analysis. The luciferase activity was significantly decreased in the TP53INP1-3′UTR-WT+ miR-3934-5p mimics-treated group (Fig. 6B). Therefore, this suggested TP53INP1 was a direct target of miR-3934-5p.
Figure 6.
TP53INP1 is a target of miR-3934-5p. The miR-3934-5p mimics and NC plasmids were transfected into TP53INP1-3′UTR-WT-and TP53INP1-3′UTR-MUT-treated A549 cells. (A) Results of TargetScan. (B) Luciferase activity in the TP53INP1-3′UTR-WT and TP53INP1-3′UTR-MUT groups. **P<0.01 vs. NC group. miR, microRNA; TP53INP1, tumor protein p53-inducible nuclear protein 1; NC, treated with negative control plasmids; 3′UTR, 3′-untranslated region; WT, wild-type; MUT, mutant.
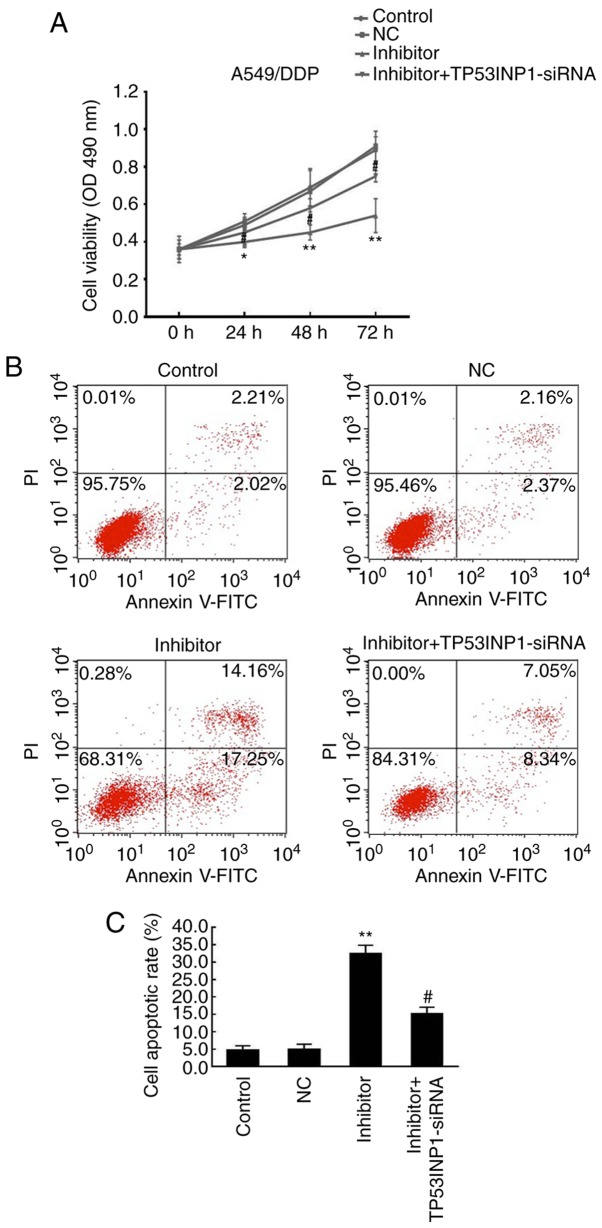
Effects of miR-3934-5p inhibitor on cell proliferation and apoptosis
The rates of cell proliferation and apoptosis of A549/DDP cells were evaluated using an MTT assay and flow cytometry following transfection with miR-3934-5p inhibitor and incubation for 48 h in RPMI-1640 medium under 5% CO2 at 37°C. As presented in Fig. 7A, these results indicated that the downregulation of miR-3934-5p significantly suppressed the proliferation rate of A549/DDP cells compared with that in the control and NC groups, which was reversed by TP53INP1 siRNA. As presented in Fig. 7B and C, cell apoptosis was significantly promoted in the miR-3934-5p inhibitor group (~30%) compared with that in the control (~5%) and NC groups (~5%), which was reversed by TP53INP1 siRNA. Therefore, the downregulation of miR-3934-5p may lead to the suppressed proliferative ability of cells and increase apoptosis by targeting TP53INP1.
Figure 7.
miR-3934-5p inhibitor causes inhibited cell proliferation and promotes cell apoptosis. The proliferation and apoptosis of A549 cells was analyzed using an MTT assay and flow cytometry, respectively. (A) Downregulation of miR-3934-5p significantly suppressed the proliferation rate of A549/DDP cells compared with that in the untreated control group and NC group, which was reversed by TP53INP1 siRNA. (B) Cell apoptosis was determined and (C) quantified. Apoptosis was promoted in the miR-3934-5p inhibitor group compared with that in the control and NC groups, which was reversed by administration of TP53INP1 siRNA. *P<0.05 and **P<0.01 vs. NC group; #P<0.05 vs. miR-3934-5p inhibitor group. miR, microRNA; TP53INP1, tumor protein p53-inducible nuclear protein 1; DDP, cisplatin; NC, treated with negative control plasmids; siRNA, small interfering RNA.
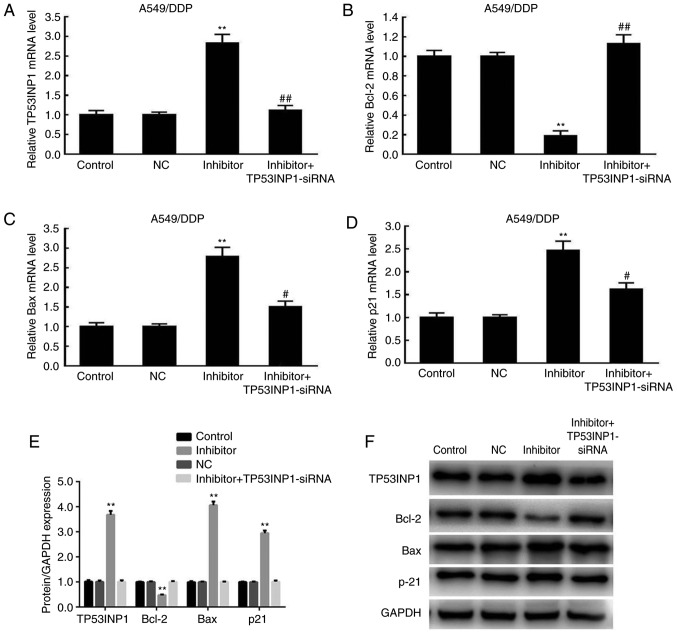
Effects of miR-3934-5p inhibitor on the expression of Bax, p21, TP53INP1 and Bcl-2
The analysis of protein expression revealed that the levels TP53INP1, Bax and p21 were significantly increased in the inhibitor group compared with those in the control and NC groups. The alterations in the mRNA expression levels of TP53INP1, Bax, Bcl-2 and p21 were similar to the changes in protein expression (Fig. 8A-D). The expression levels of the aforementioned proteins were significantly reduced by TP53INP1 siRNA transfection. By contrast, the expression of Bcl-2 was significantly decreased in the inhibitor group compared with levels in the control and NC groups, but was significantly upregulated following TP53INP1 siRNA transfection (Fig. 8E and F). These results indicated that the downregulation of miR-3934-5p resulted in the upregulated protein expression of TP53INP1, Bax and p21, whereas that of Bcl-2 was decreased.
Figure 8.
Expression levels of Bax, p21 and TP53INP1 are upregulated and expression of Bcl-2 is suppressed following miR-3934-5p inhibitor treatment. At 72 h post-miR-3934-5p inhibitor treatment, expression levels of (A) TP53INP1, (B) Bcl-2, (C) Bax and (D) p21 were examined by reverse transcription-quantitative polymerase chain reaction analysis. mRNA expression levels of TP53INP1, Bax and p21 were significantly increased in the inhibitor group compared with levels in the untreated control and NC groups, which were significantly reduced by administration of TP53INP1 siRNA. Bcl-2 was decreased in the inhibitor group compared with that in the control and NC groups, and was significantly upregulated by administration of TP53INP1 siRNA. (E) Quantification of protein levels of TP53INP1, Bax, Bcl-2 and p21 from (F) western blots showed similar changes to those of mRNA. **P<0.01 vs. NC group; #P<0.05 vs. miR-3934-5p inhibitor group; ##P<0.01 vs. miR-3934-5p inhibitor group. miR, microRNA; TP53INP1, tumor protein p53-inducible nuclear protein 1; DDP, cisplatin; NC, treated with negative control plasmids; siRNA, small interfering RNA; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X.
Discussion
The resistance of tumor cells to DDP allows cells to evade the cytotoxic effects of DDP, overcoming apoptosis (23); however, the underlying molecular mechanisms of DDP resistance require further investigation (24,25). miRNAs have been reported to be regulators of DDP resistance in NSCLC. A previous study revealed that miR-3934 serves a crucial role in the progression of lung cancer (12). However, the effects of miR-3934-5p in regulating the chemoresistance of NSCLC cells to DDP have not been elucidated.
In the present study, miR-3934-5p was downregulated in NSCLC tissues and cells, which paralleled that in A549/DDP cells. miR-3934-5p increased the IC50 of A549/DDP cells, while the downregulation of miR-3934-5p decreased the IC50 of A549/DDP cells. Therefore, miR-3934-5p induced the resistance of A549 to DDP, and this effect was reversed following treatment with the miR-3934-5p inhibitor. However, the effects of miR-3934-5p on the cell behavior of NSCLC has not been fully elucidated.
The roles of miR-3934-5p in the proliferation and apoptosis of NSCLC cells were then examined. The downregulation of miR-3934-5p significantly inhibited the proliferation and promoted the apoptosis of NSCLC cells. A previous study showed that p21 was involved in the progression of lung cancer (26). In addition, Bcl-2 has been shown to promote oncogenesis via cell death resistance and Bax is a pro-apoptotic gene in cancer (27). The present study examined the effects of miR-3934-5p on the expression of Bcl-2/Bax and p21 in NSCLC. The results showed that the suppression of miR-3934-5p significantly decreased the expression of Bcl-2 and increased the expression of Bax and p21 at the mRNA and protein levels in A549/DDP cells treated with DDP. This further demonstrated that the suppression of miR-3934-5p was important in enhancing the sensitivity of NSCLC cells to DDP. However, the underlying mechanisms remained unclear.
TP53INP1, as a tumor suppressor, can induce autophagy-dependent cell death in colorectal cancer (16). Furthermore, it has been reported that miR-182 can promote drug resistance in DDP-treated hepatocellular carcinoma cells by downregulating TP53INP1 (18). Promoting the expression of TP53INP1 in A549 cells has also been shown to enhance the sensitivity of cells to arsenic treatment (19). In the present study, TP53INP1 was predicted and confirmed to be a target of miR-3934-5p. The downregulation of miR-3934-5p increased the expression of TP53INP1. The knockdown of TP53INP1 reversed the decreased cell viability and increased apoptosis of A549/DDP induced by the miR-3934-5p inhibitor. It has been evidenced that TP53INP1 serves an important role in tumor progression via regulating downstream genes, such as Bax and p21 (14,28). Silencing TP53INP1 suppressed the expression of TP53INP1, p21 and Bax, and upregulated the expression of Bcl-2 in A549/DDP cells following transfection with the miR-3934-5p inhibitor. Taken together, the downregulation of miR-3934-5p may increase the sensitivity of A549 cells to DDP by targeting TP53INP1.
The present study had some limitations. First, the scope of the patients enrolled was limited. Second, no in vivo assay was performed. Third, a single miRNA may have various targets and a gene may be targeted by several miRNAs. These issues require further investigation.
Taken together, the present study found that miR-3934-5p was upregulated in NSCLC. Downregulated miR-3934-5p inhibited the proliferation and promoted the apoptosis of A549 cells treated with DDP, suggesting that the suppression of miR-3934-5p enhanced the sensitivity of NSCLC cells to DDP. Therefore, miR-3934-5p may be a promising therapeutic target for NSCLC.
Acknowledgements
None.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed in the present study are available from the corresponding author on reasonable request.
Authors' contributions
AR was responsible for drafting the manuscript and worked with ZW to collect, analyze and interpret the data. LZ was involved in designing the present study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the People's Hospital of Yucheng (Yucheng, China). Informed consent was obtained from all patients.
Patient consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
References
- 1.Román M, Baraibar I, López I, Nadal E, Rolfo C, Vicent S, Gil-Bazo I. KRAS oncogene in non-small cell lung cancer: Clinical perspectives on the treatment of an old target. Mol Cancer. 2018;17:33. doi: 10.1186/s12943-018-0789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 4.Peters BJM, Cramer-Vd Welle CM, Smit AAJ, Schramel FMNH, van de Garde EMW, Santeon NSCLC Study Group Trends in prescribing systemic treatment and overall survival for non-small cell lung cancer stage IIIB/IV in the Netherlands: 2008–2012. Cancer Epidemiol. 2017;51:1–6. doi: 10.1016/j.canep.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Martin P, Leighl NB. Review of the use of pretest probability for molecular testing in non-small cell lung cancer and overview of new mutations that may affect clinical practice. Ther Adv Med Oncol. 2017;9:405–414. doi: 10.1177/1758834017704329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dasari S, Tchounwou PB. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: Opportunities and challenges. Biomed Res Int. 2015;2015:125094. doi: 10.1155/2015/125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathe A, Scott RJ, Avery-Kiejda KA. miRNAs and other epigenetic changes as biomarkers in triple negative breast cancer. Int J Mol Sci. 2015;16:28347–28376. doi: 10.3390/ijms161226090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J, Zhan Y, Feng J, Luo J, Fan S. MicroRNAs associated with therapy of non-small cell lung cancer. Int J Biol Sci. 2018;14:390–397. doi: 10.7150/ijbs.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiew MSY, Cheng HP, Huang CJ, Chong KY, Cheong SK, Choo KB, Kamarul T. Incomplete cellular reprogramming of colorectal cancer cells elicits an epithelial/mesenchymal hybrid phenotype. J Biomed Sci. 2018;25:57. doi: 10.1186/s12929-018-0461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yerukala Sathipati S, Ho SY. Identifying the miRNA signature associated with survival time in patients with lung adenocarcinoma using miRNA expression profiles. Sci Rep. 2017;7:7507. doi: 10.1038/s41598-017-07739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Sun H, Zhang D, Fan D, Zhang Y, Dong X, Liu S, Yang Z, Ni C, Li Y, et al. TP53INP1 inhibits hypoxia-induced vasculogenic mimicry formation via the ROS/snail signalling axis in breast cancer. J Cell Mol Med. 2018;22:3475–3488. doi: 10.1111/jcmm.13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu SJ, Yang L, Hong Q, Kuang XY, Di GH, Shao ZM. MicroRNA-200a confers chemoresistance by antagonizing TP53INP1 and YAP1 in human breast cancer. BMC Cancer. 2018;18:74. doi: 10.1186/s12885-017-3930-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saadi H, Seillier M, Carrier A. The stress protein TP53INP1 plays a tumor suppressive role by regulating metabolic homeostasis. Biochimie. 2015;118:44–50. doi: 10.1016/j.biochi.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Liao D, Li T, Ye C, Zeng L, Li H, Pu X, Ding C, He Z, Huang GL. miR-221 inhibits autophagy and targets TP53INP1 in colorectal cancer cells. Exp Ther Med. 2018;15:1712–1717. doi: 10.3892/etm.2017.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang PH, Motoo Y, Garcia S, Iovanna JL, Pébusque MJ, Sawabu N. Down-expression of tumor protein p53-induced nuclear protein 1 in human gastric cancer. World J Gastroenterol. 2006;12:691–696. doi: 10.3748/wjg.v12.i5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q, Han Y, Wang C, Shan S, Wang Y, Zhang J, Ren T. MicroRNA-125b promotes tumor metastasis through targeting tumor protein 53-induced nuclear protein 1 in patients with non-small-cell lung cancer. Cancer Cell Int. 2015;15:84. doi: 10.1186/s12935-015-0233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin J, Luo M, Qian H, Chen W. Upregulated miR-182 increases drug resistance in cisplatin-treated HCC cell by regulating TP53INP1. Gene. 2014;538:342–347. doi: 10.1016/j.gene.2013.12.043. [DOI] [PubMed] [Google Scholar]
- 20.Luo Q, Gu S, Li Y, Zhang Z. Improving the expression of TP53INP1 in A549 cells enhances the sensitivity of cells to arsenic treatment. Wei Sheng Yan Jiu. 2017;46:120–125. [PubMed] [Google Scholar]
- 21.Lan D, Wang L, He R, Ma J, Bin Y, Chi X, Chen G, Cai Z. Exogenous glutathione contributes to cisplatin resistance in lung cancer A549 cells. Am J Transl Res. 2018;10:1295–1309. [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Sarin N, Engel F, Kalayda GV, Mannewitz M, Cinatl J, Jr, Rothweiler F, Michaelis M, Saafan H, Ritter CA, Jaehde U, Frötschl R. Cisplatin resistance in non-small cell lung cancer cells is associated with an abrogation of cisplatin-induced G2/M cell cycle arrest. PLoS One. 2017;12:e0181081. doi: 10.1371/journal.pone.0181081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong S, Li X, Zhao Y, Yang Q, Kong B. 53BP1 inhibits the migration and regulates the chemotherapy resistance of ovarian cancer cells. Oncol Lett. 2018;15:9917–9922. doi: 10.3892/ol.2018.8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Z. Miltirone-induced apoptosis in cisplatin-resistant lung cancer cells through upregulation of p53 signaling pathways. Oncol Lett. 2018;15:8841–8846. doi: 10.3892/ol.2018.8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin Y, Zhou X, Huang C, Li L, Liu H, Liang N, Chen Y, Ma D, Han Z, Xu X, et al. Lower miR-340 expression predicts poor prognosis of non-small cell lung cancer and promotes cell proliferation by targeting CDK4. Gene. 2018;675:278–284. doi: 10.1016/j.gene.2018.06.062. [DOI] [PubMed] [Google Scholar]
- 27.Campbell KJ, Tait SWG. Targeting BCL-2 regulated apoptosis in cancer. Open Biol. 2018;8(pii):180002. doi: 10.1098/rsob.180002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang CM, Zhao J, Deng HY. MiR-155 promotes proliferation of human breast cancer MCF-7 cells through targeting tumor protein 53-induced nuclear protein 1. J Biomed Sci. 2013;20:79. doi: 10.1186/1423-0127-20-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed in the present study are available from the corresponding author on reasonable request.