Abstract
Despite advancement in cancer treatment, oral cancer has a poor prognosis and is often detected at late stage. To overcome these challenges, investigators should search for early diagnostic and prognostic biomarkers. More than 700 bacterial species reside in the oral cavity. The oral microbiome population varies by saliva and different habitats of oral cavity. Tobacco, alcohol, and betel nut, which are causative factors of oral cancer, may alter the oral microbiome composition. Both pathogenic and commensal strains of bacteria have significantly contributed to oral cancer. Numerous bacterial species in the oral cavity are involved in chronic inflammation that lead to development of oral carcinogenesis. Bacterial products and its metabolic by-products may induce permanent genetic alterations in epithelial cells of the host that drive proliferation and/or survival of epithelial cells. Porphyromonas gingivalis and Fusobacterium nucleatum induce production of inflammatory cytokines, cell proliferation, and inhibition of apoptosis, cellular invasion, and migration thorough host cell genomic alterations. Recent advancement in metagenomic technologies may be useful in identifying oral cancer–related microbiome, their genomes, virulence properties, and their interaction with host immunity. It is very important to address which bacterial species is responsible for driving oral carcinogenesis. Alteration in the oral commensal microbial communities have potential application as a diagnostic tool to predict oral squamous cell carcinoma. Clinicians should be aware that the protective properties of the resident microflora are beneficial to define treatment strategies. To develop highly precise and effective therapeutic approaches, identification of specific oral microbiomes may be required. In this review, we narrate the role of microbiome in the progression of oral cancer and its role as an early diagnostic and prognostic biomarker for oral cancer.
Keywords: oral microbiome, biofilm, oral cancer, inflammation, biomarker
Introduction
Head and neck squamous cell carcinomas (HNSCC) are the major cause of cancer morbidity and mortality. Worldwide, 550 000 new cases of HNSCC and 380 000 deaths per year were reported. The majority of HNSCC are cancers of oral cavity (OC) and oropharynx.1 Oral squamous cell carcinoma (OSCC) represents 90% of all cancers. Oral squamous cell carcinoma is a common cancer occurring in the head and neck area. These tumors have very high rate of recurrence and frequently undergo lymph node metastasis. Annually 300 373 new cases of OSCC and 145 343 deaths due to this cancer was reported (http://globocan.iarc.fr.). The incidence rate of oral cancer increases among young men and women. Tobacco, betel quid, and alcohol are predominant risk factors for OSCC.2 In the Western world, tobacco smoking and alcohol consumption are the major risk factors for 74% of cases with OSCC, whereas in South Asia and the Pacific region, tobacco chewing with or without areca (betel) nut is the major risk factor for the development of OSCC. Poor oral hygiene acts synergistically to enhance the risk of oral cancer.3,4 Human papilloma virus (HPV) infection is another known risk factor for oropharyngeal cancer (OPC).5 Bacterial infection is one of the major causes of chronic inflammation which facilitates increased cell proliferation, mutagenesis, oncogene activation, and angiogenesis that lead to development of oral cancer.6 Numerous bacterial species are involved in oral cancer.7 Despite advancement in cancer treatment, OSCC has a poor prognosis with 5-year survival rates of <50%. The OSCC is often detected at late stage, and there is a high risk of having second primary tumors.8 To overcome these challenges, investigators should search for early diagnostic and prognostic biomarkers. Significant interest has been shown in the role of the microbiome showed in oral carcinogenesis.9 Human microbiome studies characterized structural and functional aspects of culturable and nonculturable bacterial communities in different regions of human body during healthy and diseased conditions using 16S rRNA-based next-generation sequencing (NGS) technology. 16SrRNA gene which is 1500 bp long is used to identify bacterial communities. The bacterial 16SrRNA gene contains 9 hypervariable regions (V1-V9). Single hypervariable region becomes unable to distinguish among all the bacteria. Among 9 hypervariable regions, the V3 to V4 region provides the maximum discriminating power for analyzing the bacterial groups. This region generates 500-bp-long amplified product by polymerase chain reaction that is normally used in metagenomic study. Human Oral Microbiome Database (14.5; www.homd.org) reported the existence of 700 bacterial strains in human OC. Alterations in oral microbiome lead to inflammation that drives OSCC through direct metabolism of carcinogens. In this review, we narrate the role of microbiome in the progression of OSCC and its role as an early diagnostic and prognostic biomarker for oral cancer. We also described the oral microbiome in healthy individuals and other oral diseases that drive progression of oral cancer.
Location of Oral Microbial Habitat and Its Ecology
The “human microbiome” represents all microorganisms and their genomes in the human body. Oral microbiome is defined as genomes of microorganisms in the OC.10 Oral microbiome is considered an ideal biomarker compared to other biomarkers from the host for oral tumor. The OC and nasopharyngeal regions maintain an ideal environment for the growth of microbiome. Aerobic bacteria create localized niche for the anaerobes. More than 700 bacterial species reside in the OC. Both pathogenic and mutualistic bacteria coevolve together to maintain homeostasis. The 37°C temperature of OC and pH 6.5 to 7.5 of saliva provide stable habitat for bacterial species. Saliva provides nutrients to the microbiome and keeps them hydrated. Aerobic and anaerobic bacteria together form oral biofilms that prevent changes in their environment.11 The oral microbiome population varies with saliva and different habitats (buccal mucosa, supragingival, and subgingival plaque) of OC. There are several microbial habitats such as periodontal pockets, surface of teeth, and tongue in the OC. Among these habitats, tongue shows the highest diversity of microbiota. Microbes in the tongue facilitate colonization of bacteria in other regions of the OC through saliva. Changes in environmental conditions increase the potential of pathogenic bacteria to create oral diseases.12
Tongue, buccal mucosa, supragingival and subgingival surfaces of the teeth, soft and hard palates, and saliva of OC may represent different ecological niches or habitats.13 The warm and moist environment and host-derived nutrients, such as saliva proteins, gingival crevicular fluid (GCF), and glycoproteins, favor the growth of microorganisms in the OC.14 Most of the bacteria in saliva are attached to exfoliated human epithelial cells.15
Microorganisms are classified based on their oxygen requirements as obligate aerobes; obligate anaerobes such as Fusobacterium and Veillonella; facultative anaerobes such as Actinomyces and Streptococci; microaerophiles that grow best at low concentrations of O2 (2%-10%); and capnophiles such as Neisseria that grow best at high CO2 concentrations (5%-10%).16,17
Bacterial species of OC manifest specific response toward different biological surfaces in the OC such as the teeth, tongue, and mucosa. Different receptors and adhesion molecules (adhesins) of bacterial species ensure colonization on different oral surfaces through “lock and key” mechanism. Different habitats have been observed in nonshedding surfaces of the teeth and continually shedding surfaces of the oral mucosa. Bacteria binds with complementary receptors on the mucosal surfaces of the host. Streptococcus salivarius, Streptococcus mitis, Streptococcus constellatus, Streptococcus oralis, Streptococcus intermedius, and Streptococcus anginosus colonize on oral soft tissues and saliva, whereas Streptococcus sanguis prefers to colonize on the teeth.18 Streptococcus is commonly found genera in the human OC, but it is highly genetically heterogeneous group.19
Veillonella atypica, P gingivalis, Selenomonas subspecies, Actinobacillus actinomycetemcomitans, Prevotella intermedia, and Capnocytophaga are observed in tongue, whereas Streptococcus pyogenes, Streptococcus pneumoniae, Haemophilus influenza, and Haemophilus parainfluenzae reside in oropharynx. Streptococcus faecalis, Eikenella corrodens, Enterobacteriaceae, Actinomyces, Lactobacilli, Veillonella, and Treponema are exclusively observed in OC but not in oropharynx (Table 1).12
Table 1.
Predominant Microbial Communities in Oral Cavity and Oropharyngeal Region.
| Different Sites in Oral Cavity and Oropharyngeal Region | Bacterial Species |
|---|---|
| Tongue |
Veillonella atypica
Porphyromonas gingivalis Selenomonas subspecies Aggregatibacter actinomycetemcomitans, Prevotella intermedia Capnocytophaga Enterococcus faecalis Eikenella corrodens |
| Oropharynx |
Streptococcus pyogenes
Streptococcus pneumoniae Haemophilus influenzae Haemophilus parainfluenzae Streptococcus mutans Streptococcus salivarius Streptococcus anginosus |
| Tooth Surface |
S mutans
Actinomyces Eubacterium Peptostreptococcus |
| Tonsil |
Streptococcus viridans
H influenzae Neisseria species Staphylococcus |
| Gingival crevice |
Fusobacterium
Prevotella Porphyromonas Streptococcus mitis Streptococcus sanguinis Propionibacterium acnes Leptotrichia buccalis Actinomyces odontolyticus Veillonella parvula |
| Dental Plaque |
Actinomyces
Rothia Microbacterium Mycobacterium Propionibacterium Corynebacterium Bifidobacterium |
Increased abundance of anaerobes such as Bacteroidaceae subspecies and Spirochetes are reported in subgingival area having less oxygen tensions. Multispecies biofilm formation on tooth surfaces occur through matrix that is made up of extracellular polymeric substances (EPS). Over exposure of fermentable carbohydrates alters the balance between commensals and pathogens. Sucrose is the combination of hexose sugars such as glucose and fructose that are used to synthesize EPS (glucans and fructans). Extracellular polymeric substances is made up of exopolysaccharides such as glucans, lipoteichoic acid, amyloid-like proteins, glycoproteins, extracellular DNA (eDNA), and host proteins. Fermentation of glucose and fructose produces lactic acid that affects the configuration and constitution of dental biofilms. Extracellular polymeric substances provide sites for the adhesion to bacterial cells. Carbohydrate-rich diet induces the formation of EPS, production of acidic metabolites, and favors accumulation of acidogenic and aciduric microorganisms in dental caries. Acids demineralize enamel that is mineralized by saliva. Streptococcus mutans, Lactobacilli, Bifidobacterium subspecies, Scardovia subspecies, and Actinomyces subspecies are associated with caries. Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, Filifactor alocis, and Peptoanaerobacter stomatis are associated with periodontal diseases. Among them P gingivalis, T forsythia, and T denticola are considered predominant pathogens in chronic periodontitis. Streptococcus sanguinis, Streptococcus gordonii, Streptococcus parasanguinis, S oralis, and S mitis are important commensals of the OC. Streptococcus gordonii enhanced bone loss along with infection of P gingivalis. Cariogenic communities such as Corynebacterium, Granulicatella, Propionibacterium, and certain strains of Leptotrichia have high saccharolytic potential and yield acids. Some aciduric bacteria such as Prevotella subspecies and Atopobium subspecies are found in cariogenic biofilms. Extracellular matrix of the cariogenic biofilms inhibits the antimicrobial activity. The extracellular matrix provides chemical or nutrient gradients, pH, and redox gradients in biofilms. The matrix can control the diffusion of charged ion, but uncharged sugars such as glucose and sucrose can readily diffuse into biofilms. Metabolism of sugars into acids makes the environment acidic. Glucans of extracellular matrix accumulate acids within biofilms by providing endogenous sugars and trapping protons. Saliva provides shelter to acidic environment of cariogenic biofilms which enhances demineralization of enamel.20
Role of Oral Microbiome in Biofilm Composition
Human Oral Microbiome Database reported that 687 species belong to 185 genera and 12 phyla such as Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria, Fusobacteria, Spirochaetes, Synergistetes, Chlamydiae, Chloroflexi, SR1, Saccharibacteria (TM7), and Gracilibacteria (GN02). Thirty-two percent of these species remain uncultivated, and 14% have not been named.21
A typical biofilm is made up of mono- or polymicrobial cells, polysaccharide, proteins, nucleic acids, and lipids.22 Oral biofilms are composed of 700 different microbial species; extracellular matrix that is composed of DNA, proteins, polysaccharides, and lipids; and salivary glycoproteins, GCF, albumin, and host cell components. Bacterial colonization, interaction between bacterial cell surface adhesins, and host receptors and extracellular matrix are the key factors for the formation, development, and maturation of oral biofilms. Pathogenic bacteria in oral biofilms contribute to the development of dental caries, periodontitis, and oral cancer. Bacteria have the capability to bind with soft and hard tissues in the OC. Adhered bacteria become unable to attach for long period of time with shedding surface. Teeth have an excellent binding surface for the bacteria. Gram-positive aerobe communities are localized in gingival margin, whereas gram-negative anaerobes are found in supragingival biofilms. Saliva is the primary nutrient source for bacteria that reside in supragingival biofilm, while GCF provides nutrient for bacteria of the subgingival biofilms.23
Formation of oral biofilms first occurs at the supragingival region and formation of subgingival biofilms second. Subgingival biofilms are mainly dominated by gram-negative obligate anaerobes.24 Gram-positive aerobic bacteria, such as Actinomyces subspecies and oral Streptococci (S intermedius and S oralis) are responsible for the initial colonization in teeth surfaces. Streptococcal AGI/II proteins induce co-aggregation between S gordonii and Actinomyces oris. Interaction between fimbriae Actinomyces naeslundii with proline-rich salivary proteins regulates interbacterial binding. Gram-negative anaerobic bacterium F nucleatum acts as a connecting link between early and late colonizers in the oral biofilms. Fusobacterium nucleatum supports the growth of P gingivalis. Metabolic products such as ammonia and organic acid of F nucleatum favor growth of acid-sensitive bacterium P gingivalis through enhancement of pH in the biofilm. Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, P intermedia, Eubacterium subspecies, T forsythia, Selenomonas flueggei, and T denticola are considered as late colonizers of the oral biofilm. Gram-positive oral bacteria such as Streptococci use 17 to 21 amino acids long competence signaling peptides and both gram-positive and gram-negative bacteria use autoinducer-2 (AI-2) to communicate with each other in the oral biofilms. Lactate of Streptococci and Lactobacilli has been utilized by A actinomycetemcomitans. Glucosyltransferases (GTFB, GTFC, and GTFD) of cariogenic bacteria S mutans are responsible for the synthesis of glucans. The surface proteins GbpA, GbpB, GbpC, and GbpD of bacteria bind with the glucans. These enzymes and proteins are involved in a sucrose-dependent pathway that induces plaque formation.25
Chemical communication among bacterial cells through expression of genes in response to high cell density is defined as quorum sensing. Periodontal pathogens such as P gingivalis, P intermedia, and F nucleatum produce highest levels of AI-2.26 The competence signaling peptide is responsible for the formation of biofilm, bacteriocins, stress response, acid tolerance, and genetic conversion by S mutans.27
Commensal bacteria live in equilibrium with the host immune response. Streptococcus is the predominant genus of healthy oral microbiome. Commensal and pathogenic bacterial species bypass the host immune response by forming biofilm. Streptococci produce oral adhesins, such as PaG, SspA, antigen I/II, amylase-binding proteins, and type 1 fimbriae-associated protein. Streptococcus gordonii and P gingivalis use AI-2 in oral biofilm. Streptococcus gordonii reduces dental plaque formation through production of hydrogen peroxide. Hydrogen peroxide of S gordonii inhibits the growth of A naeslundii. Fusobacterium nucleatum can also associate with S cristatus. Streptococcus, Actinomyces, and Lactobacillus inhibit the growth of bacterial species by making the microenvironment acidic through change in pH. Porphyromonas subspecies, Campylobacter subspecies, T forsythia, T denticola, and A actinomycetemcomitans in the biofilm are responsible for inflammation of the periodontitis.17
Porphyromonas gingivalis enhances the activity of the tyrosine kinase Ptk1 and protein tyrosine phosphorylation–dependent signaling pathway through FimA and Mfa1 fimbrial adhesins in response to streptococcal metabolite 4-amino benzoate (pABA). Interaction between Mfa1 adhesin protein and streptococcal surface proteins such as SspA or SspB activate the tyrosine phosphatase Ltp1 that dephosphorylates Ptk1 and reduces production of adhesin. The pABA of S gordonii inactivates tyrosine phosphatase Ltp1 of P gingivalis, thus reducing dephosphorylation level of tyrosine kinase Ptk1. Transcription factor CdhR is inactivated by Ptk1, and Ptk1 in turn increases the expression of FimA gene that encodes fimbrial adhesins.20
Crosstalk Between Oral Microbiome and Fungal/ Candidate Phyla Radiation organism in Oral Biofilm Formation
Candidate phyla radiation (CPR) of bacterial organisms includes 35 phyla and consists of more than 15% of the domain Bacteria. It has special characteristic features that include ultra-small cell size, 16SrRNA gene self-splicing introns, archaeal-specific RuBisCO genes, and the loss of genes that encode CRISPR/Cas bacteriophage defense system. Three CPR phyla such as Gracilibacteria (GN02), Absconditabacteria (SR1), and Saccharibacteria (TM7) are predominant in the OC. An increased abundance of TM7 was reported in periodontitis. Candida, Cladosporium, Aureobasidium, and Aspergillus are the most abundant fungal genera in human OC. The interspecies interactions between Candida albicans and S mutans or S oralis enhance the severity of dental caries. The most abundant bacteriophages identified in the OC are Siphoviridae, Myoviridae, and Podoviridae. Horizontal gene transfer in the OC provides massive diversity of organisms. It has been reported that 60% to 70% of known bacterial genomes contain prophages. Biofilm formation between C albicans and S oralis grows to a greater density. This type of interspecies biofilm formation upregulates host protease μ calpain that increases tissue invasion through degradation of cadherin junctions.28
Factors Affecting Oral Microbiota Population
Temperature, pH, atmospheric conditions, salinity, redox potential, and water of saliva affect the formation of biofilms in the OC. Saliva is used as a transporting medium of nutrients, peptides, and partially dissolved carbohydrates in the formation of oral biofilms.17
Factors such as age of host; environmental conditions such as pH, oxygen levels, and nutrition in the habitats of the OC; lifestyle of the host such as food habit, tobacco, and alcohol consumption; and oral hygiene modulate the composition of oral microbiota. Biofilm formation in dental plaque is associated with caries and periodontal diseases. Biofilm accumulation is restricted by oral epithelia. Salivary glycoproteins regulate attachment of bacteria to oral surfaces by either enhancing or preventing their adherence. The GCF contains iron that enhances probable pathogenic mechanisms in oral bacteria associated with periodontal disease.29
Salivary components such as immunoglobulin A, lactoferrin, lactoperoxidase, lysozyme, statherin, and histatins are nutritional source for the oral microbiome. Lactoperoxidase is responsible for the production of hypothiocyanite from hydrogen peroxide. Hypothiocyanite showed antimicrobial effects by suppressing bacterial glycolysis. Another salivary component with antimicrobial potential is nitrite, converted from dietary nitrates by oral bacteria. Nitrite that is produced by oral microbiome is further reduced to nitric oxide that prevents the growth of cariogenic bacteria.14,30 Veillonella subspecies metabolizes lactate into weaker acids that reduce local pH, whereas Streptococcus subspecies and Actinomyces subspecies generate the alkali, ammonia, by catabolism of arginine or urea that increases local pH.14
Oral Microbiome Signatures in Healthy Individuals
Actinomyces, Capnocytophaga, Eikenella, Eubacteria, Fusobacterium, Haemophilus, Lactobacterium, Leptotrichia, Neisseria, Porphyromonas, Prevotella, Propionibacterium, Peptostreptococcus, Streptococcus, Staphylococcus, Veillonella, and Treponema are present in OC of healthy individuals.17
Fusobacterium periodonticum, Leptotrichia trevisanii, Leptotrichia hofstadii, and Leptotrichia buccalis are abundant in saliva of control samples of Human Microbiome Project (HMP) cohorts. The predominant oral microbial communities belonged to Firmicutes (genus Streptococcus, family Veillonellaceae, genus Granulicatella), Proteobacteria (genera Neisseria, Haemophilus), Actinobacteria (genera Corynebacterium, Rothia, Actinomyces), Bacteroidetes (genera Prevotella, Capnocytophaga, and Porphyromonas), and Fusobacteria (genus Fusobacterium) are observed in healthy individuals.31,32
Comparison of Salivary Microbiome Between Indian Cohort and Western Cohort
Enrichment of Enterobacteriaceae and Proteobacteria are noticed in saliva of Indians. Bacteroidales, Paraprevotellaceae, and Spirochaetaceae were enriched in Indians. Abundance of Alphaproteobacteria and Deltaproteobacteria in saliva were higher in Indians when compared to HMP.33 Streptococcus subspecies was the commonly observed bacterial genus in saliva of Indian populations. The genera Prevotella, Fusobacterium, Veillonella, Leptotrichia, and Granulicatella were major contributors of the salivary microbiome in all populations. Abundance of Solobacterium subspecies and Alloprevotella subspecies was common in saliva samples of Indian population. Chromobacterium subspecies was most enriched in saliva of the Assamese population. Bacterial genera Atopobium, Megasphaera, and Prevotella in the saliva samples of people in Tamil Nadu, Streptobacillus and Bacillus in the saliva samples of people of Telangana, and Stenotrophomonas in the saliva samples of people of Uttarakhand and Assam showed statistically significant enrichment. Solobacterium subspecies, Alloprevotella subspecies, and Lachnoanaerobaculum subspecies represent core saliva microbiome in the Indian population.34
Beneficial Effects of Resident Oral Bacteria
Commensal bacteria can enhance the effectiveness of immunotherapy with checkpoint inhibitors. Bifidobacterium through oral administration controlled the growth of the tumor with the same efficiency with programmed cell death protein 1 ligand 1 (PD-L1)–specific antibody therapy. Combinatorial approach of both oral administration of Bifidobacterium and PD-L1-specific antibody prevented outgrowth of tumor only. Oral administration of Bifidobacterium enhances antitumor immunity and alters the efficacy of check-point inhibitors as an immunotherapeutic agent by changing the composition of intestinal microorganisms.35 Bacteriocins of Streptococcus dentisani inhibit the growth of cariogenic bacterial species.36 Antimicrobial peptides such as bacteriocins produced by S mutans and S salivarius in the OC kill other bacteria.25
Role of Oral Microbiome in Oral Lichen Planus, Periodontitis, and Dental Caries
We emphasized microbiome on caries, periodontitis, and lichen planus because oral diseases such as caries, periodontitis, and lichen planus are associated with development of SCC of tongue, OP, laryngeal, and oral cavities. Bacteria that colonize in OC and periodontitis region induce oral carcinogenesis thorough direct toxic effects, metabolism of precarcinogenic substances, inflammation, and tumor microenvironment.37
Role of Oral Microbiome in Dental Caries
Diversified microbiota contributes to the development of dental caries, gingivitis, and periodontitis. Gingivitis is the inflammation of the soft tissues of the gum surrounding the teeth that develops due to interaction between the plaque microbiota and the host tissues. The level of gram-positive Streptococci and gram-negative anaerobes such as Actinobacillus, Capnocytophaga, Campylobacter, Eikenella, Fusobacterium, and Prevotella are elevated in gingival plaque. High prevalence of L buccalis was reported in Chinese patients with gingivitis and necrotizing ulcerative gingivitis.38 Prevotella subspecies, Lactobacillus subspecies, Dialister subspecies, and Filifactor subspecies are related to pathogenesis and progression of dental caries.39 The genera Veillonella, Bifidobacterium, Selenomonas, Olsenella, Parascardovia, Scardovia, Chryseobacterium, Terrimonas, Burkholderia, Neisseria, and Sporobacter showed significantly higher abundance in patients with dental caries. Veillonella has cariogenic potential and allows acidogenic bacteria such as Streptococcus species to grow and continue to produce additional acid.40
Role of oral microbiome in periodontitis
Epidemiological studies showed a significant relationship between periodontitis and OSCC. Periodontitis enhances the risk of tongue cancer. Chronic periodontal disease is considered to be an important risk factor for oral premalignant lesions and OSCC.41 Chronic periodontitis enhances the risk of oral premalignant lesions that lead to OSCC and oropharyngeal squamous cell carcinoma (OPSCC).42 Periodontal bacteria such as P gingivalis, P intermedia, and F nucleatum are essential for developing OSCC.18,43,44 Periodontitis is associated with the subgingival microbiota such as Peptostreptococcus, Synergistes, Filifactor, Mycoplasma, and Olsenella. Among them, Peptostreptococcus is known to be a potential pathogen for periodontitis.45
Prevotella intermedia, F nucleatum, Selenomonas noxia, A actinomycetemcomitans, and Eubacterium nodatum, P gingivalis, T denticola, and T forsythia were also associated with pathogenesis of periodontal disease. These periodontal bacteria produce virulence factors such as fimbriae adhesins, lipopolysaccharides (LPSs), peptidoglycans, and lipoteichoic acids that induce release of pro-inflammatory cytokines. Among them, P gingivalis can produce LPSs that induce osteoclasts and bone resorption. Other virulence factors were produced by other periodontal bacteria such as A actinomycetemcomitans, P intermedia, T denticola, and N cinerea. A number of useful genes and metabolic pathways such as bacterial chemotaxis, flagellar assembly, and toxin biosynthesis were found to be enriched in the microbiomes of periodontal disease.46
Periodontitis-associated microorganisms induce inflammation and destruction of tissue through degradation of collagen and release nutrients such as heme-containing compounds, sources of amino acids, and iron. These nutrients are carried into the gingival crevice through GCF that favors the growth of proteolytic and asaccharolytic bacteria with iron-acquisition capacity in the subgingival region. Overexpression of genes such as proteolysis-related genes, genes for amino acid transport, and acquisition of iron as well as genes for the synthesis of LPS and genes for potassium ion transport are reported in periodontitis-associated subgingival biofilms. These meta-transcriptomic alterations drive enhancement of pro-inflammatory potential of the microbial community through production of pro-inflammatory cytokines and a decrease in the production of human β-defensin 3 in gingival epithelial cells (GECs). Aggregatibacter actinomycetemcomitans is a keystone pathogen for aggressive periodontitis where S gordonii and S parasanguinis function as an accessory pathogen. Streptococci enhances the virulence property of C albicans that increases invasion of tissue by fungi and infection in mucosa. Streptococci induces expression of β-defensins. Porphyromonas gingivalis induces Notch 1 signaling that produces PLA2-IIA in oral epithelial cells. Porphyromonas gingivalis inhibits the bactericidal activity of innate leukocytes while inducing inflammatory responses.20
Multiple host cells of myeloid and nonmyeloid origin such as neutrophil polymorphs, oral keratinocytes, monocytes, macrophages, osteoblasts, osteoclasts, and dendritic cells are involved in the development and progression of periodontal disease and oral cancer. These cells have cytosolic, membrane-associated receptors and secreted pattern recognition receptors (PRRs) as well as Toll-like receptors (TLRs), RIG-I-like receptors, Nucleotide-binding Oligomerization Domain, NLRs, and C-type lectin receptors. These receptors interact with periodontal microbial-associated molecular patterns (eg, lipoproteins, LPS, fimbriae, Bacteroides surface protein A, and nucleic acids) and damage/danger-associated molecular patterns (DAMPs; eg, fibrinogen, heat-shock proteins, and nucleic acids).47 Stratified squamous epithelium in periodontal pocket undergoes proliferation, migration, and ulcerations that provide opportunity of HPV infection.48
Role of oral microbiome in oral Lichen Planus
Oral leukoplakia, oral lichen planus (OLP), and systemic lupus erythematosus are common diseases of oral mucosa. Oral lichen planus is one of the most common chronic inflammatory autoimmune diseases. Individuals with OLP have the risk to develop oral cancer. Bacteria play an important role in these mucosal diseases.49 Higher abundance of Porphyromonas and Solobacterium and lower abundance of Haemophilus, Corynebacterium, Cellulosimicrobium, and Campylobacter were reported in patients with OLP when compared to healthy controls. Gram-negative rod-shaped anaerobes Prevotella melaninogenica showed significantly higher abundance in OLP. Treponema, Porphyromonas, Parvimonas, and Fusobacterium were considered as a core microbiota in periodontitis.50 Abundance of Bacillus, Enterococcus, Parvimonas, Peptostreptococcus, and Slackia were significantly different between epithelial precursor lesion and OSCC. Alteration in these microbial communities may be used as a predictive marker for epithelial precursor lesion–OSCC transition. Fusobacterium, Prevotella, Porphyromonas, Veillonella, Actinomyces, Clostridium, Haemophilus, Streptococcus subspecies, and Enterobacteriaceae are linked to both premalignant lesion and OSCC. Cloacibacillus, Gemmiger, Oscillospira, and Roseburia were abundant in patients with epithelial precursor lesion and OSCC when compared to normal patients. Positive correlations were reported between Prevotella and Leptotrichia in epithelial precursor lesion group and between Prevotella and Veillonella and Prevotella and Leptotrichia within both current smokers and chewers. Alistipes, Bacteroides, Blautia, Clostridium, Dorea, Escherichia, Faecalibacterium, Megamonas, and Phascolarctobacterium showed positive correlations between patients with epithelial precursor lesion and OSCC.51 Furquim et al reported about the influence of salivary microbiome for developing Fanconi anemia (FA). Patients with FA are more likely to develop HNSCC. The salivary microbiome profile of patients with FA showed similar diversity pattern with oral leukoplakia and OSCC.52
Oral Microbiome Signatures in Saliva and Tissues of Patients With OSCC
High abundance of P melaninogenica, S mitis, and Capnocytophaga gingivalis are reported in saliva of patients with OSCC. Mager et al reported the presence of F periodonticum in saliva of patients with OSCC.18 Capnocytophaga gingivalis, P melaninogenica, and S mitis were highly abundant in saliva of patients with OSCC and served as potential diagnostic markers for oral cancer.18,53 Abundance of Aggregatibacter, Lautropia, Haemophillus, Neisseria, and Leptotrichia was significantly higher in the control samples. Haemophilus, Neisseria, Gemellaceae, or Aggregatibacter were more abundant in saliva of normal individuals when compared to patients with HNSCC.54
Abundance of Lactobacillus vaginalis, Lactobacillus gasseri: Lactobacillus johnsonii, Lactobacillus fermentum, Lactobacillus salivarius, and Lactobacillus rhamnosus OTUs increased in higher TNM stage of HNSCC. Fusobacterium nucleatum, S salivarius: Streptococcus vestibularis, Prevotella oris, and Rothia mucilaginosa were highly abundant in saliva samples of patients with HNSCC. A loss in abundance of P jejuni: P melaninogenica, and Prevotella pallens was reported in HNSCC. Abundance of L gasseri/L johnsonii and L vaginalis in saliva of patients with OPC results from transportation of commensal bacterial from normal vaginal to the oral flora through oral sex. Lactobacilli prevents the binding of other bacteria to the oral epithelial cells of host. Lactic acid from Lactobacilli inhibits the growth of many other bacteria and prevents histone deacetylation that drives transcription of genes. Lactic acid enhances autophagy of epithelial cells.55
Haemophilus, Neisseria, Aggregatibacter, and Leptotrichia showed significantly higher enrichment in saliva of patients with HNSCC after surgery. Lactobacillus and Lactobacillaceae showed significantly higher abundance in saliva of patients treated with chemoradiation therapy/ surgery. The abundance of Lactobacillus was increased in saliva of patients with advanced TNM stage. Lactobacillus has been reported as a major causative agent for caries and xerostomia. Neisseria is significantly more abundant in HPV-negative individuals. Haemophilus, Neisseria, Gemellaceae, and Aggregatibacter showed overrepresentation in healthy individuals when compared to patients with oral cancer. Lactobacillus and Weeksellaceae showed more abundance in HPV-positive HNSCC tumors, whereas Eikenella, Neisseria, and Leptotrichia showed more abundance in HPV-negative HNSCC cancer (Table 2).54
Table 2.
Alterations in Predominant Bacteria Identified in Oral Cancer.
| Name of the Bacteria | Name of the Phylum | Absolute Quantitative Status in Oral Cancer | Type of Sample | References No. |
|---|---|---|---|---|
| Fusobacterium | Fusobacteria | Elevated | Oral swab Saliva | 50 |
| Fusobacterium naviforme | Fusobacteria | Elevated | Oral swabs | 56 |
| Leptotrichia buccalis | Fusobacteria | Elevated | Saliva | 51 |
| Leptotrichia | Fusobacteria | Reduced | Saliva | 50 |
| Fusobacterium nucleatum | Fusobacteria | Elevated | Saliva | 54 |
| Fusobacterium periodonticum | Fusobacteria | highly abundant in HNSCC | Tissue biopsy | 57 |
| Rothia | Actinobacteria | Reduced | Oral swab | 58 |
| Actinobacteria | Actinobacteria | Reduced | Tissue | 57 |
| Atopobium | Actinobacteria | Elevated | Tissue biopsy | 59 |
| Actinomyces odontolyticus | Actinobacteria | Reduced | Tissue biopsy | 57 |
| Rothia mucilaginosa | Actinobacteria | Reduced | Tissue biopsy | 60 |
| Rothia | Actinobacteria | Elevated | Saliva | 61 |
| Streptococcus | Firmicutes | Reduced | Saliva | 50 |
| Firmicutes | Firmicutes | Elevated | Oral rinse | 62 |
| Streptococcus oralis | Firmicutes | Elevated | Tissue biopsies and Saliva | 63 |
| Streptococcus mitis | Firmicutes | Reduced | Tissue biopsies | 57 |
| Veillonella dispar | Firmicutes | Elevated | Oral rinse | 64 |
| Streptococcus parasanguinis | Firmicutes | Elevated | Oral swab and Saliva | 61 |
| Streptococcus salivarius | Firmicutes | Elevated | Tissue biopsy and Saliva | 55,61 |
| Veillonella parvula | Firmicutes | Reduced | Oral rinse | 62 |
| Staphylococcus aureus | Firmicutes | Elevated | Tissue biopsy | 12 |
| S mitis | Firmicutes | Reduced | Saliva and tissue | 62,65 |
| Streptococcus | Firmicutes | Elevated | Saliva and tongue tumor tissue | 51,57 |
| Gemella | Firmicutes | Elevated | Oral rinse | 66 |
| Peptostreptococcus | Firmicutes | Elevated | Oral swabs | 56 |
| Filifactor alocis | Firmicutes | highly abundant in OSCC | Oral rinse | 62 |
| Oribacterium | Firmicutes | Elevated | Oral rinse | 62,64,66 |
| Streptococcus anginosus | Firmicutes | Elevated | Oral rinse and saliva | 61,64 |
| Dialister | Firmicutes | Elevated in laryngeal carcinoma | Oral rinse | 62 |
| Dialister pneumosintes | Firmicutes | highly abundant in OSCC with stage IV | Oral rinse | 62 |
| Lactobacillus gasseri | Firmicutes | highly abundant in HNSCC | Tissue biopsy and saliva | 55 |
| Lactobacillus fermentum | Firmicutes | highly abundant in oropharyngeal cancer patients | Tissue biopsy and saliva | 55 |
| Lactobacillus rhamnosus | Firmicutes | highly abundant in oropharyngeal cancer patients | Tissue biopsy and saliva | 55 |
| Porphyromonas gingivalis | Bacteroidetes | Elevated | Oral mouth wash | 67 |
| Prevotella melaninogenica | Bacteroidetes | Elevated | Tissue biopsy and saliva | 55 |
| Porphyromonas | Bacteroidetes | Elevated | Saliva | 51 |
| Prevotella | Bacteroidetes | Elevated | Saliva | 51 |
| Capnocytophaga | Bacteroidetes | Reduced | Oral mouth wash | 67 |
| Capnocytophaga gingivalis | Bacteroidetes | Elevated | Oral swabs | 56 |
| Tannerella forsythia | Bacteroidetes | highly abundant in OSCC with stage IV | Oral rinse | 62,67 |
| Prevotella salivae | Bacteroidetes | highly abundant | Tissue | 59 |
| Haemophilus | Proteobacteria | Elevated | Saliva and swab | 62 |
| Pseudomonas aeruginosa | Proteobacteria | Elevated | Tissue biopsy | 68 |
| Aggregatibacter | Proteobacteria | Highly abundant | Oral mouth wash | 67 |
| Pseudomonas | Proteobacteria | Highly abundant | Tissue | 59 |
| Haemophilus parainfluenza | Proteobacteria | highly abundant in HNSCC | Oral rinse | 62 |
| Neisseria elongata | Proteobacteria | highly abundant in OSCC | Oral rinse | 62 |
| Eikenella corrodens | Proteobacteria | highly abundant in OSCC with stage IV | Oral rinse | 62 |
| Campylobacter concisus | Epsilonbacteraeota | Highly abundant | Tissue | 59 |
Abbreviations: HNSCC, head and neck squamous cell carcinomas; OSCC, oral squamous cell carcinoma.
The existence of viable bacteria in deep parts of OSCC favored the hypothesis that bacteria survived in the tumor microenvironment.69,70 The bacterial taxa Veillonella, Fusobacterium, Prevotella, Porphyromonas, Actinomyces, and Clostridium belonging anaerobes, and Haemophilus, Enterobacteriaceae, and Streptococcus subspecies belonging to aerobes were highly abundant in tumor sites of patients with OSCC.43,58 Capnocytophaga, Pseudomonas, and Atopobium at the genus level and Campylobacter concisus, Prevotella salivae, Prevotella loeschii, and Fusobacterium oral taxon 204 at the species level were reported to be highly abundant in OSCC. Streptococcus mitis, Streptococcus oral taxon 070, Lautropia mirabilis, and Rothia dentocariosa were more abundant in fibroepithelial polyp (FEP) controls. Virulence factors, such as LPS, flagella, and exotoxin U in Pseudomonas aeruginosa played a significant role in inflammation in carcinogenesis. In OSCC, bacterial genes that encode enzymes such as transketolase, pyruvate formate lyase-activating enzyme, formate C-acetyltransferase, aspartokinase/homoserine dehydrogenase, and nitroreductase/dihydropteridine reductase were significantly overrepresented, whereas bacterial genes that encode iron complex transport system proteins, aspartyl-tRNA(Asn)/glutamyl-tRNA(Gln) amidotransferase subunits A and B were more abundant in FEP.59
Abundance of genus Streptococcus, Rothia, Gemella, Peptostreptococcus, Porphyromonas, Micromonas, Dialister, Tannerella, Veillonella, and Lactobacillus was reported in tumor tissues of patients with OSCC, whereas Prevotella, Neisseria, Leptotrichia, Capnocytophaga, Actinobacillus, and Oribacterium were more abundant in healthy individuals.9 Presence of Clavibacter michiganensis, Plantibacter flavus, Tepidimonas aquatica, and Thermus scotoductus were predominant in deep tumor tissues.71
Exiguobacterium oxidotolerans, C gingivalis, P melaninogenica, Staphylococcus aureus, Veillonella parvula, Capnocytophaga ochracea, Eubacterium saburreum, L buccalis, and S mitis were significantly abundant in tumor tissues of patients with oral cancer. DNA of S anginosus has been reported in tissue samples of HNSCC.18,32,72 Pseudomonas aeruginosa, Campylobacter subspecies. Oral taxon 44, and F nucleatum subspecies polymorphum are associated with OSCC.68 Moraxella osloensis, Prevotella veroralis, and species of Actinomyces were only detected in normal tissues.73 The level of Parvimonas was enhanced and the level of Actinomyces was reduced in tumors relative to normal tissues. The abundance level of genus Parvimonas was significantly reduced in low-grade (T0-T2) tumors compared to high-grade tumor (T3-T4). The abundance level of the genus Actinomyces was significantly increased in low-grade (T0-T2) tumor when compared to high-grade tumor (T3-T4). The relative abundances of Actinomyces and Parvimonas were reduced at hypopharyngeal/laryngeal region when compared to the OC/oropharynx. Actinomyces subspecies may inhibit tumorigenesis through the secretion protease inhibitors.60
Genus Streptococcus, Dialister, and Veillonella can be used to segregate HNSCC from healthy individuals. Lactobacillus gasseri: L johnsonii was overrepresented in patients with HNSCC compared to controls. Streptococcus mutants, L fermentum, L salivarius, and L rhamnosus were reported to be higher abundant in patients with OPC. Lactobacillus gasseri/L johnsonii, H parainfluenza, L fermentum, and F periodonticum can be used as a diagnostic biomarker for HNSCC.55 Reduction in levels of Lactobacillus subspecies induces microbial dysbiosis in patients with tongue cancer. Streptococcus, Actinomyces, Corynebacterium, Enterococcus, Micrococcus, and R mucilaginosa were significantly increased in the tongue tumor tissues when compared to normal.57
Prevotella jejuni: P melaninogenica and P pallens were less abundant in HNSCC. Fusobacterium nucleatum, Fusobacterium naviforme, and Fusobacterium canifelium were more enriched in HNSCC samples. Increased abundance of Lactobacillus species was reported in saliva of advanced stage of the tumor. Parvimonas and Actinobacteria were highly enriched in advanced stage of oral and OPC tissue samples.55
Firmicutes was the abundant phylum in oral rinse and saliva of patients with OSCC. The oral saliva contains bacteria released from diverse habitat of OC that may not be true representation of microbiome signatures of tumor lesions. The level of Fusobacteria was significantly increased and the level of Streptococcus, Haemophilus, Porphyromonas, and Actinomyces was significantly decreased with the progression of OSCC. Abundant level of F periodonticum, Parvimonas micra, S constellatus, H influenza, and F alocis were progressively increased from stage 1 to stage 4 of patients with OSCC. Higher levels of these bacteria could be used as biomarkers for early detection and tracking for the development of OSCC. The higher abundance of F periodonticum, P micra, S constellatus, H influenza, and F alocis was reported in all stages of OSCC. Neisseria elongata, E corrodens, Oribacterium sp._oral_taxon_102, and Dialister pneumosintes were highly enriched only in patients with OSCC stage 4. Filifactor alocis was significantly associated with smoking. The level of gram-positive Firmicutes bacteria F alocis was increased in periodontal disease. It was significantly increased in advanced stage of OSCC and associated with smoking. Filifactor alocis has synergistic relationship with other common periodontal bacteria. This leads to tumor progression through colonization with other pathogenic periodontal communities. Streptococcus mitis, H parainfluenzae, Porphyromonas pasteri, V parvula, and Actinomyces odontolyticus were highly abundant in the healthy controls. They were significantly decreased with the development of OSCC. Porphyromonas pasteri may be a useful bacteria marker for OSCC diagnosis. The microenvironment of the OC of patients with advanced OSCC may induce the growth of periodontal gram-negative anaerobic pathogenic species T forsythia. Bacterial genes involved in carbohydrate-related metabolism were positively associated, and bacterial genes involved in protein and amino acid metabolism such as leucine, isoleucine, and valine, phenylalanine, tyrosine, and tryptophan biosynthesis, and folate biosynthesis were inversely associated with the development of OSCC. Functional prediction showed that methane metabolism, oxidative phosphorylation, and carbon fixation were significantly higher in patients with stage 4 OSCC (Table 2).62
Filifactor alocis has genes that encode bisulfur proteins and a ferrous iron transport system that is efflux of reactive oxygen species. Filifactor alocis becomes more resistant to hydrogen peroxide-induced oxidative stress that favors its presence in periodontal pockets. Filifactor alocis induces secretion of pro-inflammatory cytokine and induces apoptosis of GECs through activation of MEK ½ and caspase 3. It modulates host cell response through activation of oncogenes. Synergistes are responsible for microecological changes and anaerobiosis in periodontal pocket, inflammation, and destruction of gingival tissue that may drive periodontitis-mediated oral carcinogenesis.74
Mycoplasma, Treponema, Campylobacter, Eikenella, Fusobacterium, Selenomonas, Dialister, Peptostreptococcus, Filifactor, Capnocytophaga, Centipeda, Lachnospiraceae_G_7, Alloprevotella, Peptococcus, Catonella, Parvimonas, and Peptostreptococcaceae_ XI_G_7 showed significantly higher abundances in oral swabs of patients with oral cancer. Among them, Fusobacterium, Dialister, Peptostreptococcus, Filifactor, Peptococcus, Catonella, and Parvimonas were also involved in periodontitis. Fusobacterium periodonticum, F naviforme, F nucleatum_subspecies, P stomatis, P micra, and E corrodens were significantly enriched in oral cancer. The genus Eikenella is associated with HPV-negative HNSCC. The taxa Megasphaera, Stomatobaculum, Granulicatella, Lautropia, Veillonella, Streptococcus, Scardovia, Rothia, and Actinomyces were significantly abundant in oral swabs of controls.56
The abundance of Rothia, Haemophilus, Corynebacterium, Paludibacter, Porphyromonas, and Capnocytophaga significantly reduced and the abundance of Oribacterium significantly increased in oral rinse of OC and OPC. Actinomyces, Parvimonas, Selenomonas, and Prevotella showed higher abundance in OCC when compared to OPC. A positive correlation between HPV infection and the abundance of Haemophilus and Gemella has also been reported in OP cancer.66 The abundance of Capnocytophaga was significantly higher in patients with recurrent OSCC when compared to controls. Abundance of periodontal pathogens Fusobacterium, Prevotella, and Alloprevotella were progressively increased, whereas the abundance of Streptococcus was progressively decreased in progression of OSCC.75 Numerous bacterial insertion sites were reported in host genes such as mycobacterium genomic element integrations in exonic regions of ADAMTS1 gene on chromosome 9, Aeromonas genomic element insertions in the exon of the RASSF5 on 1q32.1, sphingomonas genomic elements insertions in exon of chromatin remodeling gene SRCAP on chromosome 16, and Bordetella genomic insertion within the exon of the proto-oncogene WNT3 on chromosome 17. These may drive tumorigenesis by altering gene expression.76
Role of Oral Microbiome in Xenobiotic Metabolism
Tobacco and alcohol are primary causes of oral cancers. Betel nut chewing with or without tobacco has been shown to be independently associated with the development of OSCC. Alcohol itself is not carcinogenic; acetaldehyde (ALD) which is the metabolic end product of ethanol is highly carcinogenic. Tobacco, alcohol, and betel nut may alter oral microbiome. Microbiome of oral mucosa are involved in production of genotoxic metabolite ALD by oxidation of ethanol that produces DNA damage by forming DNA adducts in oral epithelial cell. Chronic smoking transforms oral microbiome to produce more ALD from ethanol. Oral microbiome may function as synergistic risk factors along with primary risk factors such as alcohol and tobacco use.77
Leukoplakia, erythroplakia, and oral submucous fibrosis are precursor lesion of oral cancer in betel nut chewers. Microbiome have been identified for the development of oral submucous fibrosis. Prolonged use of betel nut in particular, tannic acid, inhibits the growth of commensal bacteria. Cigarette smoking also increases and decreases in the abundance of common taxa of oral microbiome.78 α and β bacterial diversity showed significant difference between betel nut users with and without chewing of tobacco. Chewing of betel nut with alcohol consumption may alter oral microbiome signatures. It has been reported that β diversity significantly differed between chewers and nonchewers with excessive alcohol consumption.61 Smokers have more bacterial diversity in OP region when compared to nonsmokers.79
No significant differences in microbial diversity had been observed between smokers and nonsmokers groups with no history of betel nut use. Significantly higher abundance of Streptococcus infantis was reported in current chewers. Reduced levels of Parascardovia and Streptococcus was reported in long-term chewers. Oribacterium, Actinomyces, and S anginosus showed significant elevation in chewers with oral lesions. Streptococcus anginosus is responsible to induce the synthesis of NO and inflammatory cytokines that drive carcinogenesis. Higher abundance of S salivarius, S gordonii, and S parasanguinis have been reported in OSCC tumor tissue.61
Streptococcus salivarius, S intermedius, S mitis, and nonpathogenic Neisseria subspecies possess alcohol dehydrogenase (ADH) that is responsible for the production of carcinogenic compounds such as ALD, hydroxyl ethyl radicals, and hydroxyl radicals from ethanol.80,81 Streptococcus anginosus was frequently reported in OSCC and caused DNA damage in oral mucosa due increased synthesis of NO and cyclooxygenase 2.82
Overrepresentation of the commensal bacterial genera, Corynebacterium and Kingella, showed reduced risk of HNSCC. Corynebacterium and Kingella are functionally related to xenobiotic biodegradation and metabolism pathways that are capable to metabolize several toxicants found in cigarette smoke. Greater abundance of Neisseria, Abiotrophia, Actinomyces, Veillonella, and Capnocytophaga has been related to good oral health. Prevotella nanceiensis, Capnocytophaga leadbetteri, and Selenomonas sputigena were inversely related to HNSCC. Porphyromonas gingivalis, T forsythia, and A actinomycetemcomitans and S mutans were positively related to risk of HNSCC. Kingella dentificans and Streptococcus sanguinis were associated with reduced risk of larynx cancer; Actinomyces oris and V denticariosi were associated with reduced risk of pharynx cancer, whereas P micra and Neisseria sicca were associated with reduced risk of oral cancer.67
Tobacco smoking induces loss of microbial diversity and enhances metabolism of glutamate in the oral microbiome of patients with OSCC. Lactobacillus, Bifidobacterium, Atopobium, Prevotella, Streptococcus, and Veillonella were abundant in smokers, whereas level of Rothia, Neisseria, and Lautropia were reduced in regular smokers. Smoking lowers α-diversity and increases β-diversity. Members of the Actinomyces, Granulicatella, Oribacterium, and Campylobacter genera as well as Veillonella dispar, R mucilaginosa, and H parainfluenzae were significantly enriched in patients with HPV-positive cancer. Abundance level of S anginosus, Peptoniphilus, and Mycoplasma were significantly decreased in HPV-positive cancers.64 Smokers have unstable microbial colonization and are more susceptible to bacterial infection through alterations in innate and adaptive immune response.83,84 Higher abundance of periodontopathogenic bacteria such as Prevotella tannerae, F nucleatum, and P intermedia and lower abundance of Streptococcus tigurinus were reported in OSCC. Higher percentage of P intermedia was reported in betel quid chewers and alcohol users, whereas higher abundance of F nucleatum was reported in smokers.85 The relative abundances of Periodontopathogenic genera Porphyromonas, Tannerella, and Treponema showed no significant positive correlations with cytokine concentrations.86
Porphyromonas gingivalis, P intermedia, A actinomycetemcomitans, and F nucleatum are responsible to produce volatile sulfur compounds such as genotoxic and mutagenic agent hydrogen sulfide (H2S) in OC and methyl mercaptan in the gingival pockets that induce chronic inflammation, cell proliferation, migration, invasion, and tumor angiogenesis. Several oral microbial species such as S gordonii, S mitis, S oralis, S salivarius, and S sanguinis possess the enzyme ADH, which metabolizes alcohol to carcinogenic ALD that induces development of oral cancer. Due to high ADH activity, Neisseria produce more ALD than Streptococcus subspecies, Stomatococcus subspecies, or Moraxella subspecies Neisseria may play an essential role in alcohol-related oral carcinogenesis.63 Terrimonas is involved in tumorigenic anthracene degradation.87
Role of Oral Microbiome in Inflammation
Inflammation due to infections, environmental factors, and therapy induces angiogenesis, tumor progression, and metastasis. Bacterial infection induces initiation and progression of oncogenic processes. Host cells have PRRs such as TLR family, which recognize pathogen-associated molecular patterns or DAMPs that activate the innate immune response. Bacterial endotoxins (LPS), metabolic byproducts of bacterial infection, and increased enzymatic activity because of bacterial infection can induce somatic mutations in host genomes and alters the signaling pathway.55
Activation of transcription factor nuclear factor kB (NF-kB) is an essential feature of bacteria-associated tumor development. During infection, gram-negative bacteria release endotoxins such as LPS from their outer membrane. Bacterial LPS binds highly sensitive PRRs such as TLRs, particularly TLR4, which in turn activates inflammatory-associated cytokine production via NF-KB signaling pathway. This signaling event is one of the major factors in bacteria-induced inflammation as well as the contributor to carcinogenesis.88,89 Lipopolysaccharide from a potential pathogen such as P gingivalis and F nucleatum is responsible for the activation immune system at the cellular level in periodontal diseases. Bacterial endotoxin enhances the production of tumor necrosis factor α (TNF-α) from macrophages. Inflammatory cytokines such as interleukin (IL)-1β, IL-6, and TNF- α are responsible for periodontal tissue damage. Interleukin-1β may be involved in bone resorption and the attachment loss that are characteristic properties of periodontitis. TNF-α is responsible for the generation of free radicals during sepsis.90
Bacterial products such as endotoxins (LPS), enzymes (eg, proteases, collagenases, fibrinolysin, and phospholipase), and metabolic byproducts (eg, H2S, ammonia, and fatty acids) may induce permanent genetic alterations in epithelial cells of the host that drive proliferation and/or survival of epithelial cells.91 Microorganisms induce inflammation by activating neutrophils, macrophages, monocytes, lymphocytes, fibroblasts, and epithelial cells that drive secretion of cytokines and matrix metalloproteases. Bacteria generate reactive oxygen species (eg, hydrogen peroxide and oxygen radicals), reactive nitrogen species (nitric oxides), reactive lipids, and metabolites (eg, malondialdehyde, 4-hydroxy-2-nonenal) in epithelial cells that drive DNA damage in epithelial cells contributing to disease phenotype.71 Bacterial flagella were considered as key inflammatory structures in regulating OSCC-related inflammation.92
It has been reported that F nucleatum subspecies polymorphum, Campylobacter subspecies, and P aeruginosa showed significant association with OSCC, whereas S mitis, R mucilaginosa, and H parainfluenzae were the most significantly abundant genus in the healthy individuals. Genes involved in bacterial mobility, flagellar assembly, bacterial chemotaxis, and LPS synthesis were significantly associated with OSCC. Functional prediction also revealed that genes involved in DNA repair and combination, purine metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, ribosome biogenesis, and glycolysis/gluconeogenesis were enriched in healthy individuals. Pseudomonas aeruginosa induces DNA breaks in epithelial cells that drive chromosomal instability. Lipopolysaccharide, flagella, and cytotoxins (eg, ExoU) of P aeruginosa have potent inflammatory activity that drives carcinogenesis. This activates NF-κB signaling pathway through the recruitment of neutrophils. LasI factor, secreted from P aeruginosa, downregulates the expression of E-cadherin that induces invasion and metastasis (Table 3).65
Table 3.
Oral Bacteria and its Association With Diagnosis and Prognosis of HNSCC.
| Bacterial Phylum/Genus/Species | Association in Diagnosis and Prognosis of Oral Cancer (Reference No.) |
|---|---|
| Veillonella, Fusobacterium, Prevotella, Porphyromonas, Actinomyces, Clostridium, Haemophilus, Enterobacteriaceae, and Streptococcus subspecies | Predictive diagnostic marker for premalignant lesion and OSCC51 |
| Lactobacillus gasseri: Lactobacillus johnsonii, Lactobacillus vaginalis, Lactobacillus fermentum, Lactobacillus salivarius, and Lactobacillus rhamnosus OTUs | Increased in higher TNM stage of HNSCC55 |
|
Streptococcus salivarius: Streptococcus vestibularis, Fusobacterium nucleatum, Prevotella oris, and Rothia mucilaginosa were highly abundant in saliva samples of HNSCC. Porphyromonas gingivalis, Tannerella forsythia, Aggregatibacter actinomycetemcomitans, and Streptococcus mutans were positively related with risk of HNSCC. Fusobacterium periodonticum, Fusobacterium naviforme, F nucleatum_subspecies, Peptostreptococcus stomatis, Parvimonas micra and Eikenella corrodens were significantly enriched in oral cancer |
Predictive diagnostic marker for HNSCC. P gingivalis, Prevotella intermedia, A actinomycetemcomitans, and F nucleatum are responsible produce volatile sulfur compounds VSCs such as genotoxic and mutagenic agent hydrogen sulfide (H2 S) in oral cavity and methyl mercaptan (CH3SH) in the gingival pockets that induce chronic inflammation, cell proliferation, migration, invasion and tumor angiogenesis63 |
| Pseudomonas aeruginosa | Inflammation in carcinogenesis induces DNA breaks in epithelial cells that drive chromosomal instability, LasI factor, secreted from P aeruginosa, down regulates the expression of E-cadherin that induces invasion and metastasis65 |
| Abundance level of genus Parvimonas | Significantly reduced in low grade (T0-T2) tumor as compared to high grade tumor (T3-T4)60 |
| Abundance level of the genus Actinomyces | Significantly increased in low grade (T0-T2) tumor as compared to high grade tumor (T3-T4)60 |
| Streptococcus, Actinomyces, Corynebacterium, Enterococcus, Micrococcus and R mucilaginosa | Significantly increased in the tongue tumor tissues57 |
| Level of Streptococcus, Haemophilus, Porphyromonas, and Actinomyces | Significantly decreased with the progression of OSCC57 |
| Abundant level of F periodonticum, P micra, Streptococcus constellatus, Haemophilus influenza, and Filifactor alocis | Progressively increased from stage 1 to stage 4 of OSCC patients; used as biomarkers for early detection and tracking for the development of OSCC. F alocis induces secretion of pro-inflammatory cytokine and induces apoptosis of gingival epithelial cells through activation of MEK ½ and caspase 3. F alocis modulates host cell response through activation of oncogenes74 |
| Neisseria elongata, E corrodens, Oribacterium sp. oral taxon 102, and Dialister pneumosintes | Biomarker for stage 4 of OSCC patients. Due to high alcohol dehydrogenase (ADH) activity, Neisseria produce more acetaldehyde than Streptococcus subspecies, Stomatococcus subspecies, or Moraxella subspecies. Neisseria may play an essential role in alcohol-related oral carcinogenesis62 |
| Corynebacterium and Kingella | Involved in xenobiotic biodegradation and metabolism pathways that are capable to metabolize several toxicants found in cigarette smoke67 |
| Actinomyces, Granulicatella, Oribacterium, and Campylobacter genera as well as Veillonella dispar, R mucilaginosa, and Haemophilus parainfluenzae | Enriched in patients with HPV-positive cancer64 |
| Streptococcus anginosus, Peptoniphilus, and Mycoplasma | Decreased in HPV-positive cancers64 |
| Streptococci such as S gordonii, S mitis, S oralis, S salivarius, S sanguinis | Possess the enzyme alcohol dehydrogenase (ADH), which metabolizes alcohol to carcinogenic acetaldehyde which induces development of oral cancer63 |
Abbreviations: HNSCC, head and neck squamous cell carcinoma; HPV, human papilloma virus; OSCC, oral squamous cell carcinoma; VSC, volatile sulfur compounds.
The periodontal pathogens Fusobacterium, Porphyromonas, and Campylobacter (common in GI infections) are considered as “mobile microbiome” that originates in the OC but also associated with extra-oral infections and inflammation. Rothia, Streptococcus, and Prevotella produce oral ALD that promote oral carcinogenesis (Table 3).57 Streptococcus, Neisseria, and Veillonella are considered as anti-inflammatory mediators, whereas Selenomonas, Parvimonas, and Campylobacter are considered as pro-inflammatory mediators.93
Role of P gingivalis and F nucleatum in Progression of OSCC
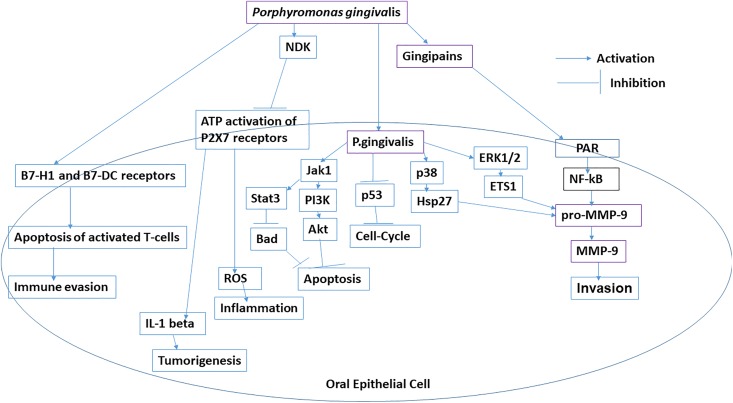
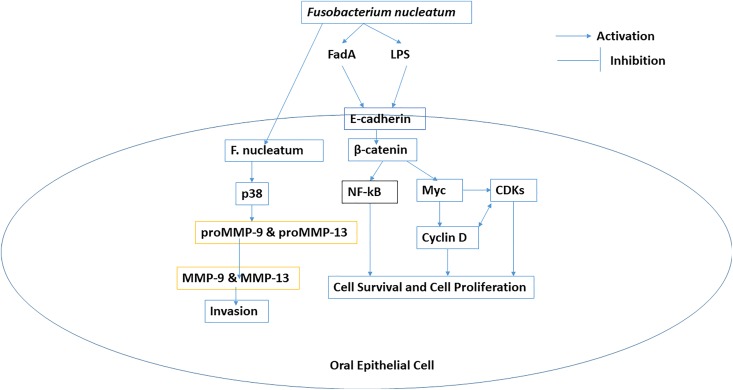
Porphyromonas gingivalis induces the overexpression of B7-H1 and B7-DC receptors in oral epithelial cells that are responsible for the development of chronic inflammation thorough the increased production of IL-1, IL-6, IL-8, and TNF-α. Porphyromonas gingivalis and F nucleatum are responsible for cellular invasion in OSCC. Porphyromonas gingivalis induces overexpression of pro-matrix metalloproteinase-9 (pro-MMP-9) through upregulation of ERK1/2-ETS1, p38/HSP27, and PAR/NF-kB pathways (Figure 1). Porphyromonas gingivalis is also responsible for the epithelial to mesenchymal transition (EMT) transition and enhances the production of MMP-1 and MMP-10. Fusobacterium nucleatum induces the overproduction of MMP-13 (collagenase 3) through upregulation of mitogen-activated protein kinase p38 and Etk/BMX, S6 kinase p70, and RhoA kinase that drive cellular invasion and migration.9 Porphyromonas gingivalis induces cell proliferation through activation and phosphorylation of cyclin-dependent kinases and reduces the expression level of TP53 through the possession of fimbriae (FimA adhesin).94 Porphyromonas gingivalis induces proliferation of buccal epithelial cells through upregulation of β-catenin and gingipain-dependent proteolytic degradation. Fusobacterium nucleatum induces proliferation of oral epithelial cell through activation of 12 kinases. Fusobacterial adhesion FadA of F nucleatum bind to E-cadherin and in turn activate the β-catenin. Porphyromonas gingivalis inhibits chemically induced intrinsic mitochondrial apoptosis in GECs through activation of JAK1/signal transducer and activator of transcription 3 (STAT3) and PI3K/Akt signaling. Porphyromonas gingivalis induces the overexpression of miR-203 which downregulates suppressor of cytokine signaling 3 that inhibits apoptosis through activation of STAT3. Nucleoside diphosphate kinase from P gingivalis inhibits ATP-dependent apoptosis through purinergic receptor P2X7 in GEC.9
Figure 1.
Role of Porphyromonas gingivalis in progression of oral squamous cell carcinoma (OSCC).
Porphyromonas gingivalis infection activates several antiapoptotic pathways such as Jak1/Akt/Stat3 signaling. The composition of LPS of P gingivalis contains phosphorylated 2-keto-3-deoxyoctonate which inhibits intrinsic mitochondrial apoptosis of epithelial cells. Porphyromonas gingivalis enhances Bcl2 (antiapoptotic); Bax (pro-apoptotic) ratio and inhibits release of cytochrome c from mitochondria.95,96 Porphyromonas gingivalis colonizes inside GEC of oral tissues and prevents apoptosis by inducing ligation of ATP with purinergic receptor P2X7 receptors. Nucleoside diphosphate kinase of P gingivalis inhibits apoptosis and promotes survival of host epithelial cells.97 Porphyromonas gingivalis reduces ATP activation of P2X7 receptors on dendritic cells that disrupt activation of the NLRP3/ASC/caspase-1 inflammasome. It prevents secretion of IL-1β and IFN-γ from CD8+ T cells.96-99
Porphyromonas gingivalis accelerates progression through the S-phase of the cell cycle by preventing activity of p53 tumor suppressor gene.100 Induced expression of B7-H1 and B7-DC receptors on OSCC cells and primary GECs by P gingivalis has been reported earlier. Expression of B7-H1 receptor inhibits effector T cells through inducing regulatory T cells. Expression of B7-H1 receptor induces immune evasion in oral cancers.101 ERK1/2-Ets1, p38/HSP27, and PAR2/NF-KB pathways are activated by P gingivalis infection to induce the expression of pro-MMP-9. Gingipains (cysteine proteinases) from P gingivalis converts pro-MMP-9 into MMP-9 that promotes cell migration invasion and metastasis in OSCC.102
Bacterial flagella and LPS are potent inflammatory structures. Lipopolysaccharide induces cancer-promoting inflammatory reactions.65 Lipopolysaccharide of F nucleatum contains 2-keto-3-deoxyoctonate and heptose that may inhibit intrinsic apoptotic pathway of oral epithelial cells. Infection of human epithelial cells by F nucleatum increases the production of MMP-13 (collagenase 3) through the activation of mitogen-activated protein kinase p38. This in turn promotes cellular migration through stimulation of Etk/BMX, S6 kinase p70, and RhoA kinase. Fusobacterium nucleatum activates p38 which in turn activates HSP-27 and induces secretion of MMP-9 and MMP-13 (collagenase 3) that drive tumor invasion and metastasis.9,103 Lipopolysaccharide of F nucleatum is involved in inflammation and cytokine-mediated damaging lesions of the GECs through the activation of translocation of NF-kB gene into nucleus that drive production of inflammatory cytokines such as IL-1α, IL-1β, IL-6, IL-8, and MMPs. Fusobacterium nucleatum in GECs activates NLRP3 inflammasome that includes HMGB1 (high-morbidity group box-1 protein), apoptosis-associated speck-like protein, and caspase-1.37
FadA adhesion molecule of F nucleatum binds to E-cadherin and activates β-catenin signaling that regulates cell proliferation and inflammatory responses in oncogenesis (Figure 2).94,104,105 Fusobacterium nucleatum synergistically with other bacteria fungi and viruses may induce pro-inflammatory microenvironment through recruitment of tumor-infiltrating immune cells that favor tumorigenesis.106
Figure 2.
Role of Fusobacterium nucleatum in progression of oral squamous cell carcinoma (OSCC).
Antigens of P gingivalis have also been detected in gingival squamous carcinoma. Expression of the B7-H1 receptor on tongue carcinoma cells is induced by infection of P gingivalis. Porphyromonas gingivalis induces production of pro-MMP-9 through ERK1/2-Ets1, p38/HSP27, and PAR2/NFκB pathways. Porphyromonas gingivalis induces expression of pro-MMP-9 in oral mucosa, dendritic cells, and monocytes. It also activates PAR2 that activates NF-κB which leads overexpression of pro-MMP-9. Proenzyme pro-MMP-9 is activated by gingipains (arginine-X [Arg-gingipain A and B (RgpA and RgpB)]- and lysine-X [Lys-gingipain (Kgp)]-specific cysteine proteinases) of P gingivalis that drive invasion. Matrix metalloproteinase-9 degrades collagen IV from basement membranes and extracellular matrix that initiates progression of tumor.102,103
Numerous studies revealed that microbial infections were potential causative agents for inflammation in cancer. Porphyromonas gingivalis is considered as a potential risk factor for oral cancer.107 Infection of P gingivalis induces EMT by downregulation of E-cadherin and nucleocytoplasmic accumulation of β-catenin that drive aggressiveness and/or metastatic potential in OSCC. Porphyromonas gingivalis increases the level of PI3K/Akt that inactivates GSK3β which increases expression of Snail and Slug transcription factors. Snail, Slug, and β-catenin enhances expressions of Zeb1, Vimentin, and MMP-2, -7, and -9.108
Microbial organisms induce immune inflammation response in host cells that enhances mutation rate in normal cells and triggers malignant transformation of normal cells of host.109 The staining intensity of P gingivalis in gingival carcinoma tissues was significantly enhanced when compared to noninvasive bacteria such as S gordonii.110 Porphyromonas gingivalis is responsible for damaging local periodontal tissues and evade host immune system. Long-term exposure of P gingivalis induces cell proliferation, migration, and invasion through regulation of tumor-related genes such as FLI1, GAS6, PDCD1LG2, CD274, and lncRNACCAT1.111 Whether profiling of expression of these genes could be used as biomarkers remain to be investigated.
Porphyromonas gingivalis induces expression of cell surface molecules that activate the TLR2-TLR1 complex, and this induces secretion of enzymes such as (HRgpA and RgpB gingipains) which has effect on complement component C5 to generate high concentration of ligand C5a for C5a receptor 1 (C5aR1). Porphyromonas gingivalis induces C5aR1-TLR2 crosstalk signaling pathway in neutrophils and macrophages that separates a host-protective TLR2–MyD88 pathway from a TLR2-MyD88-adaptor-like (MAL; also known as TIRAP)–PI3 K pathway, which blocks phagocytosis and promotes inflammation. Porphyromonas gingivalis inhibits expression of IL-8 and T helper 1 cell-biasing chemokines (CXCL9, CXCL10, and CXCL11) even in the presence of F nucleatum. Porphyromonas gingivalis inhibits MyD88-dependent inflammation, but it induces PI3K-dependent inflammatory cytokines in both neutrophils and macrophages. Toll-like receptors on epithelial cell surfaces recognizes F nucleatum which induces pro-inflammatory signaling pathways. Porphyromonas gingivalis alters these pathways by reducing expression of CXCL10 through inactivation of STAT1 and IRF1 in epithelial cells, neutrophils, and monocytes. Porphyromonas gingivalis secretes serine phosphatase SerB in epithelial cells that activate NF-κB through dephosphorylation at serine 536 residue of the p65 subunit of NF-κB. Activated p65 subunit of NF-κB inhibits transcriptional activation of IL-8.20
Role of Oral microbiota in Chemotherapy and Radiation-Induced Mucositis
Mucositis in oral or OP region develops due to chemotherapy and radiation therapy in patients with head and neck cancer.112 Tissue inflammation and cell apoptosis are induced either by radiation or by chemotherapeutic agents. This event results in mucosal ulcerations that drive secondary bacterial colonization. Cell wall products of colonized bacteria activate macrophages to produce additional inflammatory cytokines. Four genera Fusobacterium, Porphyromonas, Treponema, and Prevotella showed dynamic synchronous variations in their relative abundances during treatment.113 Oral microbial community is altered in the patients with nasopharyngeal carcinoma (NPC) who develop mucositis due to radiotherapy (RT). Streptococcus mitis was increased significantly in patients with NPC after irradiation. Streptococcus mitis induces the breakdown of mucus in the human OC that drive translocation of pathogen into the lamina propria and recruitment of inflammatory cells.114
Radiotherapy is used for the treatment of head and neck cancer. Oral mucositis and xerostomia develop in patients with HNSCC receiving RT. Xerostomia is responsible for the alterations of biofilm composition in the supragingival environment. In irradiated tissues, reduction in blood supply, delays in repairing of wound, and reduction in immune response occur that drive proliferation of anaerobic and microaerophilic bacteria in the supra or subgingival biofilms. Streptococcus anginosus, S mitis, S mutans, S oralis, S sanguinis, and Streptococcus sobrinus are responsible for the development of oral mucositis. The abundance of Porphyromonas and Prevotella genera was decreased in the supragingival biofilm and increased in the gingival sulcus of gingivitis or chronic periodontitis patients. The presence of these microorganisms in the microbial biofilm is associated with poor hygiene and oral conditions as well as alcohol and tobacco consumption which induces adhesion and oral colonization by enteric microorganisms in irradiated patients. The most significant alterations in microbial populations was observed in members of the family Enterobacteriaceae. The fimbriae of these microorganisms may facilitate adhesion of microbiota to tumor cells that enhances the colonization of microbial organisms in irradiated patients with xerostomia. The members of the family Enterobacteriaceae enhances microbial proteolytic activity on fibronectin in the surface of oral soft tissues through adhesion receptors. This induces inflammation through the release of IL-1 and IL-6 in RT-induced mucositis.115
Concluding Remarks
Oral cancer is a multifactorial disease. Environmental factors such as tobacco smoking and chewing, betel quid chewing, alcohol consumption, and HPV are major risk factors for the development oral cancer. Despite the advancement in radiation and chemotherapy, the 5-year survival rate remains below 50% and patients frequently develop secondary and recurrent tumor. Recent research focused the role of bacteria in oral carcinogenesis. Several niches of OC have predominately occupied by bacteria. A 16 S rRNA metagenomics with NGS has contributed significantly to oral microbiome signatures that drive oral cancer. This technology needs to standardize in the context of DNA isolation, selection of primers specific for amplification of hypervariable region, and bioinformatics. The 16S rRNA microbial profiling does not provide information about the microbial gene expression. Advancement of omics technologies such as improvement in bioinformatics will be useful for the identification of oral microbiome and their genomes, proteome, and metabolome. These omics data may be useful in identifying HNSCC-related microbiome, their virulence properties, and their interaction with host immunity.
The role of L gasseri/L johnsonii, L vaginalis, and F nucleatum is essential for screening, diagnosis, and chemoprevention strategies in OPSCC and OSCC. Alteration in the oral commensal microbial communities have potential application as a diagnostic tool to predict OSCC. Both pathogenic and the commensal strains of bacteria have significant contribution in OSCC. Alteration in normal resident oral bacterial flora into nonresident pathogenic microbes in the presence of known etiological factors, such as smoking and alcohol consumption, contributes to oral carcinogenesis. Clinicians should be aware of the beneficial protective properties of the resident microflora to define treatment strategies. Avoiding alcohol and tobacco consumption reduce exposure to aldehyde in the oral mucosa. Identification of oral microbiota and its functional role by NGS will be useful for therapeutic application in OSCC. The carcinogenic potential of Porphyromonas gingivalis and F nucleatum in oral cancer has been well established in both in vitro and in vivo model. Porphyromonas gingivalis and F nucleatum can be used as biomarker for oral cancer. Virulence factors such as FimA of P gingivalis and FadA of F nucleatum may also serve as therapeutic target for the prevention of oral cancer. It is very essential to address which bacterial species is responsible for driving oral carcinogenesis. Identification of phages in the OC that infect the F nucleatum, S mutans, and N meningitidis would open the possibility to develop phage therapy against these oral pathogens.
Multitargeted therapeutic strategies may be required to block pathogenic biofilm formation and disrupt established biofilms. These strategies should also target acidic pH microenvironment in biofilm by alternating pH. Oral microbiomes such as S gordonii and Actinomyces subspecies are involved in the production of alkali through arginine metabolism. These species may modulate acidic pH within oral biofilms that inhibit the growth acidogenic–aciduric bacteria. Streptococcus dentisani or Streptococcus A12 have probiotic action either through preventing the growth of cariogenic species and alteration in acidic pH through arginolytic activities. Antimicrobial peptides that consist of novispirin-derived “killing” region conjugated with species-specific peptide pheromone are involved in targeting against pathogens involved in dental caries. Antimicrobial compounds that are involved in the degradation biofilm matrix, surface modification, and antibiofilm coatings may be used in the treatment of OSCC. Nanotechnology-based drug delivery system may be used in the penetration of cariogenic oral biofilms effectively. To develop highly precise and effective therapeutic approaches, identification of specific oral microbiomes may be required. This review serves to focus the active role of oral microbiome in biofilm formation and its role in oral cancer progression through alteration in host physiology.
Abbreviations
- ADH
alcohol dehydrogenase
- AI
autoinducer
- DAMP
damage/danger-associated molecular patterns
- EMT
epithelial to mesenchymal transition
- EPS
extracellular polymeric substances
- FA
Fanconi anemia
- FEP
fibroepithelial polyp
- GCF
gingival crevicular fluid
- GEC
gingival epithelial cell
- HNSCC
head and neck squamous cell carcinomas
- HPV
human papilloma virus
- IL
interleukin
- LPS
lipopolysaccharides
- MMP
matrix metalloproteinase
- NGS
next-generation sequencing
- NF-kB
nuclear factor kB
- OC
oral cavity
- OLP
oral lichen planus
- OP
oropharyngeal
- OPSCC
oropharyngeal squamous cell carcinoma
- OSCC
oral squamous cell carcinoma
- PD-L1
death protein 1 ligand 1
- PRRs
pattern recognition receptors
- STAT
signal transducer and activator of transcription 3
- TNF
tumor necrosis factor
- TNF-α
tumor necrosis factor α
- TLRs
toll-like receptors
- RT
radiotherapy.
Footnotes
Authors’ Note: Indranil Chattopadhyay and Mukesh Verma contributed equally.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Indranil Chattopadhyay, PhD  https://orcid.org/0000-0002-0191-2621
https://orcid.org/0000-0002-0191-2621
References
- 1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. [DOI] [PubMed] [Google Scholar]
- 3. Petersen PE. Oral cancer prevention and control—the approach of the World Health Organization. Oral Oncol. 2009;45(4-5):454–460. [DOI] [PubMed] [Google Scholar]
- 4. Gupta B, Johnson NW. Systematic review and meta-analysis of association of smokeless tobacco and of betel quid without tobacco with incidence of oral cancer in South Asia and the Pacific. PLoS One. 2014;9(11):e113385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shaikh MH, McMillan NA, Johnson NW. HPV-associated head and neck cancers in the Asia Pacific: a critical literature review and meta-analysis. Cancer Epidemiol. 2015;39(6):923–938. [DOI] [PubMed] [Google Scholar]
- 6. Multhoff G, Molls M, Radons J. Chronic inflammation in cancer development. Front Immunol. 2011;2:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lax AJ, Thomas W. How bacteria could cause cancer: one step at a time. Trends Microbiol. 2002;10(6):293–299. [DOI] [PubMed] [Google Scholar]
- 8. Sklenicka S, Gardiner S, Dierks EJ, Potter BE, Bell RB. Survival analysis and risk factors for recurrence in oral squamous cell carcinoma: does surgical salvage affect outcome? J Oral Maxillofac Surg. 2010;68(6):1270–1275. [DOI] [PubMed] [Google Scholar]
- 9. Perera M, Al-hebshi NN, Speicher DJ, Perera I, Johnson NW. Emerging role of bacteria in oral carcinogenesis: a review with special reference to perio-pathogenic bacteria. J Oral Microbiol. 2016; 8(1):32762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dewhirst FE, Chen T, Izard J. et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takahashi N. Microbial ecosystem in the oral cavity: metabolic diversity in an ecological niche and its relationship with oral diseases. Int Congr Ser. 2005;1284:103–112. [Google Scholar]
- 12. Lim Y, Totsika M, Morrison M, Punyadeera C. Oral microbiome: a new biomarker reservoir for oral and oropharyngeal cancers. Theranostics. 2017;7(17):4313–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van’t Hof W, Veerman EC, Nieuw Amerongen AV, Ligtenberg AJ. Antimicrobial defense systems in saliva. Monogr Oral Sci. 2014;24:40–51. [DOI] [PubMed] [Google Scholar]
- 15. Dawes C. Estimates, from salivary analyses, of the turnover time of the oral mucosal epithelium in humans and the number of bacteria in an edentulous mouth. Arch Oral Biol. 2003;48(5):329–336. [DOI] [PubMed] [Google Scholar]
- 16. Rosan B, Lamont RJ. Dental plaque formation. Microbes Infect. 2000;2(13):1599–1607. [DOI] [PubMed] [Google Scholar]
- 17. Avila M, Ojcius DM, Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol. 2009;28(8):405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mager D, Haffajee A, Devlin P, Norris C, Posner M, Goodson J. The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, nonrandomized study of cancer-free and oral squamous cell carcinoma subjects. J Transl Med. 2005;3(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Itzek A, Gillen CM, Fulde M. et al. Contribution of plasminogen activation towards the pathogenic potential of oral Streptococci. PLoS One. 2010;5(11):e13826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018;16(12):745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The human oral microbiome database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford). 2010;2010:baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. [DOI] [PubMed] [Google Scholar]
- 23. Jakubovics NS, Kolenbrander PE. The road to ruin: the formation of disease-associated oral biofilms. Oral Dis. 2010;16(8):729–739. [DOI] [PubMed] [Google Scholar]
- 24. Aruni AW, Dou Y, Mishra A, Fletcher HM. The biofilm community—Rebels with a cause. Curr Oral Health Rep. 2015;2(1):48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sintim HO, Gürsoy UK. Biofilms as “Connectors” for oral and systems medicine: a new opportunity for biomarkers, molecular targets, and bacterial eradication. OMICS. 2016;20(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frias J, Olle E, Alsina M. Periodontal pathogens produce quorum sensing signal molecules. Infect Immun. 2001;69(5):3431–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang K, Ou M, Wang W, Ling J. Effects of quorum sensing on cell viability in Streptococcus mutans biofilm formation. Biochem Biophys Res Commun. 2009;379(4):933–938. [DOI] [PubMed] [Google Scholar]
- 28. Baker JL, Bor B, Agnello M, Shi W, He X. Ecology of the oral microbiome: beyond bacteria. Trends Microbiol. 2017;25(5):362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rosier BT, Marsh PD, Mira A. Resilience of the oral microbiota in Health: mechanisms that prevent dysbiosis. J Dent Res. 2018;97(4):371–380. [DOI] [PubMed] [Google Scholar]
- 30. Doel JJ, Hector MP, Amirtham CV, Al-Anzan LA, Benjamin N, Allaker RP. Protective effect of salivary nitrate and microbial nitrate reductase activity against caries. Eur J Oral Sci. 2004;112(5):424–428. [DOI] [PubMed] [Google Scholar]
- 31. Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009; 9:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Verma M. Mechanistic and Technical Challenges in Studying the Human Microbiome and Cancer Epidemiology. Technol Cancer Res Treat. 2017;16(2):150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Acharya A, Chan Y, Kheur S. et al. Salivary microbiome of an urban Indian cohort and patterns linked to subclinical inflammation. Oral Dis. 2017;23(7):926–940. doi: 10.1111/odi.12676 [DOI] [PubMed] [Google Scholar]
- 34. Sarkar A, Stoneking M, Nandineni MR. Unraveling the human salivary microbiome diversity in Indian populations. PLoS One. 2017;12(9):e0184515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sivan A, Corrales L, Hubert N. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. López-López A, Camelo-Castillo AJ, Ferrer García MD, Simon-Soro A, Mira A. Health-associated niche inhabitants as oral probiotics: the case of Streptococcus dentisani. Front Microbiol. 2017;8:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gholizadeh P, Eslami H, Kafil HS. Carcinogenesis mechanisms of Fusobacterium nucleatum. Biomed Pharmacother. 2017;89:918–925. [DOI] [PubMed] [Google Scholar]
- 38. Huang S, Yang F, Zeng X. et al. Preliminary characterization of the oral microbiota of Chinese adults with and without gingivitis. BMC Oral Health. 2011;11:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y, Zhang J, Chen X. et al. Profiling of oral microbiota in early childhood caries using single-molecule real-time sequencing. Front Microbiol. 2017;8:2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou J, Jiang N, Wang S. et al. Exploration of human salivary microbiomes--insights into the novel characteristics of microbial community structure in caries and caries-free subjects. PLoS One. 2016;11(1):e0147039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tezal M, Sullivan MA, Hyland A. et al. Chronic periodontitis and the incidence of head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2009;18(9):2406–2412. [DOI] [PubMed] [Google Scholar]
- 42. Laprise C, Shahul HP, Madathil SA. et al. Periodontal diseases and risk of oral cancer in Southern India: results from the HeNCe life study. Int J Cancer. 2016;139(7):1512–1519. [DOI] [PubMed] [Google Scholar]
- 43. Nagy KN, Sonkodi I, Szöke I, Nagy E, Newman HN. The microflora associated with human oral carcinomas. Oral Oncol. 1998;34(4):304–308. [PubMed] [Google Scholar]
- 44. Yao QW, Zhou DS, Peng HJ, Ji P, Liu DS. Association of periodontal disease with oral cancer: a meta-analysis. Tumor Biol. 2014;35(7):7073–7077. [DOI] [PubMed] [Google Scholar]
- 45. Shi B, Chang M, Martin J. et al. Dynamic changes in the subgingival microbiome and their potential for diagnosis and prognosis of periodontitis. MBio. 2015;6(1):e01926–14. doi: 10.1128/mBio.01926-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang J, Qi J, Zhao H. et al. Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci Rep. 2013;3:1843 doi: 10.1038/srep01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 2014;35(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tezal M, Sullivan Nasca M, Stoler DL. et al. Chronic periodontitis-human papillomavirus synergy in base of tongue cancers. Arch Otolaryngol Head Neck Surg. 2009;135(4):391–396. [DOI] [PubMed] [Google Scholar]
- 49. Gao L, Xu T, Huang G, Jiang S, Gu Y, Chen F. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018;9(5):488–500. doi: 10.1007/s13238-018-0548 -1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang K, Lu W, Tu Q. et al. Preliminary analysis of salivary microbiome and their potential roles in oral lichen planus. Sci Rep. 2016; 6:22943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee WH, Chen HM, Yang SF. et al. Bacterial alterations in salivary microbiota and their association in oral cancer. Sci Rep. 2017;7(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Furquim CP, Soares GM, Ribeiro LL. et al. The salivary microbiome and oral cancer risk: a pilot study in Fanconi anemia. J Dent Res. 2017;96(3):292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mager DL. Bacteria and cancer: cause, coincidence or cure? A review. J Transl Med. 2006;4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guerrero-Preston R, Godoy-Vitorino F, Jedlicka A. et al. 16 S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment. Oncotarget. 2016;7(32):51320–51334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guerrero-Preston R, White JR, Godoy-Vitorino F. et al. High-resolution microbiome profiling uncovers Fusobacterium nucleatum, Lactobacillus gasseri/johnsonii, and Lactobacillus vaginalis associated to oral and oropharyngeal cancer in saliva from HPV positive and HPV negative patients treated with surgery and chemo-radiation. Oncotarget. 2017;8(67):110931–110948. doi: 10.18632/oncotarget.20677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao H, Chu M, Huang Z. et al. Variations in oral microbiota associated with oral cancer. Sci Rep. 2017;7(1):11773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mukherjee PK, Wang H, Retuerto M. et al. Bacteriome and mycobiome associations in oral tongue cancer. Oncotarget. 2017;8(57):97273–97289. doi: 10.18632/oncotarget.21921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schmidt BL, Kuczynski J, Bhattacharya A. et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS One. 2014;9:e98741 doi: 10.1371/journal.pone. 0098741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Perera M, Al-Hebshi NN, Perera I. et al. Inflammatory bacteriome and oral squamous cell carcinoma. J Dent Res. 2018;97(6):725–732. doi: 10.1177/0022034518767118 [DOI] [PubMed] [Google Scholar]
- 60. Wang H, Funchain P, Bebek G. et al. Microbiomic differences in tumor and paired-normal tissue in head and neck squamous cell carcinomas. Genome Med. 2017;9(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hernandez BY, Zhu X, Goodman MT. et al. Betel nut chewing, oral premalignant lesions, and the oral microbiome. PLoS One. 2017;12(2):e0172196 doi: 10.1371/journal.pone.0172196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang CY, Yeh YM, Yu HY. et al. Oral microbiota community dynamics associated with oral squamous cell carcinoma staging. Front Microbiol. 2018;9:862 doi: 10.3389/fmicb.2018.00862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Karpiński TM. Role of Oral microbiota in cancer development. Microorganisms. 2019;7(1):pii E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Börnigen D, Ren B, Pickard R. et al. Alterations in oral bacterial communities are associated with risk factors for oral and oropharyngeal cancer. Sci Rep. 2017;7(1):17686 doi:10.1038/s41598-017-17795-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Al-Hebshi NN, Nasher AT, Maryoud MY. et al. Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci Rep. 2017;7(1):1834 doi: 10.1038/s41598-017-02079-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lim Y, Fukuma N, Totsika M, Kenny L, Morrison M, Punyadeera C. The performance of an oral microbiome biomarker panel in predicting oral cavity and oropharyngeal cancers. Front Cell Infect Microbiol. 2018;8:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hayes RB, Ahn J, Fan X. et al. Association of oral microbiome with risk for incident head and neck squamous cell cancer. JAMA Oncol. 2018;4(3):358–365. doi: 10.1001/jamaoncol.2017.4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Al-Hebshi NN, Alharbi FA, Mahri M, Chen T. Differences in the bacteriome of smokeless tobacco products with different oral carcinogenicity: compositional and predicted functional analysis. Genes. 2017. a;8:106 doi: 10.3390/genes8040106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hooper SJ, Crean SJ, Lewis MAO, Spratt DA, Wade WG, Wilson MJ. Viable bacteria present within oral squamous cell carcinoma tissue. J Clin Microbiol. 2006;44(5):1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hooper SJ, Crean SJ, Fardy MJ. et al. A molecular analysis of the bacteria present within oral squamous cell carcinoma. J Med Microbiol. 2007;56(pt 12):1651–1659. [DOI] [PubMed] [Google Scholar]
- 71. Pushalkar S, Mane SP, Ji X. et al. Microbial diversity in saliva of oral squamous cell carcinoma. FEMS Immunol Med Microbiol. 2011; 61(3):269–277. doi: 10.1111/j.1574-695X.2010.00773.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Meurman JH. Oral microbiota and cancer. J Oral Microbiol. 2010;2 doi: 10.3402/jom.v2i0.5195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kang M, Oh J, Kim H. et al. Prevalence of oral microbes in the saliva of oncological patients. J Bacteriol Virol. 2009; 39(4):277–285. [Google Scholar]
- 74. Hiranmayi KV, Sirisha K, Ramoji Rao MV, Sudhakar P. Novel pathogens in periodontal microbiology. J Pharm Bioallied Sci. 2017;9(3):155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ganly I, Yang L, Giese RA. et al. Periodontal pathogens are a risk factor of oral cavity squamous cell carcinoma, independent of tobacco and alcohol and human papillomavirus. Int J Cancer. 2019;145(3):775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Banerjee S, Tian T, Wei Z. et al. Microbial signatures associated with oropharyngeal and oral squamous cell carcinomas. Sci Rep. 2017;7(1):4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gaonkar PP, Patankar SR, Tripathi N, Sridharan G. Oral bacterial flora and oral cancer: The possible link? J Oral Maxillofac Pathol. 2018;22(2):234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wu J, Peters BA, Dominianni C. et al. Cigarette smoking and the oral microbiome in a large study of American adults. ISME journal. 2016;10(10):2435–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Charlson ES, Chen J, Custers-Allen R. et al. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One. 2010;5(12):e15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kurkivuori J, Salaspuro V, Kaihovaara P. et al. Acetaldehyde production from ethanol by oral Streptococci. Oral Oncol. 2007;43(2):181–186. [DOI] [PubMed] [Google Scholar]
- 81. Muto M, Hitomi Y, Ohtsu A. et al. Acetaldehyde production by non-pathogenic Neisseria in human oral microflora: implications for carcinogenesis in upper aerodigestive tract. Int J Cancer. 2000;88(3):342–350. [PubMed] [Google Scholar]
- 82. Sasaki M, Yamaura C, Ohara-Nemoto Y. et al. Streptococcus anginosus infection in oral cancer and its infection route. Oral Dis. 2005;11(3):151–156. [DOI] [PubMed] [Google Scholar]
- 83. Kumar PS, Matthews CR, Joshi V, de Jager M, Aspiras M. Tobacco smoking affects bacterial acquisition and colonization in oral biofilms. Infect Immun. 2011;79(11):4730–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bagaitkar J, Demuth DR, Scott DA. Tobacco use increases susceptibility to bacterial infection. Tob Induc Dis. 2008;4(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hsiao JR, Chang CC, Lee WT. et al. The interplay between oral microbiome, lifestyle factors and genetic polymorphisms in the risk of oral squamous cell carcinoma. Carcinogenesis. 2018;39(6):778–787. [DOI] [PubMed] [Google Scholar]
- 86. Vesty A, Gear K, Biswas K, Radcliff FJ, Taylor MW, Douglas RG. Microbial and inflammatory-based salivary biomarkers of head and neck squamous cell carcinoma. Clin Exp Dent Res. 2018;4(6):255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Singleton DR, Adrion AC, Aitken MD. Surfactant-induced bacterial community changes correlated with increased polycyclic aromatic hydrocarbon degradation in contaminated soil. Appl Microbiol Biotechnol. 2016;100(23):10165–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Vogelmann R, Amieva MR. The role of bacterial pathogens in cancer. Curr Opin Microbiol. 2007;10(1):76–81. [DOI] [PubMed] [Google Scholar]
- 89. Zhang G, Ghosh S. Molecular mechanisms of NF-kappaB activation induced by bacterial lipopolysaccharide through Toll-like receptors. J Endotoxin Res. 2000;6(6):453–457. [DOI] [PubMed] [Google Scholar]
- 90. Gholizadeh P, Eslami H, Yousefi M, Asgharzadeh M, Aghazadeh M, Kafil HS. Role of oral microbiome on oral cancers, a review. Biomed Pharmacother. 2016;84:552–558. doi: 10.1016/j.biopha.2016.09.082 [DOI] [PubMed] [Google Scholar]
- 91. Lax AJ. Bacterial toxins and cancer – a case to answer? Nat Rev Microbiol. 2005;3(4):343–349. [DOI] [PubMed] [Google Scholar]
- 92. Yang SF, Huang HD, Fan WL. et al. Compositional and functional variations of oral microbiota associated with the mutational changes in oral cancer. Oral Oncol. 2018;77:1–8.doi: 10.1016/j.oraloncology.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 93. Joshi V, Matthews C, Aspiras M, de Jager M, Ward M, Kumar P. Smoking decreases structural and functional resilience in the subgingival ecosystem. J Clin Periodontol. 2014;41(11):1037–1047. [DOI] [PubMed] [Google Scholar]
- 94. Pan C, Xu X, Tan L, Lin L, Pan Y. The effects of Porphyromonas gingivalis on the cell cycle progression of human gingival epithelial cells. Oral Dis. 2014;20(1):100–108. [DOI] [PubMed] [Google Scholar]
- 95. Mao S, Park Y, Hasegawa Y. et al. Lamont, Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007;9(8):1997–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yao L, Jermanus C, Barbetta B. et al. Porphyromonas gingivalis infection sequesters pro-apoptotic Bad through Akt in primary gingival epithelial cells. Mol Oral Microbiol. 2010;25(2):89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yilmaz Ö, Yao L, Maeda K. et al. ATP scavenging by the intracellular pathogen Porphyromonas gingivalis inhibits P2X7-mediated host-cell apoptosis. Cell Microbiol. 2008;10(4):863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Aymeric L, Apetoh L, Ghiringhelli F. et al. Tumor cell death and ATP release prime dendritic cells and efficient anticancer immunity. Cancer Res. 2010;70(3):855–858. [DOI] [PubMed] [Google Scholar]
- 99. Almeida-da-Silva CLC, Morandini AC, Ulrich H, Ojcius DM, Coutinho-Silva R. Purinergic signaling during Porphyromonas gingivalis infection. Biomed J. 2016;39(4):251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kuboniwa M, Hasegawa Y, Mao S. et al. Gingivalis accelerates gingival epithelial cell progression through the cell cycle. Microbes Infect. 2008;10(2):122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Groeger S, Domann E, Gonzales JR, Chakraborty T, Meyle J. B7-H1 and B7-DC receptors of oral squamous carcinoma cells are upregulated by Porphyromonas gingivalis. Immunobiology. 2011;216(12):1302–1310. [DOI] [PubMed] [Google Scholar]
- 102. Inaba H, Sugita H, Kuboniwa M. et al. Amano. Porphyromonas gingivalis promotes invasion of oral squamous cell carcinoma through induction of proMMP9 and its activation. Cell Microbiol. 2014;16(1):131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Fischman S, Revach B, Bulvik R. et al. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget. 2015;6(26):22613–22623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17(3):156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bashir A, Miskeen AY, Bhat A, Fazili KM, Ganai BA. Fusobacterium nucleatum: an emerging bug in colorectal tumorigenesis. Eur J Cancer Prev. 2015;24(5):373–385. [DOI] [PubMed] [Google Scholar]
- 107. Yuan X, Liu Y, Kong J. et al. Different frequencies of Porphyromonas gingivalis infection in cancers of the upper digestive tract. Cancer Lett. 2017;404:1–7. doi: 10.1016/j.canlet.2017.07.003 [DOI] [PubMed] [Google Scholar]
- 108. Lee J, Roberts JS, Atanasova KR, Chowdhury N, Han K, Yilmaz Ö. Human primary epithelial cells acquire an epithelial-mesenchymal-transition phenotype during long-term infection by the oral opportunistic pathogen, Porphyromonas gingivalis. Front Cell Infect Microbiol. 2017;7:493 doi: 10.3389/fcimb.2017.00493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Tribble GD, Kerr JE, Wang BY. Genetic diversity in the oral pathogen Porphyromonas gingivalis: molecular mechanisms and biological consequences. Future Microbiol. 2013;8:607–620. doi: 10.2217/fmb.13.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Katz J, Onate MD, Pauley KM, Bhattacharyya I, Cha S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int J Oral Sci. 2011;3:209–215. doi: 10.4248/IJOS11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Geng F, Liu J, Guo Y. et al. Persistent exposure to Porphyromonas gingivalis promotes proliferative and invasion capabilities, and tumorigenic properties of human immortalized oral epithelial cells. Front Cell Infect Microbiol. 2017;7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Scarantino C, Leveque F, Swann RS. et al. Effect of pilocarpine during radiation therapy: Results of RTOG 97-09, a phase III randomized study in head and neck cancer patients. J Support Oncol. 2006;4(5):252–258. [PubMed] [Google Scholar]
- 113. Hou J, Zheng H, Li P, Liu H, Zhou H, Yang X. Distinct shifts in the oral microbiota are associated with the progression and aggravation of mucositis during radiotherapy. Radiother Oncol. 2018;129(1):44–51. doi: 10.1016/j.radonc.2018.04.023 [DOI] [PubMed] [Google Scholar]
- 114. Zhu XX, Yang XJ, Chao YL. et al. The potential effect of oral microbiota in the prediction of mucositis during radiotherapy for nasopharyngeal carcinoma. EBioMed. 2017;18:23–31. doi: 10.1016/j.ebiom.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Gaetti-Jardim E, Jr, Jardim ECG, Schweitzer CM. et al. Supragingival and subgingival microbiota from patients with poor oral hygiene submitted to radiotherapy for head and neck cancer treatment. Arch Oral Biol. 2018;90:45–52. [DOI] [PubMed] [Google Scholar]