Abstract
Background:
Clinical treatment of non-small cell lung carcinoma (NSCLC) by cisplatin eventually results in drug resistance, which cancer stem cells and autophagy are believed to be involved in. In the present study, we aimed to explore the effect of autophagy-inhibited cancer stem cells in NSCLC.
Methods:
Cancer stem cells were identified by CD133 expression levels detected by immunochemistry, real-time polymerase chain reaction, western blot, and flow cytometry. Stemness was detected by sphere-forming assays of tumor cells. Autophagy was determined by LC3-II expression at mRNA and protein levels. The effect of chloroquine (CQ) on autophagy was detected by real-time polymerase chain reaction, western blot and sphere-forming assay in vitro, and tumor growth in male NOD/SCID mice.
Results:
Cisplatin (CDDP) treatment enhanced CD133+ cell ratios in clinical NSCLC specimens and NSCLC cell line A549. The CD133+ cells enriched by CDDP exhibited higher autophagy levels. Autophagy inhibition by CQ inhibited CD133+ stemness and promoted CDDP efficiency in A549 cells. In addition, the combination of CDDP and CQ treatment significantly inhibited autophagy levels and cancer stem cell proportions in vitro, and dramatically suppressed tumor growth compared with individual agents.
Conclusion:
Autophagy inhibition of cancer stem cells could promote the efficacy of cisplatin against NSCLC.
Keywords: autophagy, cancer stem cell, cisplatin, non-small cell lung carcinoma
Introduction
Lung cancer remains the most common cancer worldwide, and is the most common cause of cancer-related death.1,2 Patients with lung cancer display poor prognosis with less than 20% of 5-year overall survival.3 More than 50% of all lung cancers are non-small cell lung carcinoma (NSCLC), which accounts for the most common subtype.4,5 In the clinic, strategies for NSCLC treatment range from traditional surgery, radiation, chemotherapy to novel targeted therapy; however, the effect of various treatments is limited. More importantly, approximately 40% of patients with NSCLC are at a late stage on diagnosis.3 Platinum-based chemotherapy was mostly accepted for advanced NSCLC treatment,6,7 but the efficacy of chemotherapy is suppressed by increased drug resistance. Therefore, exploration of the mechanism of chemotherapy resistance and consequently increasing the efficiency of platinum-based chemotherapy to patients with NSCLC are urgently needed.
Accumulating evidence illustrated that cancer stem cells represent a crucial factor in platinum-based chemotherapy, owing to the properties of self-renewal, differentiation, high tumorigenicity and drug resistance.8–12 Chemotherapy successfully decreases tumor burden via the killing of fast-proliferating tumor cells; however, it remains ineffective at eliminating cancer stem cells. More importantly, chemotherapy could even lead to an enrichment of cancer stem cells.13–15
Autophagy is defined as the process of intracellular degradation of cytoplasmic materials in the lysosome and is the dynamic recycling system that provides the energy and necessary materials for cellular regeneration.16 The correlation between autophagy, tumorigenesis and drug resistance has been widely investigated. Autophagy supplies metabolic substrates essential for cancer cell survival that support tumor growth.17 Moreover, elevated autophagy levels enhance the drug resistance of cancer cells.18,19 In the present study, we aimed to explore the effect of autophagy inhibition of cancer stem cells in the cisplatin (CDDP)-based drug resistance of NSCLC. Cisplatin treatment enriched CD133+ cancer stem cells with a high autophagy level. Autophagy inhibition by chloroquine (CQ) dramatically suppressed the proportion and stemness of cancer stem cells, and increased the sensitivity of tumor cells to CDDP treatment. In mouse models, autophagy inhibition repressed tumor growth by decreasing the percentage of cancer stem cells.
Methods
Human samples and cell line
A total of 10 human NSCLC samples (5 before cisplatin treatment and 5 after cisplatin treatment) were collected in Liaocheng Cancer Hospital, China, between 2015 and 2017. The 10 tissues were collected by endobronchial biopsy, and further confirmed as NSCLC by a histologist. For CD133 immunochemistry, the tissues were fixed in formalin, paraffin-embedded and sectioned in 5 μm thickness. Written consents regarding the tumor samples used for this study were obtained before the experiments. The study was conducted according to the World Medical Association Declaration of Helsinki, and was approved by the ethics committees of Liaocheng Cancer Hospital (#2015LCHJW038). The human NSCLC cell line A549 was purchased from the Cell Bank of Shanghai, China. A549 cells were cultured in Roswell Park Memorial Institute (RPMI) medium 1640 (Gibco, Grand Island, NY) supplied with 10% fetal bovine serum (FBS; Gibco) in a humidified incubator with 5% (v/v) CO2.
Real-time polymerase chain reaction
RNAs from tumor tissues and cell lines were isolated using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Then, cDNA was synthesized using Prime-Script RT Kit (Takara, Dalian, China). The mRNA expression levels of CD133, Sox2, Oct4, Nanog and ABCG2 was determined by real-time polymerase chain reaction. The primers used were as follows: CD133: F 5′ GCC ACC GCT CTA GAT ACT GC3′, 5′GCT TTT CCT ATG CCA AAC CA3′; Sox2: F 5′CAT GTC CCA GCA CTA CCA GA3′, R 5′ ATG TGT GAG AGG GGC AGT GT3′; Sox4, F 5′AGT GAG AGG CAA CCT GGA GA3′, R 5′ ACA CTC GGA CCA CAT CCT TC3′; Nanog, F 5′ AAC TGG CCG AAG AAT AGC AA3′, R 5′ CAT CCC TGG TGG TAG GAA GA3′; ABCG2, F 5′ATG GAT TTA CGG CTT TGC AG3′, R 5′ TGA GTC CTG GGC AGA AGT TT3′. The relative mRNA levels were detected by the 2−ΔΔCt method. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA expression level was used for normalization.
Western blot
Tissues or cells were lysed by radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher, Waltham, MA) supplied with protease inhibitor cocktail (Abcam). The lysates were quantified by a bicinchoninic acid (BCA) protein assay kit (Thermo Fisher). Then, a total of 10 µg proteins were loaded into 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), followed by transferring onto an immobilon transfer membrane (Millipore, Billerica, MA). The membranes were incubated with primary antibodies of CD133 (Cell Signaling Technology, Danvers, MA, 5860s, 1:1,000), LC3 (Novus Biologicals, Shanghai, China, NB100–2220, 1:2,000), p62 (Abcam, ab194721, 1:1000), b-actin (Novus Biologicals, NB600-501, 1:1,000) and GAPDH (Chemicon, Billerica, MA, AB9132, 1:10,000) at 4°C overnight. Then, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling Technology). Signals were detected by a chemiluminescence reaction. The signal intensities were quantified using the software, ImageJ (version 1.47, NIH, Bethesda, MD).
MTT assay
Cell viability was detected by MTT assay. A total of 5,000 cells were seeded into 96-well plates, following culture for 24 h. Then, different doses of CDDP (0, 20, 40, 60, 80, 100 µM) were added into wells and incubated for 48 h. Finally, cell viability was detected using an MTT kit (Thermo Fisher Scientific, V13154, Waltham, MA, USA) according to the manufacturer’s protocol. Briefly, 3–5 × 104 cells/well were cultured in 96-well plates for 48 h. At the end of culture, 50 μl of the MTT reagent (5 mg/ml) was added to each well and incubated for 4 h, followed by adding 200 μl of dimethyl sulfoxide to each well and incubating for 15 mins. The absorbance was read using a microplate reader (SpectraMAX Plus, Molecular Devices, Sunnyvale, CA, USA) at a wavelength of 570 nm.
Immunohistochemistry
Paraffin sections in 5 μm thickness were deparaffinated and rehydrated. Then, 3% H2O2 in methanol was used to quench endogenous peroxidase activity at room temperature for 30 mins, followed by incubation with 10% bovine serum albumin at room temperature for 60 mins. The sections were then incubated with primary antibodies of CD133 (Cell Signaling Technology, 5860s, 1:1,000) or PCNA (Cell Signaling Technology, 13110S, 1:1,000) at 4°C overnight. The positive immune signals were visualized by an envision and peroxidase system. Then, DAB-chromogen (Biocare Medical, SKU: DB801) was carried out according to the manufacturer’s procedures.
Immunofluorescence
For tissues: tissues were fixed by 4% paraformaldehyde (Sigma, St. Louis, MO, 30525-89-4), followed by embedding in paraffin. The paraffin sections in 5 μm thickness were deparaffinized and rehydrated, followed by incubation with Alexa Fluor® 647-conjunted p62 primary antibody (Abcam, ab194721, 1:1,000). DAPI (4′,6-diamidino-2-phenylindole) was used for nuclear staining. The signals were detected using confocal microscopy.
For GFP-LC3-A549 cells: GFP-LC3-A549 cells were transfected with GFP-LC3pcDNA3.1/hygro (+) plasmid (Invitrogen, Waltham, MA, USA) using lipofectamine 2000 (Thermo Fisher Scientific, 11668019) following the manufacturer’s procedures. Then, the cells were fixed in sections and the signals were detected using confocal microscopy.
Colony-forming assay and sphere-forming assay
For colony-forming assays, a total of 500 cells were seeded onto six-well plate and cultured in RPMI 1640 medium (Gibco, Grand Island, NY) supplied with 10% FBS (Gibco) for 10 days. Then, colony formation was visualized by purple crystal staining. For the sphere-forming assay, a total of 100 cells were seeded onto a low adhesion six-well plate containing Dulbecco’s modified Eagle’s medium (DMEM)/F-12 with N2 supplement (Invitrogen, Grand Island, NY), 20 ng/ml epidermal growth factor (EGF), and 20 ng/ml basic fibroblast growth factor (PeproTech, Rocky Hill, CT) and referred to stem cell medium for 10 days. Then, sphere formation was detected and photographed using microscopy.
Flow cytometry
Single cell suspensions of A549 cells or those dissociated from tumor tissues were used for flow cytometry analysis. A total of 1 × 106 cells from each sample were stained with CD133-PE (phycoerythrin) antibody (BD Biosciences, Franklin Lakes, NJ, 566593, clone W6B3C1) at a dose of 1 µl/200 µl phosphate-buffered saline (PBS) for 15 mins at 4°C. Then, cells were washed with cold PBS and flow cytometric analysis was performed using FACS Calibur (BD Biosciences).
In vivo xenograft model
A total of 3 × 106 A549 cells were subcutaneously injected into the right flanks of male NOD/SCID mice aged 6–8 weeks and weighing 17.8–24.1 g. Mice were purchased from Beijing Vital River Company (Beijing, China). A total of four mice were housed per cage with free access to food and water at 22–25°C with humidity of 50 ± 2%. A total of 168 mice were randomly divided into four groups: PBS, CDDP, CQ and CDDP + CQ, and were treated every 3 days five times when the tumor volume was 50 mm3. The administration dosage of CDDP and CQ were 2.5 mg/kg intravenously (i.v.) and 10.0 mg/kg (i.v.), respectively. The tumor volumes of different groups were measured every 3 days after implantation using the formula: length × width2 × 0.5. At 21 days after implantation, the mice were euthanized by carbon monoxide and the tumors weighed. The protocols for animal experiments were in accordance with the Guide for the Care and Use of Laboratory Animals published by National Institutes of Health. Animal study was with the oversight of the animal facility in Liaocheng People’s Hospital, and was reviewed and approved by the Ethics Committee of Liaocheng People’s Hospital (#LPH-5P36).
Statistical analysis
All data were detected as the mean ± standard deviation (SD) from at least three separate experiments. The levels of significance were: *p < 0.05, **p < 0.005 and ***p < 0.001. A Student’s t-test or analysis of variance was used to determine statistical significance. In all cases, p < 0.05 was considered significant.
Results
CDDP treatment enhanced CD133+ cancer stem cells
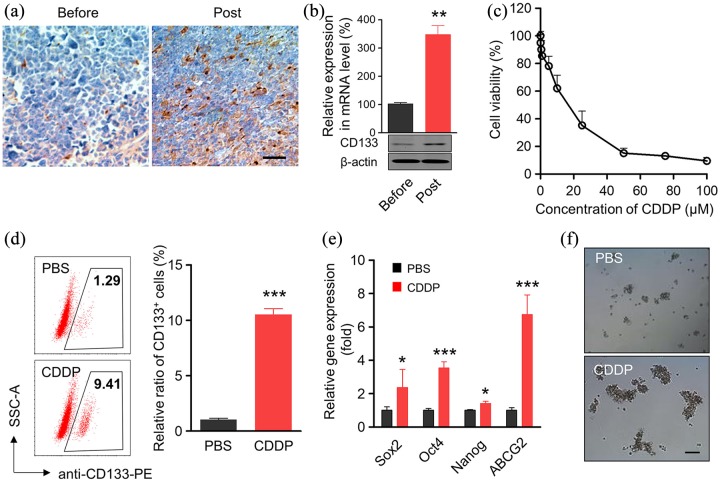
Cancer stem cells were believed to be closely involved in drug resistance,20 which promoted us to determine cancer stem cell alteration after CDDP treatment. CD133 was commonly used as a cancer stem cell marker.21 As shown in Figure 1(a), enhanced CD133 staining by immunochemistry was observed in clinical NSCLC samples after CDDP treatment. The percentage of CD133+ cells in the high-power field dramatically increased after CDDP treatment (Figure S1). Consistently, CD133 mRNA and protein levels were dramatically upregulated in CDDP-treated clinical NSCLC samples [Figure 1(b)].
Figure 1.
CDDP treatment elevates the ratio of CD133+ cells within specimens and cell line of NSCLC. (a) Representative images showed the immunohistochemistry of CD133 staining in the clinical specimens before and post CDDP treatment from patient with NSCLC. Scale bar, 20 μm. (b) mRNA and protein level of CD133 in the clinical specimens before and post CDDP treatment from patients with NSCLC. Data represent means ± SD. **p < 0.005, n = 6. (c) A549 cells were treated with different concentrations of CDDP for 48 h. The cell viability was determined by an MTT assay. (d) Percentages of CD133+ CSCs in A549 cells with or without the treatment of low dose of cisplatin for 48 h, respectively. The CDDP concentration was IC20, about 2.5 μM. CSCs were stained with anti-CD133-PE antibody and analyzed by flow cytometer. Data are shown as mean ± SD (n = 6). (e) mRNA levels of ‘stemness’-associated genes in A549 cells after CDDP treatment. A549 cells were treated with 2.5 μM CDDP for 48 h. The mRNA levels of ‘stemness’-associated genes were analyzed by qPCR, the mRNA levels of genes were normalized against the expression level of housekeeping gene GAPDH, *p < 0.05, **p < 0.005, ***p < 0.001, n = 6. (f) CDDP-treated A549 cells were subjected to the tumor sphere-forming assay. Scale bar, 100 μm.
CDDP, cisplatin; CSC, cancer stem cell; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; NSCLC, non-small cell lung cancer; qPCR, quantitative polymerase chain reaction; SD, standard deviation.
The effect of CDDP in NSCLC cell line A549 was also assessed. CDDP inhibited cell viability of A549 in a dose-dependent manner [Figure 1(c)]. Consistent with the clinical results, CDDP treatment greatly enhanced the CD133+ cancer stem cell proportion in the A549 cell line [Figure 1(d)]. To further confirm the effect of CDDP in cancer stem cells, the expression levels of stemness-associated genes were determined by RT-PCR. As shown in Figure 1(e), CDDP treatment significantly enhanced the selected stemness-associated gene (Sox2, Oct4, Nanog and ABCG2) expression levels. In addition, the tumor sphere formation ability of A549 cells was greatly improved after CDDP treatment [Figure 1(f)]. These results demonstrated that CDDP treatment enhanced CD133+ cancer stem cells.
CD133+ cancer stem cells exhibit higher autophagy levels
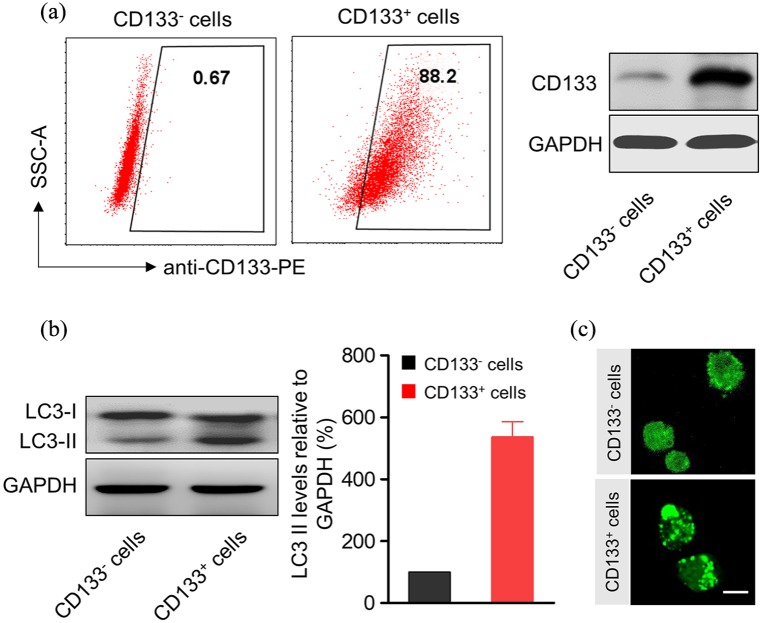
Accumulating reports indicated the correlation of autophagy with drug resistance.19 Therefore, we inspected autophagy levels of CDDP-enriched CD133+ cancer stem cells. Enrichment of CD133+ cells was first confirmed by flow cytometry and western blot [Figure 2(a)]. Comparing with CD133– cells, CD133+ exhibited much higher LC3-II protein levels [Figure 2(b)], which is commonly used as an autophagy marker.22 Enhanced autophagy levels in CD133+ were further verified by fluorescence of EGFP-LC3 in A549 cells [Figure 2(c)]. These results indicated that CDDP-enriched CD133+ cancer stem cells exhibit higher autophagy levels.
Figure 2.
CDDP-enriched CD133+ cancer stem cells exhibit enhanced levels of autophagy. (a) CD133 and CD133+ cells were sorted from A549 cells were treated with 2.5 μM CDDP for 48 h. The purities of CD133– and CD133+ cells were evaluated by flow cytometry and western blotting. (b) Western blotting and densitometric analysis of LC3-II levels in CD133 and CD133+ A549 cells. GAPDH was used as a loading control. The signal intensities were quantified using software ImageJ (version 1.47) (n = 6). (c) Fluorescent microscopy images of CD133 and CD133+ EGFP-LC3/A549 cells. Bright punctate dots indicated the induction of autophagy. Scale bar, 10 µm.
CDDP, cisplatin; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Autophagy inhibition suppressed stemness of CD133+ cells and enhanced CDDP efficiency in A549 cells
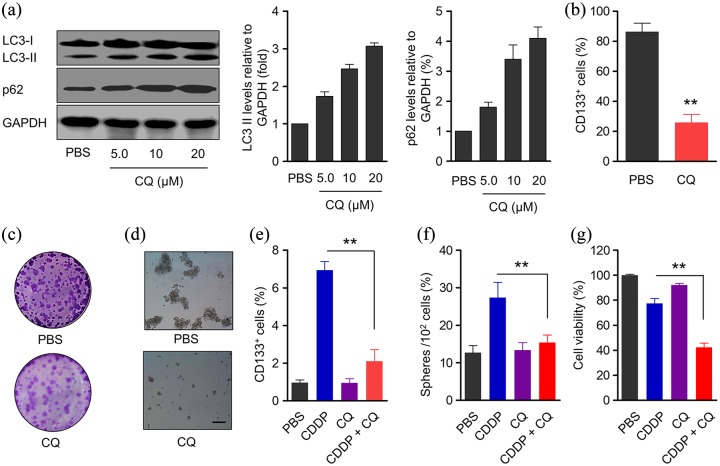
Results shown in Figure 2 demonstrated higher autophagy levels in CD133+ cancer stem cells, which caused us to explore the influence of the autophagy inhibitor chloroquine (CQ) on the biological functions of CD133+ cancer stem cells. The effect of CQ in inhibiting autophagy was affirmed by western blot, as CQ administration elevated LC3-II and p62 expression levels in a dose-dependent manner [Figure 3(a)]. Meanwhile, the proportion of CD133+ cells was dramatically suppressed after CQ treatment [Figure 3(b)]. CQ administration significantly inhibited CD133+ cell viability of CD133− and CD133+ cells [Figure S2(a and b)]. In addition, colony formation was much less in CQ-treated CD133+ cells [Figure 3(c)]. Decreased stemness of CD133+ cancer stem cells indicated by sphere-forming assay was observed after CQ treatment [Figure 3(d)].
Figure 3.
CQ-mediated inhibition of autophagy reduces the ‘stemness’ of CD133+ A549 cells and promotes efficiency of CDDP in A549 cells. (a) Protein expression of LC3 and p62 in CD133+ A549 cells after incubation with different concentration of CQ for 48 h and densitometric analysis, GAPDH was used as a loading control. The signal intensities were quantified using software ImageJ (version 1.47) (n = 6). (b) The proportion of CD133+ A549 cells in sorted cells after treatment with CQ (10 μM) for 48 h. Data represent means ± SD. **p < 0.005, n = 6. (c) CQ-treated sorted CD133+ A549 cells were subjected to the colony-forming assay. (d) CQ-treated sorted CD133+ A549 cells were subjected to sphere-forming assay. Scale bar, 100 μm. (e) Percentage of CD133+ cells in A549 cells after co-incubation with CDDP and CQ for 48 h, the concentrations of CDDP and CQ were 2.5 μM and 10 μM, respectively. Data represent means ± SD. **p < 0.005, n = 6. (f) The ability of A549 cells to regenerate spheres. A549 cells were treated with CDDP and CQ for 48 h, and then cells were seeded onto a six-well ultra-low attachment plate with complete spheres medium (100 cells per well). After 10 days, the number of newly formed spheres per well was determined under the microscope. Data represent means ± SD. **p < 0.005, (CDDP + CQ versus CDDP), n = 6. (g) MTT analyses of the viability of A549 cell after treatment with CDDP, CQ or a combination for 48 h. Cell viability was normalized to that of PBS-treated cells which served as the indicator of 100% cell viability. Data represent means ± SD. **p < 0.005 (CDDP + CQ versus CDDP), n = 6. The concentrations of CDDP and CQ were 2.5 μM and 10 μM, respectively.
CDDP, cisplatin; CQ, chloroquine; PBS, phosphate-buffered saline; SD, standard deviation.
The effect of autophagy inhibition on CDDP efficiency in A549 cells was further evaluated. Combined administration of CDDP and CQ greatly enhanced the ability of CDDP to decrease CD133+ cancer stem cells [Figure 3(e)], suppress sphere formation [Figure 3(f)] and cell viability [Figure 3(g)] of A549 cells, suggested that CQ administration improved the efficiency of CDDP in A549 cells.
Autophagy inhibition elevated CDDP efficiency in tumor growth
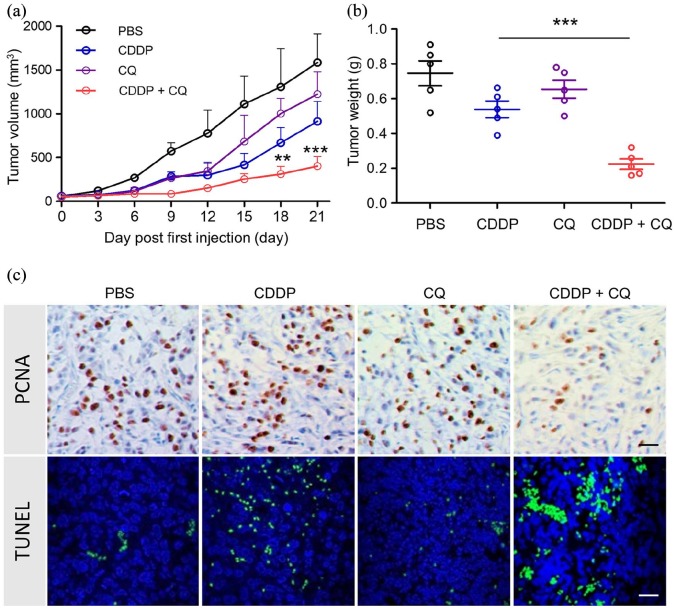
Next, we explored the effect of CQ-mediated autophagy inhibition in tumor growth after CDDP treatment. Subcutaneous A549 tumors were first established. Individual CDDP or CQ treatment displayed a trend of inhibiting tumor growth, but without significance. The combination of CDDP and CQ significantly suppressed tumor growth [Figure 4(a)]. In addition, the tumor weight in the CDDP + CQ group was much lower than the CDDP group [Figure 4(b)].
Figure 4.
In vivo anti-tumor effect of CDDP, CQ and a combination of two agents. (a) Inhibition of tumor growth by various treatments in A549 tumor-bearing NOD/SCID mice (n = 6). Male NOD/SCID mice bearing A549 tumors of ~50 mm3 received PBS, CDDP, CQ or combination of CDDP and CQ every 3 days five times, the administration dosage of CDDP and CQ were 2.5 mg/kg (i.v.) and 10.0 mg/kg (i.v.), respectively. **p < 0.005, ***p < 0.001 (versus CDDP group). (b) Tumor weights of different groups at the end of treatments. ***p < 0.001 (versus CDDP group); (c) PCNA analysis (immunohistochemistry) and TUNEL analysis (immunofluorescence) of tumor tissues after treatment with various therapeutic agents. The PCNA-positive proliferating cells are stained brown, TUNEL-positive cells are green. Scale bar was 20 μm.
CDDP, cisplatin; CQ, chloroquine; i.v., intravenously; PBS, phosphate-buffered saline; PCNA, Proliferating cell nuclear antigen; SD, standard deviation; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling.
We further detected in detail what happened in tumors after different treatments. Combination treatments dramatically suppressed tumor cell proliferation, which was indicated by immunochemistry of PCNA [Figure 4(c)]. Meanwhile, increased terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)-positive cells demonstrated that the combination treatment enhanced tumor cell apoptosis [Figure 4(c)]. These results indicate that CQ-mediated autophagy inhibition elevated CDDP efficiency in tumor growth at least partially via inhibiting tumor cell proliferation or promoting tumor cell apoptosis.
Combination therapy inhibits autophagy and suppresses cancer stem cell subpopulation in vivo
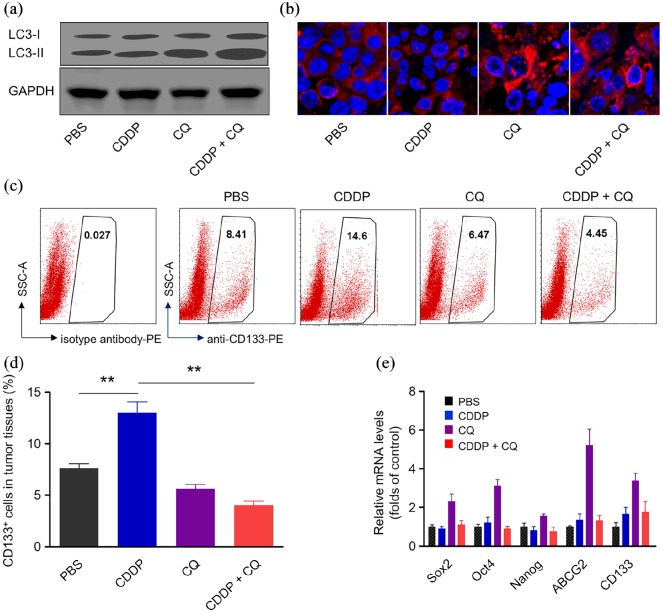
We next detected alteration of autophagy and cancer stem cells in tumor tissues after different treatments. A combination of CDDP with CQ greatly inhibited autophagy compared with CDDP or CQ alone, as evidenced by increased LC3-II expression levels [Figure 5(a)]. Immunofluorescence of the p62 protein further confirmed the decreased autophagy levels after combination therapy [Figure 5(b)]. More importantly, combination therapy dramatically suppressed the percentage of CD133+ cancer stem cells, which was elevated by CDDP treatment [Figure 5(c and d)]. Consistently, the mRNA expression levels of stemness markers, such as Sox2, Oct4, Nanog, ABCG2 and CD133 were significantly inhibited by CDDP and the CQ combination treatment [Figure 5(e)].
Figure 5.
Combination therapy inhibits autophagy and suppresses CSC subpopulation in vivo. (a) Protein expressions of LC3-I and LC3-II in tumor tissues after treatment with CDDP, CQ or a combination of two agents. GAPDH was used as a loading control. (b) The immunofluorescence of p62 protein in tumor cells after treatment with various agents. The nuclei were stained with DAPI (blue), p62 protein was stained with anti-p62 antibody labeled with Alexa Fluor 647. (c) Xenografted tumor at the end of therapy were dissociated into single cells and stained with anti-CD133 labeled with PE for flow cytometry analysis. (d) Quantitative analysis of CD133+ cells in tumor tissues. CDDP treatment significantly increase CD133+ cells and cotreatment of autophagy inhibitor CQ blocked the induction effect of CDDP on the percentage of CD133+ cells. Data represent means ± SD. n = 6, **p < 0.005. (e) mRNA expression levels of stemness-associated genes (including Sox2, Oct4, Nanog, ABCG2 and CD133) in A549 tumor tissue at the end-point of treatment.
CDDP, Cisplatin; DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PE, phycoerythrin.
To further confirm the effect of autophagy inhibition on cancer stem cells, we performed a knockdown of ATG5 (a key autophagy gene) using shRNA in A549 cell lines [Figure S3(a)]. ATG5 downregulation counteracts the increase in CD133+ cells induced by CDDP [Figure S3(b)]. These results demonstrated that a combination of CDDP and autophagy inhibition suppresses cancer stem cell subpopulation in vivo.
Discussion
NSCLC is a major type of lung cancer that is the most common cancer worldwide and is great threat for public health. CDDP was commonly used for NSCLC treatment, while drug resistance remains as the crucial challenge for CDDP-based chemotherapy of NSCLC treatment. Cancer stem cells were believed to be associated with drug resistance. In human and mouse small cell lung cancer (SCLC), CD133 expression levels increased after chemotherapy. Notably, CD133+ cancer stem cells showed increased tumorigenicity with higher Akt/PKB and Bcl-2 expression levels.11 The results prompted us to explore the effect of cancer stem cells in CDDP-based chemotherapy of NSCLC treatment. In the present study, we found that in clinical NSCLC samples, CD133+ cancer stem cells were enriched after CDDP treatment, as evidenced by elevated immunochemistry of CD133, enhanced CD133 expression levels at both mRNA and protein levels. The increased proportion of CD133+ cells after CDDP treatment was attributed to the death of bulk tumor cells (CD133− cells), or CDDP treatment itself triggers CD133 expression. We discuss the results in the revised manuscript. CDDP treatment dramatically inhibited A549 cell viability in a dose-dependent manner. However, CD133+ cancer stem cell proportions increased in the CDDP group, along with the enhanced expression of stemness markers, which is consistent with previous studies.13,15 Therefore, chemotherapy such as CDDP treatment disappointingly leads to cancer stem cell enrichment, which may be the crucial reason for drug resistance.
Autophagy is necessary for various biological processes, including tumorigenesis and tumor development; however, the correlation between autophagy and cancer is still confused. The absence of autophagy leads to abnormal accumulations of destroyed organelles and biomacromolecules that further induce oxidative stress, DNA damage and other cellular damage, and finally cellular carcinogenesis.23,24 Doxorubicin induced autophagy in wild-type cells, but not PARP-1 deficiency (parp-1-/-) or PARP inhibitor-treated cells. Absence of autophagy in parp-1-/- cells prevented ATP and NAD+ depletion, indicating the protecting role of autophagy in DNA damage.25 Through attenuating metabolic stress and preventing necrosis-induced cell death, autophagy functions as a tumor suppressor in solid tumors.26 On the other hand, materials and energy after autophagy are essential for the highly proliferating tumor cells.17 The receptor for advanced glycation end products (RAGE) downregulation in pancreatic tumor cells decreased autophagy levels and suppressed tumor cell survival, indicating the positive correlation of autophagy and tumor cell survival in RAGE-mediated cell response to stress.27 Beclin1 deficiency-mediated autophagy impairment suppressed mammary cancer development in a PALB2 loss-associated hereditary breast cancer model,28 indicating autophagy accelerated mammary cancer growth. In our study, enriched CD133+ A549 cancer stem cells displayed higher autophagy levels than CD133− A549 cells, indicating the correlation of autophagy with NSCLC cancer stem cells. Autophagy inhibition by CQ greatly suppressed CD133+ cancer stem cells in sorted cells. The inhibition of CQ-mediated autophagy dramatically decreased the ability of colony formation and sphere formation of CD133+ cancer stem cells. Therefore, autophagy levels are positively associated with the biological properties of NSCLC cancer stem cells.
Another important influence of autophagy on the effect of CDDP was determined based on the fact that increased autophagy levels enhanced drug resistance of CDDP-based chemotherapy in various types of cancer.18,19 CDDP treatment promoted the autophagy of ovarian cancer cells by activating ERK signaling. ERK inhibition by MEK inhibitors or siRNA-mediated ERK downregulation suppressed autophagy and sensitized tumor cells to CDDP challenge. More importantly, in established CDDP-resistant tumor cells, autophagy inhibition enhanced CDDP-induced cell apoptosis.18 Disruption of autophagy by CQ dramatically enhanced the effect of suberanilohydroxamic acid in imatinib-resistant chronic myelogenous leukemia cells.29 Our results demonstrated that CDDP treatment alone showed no significant influence on cell viability of A549 cells, while a combination of CDDP and CQ-mediated autophagy inhibition significantly suppressed cell viability of A549 cells. In xenograft tumors, combination treatment displayed a more effective role in inhibiting tumor growth and tumor cell proliferation and promoting cell apoptosis. More importantly, CD133+ cancer stem cells were significantly inhibited in tumor tissues.
It is important to note the limitations of this study. First, only one human NSCLC cell line (A549) was used in this study. It will be more convincing if we found similar results using another cell line. Second, the in vivo experiments were performed in NOD/SCID mice, which is somewhat limiting because of the inefficiency of the immune system. Spontaneous NSCLC mouse models or mouse NSCLC cell lines with injection into the same background mice would be a better choice to detect the effect of CDDP + CQ treatment. Third, the correlation between autophagy levels and clinical NSCLC progression may need better evidence to support the conclusion.
In summary, chemotherapy enhanced CD133+ cells in clinical NSCLC specimens and NSCLC cell lines. The enriched CD133+ cells exhibited higher autophagy levels. Autophagy inhibition by CQ inhibited CD133+ cell stemness and promoted CDDP efficiency in A549 cells. More importantly, a combination of CDDP and CQ treatment significantly inhibited cancer stem cell proportions in vitro, and dramatically suppressed tumor growth in vivo. All the results indicated that autophagy inhibition of cancer stem cells promotes the efficacy of CDDP against NSCLC.
Conclusion
Autophagy inhibition of cancer stem cells promotes the efficacy of CDDP against NSCLC. Autophagy inhibition could serve as a promising strategy for NSCLC treatment.
Supplemental Material
Supplemental material, Author_Response_1_(2) for Autophagy inhibition of cancer stem cells promotes the efficacy of cisplatin against non-small cell lung carcinoma by Chengcheng Hao, Guiping Liu and Guangliang Tian in Therapeutic Advances in Respiratory Disease
Supplemental Material
Supplemental material, Reviewer_2_v.1_(1) for Autophagy inhibition of cancer stem cells promotes the efficacy of cisplatin against non-small cell lung carcinoma by Chengcheng Hao, Guiping Liu and Guangliang Tian in Therapeutic Advances in Respiratory Disease
Supplemental Material
Supplemental material, Supplemental_information for Autophagy inhibition of cancer stem cells promotes the efficacy of cisplatin against non-small cell lung carcinoma by Chengcheng Hao, Guiping Liu and Guangliang Tian in Therapeutic Advances in Respiratory Disease
Footnotes
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
Conflict of interest statement: The authors declare that there is no conflict of interest.
ORCID iD: Guangliang Tian  https://orcid.org/0000-0001-9464-8307
https://orcid.org/0000-0001-9464-8307
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Chengcheng Hao, Department of Radiation Oncology, Liaocheng People’s Hospital, Liaocheng, Shandong, China.
Guiping Liu, Department of Radiation Oncology, Liaocheng People’s Hospital, Liaocheng, Shandong, China.
Guangliang Tian, Department of Radiation Oncology, Liaocheng Cancer Hospital, No 45 Jianshe East Road, Liaocheng, Shandong, 252000, China.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017; 67: 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 3. Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res 2016; 5: 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Doroshow DB, Herbst RS. Treatment of advanced non-small cell lung cancer in 2018. JAMA Oncol 2018; 4: 569–570. [DOI] [PubMed] [Google Scholar]
- 5. Tilz GP, Becker H. Antigen-antibody complexes: physiology and pathology. Wien Med Wochenschr Suppl 1990; 107: 2–3. [PubMed] [Google Scholar]
- 6. Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer 2007; 7: 573–584. [DOI] [PubMed] [Google Scholar]
- 7. Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 2002; 346: 92–98. [DOI] [PubMed] [Google Scholar]
- 8. Shackleton M, Quintana E, Fearon ER, et al. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell 2009; 138: 822–829. [DOI] [PubMed] [Google Scholar]
- 9. Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell stem cell 2012; 10: 717–728. [DOI] [PubMed] [Google Scholar]
- 10. Gottschling S, Schnabel PA, Herth FJ, et al. Are we missing the target? cancer stem cells and drug resistance in non-small cell lung cancer. Cancer Genomics Proteomics 2012; 9: 275–286. [PubMed] [Google Scholar]
- 11. Sarvi S, Mackinnon AC, Avlonitis N, et al. CD133+ cancer stem-like cells in small cell lung cancer are highly tumorigenic and chemoresistant but sensitive to a novel neuropeptide antagonist. Cancer Res 2014; 74: 1554–1565. [DOI] [PubMed] [Google Scholar]
- 12. Akunuru S, James Zhai Q, Zheng Y. Non-small cell lung cancer stem/progenitor cells are enriched in multiple distinct phenotypic subpopulations and exhibit plasticity. Cell Death Dis 2012; 3: e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu YP, Yang CJ, Huang MS, et al. Cisplatin selects for multidrug-resistant CD133+ cells in lung adenocarcinoma by activating Notch signaling. Cancer Res 2013; 73: 406–416. [DOI] [PubMed] [Google Scholar]
- 14. Ma S, Lee TK, Zheng BJ, et al. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene 2008; 27: 1749–1758. [DOI] [PubMed] [Google Scholar]
- 15. Bertolini G, Roz L, Perego P, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci USA 2009; 106: 16281–16286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147: 728–741. [DOI] [PubMed] [Google Scholar]
- 17. Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell 2013; 155: 1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang J, Wu GS. Role of autophagy in cisplatin resistance in ovarian cancer cells. J Biol Chem 2014; 289: 17163–17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen S, Rehman SK, Zhang W, et al. Autophagy is a therapeutic target in anticancer drug resistance. Biochim Biophys Acta 2010; 1806: 220–229. [DOI] [PubMed] [Google Scholar]
- 20. Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer 2005; 5: 275–284. [DOI] [PubMed] [Google Scholar]
- 21. Zeppernick F, Ahmadi R, Campos B, et al. Stem cell marker CD133 affects clinical outcome in glioma patients. Clin Cancer Res 2008; 14: 123–129. [DOI] [PubMed] [Google Scholar]
- 22. Mizushima N. Methods for monitoring autophagy. Int J Biochem 2004; 36: 2491–2502. [DOI] [PubMed] [Google Scholar]
- 23. Robert T, Vanoli F, Chiolo I, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature 2011; 471: 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature 2008; 451: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Munoz-Gamez JA, Rodriguez-Vargas JM, Quiles-Perez R, et al. PARP-1 is involved in autophagy induced by DNA damage. Autophagy 2009; 5: 61–74. [DOI] [PubMed] [Google Scholar]
- 26. Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006; 10: 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kang R, Tang D, Schapiro NE, et al. The receptor for advanced glycation end products (RAGE) sustains autophagy and limits apoptosis, promoting pancreatic tumor cell survival. Cell Death Differ 2010; 17: 666–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huo Y, Cai H, Teplova I, et al. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer Discov 2013; 3: 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carew JS, Nawrocki ST, Kahue CN, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood 2007; 110: 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Author_Response_1_(2) for Autophagy inhibition of cancer stem cells promotes the efficacy of cisplatin against non-small cell lung carcinoma by Chengcheng Hao, Guiping Liu and Guangliang Tian in Therapeutic Advances in Respiratory Disease
Supplemental material, Reviewer_2_v.1_(1) for Autophagy inhibition of cancer stem cells promotes the efficacy of cisplatin against non-small cell lung carcinoma by Chengcheng Hao, Guiping Liu and Guangliang Tian in Therapeutic Advances in Respiratory Disease
Supplemental material, Supplemental_information for Autophagy inhibition of cancer stem cells promotes the efficacy of cisplatin against non-small cell lung carcinoma by Chengcheng Hao, Guiping Liu and Guangliang Tian in Therapeutic Advances in Respiratory Disease