Abstract
Aims
We aimed to investigate whether metabolomic profiling of blood can lead to novel insights into heart failure pathogenesis or improved risk prediction.
Methods and results
Mass spectrometry‐based metabolomic profiling was performed in plasma or serum samples from three community‐based cohorts without heart failure at baseline (total n = 3924; 341 incident heart failure events; median follow‐up ranging from 4.6 to 13.9 years). Cox proportional hazard models were applied to assess the association of each of the 206 identified metabolites with incident heart failure in the discovery cohorts Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) (n = 920) and Uppsala Longitudinal Study of Adult Men (ULSAM) (n = 1121). Replication was undertaken in the independent cohort TwinGene (n = 1797). We also assessed whether metabolites could improve the prediction of heart failure beyond established risk factors (age, sex, body mass index, low‐density and high‐density lipoprotein cholesterol, triglycerides, lipid medication, diabetes, systolic and diastolic blood pressure, blood pressure medication, glomerular filtration rate, smoking status, and myocardial infarction prior to or during follow‐up). Higher circulating urobilin and lower sphingomyelin (30:1) were associated with incident heart failure in age‐adjusted and sex‐adjusted models in the discovery and replication sample. The hazard ratio for urobilin in the replication cohort was estimated to 1.29 per standard deviation unit, 95% confidence interval (CI 1.03–1.63), and for sphingomyelin (30:1) to 0.72 (95% CI 0.58–0.89). Results remained similar after further adjustment for established heart failure risk factors in meta‐analyses of all three cohorts. Urobilin concentrations were inversely associated with left ventricular ejection fraction at baseline in the PIVUS cohort (β = −0.70, 95% CI −1.03 to −0.38). No major improvement in risk prediction was observed when adding the top 2 metabolites (C‐index 0.787, 95% CI 0.752–0.823) or nine Lasso‐selected metabolites (0.790, 95% CI 0.754–0.826) to a modified Atherosclerosis Risk in Communities heart failure risk score model (0.780, 95% CI 0.745–0.816).
Conclusions
Our metabolomic profiling of three community‐based cohorts study identified associations of circulating levels of the haem breakdown product urobilin, and sphingomyelin (30:1), a cell membrane component involved in signal transduction and apoptosis, with incident heart failure.
Keywords: Biomarkers, Metabolomics, Heart failure, Risk prediction, Epidemiology
Introduction
Heart failure is a major global health problem that needs to be addressed in terms of better identification of high‐risk individuals, as well as new therapeutic and preventive strategies.1 New biomarkers for heart failure could improve risk assessment and provide insights into underlying pathophysiology, thereby improving quality of life for patients and reducing the burden on the health system.2
Metabolomic profiling, or metabolomics, is the study of low‐weight molecules (<1.5 kDa), derived either from the environment (exogenous metabolites) or from metabolism (endogenous metabolites).3 Recent advances in high‐throughput technologies have enabled metabolic profiling in large cohorts, which has provided novel possibilities for biomarker discovery. Yet few studies have investigated the association between metabolomics profiles and heart failure risk.4, 5
The objective of this study was to investigate the association between 206 metabolites measured by ultra‐performance liquid chromatography coupled with time‐of‐flight tandem mass spectrometry and heart failure incidence in three independent community‐based cohorts of middle‐aged to elderly individuals using a discovery/replication approach. Secondary aims were to assess whether the metabolites could improve the prediction of heart failure beyond established heart failure risk factors and to investigate the association between the metabolites and the baseline echocardiographic measurements of left ventricular function.
Methods
Cohorts
In the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS), all 70‐year‐old residents of Uppsala County, Sweden, were invited to participate in a health survey and detailed clinical assessment between 2001 and 2004, described in detail previously.6 Of 2025 invited, 1016 (50.2%) participated in the baseline assessment, which occurred within 1 month of their 70th birthday.
In the Uppsala Longitudinal Study of Adult Men (ULSAM), all men born in Sweden between 1920 and 1924 and living in Uppsala were invited to participate in a health assessment between 1970 and 1973, described in detail previously.7 Of 1681 invited, 1221 (72.6%) participated in the follow‐up assessment at age 71 between 1991 and 1995 that serves as the baseline examination for the present study.
The Swedish Twin Register is a population‐based study of 194 000 Swedish twins born between 1886 and 2008.8 TwinGene is a longitudinal study of men and women nested within the Swedish Twin Register. All twins born before 1958 who also participated in the Screening Across the Lifespan Twin telephone screening between 1998 and 2002 were re‐contacted between 2004 and 2008, and 12 591 individuals (55% women) participated in the study.8 Metabolomic analyses were performed in a subset of TwinGene using a case–cohort design, including all incident cases of type 2 diabetes (n = 218), coronary artery disease (n = 282), ischaemic stroke (n = 186), and dementia (n = 114) prior to 31 December 2010, and a sub‐cohort (controls) of 1643 individuals (43% women) stratified on age and sex were included.9 individuals in all these groups were eligible for inclusion in this study.
Exclusion criteria
Individuals with (i) prevalent heart failure, (ii) missing metabolite measurements, (iii) missing model covariates, or (iv) non‐fasting individuals (106 individuals in TwinGene) were excluded. The final number of participants included in the analysis was 920 in PIVUS, 1121 in ULSAM, and 1797 in TwinGene (Figure 1). The regional ethics review boards in Uppsala (PIVUS and ULSAM) and Stockholm (TwinGene) approved the studies, and all participants gave written informed consent.
Figure 1.
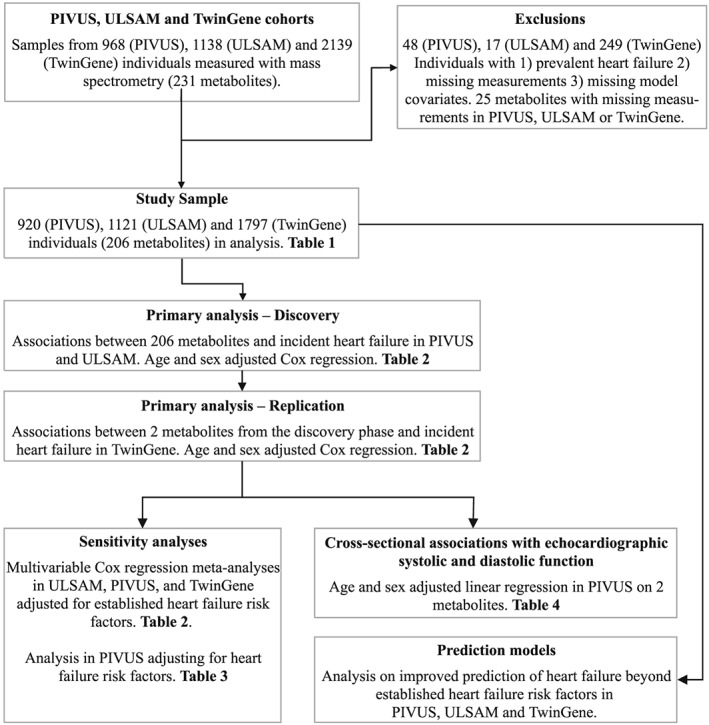
Flow chart illustrating the design of the study, including inclusion and exclusion criteria.
Outcome
In PIVUS and ULSAM, medical records for all individuals hospitalized for heart failure [International Classification of Diseases, Ninth Revision (ICD‐9) 428 and ICD‐10 I50], or hypertensive heart disease with heart failure (ICD‐9 402 and ICD‐10 I11), in the National Patient Register (inpatient) during follow‐up were reviewed by physicians blinded to the baseline data.10 Heart failure events were classified as definite, questionable, or miscoded according to the European Society of Cardiology definitions.11 Thus, to be classified as a definite heart failure case, there had to be symptoms and signs of heart failure and ‘objective evidence’ of cardiac dysfunction at rest. In cases of doubt, the response to heart failure treatment was a useful check of the diagnosis. The required ‘objective evidence’ was echocardiography; however, because the study commenced prior to the widespread availability of echocardiography, electrocardiography and X‐ray were also considered acceptable when an echocardiography report was not available. We used definite cases of heart failure in our analyses for definition of the outcome. In TwinGene, heart failure diagnosis was obtained from the National Patient Register (ICD‐9 428 and ICD‐10 I50).
Individuals were followed for up to 17 years from baseline until heart failure diagnosis, death, or end of follow‐up (10 June 2014 in PIVUS, 31 December 2008 in ULSAM, and 31 December 2010 in TwinGene).
Clinical characteristics
In PIVUS and ULSAM, participants were investigated in the morning after an overnight fast. Venous blood samples were frozen as plasma (ULSAM), or as serum (PIVUS), and stored at −70°C until analysis. Participants in TwinGene went to their local healthcare centre for blood sampling and health check‐up. Participants were instructed to perform the sample collection in the morning after an overnight fast. Serum samples were sent by overnight mail to the Karolinska Biobank where they were frozen at −80° C until analysis.
In all cohorts, information on lifestyle and medication at baseline was collected through questionnaires. Data on myocardial infarction prior to baseline or during follow‐up were retrieved from the Swedish hospital discharge register (ICD‐9 410 and ICD‐10 I21 and I22). Diabetes was defined as having a fasting plasma glucose ≥7.0 mmol/L, taking antidiabetic medication, or having an HbA1c ≥ 6.5% (48 mmol/mol, TwinGene only). Glomerular filtration rate was estimated from creatinine using the CKD‐EPI formula.12
In PIVUS, the left ventricular volume was calculated according to the Teichholz M‐mode formula.13 Left ventricular systolic function was assessed by ejection fraction, calculated as (left ventricular diastolic volume − left ventricular systolic volume)∕left ventricular diastolic volume. Left ventricular diastolic function was assessed by the isovolumic relaxation time (the interval between aortic valve closure and the onset of mitral flow) using the Doppler signal from the area between the left ventricular outflow tract and the mitral flow.13
Metabolomic profiling
The analytical procedure for metabolomics has been previously described,14 and the code to process the data is publicly available (https://github.com/andgan/metabolomics_pipeline). In brief, metabolomics profiling in PIVUS, ULSAM, and TwinGene was performed using a Waters Acquity UPLC system coupled to a Waters Xevo G2‐Time‐Of‐Flight‐Mass Spectrometry platform at Colorado State University (Fort Collins, CO, USA). Data acquisition using positive electrospray ion mode with a mass‐to‐charge ratio range of 50–1200 at five scans per second was alternately performed at collision energies of 6 and 15–30 V. Data were processed by XCMS in R.15 In total, 10 162 (ULSAM), 9755 (TwinGene), and 7522 (PIVUS) features were detected and adjusted for factors of external variability (plate effect, analysis date, retention time drift, and sample collection) by analysis of variance‐type standardization and/or log transformation and by removal of spectra with abnormal intensities and/or low interduplicate correlations and/or retention times. Features in common between PIVUS, ULSAM, and TwinGene were identified by matching m/z and retention time, followed by manual inspection. For each feature, retention time, m/z, and fragmentation pattern were compared with in‐house and public database reference libraries and matched according to Metabolomics Standard Initiative guidelines to annotate spectra to metabolite names.16 We confirmed the annotation of sphingomyelin (30:1) and urobilin at the highest level of confidence by spectral comparison of standards prepared in methanol and injected at a concentration of 5 μg/mL using the same method as described earlier (Supporting Information, Figures S2–S4 ). Full metabolomics data are available in the MetaboLights archive (study identifiers MTBLS90 for PIVUS, MTBLS124 for ULSAM, and MTBLS93 for TwinGene; http://www.ebi.ac.uk/metabolights/).
Statistical analyses
We fitted Cox proportional hazard regression models for new‐onset heart failure separately for each of the 206 identified metabolites, using attained age as timescale and adjusting for sex, and meta‐analysed the discovery cohorts PIVUS and ULSAM (fixed effect model using the method of Mantel and Haenszel)17 using the Stata package metan. All metabolites were normalized (on log2 scale) prior to analysis to have a mean of 0 and a standard deviation of 1, and those associated with incident heart failure at a 15% false discovery rate (estimated using the Benjamini and Hochberg method18) were taken forward to replication in TwinGene (validation cohort), where a nominal significance threshold of P ≤ 0.05 was used. We expect the false rediscovery rate in the validation cohort to be 0.23%, implying that 0.23% of metabolites replicating at P = 0.05 are expected to be false positives (Supporting Information, Methods).19 The Cox proportional hazard assumption was assessed by visually inspecting plots of Schoenfeld residuals against time in those metabolites passing the discovery stage. In TwinGene, we applied weighted Cox proportional hazard models compensating for the case–cohort design, as described in detail previously.14
We constructed directed acyclic graphs for the assumed causal relations to guide the choice of covariates (Supporting Information, Figure S1 ).20 For the metabolites that were associated with heart failure in the replication phase, we performed additional multivariable adjustment based on the DAGs to establish models with minimal confounding in each cohort and meta‐analysed the results. In these analyses using age as time scale, we adjusted for sex, body mass index, low‐density lipoprotein (LDL) and high‐density lipoprotein cholesterol, triglycerides, lipid medication, diabetes, systolic and diastolic blood pressure, blood pressure medication, kidney function (glomerular filtration rate), smoking status, myocardial infarction prior to or during study (time updated; the covariate is updated the day individuals are hospitalized with acute myocardial infarction). In additional models, we adjusted for baseline N‐terminal prohormone of brain natriuretic peptide levels (estimated by a commercial or enzyme‐linked immunosorbent assay (ELISA), Roche Diagnostics), heart rate, haemoglobin, cardiovascular medications (angiotensin‐converting enzyme inhibitors, angiotensin II receptor antagonists, beta‐blockers, calcium inhibitors, and diuretics), and calendar year in the PIVUS cohort to determine the influence of these risk factors on our estimates. In order to investigate the influence of liver damage on the association between metabolite and heart failure, we performed additional multivariable models adjusting for alanine transaminase, albumin, γ‐glutamyltransferase in the PIVUS cohort. For the measured upstream metabolites of urobilin, we performed sensitivity analysis to investigate the association of bilirubin and its metabolites with incident heart failure. We also assessed the association of all measured sphingomyelins in the three cohorts combined to evaluate the association between sphingomyelins as a group and incident heart failure. In additional sensitivity analysis, missing covariates in TwinGene (475 data points in 258 individuals) were imputed using multiple imputation by chained equations using the mi command package in Stata 14, and results from fully adjusted models were compared with the complete case analysis. We also evaluated if the metabolites had a linear relationship by fitting restricted cubic spline models with Cox and plotting the log relative hazards, using the predict command in Stata.
We further investigated the cross‐sectional association with echocardiographic indices of left ventricular systolic and diastolic function in PIVUS using age‐adjusted and sex‐adjusted linear regression analyses.
To assess whether metabolomic data could improve heart failure risk prediction beyond the Atherosclerosis Risk in Communities (ARIC) heart failure score,21 we assessed Cox proportional hazard models with and without urobilin and sphingomyelin (30:1) in a pooled sample of all three cohorts. We adjusted both models for cohort and for available ARIC covariates (age, sex, previous or current smoker, body mass index, systolic blood pressure, history of myocardial infarction, blood pressure medication and diabetes diagnosis). Heart rate was not available.
Second, we used L1‐regularized (Lasso) Cox proportional hazard regression to select a sparse model that maximized discrimination performance, while minimizing the number of metabolites used for prediction. Using the glmnet package in R, we forced the established risk factors unpenalized into the model and selected the best performing solution retaining nine metabolites (<5% of 206 candidates), in a 75% random split of the combined cohorts.22
We evaluated discrimination performance for the two prediction models by calculating the C‐index with coxph and survConcordance function in the survival package in a pooled sample of all three cohorts.
Analyses were performed using Stata 13.1 (Stata Corp, College Station, TX, USA) or R version 3.4.3.
Results
Cohort characteristics
The baseline characteristics, number of events, and incidence rate of the final analytical dataset for PIVUS (median age 70 years, n = 920), ULSAM (median age 71 years, n = 1121), and TwinGene (median age 68 years, n = 1797) are shown in Table 1 and stratified by sex in Supporting Information, Table S1 . Seventy‐four participants in PIVUS, 183 participants in ULSAM, and 84 participants in TwinGene were diagnosed with heart failure. In total, 96 of the 324 heart failure events had a preceding myocardial infarction.
Table 1.
Baseline characteristics of the participants of the PIVUS, ULSAM, and TwinGene cohorts
| Cohort (number of individuals) | PIVUS (920) | ULSAM (1121) | TwinGene (1797) |
|---|---|---|---|
| Age (years) | 70.2 (0.2) | 71 (0.6) | 68.5 (8.1) |
| BMI (kg/m2) | 27 (4.2) | 26.3 (3.4) | 26.3 (4.0) |
| LDL‐C (mmol/L) | 3.4 (0.9) | 3.9 (0.9) | 3.7 (1.0) |
| HDL‐C (mmol/L) | 1.5 (0.4) | 1.3 (0.3) | 1.4 (0.4) |
| Triglycerides (mmol/L) | 1.3 (0.6) | 1.4 (0.7) | 1.4 (0.7) |
| Fasting glucose (mmol/L) | 5.6 (5.1–6) | 5.4 (5–5.9) | 5.4 (5–6) |
| Glomerular filtration rate (mL/min/1.73 m2) | 80 (14) | 76 (11) | 82 (14) |
| Systolic blood pressure (mmHg) | 150 (22) | 147 (19) | 142 (20) |
| Diastolic blood pressure (mmHg) | 79 (10) | 84 (9.0) | 82 (10) |
| NT‐proBNP (mmol/L) | 109 (63–175) | 105 (59–214) | NA |
| Heart rate (b.p.m.) | 61 (55–68) | 64 (60–72) | NA |
| Haemoglobin (g/L) | 137 (131–145) | NA | 143 (135–152) |
| Male | 454 (49%) | 1121 (100%) | 1013 (57%) |
| Current smoker | 94 (10%) | 226 (20%) | 259 (15%) |
| Previous smoker | 376 (41%) | 487 (43%) | 662 (37%) |
| Left ventricular hypertrophy | 102 (12%) | 246 (27%) | NA |
| Lipid‐lowering medication | 142 (15%) | 97 (9%) | 299 (17%) |
| Blood pressure‐lowering medication | 275 (30%) | 377 (34%) | 467 (26%) |
| ACE inhibitors | 69 (8%) | 59 (6%) | 65 (4%) |
| Angiotensin II receptor antagonists | 75 (8%) | NA | NA |
| Beta‐blockers | 184 (20%) | 206 (19%) | 80 (4%) |
| Diuretics | 101 (11%) | 124 (12%) | NA |
| Calcium channel blockers | 103 (11%) | 128 (12%) | NA |
| Myocardial infarction at baseline | 53 (6%) | 84 (7%) | 129 (7%) |
| Type 2 diabetes at baseline | 165 (18%) | 142 (13%) | 241 (14%) |
| Treated with insulin | 15 (2%) | 13 (1%) | NA |
| Taking diabetes medication | 55 (6%) | 48 (4%) | 115 (6%) |
| Follow‐up time (median, range) | 10.0 (0.1–10.9) | 13.9 (0.0–17.4) | 4.6 (0.0–6.6) |
| Incidence rate (per 100 person years at risk) | 0.87 | 1.40 | 0.26 |
ACE, angiotensin‐converting enzyme; BMI, body mass index; HDL‐C, high‐density lipoprotein cholesterol; LDL‐C, low‐density lipoprotein cholesterol; NA, not applicable; NT‐proBNP, N‐terminal prohormone of brain natriuretic peptide.
Data are mean (standard deviation) for normally distributed variables, median (interquartile range) for skewed variables, and n (%) for dichotomous variables, unless specified.
Main results
Three metabolites, urobilin, sphingomyelin (30:1), and sphingomyelin (28:1), were associated with incident heart failure after adjusting for age and sex at the 15% false discovery rate threshold in PIVUS and ULSAM combined (Supporting Information, Table S2 ). Urobilin and sphingomyelin (30:1) were also associated with incident heart failure at P < 0.05 with consistent effect direction in TwinGene (Table 2 and Supporting Information, Table S3 ).
Table 2.
Assessment of the association between urobilin and sphingomyelin (30:1) and incident heart failure in the PIVUS, ULSAM, and TwinGene cohorts
| Metabolite | PIVUS and ULSAMa | TwinGenea | Meta‐analysisa | Meta‐analysis, adjustedb |
|---|---|---|---|---|
| Urobilin | 1.45 (1.19–1.76) *** | 1.29 (1.03–1.63)* | 1.38 (1.19–1.60) *** | 1.30 (1.10–1.52)** |
| Sphingomyelin (30:1) | 0.80 (0.72–0.90) *** | 0.72 (0.58–0.89)* | 0.78 (0.71–0.87) *** | 0.85 (0.75–0.95)** |
Data are hazard ratio with 95% confidence intervals expressed per standard deviation increase of metabolite levels.
Age and sex adjusted.
Established heart failure risk factors: age, sex, body mass index, low‐density and high‐density lipoprotein cholesterol, triglycerides, lipid medication, diabetes, systolic and diastolic blood pressure, blood pressure medication, kidney function (glomerular filtration rate), smoking status, and myocardial infarction prior to or during study (time updated).
P‐value <0.05.
P‐value <0.01.
P‐value <0.001.
We performed multivariable adjustment for established heart failure risk factors in each cohort and meta‐analysed the results. As seen in Table 2, adjustment for heart failure risk factors had a modest influence on these associations. Urobilin (hazard ratio 1.27 per standard deviation unit increase, 95% confidence interval CI 1.08–1.49) and sphingomyelin (30:1) (hazard ratio 0.85, 95% CI 0.75–0.95) were associated with incident heart failure also after adjustment for established heart failure risk factors.
In PIVUS, we added further adjustments for N‐terminal prohormone of brain natriuretic peptide, heart rate, haemoglobin, cardiovascular medication, or calendar year to the model with established heart failure risk factors. These additional adjustments had a modest influence on the hazard ratio estimates for the two metabolites and heart failure risk (Table 3).
Table 3.
Assessment of potential confounders of the association between urobilin and sphingomyelin (30:1) and incident heart failure, respectively, in the PIVUS cohort (n = 829, number of events = 58)
| Metabolite | Model | Hazard ratio (95% CI) | P‐value |
|---|---|---|---|
| Urobilin | Age and sex | 1.36 (1.07–1.72) | 0.01 |
| Established risk factorsa | 1.24 (0.97–1.60) | 0.08 | |
| Established risk factors + NT‐proBNP | 1.27 (0.99–1.63) | 0.06 | |
| Established risk factors + heart rate | 1.24 (0.96–1.59) | 0.09 | |
| Established risk factors + haemoglobin | 1.24 (0.97–1.59) | 0.08 | |
| Established risk factors + calendar year | 1.25 (0.98–1.61) | 0.08 | |
| Established risk factors + cardiovascular medications | 1.24 (0.97–1.60) | 0.09 | |
| Sphingomyelin (30:1) | Age and sex | 0.72 (0.58–0.88) | 1.4 × 10−3 |
| Established risk factorsa | 0.70 (0.56–0.88) | 2.0 × 10−3 | |
| Established risk factors + NT‐proBNP | 0.61 (0.50–0.74) | 6.8 × 10−7 | |
| Established risk factors + heart rate | 0.70 (0.56–0.88) | 2.1 × 10−3 | |
| Established risk factors + haemoglobin | 0.70 (0.56–0.87) | 1.7 × 10−3 | |
| Established risk factors + calendar year | 0.70 (0.56–0.89) | 3.1 × 10−3 | |
| Established risk factors + cardiovascular medications | 0.69 (0.54–0.87) | 2.2 × 10−3 |
CI, confidence interval; NT‐proBNP, N‐terminal prohormone of brain natriuretic peptide.
Data are hazard ratio with 95% CI adjusted for age and gender expressed per standard deviation increase of metabolite levels.
Established heart failure risk factors: age, sex, previous or current smoker, body mass index, systolic and diastolic blood pressure, blood pressure medication, diabetes, low‐density lipoprotein cholesterol, high‐density lipoprotein cholesterol, triglycerides, glucose, diastolic blood pressure, lipid‐lowering medication, blood pressure, myocardial infarction before/during study, glomerular filtration rate, and insulin/oral antidiabetics treatment. Cardiovascular medications: angiotensin‐converting enzyme inhibitors, angiotensin II receptor antagonists, beta‐blockers, calcium inhibitors, and diuretics.
Sensitivity analysis
Adding liver markers (alanine transaminase, alkaline phosphatase, bilirubin, albumin, and γ‐glutamyltransferase) to the fully adjusted model had no effect on the association between each metabolite and heart failure (Supporting Information, Table S4 ), and the measured upstream metabolites of urobilin were not associated with incident heart failure (Supporting Information, Table S5 ).
For all of the 15 sphingomyelins measured in PIVUS, ULSAM, and TwinGene, all the associations went in the same direction as for sphingomyelin (30:1), and four of them were nominally significant (Supporting Information, Table S6 ).
Results from the analysis based on individuals with imputed missing covariates in TwinGene yielded essentially identical results as the complete case analysis (Supporting Information, Table S7 ).
We assessed deviance from a log‐linear relationship between urobilin and sphingomyelin (30:1) and incident heart failure using restricted cubic spline models. No evidence of non‐linearity was observed when plotting the log relative hazard (data not shown).
Left ventricular function
Urobilin concentration was inversely associated with left ventricular ejection fraction at baseline in the PIVUS cohort (regression coefficient per standard deviation increase −0.70, P = 3.1 × 10−5), while neither metabolite was associated with a measure of early diastolic function (isovolumic relaxation time) (Table 4).
Table 4.
The association between urobilin, sphingomyelin (30:1), and systolic and diastolic function, as evaluated by left ventricular ejection fraction and isovolumic relaxation time, respectively
| Metabolite | Systolic function (left ventricular ejection fraction, n = 755) | Diastolic function (isovolumic relaxation time, n = 824) | ||
|---|---|---|---|---|
| β valuesa, 95% CI | P‐value | β valuesb, 95% CI | P‐value | |
| Urobilin | −0.70 (−1.03 to −0.38) | 3 × 10−5 | 47 (−46 to 141) | 0.32 |
| Sphingomyelin (30:1) | 0.41 (−0.48 to 1.31) | 0.37 | 6 (−255 to 268) | 0.96 |
β values can be interpreted as the change in ejection fraction (%) for each standard deviation unit increase in the metabolite measurement. Lower ejection fraction implies worsened systolic function.
β values can be interpreted as the change in isovolumic relaxation time (milliseconds) per standard deviation unit increase in the metabolite measurement. Higher isovolumic relaxation time implies worsened diastolic function.
Risk prediction
No major improvement in risk prediction was found when adding (i) the top 2 metabolites (C‐index 0.787, 95% CI 0.752–0.823) or (ii) nine Lasso‐selected metabolites (0.790, 95% CI 0.754–0.826) to the ARIC model (0.780, 95% CI 0.745–0.816) in the three cohorts combined. The corresponding C‐index for the nine Lasso‐selected metabolites and ARIC covariates in the 25% validation set was 0.793 (95% CI 0.717‐0.869) compared to ARIC only 0.775 (0.689‐0.862).
Discussion
Principal findings
In this large well‐characterized sample, we identified novel associations of urobilin and sphingomyelin (30:1) with incident heart failure, independently of established risk factors. Furthermore, higher urobilin was associated with worse echocardiographic left ventricular systolic function.
Comparison with other studies
We are aware of two previous studies investigating the longitudinal associations between untargeted metabolomics and heart failure risk. Zheng et al.4 investigated 1744 African‐Americans in the ARIC cohort. Six named metabolites (erythritol, N‐acetylalanine, p‐cresol sulfate, phenylacetylglutamine, prolylhydroxyproline, and pyroglutamine) and 10 unnamed metabolites were independently associated with incident heart failure, adjusting for established heart failure risk factors. None of the named but two unnamed metabolites remained associated after additional adjustment for kidney function. Moreover, in an untargeted metabolomics study measuring 308 metabolites, also in the ARIC cohort, the metabolite hexadecanedioate was independently associated with heart failure risk.5
To our knowledge, the specific positive association between urobilin and heart failure risk has not been reported previously. However, metabolites related to urobilin, such as bilirubin,23 have been reported to be associated with heart disease in humans including inverse associations with coronary artery disease,24 stroke,25 atherosclerosis,26 and one study showing a U‐shaped relationship with ischaemic heart disease.27
We found a consistent inverse association of measured sphingomyelins with heart failure risk. In line with our findings, cross‐sectional studies have reported an inverse association between sphingomyelins and prevalent heart failure.28, 29 Further, a recent investigation in the ULSAM, PIVUS, and TwinGene cohorts found that sphingomyelin (28:1) was associated with decreased risk of coronary heart disease.9 In the current study, sphingomyelin (28:1) was associated with decreased risk of incident heart failure in the discovery sample but not in the replication sample. This might be due to the lower power or the younger age of participants in TwinGene. Total plasma sphingomyelin has also been associated with lower 5 year coronary heart disease incidence in the prospective Multi‐Ethnic Study of Atherosclerosis cohort, consisting of 6809 individuals between 45 and 84 years of age.30
Potential mechanisms
Because of the observational study design, it is not possible to draw conclusions regarding causality between metabolites and heart failure incidence. Our findings warrant additional experimental studies to clarify the underlying mechanisms for the present associations.
Urobilin is a degradation product of haem, through conversion of haem to biliverdin, and then to bilirubin, which is excreted as bile and further degraded by microbes present in the large intestine to urobilinogen. Approximately half of the urobilinogen is reabsorbed and oxidized to urobilin by the liver, which is then excreted by the kidneys, causing the yellow colour of urine.23, 31 Already in 1929, it was observed that patients with heart failure had increased haemolysis, bilirubin formation, and urinary urobilinogen, thought to arise from liver dysfunction.32 Abnormal liver function is commonly seen in heart failure patients and is linked to worse prognosis.33
The association between urobilin and incident heart failure could be explained by an increased breakdown of the upstream compounds haem and bilirubin, a larger reabsorption of urobilinogen in the large intestine into the blood, or slower clearance of urobilin from the blood by the kidneys. We saw no association with incident heart failure between bilirubin, biliverdin A, biliverdin B, or unconjugated bilirubin in the present study (Supporting Information, Tables S2 , S3 , and S5 ), and the association with incident heart failure was not attenuated after adjustment for kidney function or haemoglobin (Table 3). Taken together, it seems more likely that the observed association would be due to increased reabsorption of urobilinogen into the blood from the large intestine than a decreased clearance of urobilin by the kidney.
Sphingomyelin is an abundant constituent of cell membranes, and its involvement in signal transduction,34 apoptosis,35 atherosclerosis,36 and plasma lipid metabolism37 may explain part of these associations, although further studies are needed to investigate the mechanism behind the association with incident heart failure.
Plasma sphingomyelin is not degraded in plasma; instead, it accumulates in lipoproteins,38 a mechanism that has been proposed to be directly involved in the early stages of atherosclerotic development.36 Knockout mice for the LDL receptor gene fed on a sphingolipid‐rich (1%) diet had increased plasma sphingomyelin, LDL aggregation, and atherosclerotic lesions, indicating that accumulation of circulating sphingomyelin has a direct effect on plaque development.39 Thus, speculatively, higher circulating levels of sphingomyelins could indicate a reduced accumulation of sphingomyelins in atherosclerotic plaques and consequently a lower atherosclerotic load that might explain the inverse association with heart failure risk. In the current study the association with incident heart failure was independent of LDL‐C. There may also be some other, currently unidentified cardioprotective roles of sphingomyelin (30:1) and other sphingomyelins. Additional mechanistic studies are highly warranted to shed light into the underlying pathophysiology of our findings.
Clinical implications
The addition of metabolomics to a model of established heart failure risk factors did not improve the prediction of heart failure in our study to any large extent. Thus, our study does not provide support that circulating metabolomics could be useful in identifying individuals at increased risk for heart failure in clinical practice. However, other metabolites not captured on our platform may be more informative.
Strengths and limitations
Strengths of the study include the longitudinal study design, up to 17 years of follow‐up, detailed assessment of participants, use of a state‐of‐the‐art mass spectrometry platform, rigorous annotation and data processing, with manual validation, the discovery and replication approach in independent cohorts, and the balanced multiple testing correction. Limitations include generalizability, as participants were middle‐aged to elderly European residents of a geographically defined part of Sweden and that the ULSAM cohort included only men. The replication cohort however was recruited nationwide in Sweden and consisted of 43% women. We excluded persons previously hospitalized for heart failure, but it is possible that some participants had subclinical heart failure at baseline. We did not have data on primary care diagnoses or the incidence of subtypes of heart failure. We could not differentiate between heart failure with reduced ejection fraction and heart failure with preserved ejection fraction as echocardiographic data from the heart failure event were not available, nor were data on heart failure severity according to New York Heart Association functional classification available. The estimated hazard ratios should be interpreted with caution because the storage time may decrease the precision of the estimates. The mass spectrometry platform does not provide standard concentration units, making comparisons with clinically applied cut‐offs difficult. Our observational study cannot establish causality but may help in giving future directions for experimental studies on the causal mechanisms leading to heart failure.
Conclusions
We identified two novel associations between circulating metabolites and incident heart failure, independent of traditional risk factors. Our data suggest that circulating metabolomics is a promising technique for discovering novel risk markers for incident heart failure and encourage future large‐scale efforts to evaluate the utility of metabolomic profiling in clinical practice.
Conflict of interest
E.I. is a scientific advisor for Precision Wellness, Inc., and has received consulting fees from Olink Proteomics for work unrelated to the present project. J.S. has an advisory board membership for Itrim. J.Ä. has received lecturing fees from AstraZeneca. The remaining authors declare no conflicts of interest.
Funding
The Swedish Twin Registry is managed by Karolinska Institutet and receives funding through the Swedish Research Council under the grant no. 2017‐00641. J.Ä. is supported by the Swedish Research Council (2012‐02215), Swedish Heart‐Lung Foundation, and the European Union Horizon 2020 (grant no. 634869).
Author contributions
T.F. and J.Ä. are joint senior authors. J.Ä. had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. M.S., E.I., P.K.E.M., T.F., and J.Ä. contributed in the study concept and design; M.S., C.N., S.S., P.S., J.S., V.G., L.L., P.K.E.M., E.I., T.F., C.D.B., J.E.P., A.G., and J.Ä. in the acquisition, analysis, or interpretation of the data; M.S. in drafting of the manuscript; C.N., S.S., P.S., J.S., V.G., L.L., P.K.E.M., C.D.B., E.I., T.F., and J.Ä. in the critical revision of the manuscript for important intellectual content; M.S., C.N., T.F., and J.Ä. in the statistical analysis; and E.I., T.F., and J.Ä. in the study supervision.
Declaration of Helsinki
This study complies with the Declaration of Helsinki, the locally appointed ethics committee that has approved the research protocol, and informed consent has been obtained from the subjects (or their legally authorized representative).
Supporting information
Figure S1. Proposed causal diagram for the association between circulating biomarkers and incident heart failure – Directed Acyclic Graph.
Figure S2. Validation of sphingomyelin(30:1) annotation.
Figure S3. Validation of urobilin annotation
Figure S4. Validation of sphingomyelin(28:1) annotation
Table S1. Sex‐specific baseline characteristics in participants of the PIVUS, ULSAM, and TwinGene cohorts.
Table S2. Hazard ratio estimates for all metabolites and incident heart failure in PIVUS and ULSAM – age and sex adjusted.
Table S3. Hazard ratio estimates for all metabolites and incident heart failure in TwinGene – age and sex adjusted.
Table S4. Assessment of potential confounders of the association between urobilin and sphingomyelin (30:1), and incident heart failure, respectively, in the PIVUS cohort (n = 829, number of events = 58).
Table S5. Association of each urobilin metabolite in ULSAM, PIVUS and TwinGene combined – age and sex adjusted.
Table S6. Association of each sphingomyelin in ULSAM, PIVUS and TwinGene combined – age and sex adjusted.
Table S7. Assessment of the association between urobilin and sphingomyelin(30:1), and incident heart failure in the PIVUS, ULSAM and TwinGene cohorts before and after multiple imputation in TwinGene.
Acknowledgement
We acknowledge the Swedish Twin Registry for access to data.
Stenemo M., Ganna A., Salihovic S., Nowak C., Sundström J., Giedraitis V., Broeckling C. D., Prenni J. E., Svensson P., Magnusson P. K. E., Lind L., Ingelsson E., Ärnlöv J., and Fall T. (2019) The metabolites urobilin and sphingomyelin (30:1) are associated with incident heart failure in the general population, ESC Heart Failure, 6, 764–773. 10.1002/ehf2.12453.
References
- 1. McMurray JJ, Petrie MC, Murdoch DR, Davie AP. Clinical epidemiology of heart failure: public and private health burden. Eur Heart J 1998; 19: P9–P16. [PubMed] [Google Scholar]
- 2. Bahrami H, Bluemke DA, Kronmal R, Bertoni AG, Lloyd‐Jones DM, Shahar E, Szklo M, Lima JAC. Novel metabolic risk factors for incident heart failure and their relationship with obesity. J Am Coll Cardiol 2008; 51: 1775–1783. [DOI] [PubMed] [Google Scholar]
- 3. Wishart DS. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov 2016; 15: 473–484. [DOI] [PubMed] [Google Scholar]
- 4. Zheng Y, Yu B, Alexander D, Manolio TA, Aguilar D, Coresh J, Heiss G, Boerwinkle E, Nettleton JA. Associations between metabolomic compounds and incident heart failure among African Americans: the ARIC Study. Am J Epidemiol 2013; 178: 534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu B, Li AH, Metcalf GA, Muzny DM, Morrison AC, White S, Mosley TH, Gibbs RA, Boerwinkle E. Loss‐of‐function variants influence the human serum metabolome. Sci Adv 2016; 2: e1600800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lind L, Fors N, Hall J, Marttala K, Stenborg A. A comparison of three different methods to evaluate endothelium‐dependent vasodilation in the elderly: the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study. Arterioscler Thromb Vasc Biol 2005; 25: 2368–2375. [DOI] [PubMed] [Google Scholar]
- 7. Hedstrand H. A study of middle‐aged men with particular reference to risk factors for cardiovascular disease. Ups J Med Sci Suppl 1975; 19: 1–61. [PubMed] [Google Scholar]
- 8. Magnusson PKE, Almqvist C, Rahman I, Ganna A, Viktorin A, Walum H, Halldner L, Lundstrom S, Ullen F, Langstrom N, Larsson H, Nyman A, Gumpert CH, Rastam M, Anckarsater H, Cnattingius S, Johannesson M, Ingelsson E, Klareskog L, de Faire U, Pedersen NL, Lichtenstein P. The Swedish Twin Registry: establishment of a biobank and other recent developments. Twin Res Hum Genet 2013; 16: 317–329. [DOI] [PubMed] [Google Scholar]
- 9. Ganna A, Salihovic S, Sundstrom J, Broeckling CD, Hedman AK, Magnusson PK, Pedersen NL, Larsson A, Siegbahn A, Zilmer M, Prenni J, Arnlov J, Lind L, Fall T, Ingelsson E. Large‐scale metabolomic profiling identifies novel biomarkers for incident coronary heart disease. PLoS Genet 2014; 10: e1004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ingelsson E, Arnlov J, Sundstrom J, Lind L. The validity of a diagnosis of heart failure in a hospital discharge register. Eur J Heart Fail 2005; 7: 787–791. [DOI] [PubMed] [Google Scholar]
- 11. The Task Force on Heart Failure of the European Society of Cardiology . Guidelines for the diagnosis of heart failure. Eur Heart J 1995; 16: 741–751. [PubMed] [Google Scholar]
- 12. Levey AS, Stevens LA, Schmid CH, Zhang YP, Castro AF, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J, Co CKDE. A new equation to estimate glomerular filtration rate. Ann Intern Med 2009; 150: 604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nerpin E, Ingelsson E, Riserus U, Sundstrom J, Andren B, Jobs E, Larsson A, Lars L, Arnlov J. The association between glomerular filtration rate and left ventricular function in two independent community‐based cohorts of elderly. Nephrol Dial Transplant 2014; 29: 2069–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ganna A, Fall T, Salihovic S, Lee W, Broeckling CD, Kumar J, Hagg S, Stenemo M, Magnusson PKE, Prenni JE, Lind L, Pawitan Y, Ingelsson E. Large‐scale non‐targeted metabolomic profiling in three human population‐based studies. Metabolomics 2016; 12. [Google Scholar]
- 15. Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 2006; 78: 779–787. [DOI] [PubMed] [Google Scholar]
- 16. Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TWM, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007; 3: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. Biometrics 1959; 15: 639–640. [PubMed] [Google Scholar]
- 18. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Methodol 1995; 57: 289–300. [Google Scholar]
- 19. Ganna A, Lee D, Ingelsson E, Pawitan Y. Rediscovery rate estimation for assessing the validation of significant findings in high‐throughput studies. Brief Bioinform 2015; 16: 563–575. [DOI] [PubMed] [Google Scholar]
- 20. Textor J, Hardt J, Knüppel S. DAGitty: a graphical tool for analyzing causal diagrams. Epidemiology 2011; 22: 745. [DOI] [PubMed] [Google Scholar]
- 21. Agarwal SK, Chambless LE, Ballantyne CM, Astor B, Bertoni AG, Chang PP, Folsom AR, He M, Hoogeveen RC, Ni H, Quibrera PM, Rosamond WD, Russell SD, Shahar E, Heiss G. Prediction of incident heart failure in general practice: the Atherosclerosis Risk in Communities (ARIC) Study. Circ Heart Fail 2012; 5: 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Friedman JH, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010; 33: 22. [PMC free article] [PubMed] [Google Scholar]
- 23. Kundur AR, Singh I, Bulmer AC. Bilirubin, platelet activation and heart disease: a missing link to cardiovascular protection in Gilbert's syndrome? Atherosclerosis 2015; 239: 73–84. [DOI] [PubMed] [Google Scholar]
- 24. Schwertner HA, Jackson WG, Tolan G. Association of low serum concentration of bilirubin with increased risk of coronary artery disease. Clin Chem 1994; 40: 18–23. [PubMed] [Google Scholar]
- 25. Kimm H, Yun JE, Jo J, Jee SH. Low serum bilirubin level as an independent predictor of stroke incidence. Stroke 2009; 40: 3422–3427. [DOI] [PubMed] [Google Scholar]
- 26. Ladislav N, Libor V. Inverse relationship between serum bilirubin and atherosclerosis in men: a meta‐analysis of published studies. Exp Biol Med 2003; 228: 568–571. [DOI] [PubMed] [Google Scholar]
- 27. Breimer LH, Wannamethee G, Ebrahim S, Shaper AG. Serum bilirubin and risk of ischemic heart disease in middle‐aged British men. Clin Chem 1995; 41: 1504–1508. [PubMed] [Google Scholar]
- 28. Mueller‐Hennessen M, Dungen HD, Lutz M, Trippel TD, Kreuter M, Sigl J, Muller OJ, Tahirovic E, Witt H, Ternes P, Carvalho S, Peter E, Rein D, Schatz P, Herth F, Giannitsis E, Weis T, Frey N, Katus HA. A novel lipid biomarker panel for the detection of heart failure with reduced ejection fraction. Clin Chem 2017; 63: 267–277. [DOI] [PubMed] [Google Scholar]
- 29. Zordoky BN, Sung MM, Ezekowitz J, Mandal R, Han B, Bjorndahl TC, Bouatra S, Anderson T, Oudit GY, Wishart DS, Dyck JRB, Alberta HEART . Metabolomic fingerprint of heart failure with preserved ejection fraction. Plos One 2015; 10: e0124844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yeboah J, McNamara C, Jiang XC, Tabas I, Herrington DM, Burke GL, Shea S. Association of plasma sphingomyelin levels and incident coronary heart disease events in an adult population multi‐ethnic study of atherosclerosis. Arterioscler Thromb Vasc Biol 2010; 30: 628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kirk JM. Neonatal jaundice: a critical review of the role and practice of bilirubin analysis. Ann Clin Biochem 2008; 45: 452–462. [DOI] [PubMed] [Google Scholar]
- 32. Jolliffe N. Liver function in congestive heart failure. J Clin Investig 1930; 8: 419–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van Deursen VM, Damman K, Hillege HL, van Beek AP, van Veldhuisen DJ, Voors AA. Abnormal liver function in relation to hemodynamic profile in heart failure patients. J Card Fail 2010; 16: 84–90. [DOI] [PubMed] [Google Scholar]
- 34. Kolesnick R. Signal‐transduction through the sphingomyelin pathway. Mol Chem Neuropathol 1994; 21: 287–297. [DOI] [PubMed] [Google Scholar]
- 35. Green DR. Apoptosis and sphingomyelin hydrolysis: the flip side. J Cell Biol 2000; 150: F5–F7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tabas I. Nonoxidative modifications of lipoproteins in atherogenesis. Annu Rev Nutr 1999; 19: 123–139. [DOI] [PubMed] [Google Scholar]
- 37. Jiang XC, Yeang C, Li ZQ, Chakraborty M, Liu J, Zhang HQ, Fan YF. Sphingomyelin biosynthesis: its impact on lipid metabolism and atherosclerosis. Clin Lipidol 2009; 4: 595–609. [Google Scholar]
- 38. Jeong T, Schissel SL, Tabas I, Pownall HJ, Tall AR, Jiang X. Increased sphingomyelin content of plasma lipoproteins in apolipoprotein E knockout mice reflects combined production and catabolic defects and enhances reactivity with mammalian sphingomyelinase. J Clin Invest 1998; 101: 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Z, Basterr MJ, Hailemariam TK, Hojjati MR, Lu S, Liu J, Liu R, Zhou H, Jiang XC. The effect of dietary sphingolipids on plasma sphingomyelin metabolism and atherosclerosis. Biochim Biophys Acta 2005; 1735: 130–134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Proposed causal diagram for the association between circulating biomarkers and incident heart failure – Directed Acyclic Graph.
Figure S2. Validation of sphingomyelin(30:1) annotation.
Figure S3. Validation of urobilin annotation
Figure S4. Validation of sphingomyelin(28:1) annotation
Table S1. Sex‐specific baseline characteristics in participants of the PIVUS, ULSAM, and TwinGene cohorts.
Table S2. Hazard ratio estimates for all metabolites and incident heart failure in PIVUS and ULSAM – age and sex adjusted.
Table S3. Hazard ratio estimates for all metabolites and incident heart failure in TwinGene – age and sex adjusted.
Table S4. Assessment of potential confounders of the association between urobilin and sphingomyelin (30:1), and incident heart failure, respectively, in the PIVUS cohort (n = 829, number of events = 58).
Table S5. Association of each urobilin metabolite in ULSAM, PIVUS and TwinGene combined – age and sex adjusted.
Table S6. Association of each sphingomyelin in ULSAM, PIVUS and TwinGene combined – age and sex adjusted.
Table S7. Assessment of the association between urobilin and sphingomyelin(30:1), and incident heart failure in the PIVUS, ULSAM and TwinGene cohorts before and after multiple imputation in TwinGene.


