Abstract
Background and Aims
Most crucifer species (Brassicaceae) have small nuclear genomes (mean 1C-value 617 Mb). The species with the largest genomes occur within the monophyletic Hesperis clade (Mandáková et al., Plant Physiology174: 2062–2071; also known as Clade E or Lineage III). Whereas most chromosome numbers in the clade are 6 or 7, monoploid genome sizes vary 16-fold (256–4264 Mb). To get an insight into genome size evolution in the Hesperis clade (~350 species in ~48 genera), we aimed to identify, quantify and localize in situ the repeats from which these genomes are built. We analysed nuclear repeatomes in seven species, covering the phylogenetic and genome size breadth of the clade, by low-pass whole-genome sequencing.
Methods
Genome size was estimated by flow cytometry. Genomic DNA was sequenced on an Illumina sequencer and DNA repeats were identified and quantified using RepeatExplorer; the most abundant repeats were localized on chromosomes by fluorescence in situ hybridization. To evaluate the feasibility of bacterial artificial chromosome (BAC)-based comparative chromosome painting in Hesperis-clade species, BACs of arabidopsis were used as painting probes.
Key Results
Most biennial and perennial species of the Hesperis clade possess unusually large nuclear genomes due to the proliferation of long terminal repeat retrotransposons. The prevalent genome expansion was rarely, but repeatedly, counteracted by purging of transposable elements in ephemeral and annual species.
Conclusions
The most common ancestor of the Hesperis clade has experienced genome upsizing due to transposable element amplification. Further genome size increases, dominating diversification of all Hesperis-clade tribes, contrast with the overall stability of chromosome numbers. In some subclades and species genome downsizing occurred, presumably as an adaptive transition to an annual life cycle. The amplification versus purging of transposable elements and tandem repeats impacted the chromosomal architecture of the Hesperis-clade species.
Keywords: Genome size evolution, repetitive DNA, tandem repeats, retrotransposons, interstitial telomeric repeats (ITRs), chromosome organization, Bunias, Hesperis, Matthiola, Lineage III, Brassicaceae
INTRODUCTION
Angiosperms, flowering plants, exhibit 2440-fold variation in nuclear genome size. The smallest genome has only ~60 Mb, whereas the size of the largest angiosperm genome is almost 150 000 Mb and the mean and modal genome size equals 5020 and 587 Mb, respectively (Pellicer et al., 2018). Nuclear genomes expand as the consequence of whole-genome duplications (polyploidy) and due to the accumulation of transposable elements (TEs) and tandem repeats (e.g. Kubis et al., 1998; Macas et al., 2015; Willing et al., 2015; Gaiero et al., 2018; Pellicer et al., 2018). Genome expansion is counterbalanced by deletion-biased double-strand break repair, including transposon excision and homologous and illegitimate recombination (e.g. Devos et al., 2002; Hawkins et al., 2009; Waterworth et al., 2011; Vu et al., 2017). Large chromosome regions can be lost as the consequence of chromosomal rearrangements, such as deletions and translocations (Schubert and Lysak, 2011), and inversions moving inverted regions to more proximal chromosomal positions can increase the elimination of repetitive sequences due to higher illegitimate recombination rates in these regions (Ren et al., 2018). As genome expansion and downsizing mechanisms can be (in)active to strikingly different extents, huge genome and chromosome size variation can be encountered even in plant groups with overall constant chromosome numbers, such as grasses and the Pinaceae (Heslop-Harrison and Schwarzacher, 2011).
In comparison with the 2440-fold variation across all angiosperms, genome sizes of crucifer species (the mustard family or Brassicaceae) vary only by 52-fold (from 157 Mb in Arabidopsis thaliana to 8117 Mb in the tetraploid Hesperis matronalis; Bennett et al., 2003; Kiefer et al., 2014; https://brassibase.cos.uni-heidelberg.de), with most species having a small genome size (mean and modal genome size is 617 and 392 Mb, respectively; Lysak et al., 2009). In fact, a crucifer species, namely arabidopsis (A. thaliana) was considered to have the smallest genome (157 Mb; Bennett et al., 2003) among flowering plants until its special position was replaced by the extremely small genomes (∼60 Mb) of the bladderwort family (Lentibulariaceae; Greilhuber et al., 2006).
When analysing genome size variation across 3977 crucifer species classified in 341 genera and 52 tribes (Kiefer et al., 2014; https://brassibase.cos.uni-heidelberg.de), it becomes evident that the variation is not equally distributed among the tribes and six or so super-tribes, i.e. lineages or clades (Beilstein et al., 2006; Huang et al., 2016). With some rare exceptions, crucifer species with very large as well as the largest genome sizes (Lysak et al., 2009) belong to the Hesperis clade (Mandáková et al., 2017), also known as Lineage III (Beilstein et al., 2006) or Clade E (Huang et al., 2016). The monophyletic Hesperis clade comprises seven tribes harbouring ~350 species classified in ~48 genera (Mandáková et al., 2017; but see Chen et al., 2018; German and Al-Shehbaz, 2017, 2018 for recent taxonomic reappraisals in the clade). Among the several crucifer super-tribes, the Hesperis clade not only contains the largest genomes, but also exhibits the broadest range of genome sizes. Holoploid genome size varies by >30-fold, ranging from 265 Mb in Diptychocarpus strictus and Euclidium syriacum (Kiefer et al., 2014; https://brassibase.cos.uni-heidelberg.de; this study) to 8117 Mb in the tetraploid Hesperis matronalis (Kiefer et al., 2014; https://brassibase.cos.uni-heidelberg.de). Monoploid genome size varies 16.8-fold, ranging from 265 to 4273 Mb in H. sylvestris (this study). Interestingly, the extensive genome size variation contrasts with the evolutionary stability of chromosome numbers, with most species having rather low chromosome numbers (n = 6 or n = 7) (Mandáková et al., 2017). As noted by early scholars (Jaretzky, 1928; Manton, 1932), few chromosomes accommodating a large nuclear genome make the chromosomes of the Hesperis-clade species some of the largest chromosomes in the Brassicaceae.
In the present study, we aimed to analyse repeatomes of selected Hesperis-clade species to get a deeper insight into processes underlying genome size variation across the clade. To this end, we carried out low-pass Illumina sequencing of genomic DNA in seven diploid species representing six tribes as well as the 16-fold genome size variation within the Hesperis clade. In the context of gene-based phylogenetic hypotheses, our objective was to elucidate the directionality of repeatome evolution in the clade; in particular we aimed to analyse why genome obesity is not a universal feature of all species belonging to the apparently monophyletic super-tribe (Mandáková et al., 2017). In the case of large-genome species, we asked whether these genomes were inflated by only a few abundant repeats amplified to high copy numbers or due to the proliferation of many repeat types with fewer genomic copies. Finally, yet importantly, we aimed to compare chromosomal organization in small versus large crucifer genomes, to challenge the stereotype of the arabidopsis-type chromosomal organization being universal for all crucifer taxa.
MATERIALS AND METHODS
Plant material
Plants used in this study were grown from seeds or collected in the field (for the origins see Mandáková et al., 2017). Genomic DNA was extracted from fresh or silica-dried leaves using the NucleoSpin Plant II kit (Macherey-Nagel). Young inflorescences from several plants of the analysed species were collected and fixed in freshly prepared fixative (ethanol:acetic acid, 3:1) overnight, transferred to 70 % ethanol and stored at −20 °C until further use.
Genome size measurements
Holoploid genome sizes were estimated by flow cytometry. For each species, preferentially two intact petals or one young, intact leaf, ~1 cm in length, was prepared according to the two-step procedure of Otto (1990) in a simplified version (Doležel et al., 2007). The samples were stained (solution containing propidium iodide + RNAase IIA, both at final concentrations of 50 µg mL−1) for 5 min at room temperature and analysed using a CyFlow cytometer (Partec) equipped with a 532 nm diode-pumped solid-state laser (Cobolt Samba; Cobolt). A fluorescence intensity of 5000 particles was recorded. Pisum sativum ‘Ctirad’ (1C = 4.38 pg; Trávníček et al., 2015) served as the primary reference standard and Solanum pseudocapsicum as the secondary standard (1C = 1.29 pg recalculated against the primary reference). One individual of each species measured on three consecutive days was used for genome size estimation.
Low-pass genome sequencing
Genome sequencing of five species (Braya humilis, Bunias orientalis, Chorispora tenella, Dontostemon micranthus, Euclidium syriacum), generating 100-bp paired-end reads, was performed on an Illumina HiSeq 2000 platform at GATC Biotech (Konstanz, Germany), and genomes of two species (Hesperis sylvestris and Matthiola incana) were sequenced using an Illumina MiSeq, paired 300-bp reads, and MiSeq v3 reagents, at the sequencing core facility of the Oklahoma Medical Research Foundation (Oklahoma City, USA).
Phylogenetic analysis and ancestral genome size reconstruction
Internal transcribed spacer (ITS) sequences were obtained from BrassiBase (Kiefer et al., 2014; https://brassibase.cos.uni-heidelberg.de/) and ndhF sequences from NCBI GenBank (www.ncbi.nlm.nih.gov). Nucleotide sequences were aligned and manually checked using Geneious v11.1.5 (https://www.geneious.com;Kearse et al., 2012). Only sequences of Hesperis-clade species with known genome sizes were used for further phylogenetic analyses and reconstruction of genome size evolution. C-values of Hesperis-clade species were either estimated in the present study or adopted from Greilhuber and Obermayer (1999), Suda et al. (2005), Lysak et al. (2009), Kubešová et al. (2010) and BrassiBase (Kiefer et al., 2014; https://brassibase.cos.uni-heidelberg.de/) (Supplementary Data Table S1).
Phylogenetic unrooted trees for ITS and ndhF datasets were reconstructed using MrBayes v3.2.6 (Ronquist et al., 2012). In all Bayesian analyses, starting trees were random, four simultaneous Markov chains were run for 5 000 000 generations, burn-in values were set at 500 000 and trees were sampled every 5000 generations. Bayesian posterior probabilities were calculated using a Markov chain Monte Carlo sampling approach. The 50 % majority rule was used for constructing consensus trees. All parameters were inspected with Tracer v.1.6 (Rambaut and Drummond, 2009).
The R package GEIGER (Harmon et al., 2007) was used to estimate Pagel’s λ, measuring phylogenetic dependence of the observed trait, i.e. genome size. A λ value equal or close to 1 suggests trait evolution according a Brownian motion model. As λ values were close to 1 (0.96) for both datasets we used a Brownian motion model for further analyses.
Ancestral genome sizes were reconstructed for each node using the function ace in the R package APE (Paradis et al., 2004) using the Brownian motion-based maximum likelihood. The reconstructions were subsequently mapped onto the Bayesian phylograms using the function contMap in the package phytools (Revell, 2012).
Genome size and life forms
Information on life forms was obtained from Hohmann et al. (2015). Hesperis-clade species with known genome sizes were divided into two categories based on their life forms (annuals versus biennials and perennials). The Shapiro–Wilk normality test showed that the genome size values did not have a normal distribution. Thus, we used an unpaired two-sided Mann–Whitney test to find whether genome size differs significantly in annuals versus biennials and perennials. To test the correlation between genome size and life form we performed Spearman’s rank correlation test.
Data pre-processing and de novo identification of repetitive sequences
A quality check of paired-end reads was carried out using FastQC (Andrews, 2010). Raw sequencing data pre-processing was done before clustering analysis. Removal of reads with similarity to the PhiX was done using our custom-made script. Read-quality filtering (Phred score >20 and cutoff value 80 %), adapter trimming (removal of adapter-containing reads) and conversion of fastq to fasta were performed using the FASTX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) implemented within the Galaxy environment (Afgan et al., 2018). MiSeq reads were trimmed to 100 bp.
Repeat identification by similarity-based clustering of reads was performed using local installation of the RepeatExplorer pipeline (Novák et al., 2013) using (1) the maximum number of reads possible, and (2) the number of reads representing 0.05× genome coverage. Each species was analysed separately. The settings for each analysis were left at the default with the minimum overlap length for clustering set as 55 %, and the minimal overlap for assembly set as 40 %. Repeat clusters with genome proportions >0.01 % were annotated in detail. Both genome coverages were analysed with two or three replicates.
The detailed repeatome analysis was based on clustering with maximum reads as we aimed to capture all repetitive sequences responsible for genome size variation; a higher genome coverage (at least 0.01×) has to be used to estimate abundance of repeats with low(er) genome proportions (http://repeatexplorer.org/?page_id=179).
Clusters with known protein domains were classified by the RepeatExplorer pipeline directly. Other clusters were further analysed using similarity search tool BLAST (Altschul et al., 1990) against GenBank nucleotide and protein databases, and the software tool CENSOR (Kohany et al., 2006), which screens query sequences against a Viridiplantae reference database of repeats. Contigs of clusters classified as putative satellites were manually inspected and analysed using Tandem Repeat Finder (TRF; Benson, 1999) and Dotter (Sonnhammer and Durbin, 1995). Reconstruction of consensus monomer sequences of satellites was performed using the tandem repeat analyser TAREAN (Novák et al., 2017) pipeline; interlaced paired-end reads of individual species were used as inputs. TAREAN’s advanced option Perform cluster merging was used to merge clusters connected through paired-end reads. TAREAN is available as part of the RepeatExplorer2 pipeline (https://repeatexplorer-elixir.cerit-sc.cz/galaxy/).
Up to 14 % chloroplast DNA (cpDNA) was found in cluster analysis. It has been reported that cpDNA could be found incorporated in the nuclear genome (Roark et al., 2010). However, the significantly high proportion of cpDNA and high similarity to cpDNA of other crucifer species, verified by BLASTN to the NCBI nucleotide database, suggested that it might have come from the DNA extraction process, and thus we excluded cpDNA clusters from our analyses.
Cluster analysis of H. sylvestris data using the maximum number of reads resulted in an error due to high computation demands. Therefore, we used automatic filtering of abundant satellite repeats option as this automatic filtering tries to identify the most abundant tandem repeats and removes such sequences partially (10 % left) from analysis. Removal of abundant tandem repeats enabled us to analyse less abundant repeats and a higher number of reads in total. The modified clustering parameters helped to identify additional copies of TEs, particularly LTR retrotransposons (Ty3-gypsy/Athila and Ty1-copia/Ale elements).
Additionally, E. syriacum sequence data from the study by Jiao et al. (2017) were downloaded from the Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra). SRA archives (ERR1773712 to ERR1773714) were converted into fastq files with fastq-dump from SRA Toolkit v2.4.2. These data were submitted to the TAREAN pipeline (Novák et al., 2017). The assembled E. syriacum genome (Jiao et al., 2017) was downloaded from the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena), BioProject ID PRJEB16743. Sequence contigs were analysed using TRF (Benson, 1999). Satellite monomers obtained from the two E. syriacum datasets were compared and mapped to E. syriacum contigs by BLASTN (e-value 1e−3, identity >70 %). The Integrative Genomics Viewer (Robinson et al., 2011) was used to visualize satellite localization on assembled scaffolds (data not shown).
A comparative analysis of repetitive sequences of Hesperis-clade species was done on pooled reads of all species sampled to 0.01× genome coverage. The settings for the comparative analysis were the same as those for the individual species cluster analyses.
Correlation between genome size and repeat content
To test whether there were correlations between the amounts of different types of repeats with genome size variation in the Hesperis clade, we used the function lm for linear regression in the package stats in R software (R Development Core Team, 2013) using absolute amounts of repeats estimated for individual species.
Construction of phylogenetic tree for TE reverse transcriptase domains
Protein domain finder tool embedded in RepeatExplorer Galaxy platform (https://repeatexplorer-elixir.cerit-sc.cz/galaxy?tool_id=domains_finder&version=1.0.0) was used to find and classify all TE protein domains in concatenated contigs from the individual cluster analyses (contigs from individual species were distinguished by sample code). This tool uses the external aligning program LAST (Kiełbasa et al., 2011) and the RepeatExplorer database of TE protein domains (Viridiplantae). The Protein domain filter tool was then applied to filter out only contigs with reverse transcriptase (RT) domains. Default alignment quality criteria were used: minimum identity 35 %, minimum similarity 45 % and minimum alignment length 80 %. To extract protein sequences of RT domains, the Protein domain search tool was used. A database of protein domains derived from plant mobile elements is used in this tool for a similarity search using the fasty36 program (Pearson et al., 1997). Raw fasty36 output was filtered for minimal quality of alignment. The output consisted of protein sequences translated from query DNA and best matching sequences from the protein database. Two output datasets were created according to LTR retroelement superfamilies, for Ty1-copia-related sequences and for Ty3-gypsy sequences. Multialignment of protein domains was done in MAFFT v7.017 (Katoh and Standley, 2013) in Geneious v11.1.5 (https://www.geneious.com;Kearse et al., 2012) and manually checked. The phylogenetic trees were built using a Bayesian methods algorithm by MrBayes 3.2.6 (Ronquist et al., 2012); the number of generations was set to 5 000 000 and burn-in values were set at 500 000. Parameter values of each run were checked using Tracer v.1.6 (Rambaut and Drummond, 2009).
Identification of shared tandem repeats
Putative satellite sequences from all species were compared with each other by BLASTN (e-value 1e−3, identity >70 %) to assess their sequence similarity. BLAST searching against the GenBank nucleotide database of each satellite was done to investigate whether they showed similarity hits to already known satellite sequences from Brassicaceae species.
Chromosome preparations
Chromosome spreads from fixed young flower buds containing immature anthers were prepared according to published protocols (Mandáková and Lysak, 2016a). Briefly, selected flower buds were rinsed in distilled water and citrate buffer, and digested in 0.3 % cellulase, cytohelicase and pectolyase (all from Sigma–Aldrich) in citrate buffer at 37 °C for 3 h. After digestion, individual anthers were dissected and spread in 20 μL of 60 % acetic acid on a microscope slide placed on a metal hot plate (50 °C) for ~30 s. The preparation was then fixed in freshly prepared fixative (ethanol:acetic acid, 3:1) by dropping the fixative around the remaining drop of acetic acid and into it. Chromosome spreads were dried using a hair dryer, post-fixed in freshly prepared 4 % formaldehyde in distilled water and air-dried. Preparations were kept in a dust-free box at room temperature until used.
Fluorescence in situ hybridization probes
Oligonucleotide probes were designed from consensus DNA sequences of tandem repeat sequences (Supplementary Data Table S2). Target sequences (59–82 nt) were manually selected to obtain a high level of sequence complexity to maximize probe specificity and ensure a GC content between 30 and 50 %. The sequences were checked to minimize self-annealing and formation of hairpin structures in Geneious 11.1.5 (https://www.geneious.com, Kearse et al., 2012). The double-stranded DNA probes were generated and labelled with biotin-dUTP, digoxigenin-dUTP or Cy3-dUTP by nick translation as described by Mandáková and Lysak (2016b).
For retrotransposon probes, PCR primers were designed to the gag gene of various retrotransposon families (Supplementary Data Table S3). PCR products were sequenced at Macrogen Ltd. to validate them and then labelled by nick translation according to Mandáková and Lysak (2016b).
For comparative chromosome painting (CCP), chromosome-specific bacterial artificial chromosome (BAC) clones of A. thaliana grouped into contigs according to genomic blocks Jb and M of the Ancestral Crucifer Karyotype (Lysak et al., 2016) were used and labelled with biotin-dUTP and digoxigenin-dUTP, respectively (Mandáková and Lysak, 2016b).
Fluorescence in situ hybridization and microscopy
Labelled probes were pooled, ethanol-precipitated, dried and dissolved in 20 µL of 50 % formamide and 10 % dextran sulphate in 2× saline–sodium citrate (SSC) per slide. Then 20 µL of the labelled probe was pipetted onto a suitable slide and denatured on a hotplate at 80 °C for 2 min. Hybridization was carried out in a moist chamber at 37 °C overnight. Post-hybridization washing was performed in 20 % formamide in 2× SSC at 42 °C. The immunodetection of hapten-labelled probes was performed as described by Mandáková and Lysak (2016b). Chromosomes were counterstained with 2 μg mL−1 4′,6-diamidino-2-phenylindole (DAPI) in Vectashield. The preparations were photographed using a Zeiss Axioimager Z2 epifluorescence microscope with a CoolCube camera (MetaSystems). Images were acquired separately for all four fluorochromes using appropriate excitation and emission filters (AHF Analysentechnik). At least ten chromosome figures were photographed for each probe localized; however, due to combining different probes, almost all probes were localized on several slides repeatedly. The four monochromatic images were pseudocoloured, merged, processed and cropped using Photoshop CS (Adobe Systems). The images were processed only using the software functions applying to all pixels of the image.
Quantification of selected repeats using dot-blot analysis
Four repeats were quantified using a dot-blot analysis in C. tenella and H. sylvestris. We chose one satellite (ChTe2 and HeSy1) and one LTR retrotransposon (gag domain) from the Athila lineage (ChTe_Athila and HeSy_Athila) for each species. The radioactively labelled probes (synthesized oligonucleotides for satellites as described above, and purified and cloned PCR products for retroelements) were hybridized to diluted standards of unlabelled probes (0.125, 0.25, 0.5, 1 and 2 ng) and genomic DNA (1, 5, 50, 100 and 200 ng) of the two species onto Hybond-XL membrane (GE Healthcare). The dot-blot signals were quantified using a Typhoon FLA 9500 (GE Healthcare).
RESULTS
Extensive genome size variation versus chromosome number stasis
Our initial analysis confirmed that all the seven species analysed were diploid, with either 2n = 12 (H. sylvestris) or 2n = 14 (Br. humilis, Bu. orientalis, C. tenella, D. micranthus, E. syriacum and M. incana). Flow-cytometric analysis of nuclear DNA content revealed and confirmed extensive genome size variation among the seven species (Table 1). The smallest genome sizes were estimated for E. syriacum (254 Mb) and C. tenella (342 Mb), whereas H. sylvestris had the largest genome (4264 Mb). The four remaining species had medium to large genomes ranging from 1594 to 2611 Mb. Thus, the analysed species have comparable numbers of chromosomes, while their genome sizes differ by 16-fold and average chromosome size (genome size/haploid chromosome number) varies 20-fold (Table 1).
Table 1.
Chromosome numbers and genome sizes of the analysed Hesperis-clade plants
| Species | Tribe | 2n | Genome size (1C) | Average chromosome size | ||
|---|---|---|---|---|---|---|
| (pg) | (Mb) | (pg) | (Mb) | |||
| E. syriacum | Euclidieae | 14 | 0.26 | 254.28 | 0.04 | 36.33 |
| C. tenella | Chorisporeae | 14 | 0.35 | 342.30 | 0.05 | 48.90 |
| Br. humilis | Euclidieae | 14 | 1.63 | 1594.14 | 0.23 | 227.73 |
| D. micranthus | Dontostemoneae | 14 | 1.66 | 1623.48 | 0.24 | 231.93 |
| M. incana | Anchonieae | 14 | 2.20 | 2151.60 | 0.31 | 307.37 |
| Bu. orientalis | Buniadeae | 14 | 2.67 | 2611.26 | 0.38 | 373.04 |
| H. sylvestris | Hesperideae | 12 | 4.36 | 4264.08 | 0.73 | 710.68 |
1 pg = 978 Mb (Doležel et al., 2003).
Genome size evolution
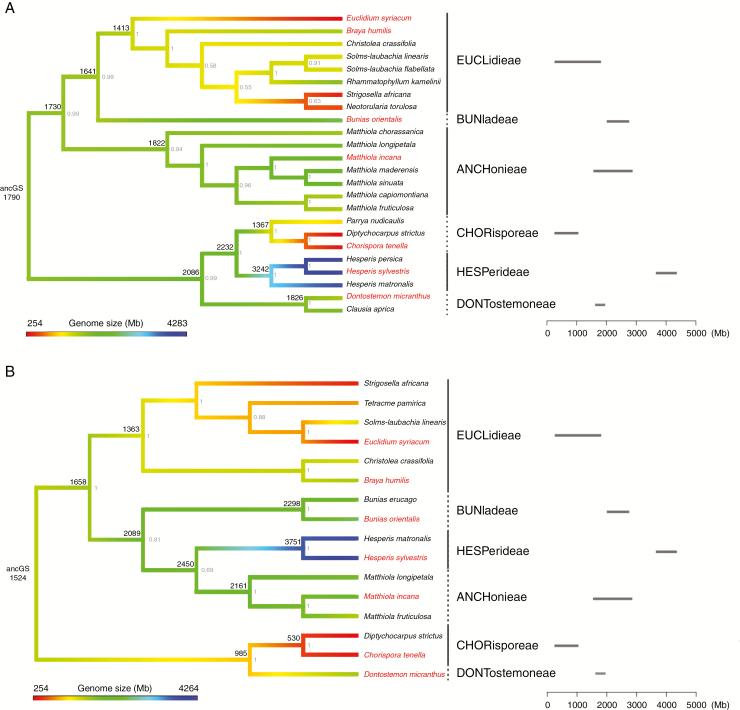
To reconstruct the evolution of genome size in the Hesperis clade, ITS and ndhF phylogenies were constructed using sequences retrieved from GenBank and BrassiBase (Kiefer et al., 2014; https://brassibase.cos.uni-heidelberg.de) for species with known C-values (Fig. 1). Although the two trees showed similar basal dichotomy, splitting the six tribes into two groups, the position of Hesperideae (HESP) was not consistent among the ITS and ndhF trees. Due to the conflicting position of HESP, both trees were used to model genome size evolution and infer ancestral genome size (ancCS) for the Hesperis clade (Fig. 1, Supplementary Data Table S4).
Fig. 1.
Bayesian phylogenetic trees of the Hesperis clade with results of ancestral genome size reconstruction. (A) ITS tree. (B) ndhF tree. See Supplementary Data Table 1 for GenBank accession numbers. The reconstructed genome sizes (Mb) are shown at the nodes; posterior probability values are shown in grey. Horizontal bars represent the range of Cx-values for each tribe (C-values are from Table 1 and Kiefer et al., 2014, https://brassibase.cos.uni-heidelberg.de). Species with an analysed repeatome are labelled in red.
For both phylogenies, Pagel’s λ was estimated to determine the phylogenetic signal of genome size variation. As the λ values (0.96) were close to 1.0 for both trees, genome size evolution should be correlated with the tree structure. In the ITS phylogeny, ancGS was estimated as 1790 Mb (Fig. 1A, Supplementary Data Table S4) and a similar value was inferred based on the ndhF tree: 1524 Mb (Fig. 1B, Supplementary Data Table S4).
While the topology of the ndhF tree (Fig. 1B) supports morphological differences between the two tribal subclades (Mandáková et al., 2017), namely between Chorisporeae (CHOR) and Dontostemoneae (DONT) on the one hand and Anchonieae (ANCH), Buniadeae (BUNI), Euclidieae (EUCL) and HESP on the other hand, the ~40 % more species used in the ITS tree provide a more realistic picture of genome size variation within the clade. In the context of the ITS tree (Fig. 1A), the inferred ancGS value points to independent genome size increases in ANCH, BUNI, DONT and HESP (note that only two C-values for DONT do not reflect the real extent of variation), accompanied by decreases in CHOR and EUCL. The maternal phylogeny (Fig. 1B) congruently suggests independent genome size increases in ANCH, BUNI and HESP, and downsizing in CHOR and EUCL (and DONT). As both inferred ancGS values are substantially bigger than the family’s mean (617 Mb) and modal (392 Mb) genome sizes (Lysak et al., 2009), the early diversification of the Hesperis clade was most likely marked by a genome size increase. The elevated ancestral genome size was subjected to stasis or further increase in ANCH, BUNI, DONT and HESP, while ~6-fold genome reductions occurred in CHOR and EUCL.
Genome size variation is correlated with life histories
In species with known genome sizes we tested whether the inter-species genome size differences are related to life-history strategies (Supplementary Data Table S1). The median and mean genome size of annual species (n = 9) was 697 and 1003 Mb, respectively. The species with prevalent perennial or biennial life history (n = 20) had median and mean genome size of 2054 and 2381 Mb, respectively. The median genome size was significantly lower in annuals than in perennials (Mann–Whitney test, P = 0.0024). We found a weak but significant positive correlation between increased genome size and perenniality (Spearman’s rank correlation test, ρ = 0.5549, P = 0.0018).
Repeatome analysis
To identify and analyse the underlying sequences responsible for genome size variation in the Hesperis clade, whole-genome shotgun sequencing was performed in the seven species (Table 1) using an Illumina platform, generating 100- (Br. humilis, Bu. orientalis, C. tenella, D. micranthus and E. syriacum) or 300-nucleotide (H. sylvestris and M. incana) paired-end reads. All the longer reads were trimmed to 100 nucleotides prior conducting analyses embedded in the RepeatExplorer pipeline (Novák et al., 2013). The cluster analysis permitting identification of reads derived from repetitive sequences was carried out for each species separately with two different samplings: (1) the maximum number of reads (genome coverage from 0.02× to 1.85×), and (2) at 0.05× genome coverage (Table 2). The detailed repeatome analysis was based on clustering with maximum reads.
Table 2.
Numbers of high-throughput sequencing reads used in the RepeatExplorer bioinformatic pipeline and clustering statistics
| Species | Maximum no. of reads | Genome coverage 0.05× | ||||||
|---|---|---|---|---|---|---|---|---|
| No. of reads | Genome coverage | Total repeats* (%) | Total repeats excluding cpDNA† (%) | No. of clusters | No. of reads | Total repeats* (%) | No. of clusters | |
| E. syriacum | 4 711 370 | 1.85 | 38.05 | 24.31 | 385 | 128 516 | 22.96 | 333 |
| C. tenella | 1 898 952 | 0.55 | 41.10 | 33.33 | 401 | 171 150 | 25.98 | 354 |
| Br. humilis | 4 235 224 | 0.27 | 53.46 | 42.40 | 454 | 796 316 | 43.28 | 444 |
| D. micranthus | 4 258 534 | 0.26 | 62.41 | 60.30 | 475 | 809 780 | 54.67 | 380 |
| M. incana | 2 589 598 | 0.12 | 65.98 | 62.40 | 467 | 1 075 800 | 61.43 | 430 |
| Bu. orientalis | 2 969 920 | 0.11 | 67.19 | 65.50 | 440 | 1 305 300 | 59.58 | 471 |
| H. sylvestris | 1 221 831 | 0.03 | 63.78‡ | 62.39‡ | 323 | 2 133 750 | No data¶ |
*Percentage of repeats in clusters constituting at least 0.01 % of the genome.
†Percentage of repeats in clusters constituting at least 0.01 % of the genome; clusters annotated as cpDNA were excluded.
‡RepeatExplorer analysis was performed with the advanced option of automatic filtering out the most abundant tandem repeats.
¶Not possible to compute due to computational resources restriction.
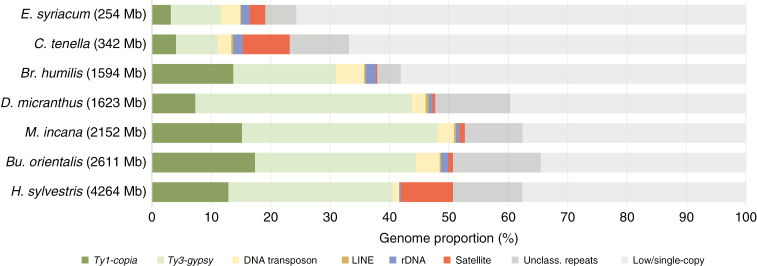
We identified main types of repetitive sequences and their genome proportions in all the seven species analysed (Table 3, Fig. 2). Small genomes, i.e. those of E. syriacum and C. tenella, exhibited the lowest proportion of repeats: 24.31 and 33.33 %, respectively. In medium-sized genomes, repetitive sequences represented at least 40 % of their genomes (Br. humilis, 42.4 %; D. micranthus, 60.3 %; M. incana, 62.4 %; Bu. orientalis, 65.5 %). Within the largest genome of H. sylvestris at first only 52.82 % of repetitive DNA (sampled at 0.01× genome coverage; data not shown) was identified. However, after filtering out the most abundant tandem repeats (see Materials and methods section for details), a new round of cluster analysis retrieved 10.96 % additional repetitive sequences, increasing the total repeat content in H. sylvestris to 63.78 % (62.39 % when excluding cpDNA reads; Table 2). Among all seven species, low- or single-copy sequences constituted 35 % (900 Mb, Bu. orientalis) to 76 % (192 Mb, E. syriacum) of the sequence data and ~4–14 % of repeats remained unclassified (Table 3).
Table 3.
Genome proportions and classification of repetitive sequences from the individual RepeatExplorer analyses performed using the maximum number of reads for clustering
| E. syriacum | C. tenella | Br. humilis | D. micranthus | M. incana | Bu. orientalis | H. sylvestris | ||
|---|---|---|---|---|---|---|---|---|
| Genome size (Mb) | 254.28 | 342.30 | 1 594.14 | 1 623.48 | 2 151.60 | 2 611.26 | 4 264.08 | |
| Repeat family | Lineage | % of the genome/Mb | ||||||
| LTR retrotransposons | 11.65/29.63 | 11.11/38.03 | 31.02/494.51 | 43.74/710.12 | 48.11/1035.14 | 44.48/1161.41 | 40.56/1729.51 | |
| Ty1-copia | Ale | 0.09/0.23 | 0.10/0.35 | 1.36/21.69 | 0/0 | 2.75/59.17 | 2.20/57.45 | 2.00/85.28 |
| Angela | 1.70/4.33 | 1.74/5.96 | 9.39/149.69 | 3.38/54.88 | 9.76/210.00 | 6.19/161.64 | 7.30/311.28 | |
| Bianca | 0.30/0.77 | 0.90/3.09 | 0.80/12.76 | 1.05/17.05 | 1.13/24.32 | 1.94/50.66 | 1.79/76.33 | |
| Ivana/Oryco | 0.14/0.36 | 0.08/0.28 | 0.20/3.19 | 0.24/3.90 | 0.06/1.30 | 0.21/5.48 | 0.05/2.13 | |
| Maximus/SIRE | 0.55/1.4 | 0.08/0.28 | 0.85/13.56 | 1.15/18.68 | 0.07/1.51 | 0.90/23.50 | 1.14/48.61 | |
| TAR | 0/0 | 0.17/0.59 | 0.06/0.96 | 0.11/1.79 | 0.35/7.54 | 0.38/9.92 | 0.22/9.38 | |
| Tork | 0.12/0.31 | 0.12/0.42 | 0.26/4.15 | 0.35/5.69 | 0.20/4.31 | 0.51/13.32 | 0.38/16.20 | |
| Unclassified | 0.30/0.77 | 0.88/3.02 | 0.79/12.60 | 1.03/16.73 | 0.84/18.08 | 5.02/131.09 | 0/0 | |
| Total | 3.20/8.14 | 4.06/13.90 | 13.71/218.56 | 7.31/118.68 | 15.16/326.19 | 17.36/453.31 | 12.88/ 549.21 | |
| Ty3-gypsy | Chromovirus | 1.27/3.23 | 0.64/2.20 | 1.79/28.54 | 1.69/27.44 | 1.63/35.08 | 1.68/43.87 | 1.75/74.62 |
| CRM | 1.23/3.13 | 0.62/2.12 | 1.53/24.39 | 1.16/18.83 | 0.98/21.09 | 1.04/27.16 | 0.75/31.98 | |
| Tekay | 0.04/3.10 | 0.02/0.08 | 0.26/4.15 | 0.53/8.61 | 0.65/13.99 | 0.64/16.71 | 1.00/42.64 | |
| Athila | 2.19/5.57 | 3.25/11.13 | 11.29/179.98 | 22.62/367.24 | 22.56/485.41 | 10.73/280.19 | 20.54/875.84 | |
| Ogre/Tat | 2.42/6.16 | 0.16/0.55 | 0.35/5.58 | 8.05/130.7 | 6.57/141.37 | 1.33/34.73 | 0.75/31.98 | |
| Unclassified | 2.57/6.54 | 3.00/10.27 | 3.88/61.86 | 4.07/66.08 | 2.20/47.34 | 13.38/349.39 | 4.64/197.85 | |
| Total | 8.45/21.49 | 7.06/24.17 | 17.31/275.95 | 36.43/591.44 | 32.95/708.96 | 27.12/708.17 | 27.68/1180.30 | |
| DNA transposons | CACTA | 0.70/1.78 | 0.59/2.02 | 1.62/25.83 | 1.81/29.39 | 1.71/36.80 | 2.09/54.58 | 0.63/26.86 |
| Helitron | 0.62/1.58 | 0.36/1.24 | 0.22/3.51 | 0.03/0.49 | 0.22/4.74 | 0.02/0.52 | 0.05/2.13 | |
| Mutator | 0.37/0.95 | 0.87/2.98 | 1.27/20.25 | 0.40/6.50 | 0.38/8.18 | 1.38/36.04 | 0.36/15.35 | |
| Unclassified | 1.56/3.97 | 0.45/1.55 | 1.61/25.67 | 0.08/1.30 | 0.46/9.90 | 0.50/13.06 | 0/0 | |
| Total | 3.25/8.27 | 2.27/7.78 | 4.72/75.25 | 2.32/37.67 | 2.77/59.60 | 3.99/104.19 | 1.04/44.35 | |
| LINE | 0.14/0.36 | 0.31/1.07 | 0.26/4.15 | 0.51/8.28 | 0.32/6.89 | 0.23/6.01 | 0.08/3.41 | |
| rDNA | 1.38/3.51 | 1.64/5.62 | 1.69/26.95 | 0.68/11.04 | 0.63/13.56 | 1.20/31.36 | 0.25/10.66 | |
| Satellites | 2.69/6.85 | 7.88/26.98 | 0.26/4.15 | 0.46/7.47 | 0.87/18.72 | 0.80/20.89 | 8.77/373.96 | |
| Unclassified repeats | 5.20/13.23 | 9.98/34.17 | 3.96/63.13 | 12.53/203.43 | 9.70/208.71 | 13.90/362.97 | 11.68/498.04 | |
| Low/single-copy sequences | 75.69/192.47 | 66.67/228.22 | 57.60/918.23 | 39.70/644.53 | 37.60/809.01 | 34.50/900.88 | 37.61/1603.72 | |
| All repeats total | 24.31/61.82 | 33.33/114.09 | 42.40/675.92 | 60.30/978.96 | 62.40/1342.60 | 65.50/1710.38 | 62.39/2660.36 |
Fig. 2.
Relative abundances of repeat families and low/single-copy sequences identified in genomes of the seven Hesperis-clade species analysed.
To determine how reliable our in silico estimates of repeat abundances were, we quantified the number of genomic copies for one tandem repeat and Athila retrotransposon (gag domain) by a dot-blot analysis in two species with contrasting genome sizes. The dot-blot and in silico estimates were largely congruent for C. tenella (1C = 342 Mb) and H. sylvestris (1C = 4264 Mb), except for the ChTe2 tandem repeat in C. tenella, being 1.75-fold more abundant in the dot-blot analysis (Supplementary Data Table S5). This discrepancy suggests that in silico-estimated abundances of tandem repeats can be somewhat underestimated compared with dot-blot or Southern blot analyses due to G/C bias in Illumina reads (Chen et al., 2013) and tandem repeats usually being A/T-rich.
Retrotransposon diversity and abundances
In all seven genomes, LTR retrotransposons made up the majority of repeatomes, ranging from 11.11 % in C. tenella to nearly 48.11 % in M. incana (Table 3). Although H. sylvestris has the largest genome among the species analysed (Table 1), only 40.56 % (1 729.51 Mb) of its genome was identified to be built from LTR retrotransposons. The identified Ty1-copia elements belonged to seven lineages (Ale, Angela, Bianca, Ivana/Oryco, Maximus/SIRE, TAR and Tork; Table 3) out of the 16 known lineages (Neumann et al., 2019). The identified Ty3-gypsy elements belonged to two major lineages (Neumann et al., 2019): Chromovirus (represented by CRM and Tekay clades) and non-Chromovirus (Athila and Ogre/Tat clades; Table 3).
In all genomes, LTR retroelements of the Ty3-gypsy superfamily prevailed and were mainly represented by the Athila clade, followed by Ogre/Tat (Table 3). The abundance of Athila elements ranged from 2.19 % in E. syriacum to 22.62 % in D. micranthus. In the smallest genome, that of E. syriacum, the Ogre/Tat element was the most abundant Ty3-gypsy element (2.42 %), followed by Athila (2.19 %) and Chromovirus (1.27 %, mainly CRM lineage). However, the Ogre/Tat clade was most amplified in genomes of M. incana (6.57 %) and D. micranthus (8.05 %). Some Ty3-gypsy elements remained unclassified, as we were not able to assign them clearly to any lineage; the highest proportion of unclassified Ty3-gypsy elements was identified in the larger genomes of H. sylvestris (~10 %) and Bu. orientalis (~13 %). In all but one species, the Chromovirus lineage was represented by the CRM and Tekay clades; in H. sylvestris, the Tekay clade was more abundant than CRM.
Ty1-copia retroelements, represented mainly by the Angela lineage, were 2- to 5-fold less abundant than Ty3-gypsy elements (Table 3). Angela retroelements occupied 1.70 % (E. syriacum) to 9.76 % (M. incana) of the genome. Other common lineages in medium- and large-sized genomes were Ale (from 1.36 % in Br. humilis to 2.75 % in M. incana), Bianca (from 0.80 % in Br. humilis to 1.94 % in Bu. orientalis) and Maximus (from 0.07 % in M. incana to 1.15 % in D. micranthus). The representation of Ty1-copia retroelements in D. micranthus was significantly lower than in other medium-sized genomes (e.g. Ale was not identified and Angela elements occupied only 3.38 % of the genome). In small-sized genomes, after Angela, the second most abundant Ty1-copia element was Maximus in E. syriacum (0.55 %) and Bianca in C. tenella (0.80 %). Other Ty1-copia lineages, such as Ivana/Oryco, TAR and Tork, were found only in low amounts or were absent in repeat clusters constituting at least 0.01 % of a genome (Table 3).
From non-LTR retrotransposons, LINE elements were identified only at very low genome proportions in the analysed species: 0.08 % in H. sylvestris to 0.51 % in D. micranthus (Table 3). MITE and SINE elements were not detected in clusters constituting at least 0.01 % of a genome. DNA transposons were represented by abundances ranging from 1.04 % in H. sylvestris up to 4.72 % in Br. humilis; the most abundant of these were CACTA and Mutator elements (Table 3).
With the exception of H. sylvestris, tandem repeats did not contribute significantly to genome expansion in the Hesperis-clade species
Tandem repeats were found in different abundances, from a very low (0.26 % in Br. humilis and 0.46 % in D. micranthus) to a high genome proportion in H. sylvestris (8.77 %) (Table 3 and Fig. 2). Results of tandem repeat analysis are summarized in Table 4. The identified monomer sizes were variable among the seven species, ranging from 20 to ~350 bp. The 825-bp ChTe2 satellite identified in C. tenella had an exceptional monomer length.
Table 4.
Tandem repeats identified by RepeatExplorer and TAREAN analyses. Only repeats with genome proportion >0.01 % were analysed and are listed
| Species | Tandem repeat | Monomer length (bp) | Genome proportion (%) |
|---|---|---|---|
| E. syriacum | EuSy1 (EuSy1A, EuSy1B) | 60 (motif) | 2.70 |
| EuSy2 | 20 | 0.05 | |
| EuSy3 | 354 | 0.03 | |
| EuSy4 | 132 | 0.01 | |
| C. tenella | ChTe1 | 39 | 2.27 |
| ChTe2 | 825 | 1.60 | |
| ChTe3 | 139 | 0.84 | |
| ChTe4 | 102 | 0.12 | |
| ChTe5 | 28 | 0.04 | |
| ChTe6 | 52 | 0.02 | |
| ITR and telomeric repeat | 7 | 2.72 | |
| Br. humilis | BrHu1 | 161 | 0.16 |
| BrHu2 | 295 | 0.04 | |
| BrHu3 | 87 | 0.02 | |
| BrHu4 | 338 | 0.02 | |
| BrHu5 | 345 | 0.02 | |
| telomeric repeat | 7 | 0.24 | |
| D. micranthus | DoMi1 | 36 | 0.30 |
| DoMi2 | 143 | 0.06 | |
| DoMi3* | 350 | 0.05 | |
| DoMi4 | 26 | 0.03 | |
| DoMi5 | 354 | 0.02 | |
| DoMi6 | 182 | 0.01 | |
| M. incana | MaIn1* | 352 | 0.58 |
| MaIn2 | 355 | 0.10 | |
| MaIn3 | 69 | 0.08 | |
| MaIn4 | 88 | 0.06 | |
| MaIn5 | 590 | 0.05 | |
| Bu. orientalis | BuOr1* | 352 | 0.36 |
| BuOr2 | 192 | 0.18 | |
| BuOr3 | 179 | 0.10 | |
| BuOr4 | 20 | 0.09 | |
| BuOr6 | 171 | 0.01 | |
| BuOr7 | 77 | 0.01 | |
| BuOr8 | 177 | 0.01 | |
| BuOr9 | 490 | 0.01 | |
| telomeric repeat | 7 | 0.40 | |
| H. sylvestris | HeSy1 | 91 | 7.38 |
| HeSy2 | 161 | 0.69 | |
| HeSy3 | 91 | 0.08 | |
| HeSy4 | 200 | 0.07 | |
| HeSy5 | 174 | 0.06 |
*Shared repeats.
In the small-sized genomes of E. syriacum and C. tenella, tandem repeats occupied 2.78 % (four different repeats) and 7.61 % (seven different repeats), respectively. In the E. syriacum genome, while only 0.08 % of the genome was found to consist of typical tandemly repeated DNA, a satellite family of non-homogeneous monomers containing a 60-bp repetitive motif occupied ~ 2.70 % of the genome. All contigs from the RepeatExplorer cluster analysis whose graph shapes indicated putative tandem repeats were further analysed using Dotter and TRF to create self-dot plots and to identify satellite monomer lengths, respectively. The 60-bp motif was identified by TRF using all reads (average 75 % matches) and by the TAREAN pipeline using sampled reads, which additionally identified satellites with monomer lengths of 519, 179, 60 and 40 bp. To further investigate these sequences, we analysed the sequenced E. syriacum genome (Jiao et al., 2017) by TRF and TAREAN. Whereas TAREAN identified two satellites with a monomer length of 717 and 377 bp, the TRF analysis revealed two more monomer lengths: 357 and 397 bp. All the identified monomers contained the 60-bp motif (Supplementary Data Fig. S1). In C. tenella, approximately one-third (2.72 %) of the tandem repeats identified were represented by ITRs derived from the arabidopsis-type telomeric repeat (TTTAGGG).
In species with medium-sized genomes, tandem repeats represented <0.9 % of their genomes. In M. incana and Bu. orientalis, tandem repeats constituted only 0.87 % (five different satellites) and 0.77 % (nine different satellites) of the genome, respectively. Among the five identified tandem repeats in H. sylvestris, the 91-bp HeSy1 satellite repeat occupied 7.38 % of the genome.
Chromosomal localization of the identified repeats
Chromosomal localization of the identified repeats was determined by fluorescence in situ hybrization (FISH) of fluorochrome- or hapten-labelled DNA probes to mitotic chromosomes. To localize retrotransposons, probes designed to the gag domain of Angela, Athila and Chromovirus were used.
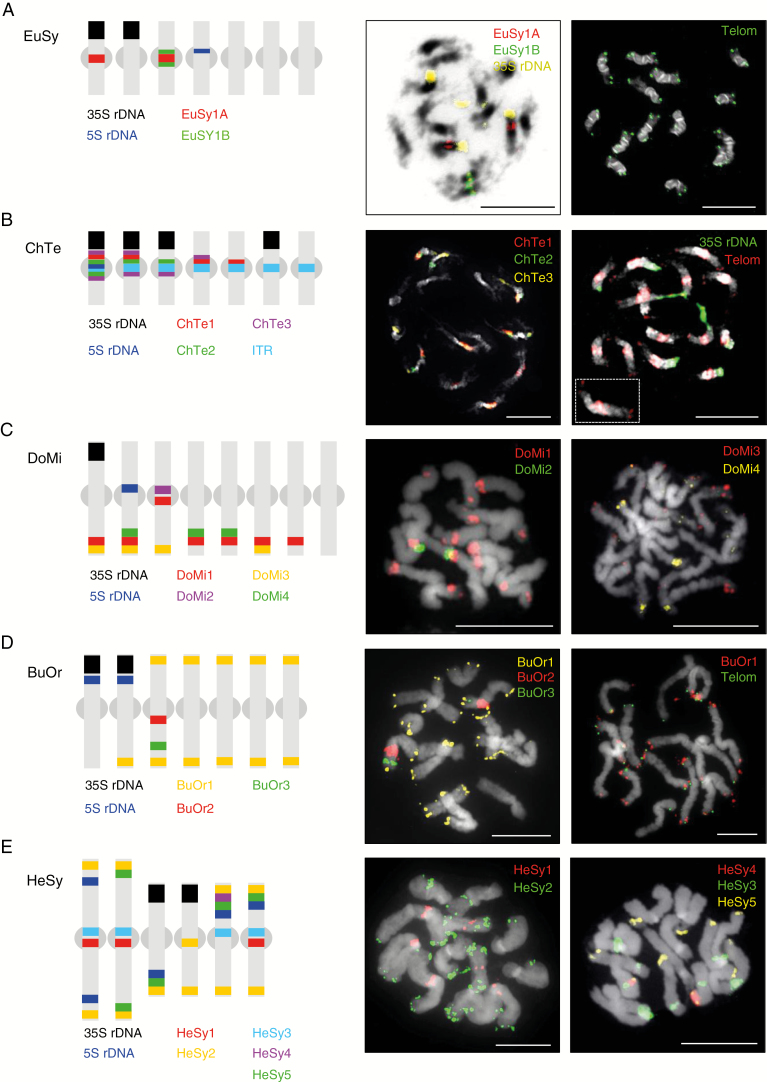
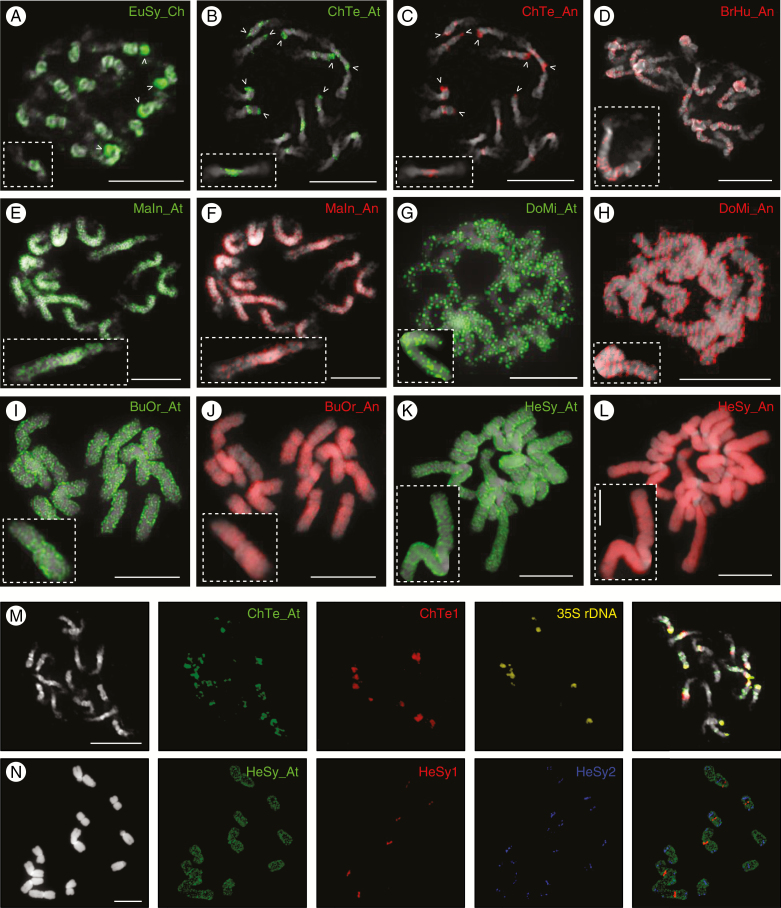
In species with a small genome size (C. tenella and E. syriacum), tandem repeats as well as retrotransposons clustered within heterochromatic pericentromere regions. In E. syriacum, FISH of DNA probes corresponding to consensus monomer sizes of 357 bp (EuSy1A) and 377 bp (EuSy1B), containing the 60-bp repetitive motif, showed that these repeats occurred on two and one chromosome pair(s), respectively (Fig. 3A). In C. tenella, three major tandem repeats formed pericentromere chromatin (Fig. 3B). The 39-bp ChTe1 satellite (2.27 % of the genome) localized to four chromosome pairs, the 825-bp ChTe2 repeat (1.60 %) provided weak hybridization signals on three chromosome pairs, and the 139-bp ChTe3 repeat (0.84 %) gave a stronger hybridization signal at the heterochromatin/euchromatin boundary of four chromosome pairs. The large blocks of ITRs (~2.7 %) were located at all pericentromeres in C. tenella (Fig. 3B), whereas telomeric repeats were localized only at chromosome ends in E. syriacum (Fig. 3A). In both species, LTR retrotransposons were present in all pericentromere regions (Fig. 4A–C), largely co-localizing with the identified tandem repeats (Fig. 4M). Apart from the pericentromeric heterochromatin, Chromovirus and Athila retroelements co-localized with four terminal nucleolar organizing regions (NORs) in E. syriacum (Fig. 4A) and eight NORs in C. tenella (Fig. 4B). The DNA probe for the Angela retrotransposon hybridized to all pericentromeres and the eight NORs in C. tenella (Fig. 4C).
Fig. 3.
Chromosomal localization of the most abundant tandem repeats and rDNA loci on mitotic metaphase chromosomes in five Hesperis-clade species. Telomeres and ITRs (B) were localized using a FISH probe for the arabidopsis-type telomeric repeat. Chromosomes were counterstained by DAPI (displayed in black and white); FISH signals are shown in colour as indicated. Grey spheroids in the schematic ideograms represent (peri)centromeric regions. EuSy, E. syriacum, ChTe, C. tenella; DoMi, D. micranthus; BuOr, Bu. orientalis; HeSy, H. sylvestris. All scale bars = 10 µm.
Fig. 4.
Chromosomal localization of dispersed repeats on mitotic metaphase chromosomes in seven Hesperis-clade species. (A) E. syriacum (EuSy), (B, C) C. tenella (ChTe), (D) Br. humilis (BrHu), (E, F) M. incana (MaIn) (G, H) D. micranthus (DoMi), (I, J) Bu. orientalis (BuOr), (K, L) H. sylvestris (HeSy) (M, N) Co-localization of tandem repeats (Fig. 3) and Athila retrotransposons in C. tenella (M) and H. sylvestris (N). Arrowheads in (A–C) point to 35S rDNA (NOR) loci. Chromosomes were counterstained by DAPI (displayed in black and white); FISH signals are shown in colour as indicated. Lineage abbreviations: An, Angela; At, Athila; Ch, Chromovirus. Scale bars (complete chromosome spreads) = 10 µm; (insets) = 5 µm (L).
In medium-sized genomes (Br. humilis, D. micranthus, Bu. orientalis and M. incana), tandem repeats predominantly constituted pericentromere and subtelomere heterochromatic regions. In D. micranthus, the 36-bp DoMi1 satellite (0.30 % of genome) localized to subtelomeric regions of six chromosome pairs and, together with the 143-bp DoMi2 satellite (0.06 %), to the pericentromere of an additional chromosome pair (Fig. 3C). The 350-bp DoMi3 satellite (0.05 %) localized to subtelomeric regions of four chromosome pairs, whereas the 26-bp DoMi4 repeat (0.03 %) occurred on three chromosomes (Fig. 3C). In Bu. orientalis, the 352-bp tandem repeat BuOr1 (0.36 % of genome) showed localization at chromosome termini of six out of the seven chromosome pairs (Fig. 3D). FISH with the BuOr1 satellite and the arabidopsis-like telomeric repeat showed that the newly identified repeat occupied the most distal chromosome regions immediately adjacent to the telomeric repeats at 11 chromosome ends (Fig. 3D). The 192-bp satellite BuOr2 (0.18 %), together with the 179-bp BuOr3 (0.10 %), was localized on the same arm of a single chromosome pair (Fig. 3D).
In Br. humilis and M. incana, LTR retrotransposons co-localized with pericentromeric heterochromatin and both adjacent chromosome arms, except for the most proximal regions (Fig. 4D–F). In Bu. orientalis and D. micranthus the Athila and Angela retrotransposons showed dispersed distribution along the entire length of all chromosomes (Fig. 4G–J).
In H. sylvestris, the most abundant 91-bp tandem repeat, HeSy1 (7.38 % of the genome), was localized at pericentromeres of only three chromosome pairs (Fig. 3E). The 91-bp HeSy3 (0.08 %), showing 80 % sequence identity to HeSy1, co-localized with HeSy1 on three chromosome pairs, in addition to a solo localization on a fourth chromosome. The 161-bp satellite HeSy2 (0.69 %) localized to ten subtelomeric regions; the 174-bp HeSy5 (0.06 %) had a similar localization, with signals on four chromosome pairs, and the 200-bp HeSy4 (0.07 %) localized to one arm of a single chromosome pair (Fig. 3E). In H. sylvestris, retrotransposon probes hybridized along the entire length of all chromosome pairs, except for (peri)centromeric regions (Fig. 4K, L). FISH with DNA probes for HeSy1 and Athila and Angela (not shown) retrotransposons confirmed that the (peri)centromeric regions with a low abundance of dispersed repeats were occupied by tandem repeats (Fig. 4N).
Repeat content is positively correlated with genome size
We tested whether the estimated abundances of the identified repeats reflect the ~16-fold genome size variation among the analysed species (Supplementary Data Table S6). A strong positive correlation was found between the repeat content and genome size (R2 = 0.984, P = 1.041e−5), but also between abundances of both LTR retrotransposon superfamilies and increasing genome size (Ty1-copia plus Ty3-gypsy, R2 = 0.967, P = 4.168e−5; Ty3-gypsy, R2 = 0.940, P = 0.0003; Ty1-copia, R2 = 0.918, P = 0.0007), and in particular for both families, Athila and Angela (Athila, R2 = 0.874, P = 0.0020; Angela, R2 = 0.885, P = 0.0016). Despite a significant positive correlation between tandem repeat contents and genome size variation, the lower R2 value (0.604) indicates that tandem repeat amplification influenced genome size expansions across the Hesperis clade to a lesser extent than proliferation of LTR retrotransposons.
Phylogenetic relationships among the identified LTR retroelement lineages
Phylogenetic analyses based on the reverse transcriptase (RT) sequence of the identified LTR retrotransposons were carried out to assess whether their relationships reflect tribal relationships within the Hesperis clade. As expected, Ty1-copia and Ty3-gypsy elements clustered into major clades, namely Angela, Ale, Bianca, Ivana/Oryco, Maximus/SIRE, TAR and Tork (Supplementary Data Fig. S2), and Athila, Chromovirus and Ogre/Tat (Supplementary Data Fig. S3), respectively. No species-specific LTR retroelements were found. These analyses further supported the antiquity and ubiquity of LTR retrotransposon lineages shared among the six tribes.
Identification of shared repeats within the Hesperis clade and across the Brassicaceae
A comparative RepeatExplorer cluster analysis was performed by pooling single dataset reads used in individual analyses as random samples corresponding to 0.01× genome coverage; the total of 1 284 124 reads were analysed, the number of reads per species ranging from 25 704 (E. syriacum) to 409 540 (H. sylvestris). The detailed analysis of the first 300 clusters showed variation in the proportion of reads contributed by individual species due to a positive correlation between abundances of species-specific reads and genome size of the analysed species. The majority of the first 300 repeat clusters contained sequences of all or most species analysed and were annotated as LTR retrotransposons (Supplementary Data Fig. S4).
In contrast to LTR retrotransposons, most of the tandem repeat clusters were made up of reads originating from a single species and no tandem repeat was shared among all the species analysed. To compare the sequence identity of all the identified tandem repeats, we BLASTed these sequences against each other and against known sequences in the NCBI GenBank database. A tandem repeat with an average monomer size of 352 bp was found to be shared among Bu. orientalis (BuOr1), M. incana (MaIn1) and partly D. micranthus (DoMi3). These three satellites showed hits to Brassica oleracea (pBoSTRb) and Brassica rapa (pBrSTRb) subtelomeric satellites (Koo et al., 2011) with identities up to 79 % for BuOr1 and MaIn1, 67 % for DoMi3 (Supplementary Data Fig. S5).
The CRAMBO tandem repeat (CRAMBO7 and CRAMBO.6 both 338 bp, and CRAMBO.11 309 bp), previously found only in Cardamine species (Mandáková et al., 2013), was found in the Br. humilis genome as the 338-bp BrHu4 tandem repeat (0.02 % of the genome, Table 4). In a pairwise BLASTN comparison, the BrHu4 repeat and the three CRAMBO variants (accession numbers JQ412178, JQ412179 and JQ412180) exhibited 96–99 % sequence identity with 81–95 % query coverage. The remaining identified tandem repeats did not show a significant sequence similarity to already known repeats.
Feasibility of comparative BAC-based painting decreases with increasing genome size and repeat content
While reconstructing genome evolution in Hesperis-clade tribes by CCP based on arabidopsis BAC clones (Mandáková et al., 2017), we noticed that the method was less efficient or even not applicable to species with large(r) genomes. Here we used the repeatome data and chromosomal localization of the identified repeats to reassess the feasibility of BAC painting in crucifer species with large genomes.
In six Hesperis-clade species and under identical experimental conditions, CCP with BAC contigs spanning genomic blocks Jb and M and forming chromosome 4 of CEK (ancestral karyotype of Clade E; Mandáková et al., 2017) demonstrated that chromosome specificity of painting probes and overall efficacy of CCP gradually decreased with increasing repeat content and genome size (Supplementary Data Fig. S6). Whereas in C. tenella and E. syriacum both painting probes provided highly specific and strong hybridization signals, weaker, less specific and homogeneous signals were observed in D. micranthus and M. incana. In Bu. orientalis and H. sylvestris, painting was even more compromised, with fluorescent signals hardly specific and distinguishable.
DISCUSSION
Due to the prominent role of the minute arabidopsis genome in plant research, crucifers are traditionally viewed as an angiosperm lineage harbouring species with comparably small genomes. Here we showed that species and genera of the Hesperis clade represent an exception to the rule and that these genomes followed evolutionary trajectories different from most crucifer taxa.
Genome size evolution in the Hesperis clade: genome obesity, with rare genome downsizing
Although based on a very limited dataset, our reconstruction of ancestral genome size suggested that the common ancestor of the Hesperis clade (called CEK; Mandáková et al., 2017) most likely had a genome larger (~1600 Mb) than the modal (392 Mb) and mean (617 Mb) C-values for Brassicaceae species, and that the expansion of the ancestral genome has preceded the tribal diversification within the clade. As the ancestral genome upsizing was followed by further genome size increases in all six tribes (Fig. 1), the Hesperis clade genomes must have intrinsic propensities to tolerate or benefit from further genome expansion. When plotting the available C-values on phylogenetic trees of two tribes harbouring species with small and large genomes, namely Chorisporeae and Euclidieae, the prevailing tendency for genome expansion is further supported. In Chorisporeae (~63 species in four genera), the small Chorispora/Diptychocarpus subclade (12 species), containing species with small genomes, is sister to or younger than (German et al., 2011; BrassiBase, https://brassibase.cos.uni-heidelberg.de) the species-rich Parrya/Litwinowia subclade of 43 perennial species with genomes presumably as large as that of Parrya nudicaulis. Thus, these phylogenies point to a more recent origin of Chorispora/Diptychocarpus genomes followed by genome downsizing. In the diverse and species-rich Euclidieae (28 genera and 152 species; Chen et al., 2018), large genomes of perennial species prevail (Supplementary Data Table S1), whereas small genomes have been identified so far only in the annual species E. syriacum (one species in the genus), Neotorularia torulosa (~14 species) and Strigosella africana (24 species). The dominance of large genomes and the phylogenetic position of the three genera in the tribe (Chen et al., 2018) point to genome downsizing specific for Euclidium and (some) species of Neotorularia and Strigosella.
As the genome obesity of Hesperis-clade species was caused mainly by the activity of LTR retrotransposons, particularly Ty3-gypsy elements, whole-genome duplication(s) (WGD) as a possible mechanism underlying the genome size increases can be ruled out. This was corroborated by earlier CCP analyses which failed to detect duplicated genomic regions in all Hesperis-clade species analysed (Mandáková et al., 2017). Interestingly, when comparing genome sizes in species from the 13 crucifer clades (Lysak et al., 2009; Kiefer et al., 2014; https://brassibase.cos.uni-heidelberg.de/;Hohmann et al., 2015) that have undergone a mesopolyploid WGD (Mandáková et al., 2017), it turns out that these species have usually substantially smaller genome sizes than many Hesperis-clade species. This is due to long-lasting and genome-wide post-polyploid diploidization effectively downsizing the inflated mesopolyploid genomes. The peculiar exception to this trend is the 2300-Mb genome of Physaria bellii (n = 4, Physarieae; Lysak and Lexer, 2006; Lysak et al., 2009). In this species, and potentially in some of its congeners, the tribe-specific whole-genome triplication (Mandáková et al., 2017) was followed by extensive diploidization, including descending dysploidy to only four chromosome pairs, and amplification of repetitive sequences increasing the average chromosome size in P. bellii (575 Mb) to values comparable with Hesperis-clade species (Fig. 1).
Genome expansion through amplification of TEs
Genome size variation among crucifer species with the arabidopsis-like chromosomal architecture was associated with the expansion (or contraction) of repeat-rich pericentromeres (Hall et al., 2006), as the insertion of amplified retrotransposon copies and other repeats into pericentromeres is potentially less harmful than targeting gene-rich chromosome arms. Although this has certainly occurred in some species, as evidenced by the ITR arrays at all pericentromeres in C. tenella, here we showed that the Hesperis-clade genomes expanded due to the chromosome-wide amplification of LTR retrotransposons and, to a lesser extent, the origin and amplification of tandem repeats. Whereas the diversity of TEs was comparable among all the sequenced genomes, the abundances of individual TE families differed substantially among the genomes and were positively correlated with increasing genome sizes. In all the large-genome species, Ty3-gypsy elements were identified as the key repeatome components driving the observed genome expansions. The frequently dominating role of Ty3-gypsy elements in genome size upsizing was documented in species from diverse plant families (e.g. Park et al., 2012; Macas et al., 2015; Willing et al., 2015; Dodsworth et al., 2017). Based on our partial repeatome analysis, tandem repeats represented only 0.26–0.8 % of repeatomes in four genomes >1500 Mb (Table 1). The high genome abundance (~7 %) of the HeSy1 repeat in H. sylvestris makes one notable exception. It remains unclear whether the accumulation of this repeat at three pericentromeres in H. sylvestris could have a functional role and whether this or similar high-copy tandem repeats can be found in genomes of other Hesperideae species.
The small genomes characterizing annual species of Chorisporeae (Chorispora and Diptychocarpus) and Euclidieae (Euclidium, Neotorularia and Strigosella) presumably represent independent subclade- (Chorisporeae) or species-specific (Euclidieae) genome downsizing events. Although our repeatome analysis, together with the phylogenetic position of these taxa, points to genome purging, it is difficult to pinpoint the underlying mechanism(s) using short read sequences (Macas et al., 2015). Repetitive sequences can be removed by recombination within or between repeat copies (Devos et al., 2002; Hawkins et al., 2009) or during double-strand break repair (e.g. Vu et al., 2017). However, a first prerequisite of deeper understanding of DNA purging in these tribes is more supported phylogenetic relationships with the aim of identifying species and genus pairs with and without genome contraction.
Chromosomal architecture in Brassicaceae species
Repetitive sequences in plant genomes usually show specific chromosomal organization, with tandem repeats localized in spatially separated domains, while TEs have more ubiquitous chromosomal distribution (e.g. Schmidt and Heslop-Harrison, 1998; Heslop-Harrison and Schwarzacher, 2011). Tandemly repeated sequences usually constitute chromosomal heterochromatic arrays, whereas TEs, despite their frequent co-localization with tandem repeats, can intersperse throughout gene-rich euchromatic regions. The angiosperm plants with small nuclear genomes, exemplified by the arabidopsis genome, show non-uniform distribution of repetitive sequences, which are preferentially localized in heterochromatic pericentromeric regions and knobs, and mostly absent on chromosome arms (Fransz et al., 1998, 2002; Lim et al., 1998; Cheng et al., 2001; Grob et al., 2013; Simon et al., 2015; Underwood et al., 2017; Morata et al., 2018). This distribution of repetitive sequences is widespread across Brassicaceae (as indirectly evidenced by dozens of CCP analyses carried out in our laboratory), as most crucifer species possess small genomes (modal 1C-value 392 Mb; Lysak et al., 2009; Hohmann et al., 2015). In the Hesperis clade, the arabidopsis-like chromosomal architecture is characteristic of species with smallest genome sizes, i.e. C. tenella, E. syriacum and Strigosella africana (390 Mb; Lysak et al., 2009). As nuclear genome size, average chromosome size and TE content increase in most Hesperis clade species (Tables 1 and 3), the longitudinal chromosomal compartmentalization disappears. In genomes larger than 1500 Mb and average chromosome sizes above 200 Mb, TEs are evenly distributed along the entire chromosome length, except for distinct subtelomeric and pericentromeric loci occupied by tandem repeats. The increasing chromosome arm lengths pose a serious challenge to centromeres to ensure a correct segregation of ‘obese’ chromosomes during cell division. It should be interesting to analyse whether the increasing chromosome arm length was reflected by a corresponding increase in centromere size and copy number of centromeric tandem repeats (Zhang and Dawe, 2012).
Genome size and life-form transition
There is a substantial body of evidence linking genome size, ecophysiological parameters and life-history strategies in plant species. Whereas species with small genomes can grow in more diverse habitats and can adopt any life form, species with larger genomes are confined to narrower ecological amplitudes and perenniality (Bennett, 1987; Knight et al., 2005; Suda et al., 2015; Pellicer et al., 2018). The Hesperis-clade species show the statistically significant tendency of ephemeral or annual species to have small genomes, whereas species with large(er) genomes are more likely to adopt a biennial or perennial life history. Scarce C-value data are not sufficient to rigorously test this causal relationship in closely related or sister species of different life forms. The inferred correlation is found, for example, in Bunias and Chorispora. Genome size of the annual Bunias erucago (2083 Mb) is 0.8-fold the C-value (2585 Mb) of the perennial Bu. orientalis (Greilhuber and Obermayer, 1999). Whereas the annual C. tenella has a 342-Mb genome, the genome size of the perennial C. bungeana, confined to high alpine environment (2000–4200 m; Song et al., 2015), is 817–830 Mb (Liu, 2017). Altogether, our data suggest that the smaller Hesperis-clade genomes could have been selected for, as genome downsizing enables short-lived ephemerals and annuals to adapt to time-limited habitats. For example, in the Asian cold deserts ephemeral crucifer species are an important component of the flora. In the Junggar Desert of northwest China, of the 24 ephemeral Brassicaceae species, ten belong to the Hesperis clade and nine are annual herbs with indehiscent or dehiscent fruits (Liu and Tan, 2007; Lu et al., 2017). Among the nine taxa, small C-values are known for four species and a comparably small genome can be predicted for the remainder of the annuals. However, most Hesperis-clade species are biennials and perennials with large genomes. Longer life cycles of perennials, associated with genome inflation, were important in the adaptation of Hesperis-clade species to extreme mountain and alpine conditions, with frequent fluctuations of temperature and precipitation, long-lasting snow cover and high solar radiation (e.g. Hughes and Atchison, 2015).
The feasibility of chromosome painting
Chromosome painting based on BAC in plants is based on hybridization and subsequent visualization of non-repetitive sequences on chromosomes (Lysak et al., 2003; Betekhtin et al., 2014). Large-scale CCP in plants takes advantage of small genomes, such as that of arabidopsis, with euchromatic gene-rich chromosome arms and most repetitive sequences clustered within heterochromatic pericentromeres. The amplification and mobility of repeats, underlying genome upsizing, transform arabidopsis-type chromosomes into the less compartmentalized chromosomes characterizing most plant genomes (Kejnovsky et al., 2009). In Hesperis-clade genomes with >40 % of repetitive sequences, CCP is significantly compromised or unfeasible (Mandáková et al., 2017; this study) due to the changed chromosomal architecture. As genome sequences of these species are not available, we may only hypothesize that painting probes, based on single-copy coding sequences, render weaker hybridization signals as the target sequences are interspersed with abundant dispersed repeats. Moreover, heterochromatinization, including DNA methylation and histone modifications, may further hinder the accessibility of target sequences for the DNA probe.
Conclusions
The Hesperis clade represents a unique crucifer lineage grouping taxa with unusually large nuclear genomes and low chromosome numbers. We demonstrated that the phylogenetically shared genome obesity has not been caused by a clade-specific WGD, but by proliferation of LTR retrotransposons, initially in the Hesperis-clade ancestor and subsequently in taxa of the six tribes. It is assumed that the predominance of genome obesity was associated with the selection for biennial or perennial life histories. Rarely, but repeatedly, genome expansion was counteracted by purging of TEs, enabling in some species an adaptive transition to the annual life strategy. Genome downsizing versus expansion significantly impacted chromosome size and architecture of the Hesperis-clade species towards small and highly compartmentalized chromosomes (e.g. C. tenella, E. syriacum) versus large and less structured chromosomes (e.g. Matthiola and Hesperis spp.).
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: summary of chromosome number, monoploid genome size, phylogenetic and life form data available for Hesperis-clade species. Table S2: nucleotide sequences of consensus satellites monomers. Table S3: sequences of PCR primers designed to amplify the gag domain in selected LTR retrotransposons. Table S4: reconstruction of ancestral genome sizes based on ITS and ndhF phylogenies. Table S5: comparison of in silico and dot-blot estimates of repeat abundances in two Hesperis-clade species with contrasting genome size. Table S6: correlation between repeat amounts and genome size variation in the analysed Hesperis-clade species. Figure S1: self dot-plot comparison of non-homogenized monomers of a satellite family with a 60-bp repetitive motif in the E. syriacum genome. Figure S2: phylogenetic analysis of Ty1-copia LTR retrotransposons based on multiple alignment of their RT domains. Figure S3: phylogenetic analysis of Ty3-gypsy LTR retrotransposons based on multiple alignment of their reverse transcriptase domains. Figure S4: repeat sequence proportions in the 50 largest clusters based on the comparative clustering analysis. Figure S5: dot plot showing sequence similarities between satellites identified in Bu. orientalis, M. incana, D. micranthus, Brassica rapa and Brassica oleracea. Figure S6: comparative chromosome painting in Hesperis-clade species.
ACKNOWLEDGEMENTS
We thank Dr Dmitry German (Heidelberg University) and Dr Jiří Macas (Institute of Plant Molecular Biology, České Budějovice) for their advice on the manuscript. This work was supported by research grants from the Czech Science Foundation (grants P501/12/G090 and 18-20134S), the CEITEC 2020 (grant LQ1601) project and by the Czech Academy of Sciences (long-term research development project RVO 67985939).
LITERATURE CITED
- Afgan E, Baker D, Batut B, et al. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research 46: W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. [Google Scholar]
- Beilstein MA, Al-Shehbaz IA, Kellogg EA. 2006. Brassicaceae phylogeny and trichome evolution. American Journal of Botany 93: 607–619. [DOI] [PubMed] [Google Scholar]
- Bennett MD. 1987. Variation in genomic form in plants and its ecological implications. New Phytologist 106: 177–200. [Google Scholar]
- Bennett MD, Leitch IJ, Price HJ, Johnston JS. 2003. Comparisons with Caenorhabditis (∼100 Mb) and Drosophila (∼175 Mb) using flow cytometry show genome size in Arabidopsis to be ∼157 Mb and thus ∼25 % larger than the Arabidopsis genome initiative estimate of ∼125 Mb. Annals of Botany 91: 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betekhtin A, Jenkins G, Hasterok R. 2014. Reconstructing the evolution of Brachypodium genomes using comparative chromosome painting. PLoS ONE 9: e115108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Al-Shehbaz IA, Yue J, Sun H. 2018. New insights into the taxonomy of tribe Euclidieae (Brassicaceae), evidence from nrITS sequence data. PhytoKeys 100: 125–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Liu T, Yu CH, Chiang TY, Hwang CC. 2013. Effects of GC bias in next-generation-sequencing data on de novo genome assembly. PLoS ONE 8: e62856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Buell CR, Wing RA, Gu M, Jiang J. 2001. Toward a cytological characterization of the rice genome. Genome Research 11: 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Brown JK, Bennetzen JL. 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Research 12: 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodsworth S, Jang TS, Struebig M, Chase MW, Weiss-Schneeweiss H, Leitch AR. 2017. Genome-wide repeat dynamics reflect phylogenetic distance in closely related allotetraploid Nicotiana (Solanaceae). Plant Systematics and Evolution 303: 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Bartoš J, Voglmayr H, Greilhuber J. 2003. Nuclear DNA content and genome size of trout and human. Cytometry 51: 127–128. [DOI] [PubMed] [Google Scholar]
- Doležel J, Greillhuber J, Suda J. 2007. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols 2: 2233–2244. [DOI] [PubMed] [Google Scholar]
- Fransz P, Armstrong S, Alonso-Blanco C, Fischer TC, Torres-Ruiz RA, Jones G. 1998. Cytogenetics for the model system Arabidopsis thaliana. Plant Journal 13: 867–876. [DOI] [PubMed] [Google Scholar]
- Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. 2002. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proceedings of the National Academy of Sciences of the USA 99: 14584–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiero P, Vaio M, Peters SA, Schranz ME, de Jong H, Speranza PR. 2018. Comparative analysis of repetitive sequences among species from the potato and the tomato clades. Annals of Botany 123: 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German DA, Al-Shehbaz IA. 2017. A taxonomic note on Sterigmostemum and related genera (Anchonieae, Cruciferae). Novosti Sistematicheski Vysshikh Rasteniĭ 48: 78–83. [Google Scholar]
- German DA, Al-Shehbaz IA. 2018. A reconsideration of Pseudofortuynia and Tchihatchewia as synonyms of Sisymbrium and Hesperis, respectively (Brassicaceae). Phytotaxa 334: 95–98. [Google Scholar]
- German DA, Grant JR, Lysak MA, Al-Shehbaz IA. 2011. Molecular phylogeny and systematics of the tribe Chorisporeae (Brassicaceae). Plant Systematics and Evolution 294: 65–86. [Google Scholar]
- Greilhuber J, Obermayer R. 1999. Cryptopolyploidy in Bunias (Brassicaceae) revisited—a flow-cytometric and densitometric study. Plant Systematics and Evolution 218: 1–4. [Google Scholar]
- Greilhuber J, Borsch T, Müller K, Worberg A, Porembski S, Barthlott W. 2006. Smallest angiosperm genomes found in Lentibulariaceae, with chromosomes of bacterial size. Plant Biology 8: 770–777. [DOI] [PubMed] [Google Scholar]
- Grob S, Schmid MW, Luedtke NW, Wicker T, Grossniklaus U. 2013. Characterization of chromosomal architecture in Arabidopsis by chromosome conformation capture. Genome Biology 14: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AE, Kettler GC, Preuss D. 2006. Dynamic evolution at pericentromeres. Genome Research 16: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. 2007. GEIGER: investigating evolutionary radiations. Bioinformatics 24: 129–131. [DOI] [PubMed] [Google Scholar]
- Hawkins JS, Proulx SR, Rapp RA, Wendel JF. 2009. Rapid DNA loss as a counterbalance to genome expansion through retrotransposon proliferation in plants. Proceedings of the National Academy of Sciences of the USA 106: 17811–17816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Schwarzacher T. 2011. Organisation of the plant genome in chromosomes. Plant Journal 66: 18–33. [DOI] [PubMed] [Google Scholar]
- Hohmann N, Wolf EM, Lysak MA, Koch MA. 2015. A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell 27: 2770–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Sun R, Hu Y, et al. 2016. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Molecular Biology and Evolution 33: 394–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CE, Atchison GW. 2015. The ubiquity of alpine plant radiations: from the Andes to the Hengduan Mountains. New Phytologist 207: 275–282. [DOI] [PubMed] [Google Scholar]
- Jaretzky R. 1928. Untersuchungen über Chromosomen und Phylogenie bei einigen Cruciferen. Jahrbücher für Wissenschaftliche Botanik 68: 1–45. [Google Scholar]
- Jiao WB, Accinelli GG, Hartwig B, et al. 2017. Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Research 27: 778–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kejnovsky E, Leitch IJ, Leitch AR. 2009. Contrasting evolutionary dynamics between angiosperm and mammalian genomes. Trends in Ecology & Evolution 24: 572–582. [DOI] [PubMed] [Google Scholar]
- Kiefer M, Schmickl R, German DA, et al. 2014. BrassiBase: introduction to a novel knowledge database on Brassicaceae evolution. Plant and Cell Physiology 55: e3. [DOI] [PubMed] [Google Scholar]
- Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Research 21: 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight CA, Molinari NA, Petrov DA. 2005. The large genome constraint hypothesis: evolution, ecology and phenotype. Annals of Botany 95: 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo DH, Hong CP, Batley J, et al. 2011. Rapid divergence of repetitive DNAs in Brassica relatives. Genomics 97: 173–185. [DOI] [PubMed] [Google Scholar]
- Kubešová M, Moravcova L, Suda J, Jarošík V, Pyšek P. 2010. Naturalized plants have smaller genomes than their non-invading relatives: a flow cytometric analysis of the Czech alien flora. Preslia 82: 81–96. [Google Scholar]
- Kubis S, Schmidt T, Heslop-Harrison JS. 1998. Repetitive DNA elements as a major component of plant genomes. Annals of Botany 82: 45–55. [Google Scholar]
- Lim KY, Leitch IJ, Leitch AR. 1998. Genomic characterisation and the detection of raspberry chromatin in polyploid Rubus. Theoretical and Applied Genetics 97: 1027–1033. [Google Scholar]
- Liu L. 2017. The epigenetic modifications of Chorispora bungeana and the function of ADH1 in cold response. Thesis retrieved from China Integrated Knowledge Resources Database. http://cdmd.cnki.com.cn/Article/CDMD-10730-1018803968.htm. [Google Scholar]
- Liu XF, Tan DY. 2007. Diaspore characteristics and dispersal strategies of 24 ephemeral species of Brassicaceae in the Junggar Desert of China. Journal of Plant Ecology 31: 1019–1027. [Google Scholar]
- Lu JJ, Tan DY, Baskin CC, Baskin JM. 2017. Role of indehiscent pericarp in formation of soil seed bank in five cold desert Brassicaceae species. Plant Ecology 218: 1187–1200. [Google Scholar]
- Lysak MA, Lexer C. 2006. Towards the era of comparative evolutionary genomics in Brassicaceae. Plant Systematics and Evolution 259: 175–198. [Google Scholar]
- Lysak MA, Mandáková T. 2013. Analysis of plant meiotic chromosomes by chromosome painting. Methods in Molecular Biology 990: 13–24. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Pecinka A, Schubert I. 2003. Recent progress in chromosome painting of Arabidopsis and related species. Chromosome Research 11: 195–204. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Koch MA, Beaulieu JM, Meister A, Leitch IJ. 2009. The dynamic ups and downs of genome size evolution in Brassicaceae. Molecular Biology and Evolution 26: 85–98. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Mandáková T, Schranz ME. 2016. Comparative paleogenomics of crucifers: ancestral genomic blocks revisited. Current Opinion in Plant Biology 30: 108–115. [DOI] [PubMed] [Google Scholar]
- Macas J, Novak P, Pellicer J, et al. 2015. In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the legume tribe Fabeae. PLoS ONE 10: e0143424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáaková T, Kovařík A, Zozomová-Lihová J, Shimizu-Inatsugi R, Shimizu KK, Mummenhoff K, Marhold K, Lysak MA. 2013. The more the merrier: recent hybridization and polyploidy in Cardamine. Plant Cell 25: 3280–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA. 2016a Chromosome preparation for cytogenetic analyses in Arabidopsis. Current Protocols in Plant Biology 1: 43–51. [DOI] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA. 2016b Painting of Arabidopsis chromosomes with chromosome-specific BAC clones. Current Protocols in Plant Biology 1: 359–371. [DOI] [PubMed] [Google Scholar]
- Mandáková T, Hloušková P, German D, Lysak MA. 2017. Monophyletic origin and evolution of the largest crucifer genomes. Plant Physiology 174: 2062–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manton I. 1932. Introduction to the general cytology of the Cruciferae. Annals of Botany 46: 509–556. [Google Scholar]
- Morata J, Tormo M, Alexiou KG, et al. 2018. The evolutionary consequences of transposon-related pericentromer expansion in melon. Genome Biology and Evolution 10: 1584–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Novák P, Hoštáková N, Macas J. 2019. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mobile DNA 10: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Pech J, Steinhaisl J, Macas J. 2013. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29: 792–793. [DOI] [PubMed] [Google Scholar]
- Novák P, Ávila Robledillo L, Koblížková A, Vrbová I, Neumann P, Macas J. 2017. TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Research 45: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F. 1990. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In: Darzynkiewicz Z, Crissman HA, eds. Methods in Cell Biology, Vol. 33 New York: Academic Press, 105–110. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Park M, Park J, Kim S, et al. 2012. Evolution of the large genome in Capsicum annuum occurred through accumulation of single-type long terminal repeat retrotransposons and their derivatives. Plant Journal 69: 1018–1029. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Wood T, Zhang Z, Miller W. 1997. Comparison of DNA sequences with protein sequences. Genomics 46: 24–36. [DOI] [PubMed] [Google Scholar]
- Pellicer J, Hidalgo O, Dodsworth S, Leitch I. 2018. Genome size diversity and its impact on the evolution of land plants. Genes 9: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2013. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; http://www.R-project.org. [Google Scholar]
- Rambaut A, Drummond A. 2009. Tracer v1.6. http://tree.bio.ed.ac.uk/software/tracer. [Google Scholar]
- Ren L, Huang W, Cannon EK, Bertioli DJ, Cannon SB. 2018. A mechanism for genome size reduction following genomic rearrangements. Frontiers in Genetics 9: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. [Google Scholar]
- Roark LM, Hui AY, Donnelly L, Birchler JA, Newton KJ. 2010. Recent and frequent insertions of chloroplast DNA into maize nuclear chromosomes. Cytogenetic and Genome Research 129: 17–23. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, et al. 2011. Integrative genomics viewer. Nature Biotechnology 29: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T, Heslop-Harrison JS. 1998. Genomes, genes and junk: the large-scale organization of plant chromosomes. Trends in Plant Science 3: 195–199. [Google Scholar]
- Schubert I, Lysak MA. 2011. Interpretation of karyotype evolution should consider chromosome structural constraints. Trends in Genetics 27: 207–216. [DOI] [PubMed] [Google Scholar]
- Simon L, Voisin M, Tatout C, Probst AV. 2015. Structure and function of centromeric and pericentromeric heterochromatin in Arabidopsis thaliana. Frontiers in Plant Science 6: 1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Liu L, Feng Y, et al. 2015. Chilling- and freezing-induced alterations in cytosine methylation and its association with the cold tolerance of an alpine subnival plant, Chorispora bungeana. PLoS ONE 10: e0135485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: GC1–GC10. [DOI] [PubMed] [Google Scholar]
- Suda J, Kyncl T, Jarolímová V. 2005. Genome size variation in Macaronesian angiosperms: forty percent of the Canarian endemic flora completed. Plant Systematics and Evolution 252: 215–238. [Google Scholar]
- Suda J, Meyerson LA, Leitch IJ, Pyšek P. 2015. The hidden side of plant invasions: the role of genome size. New Phytologist 205: 994–1007. [DOI] [PubMed] [Google Scholar]
- Trávníček P, Ponert J, Urfus T, et al. 2015. Challenges of flow-cytometric estimation of nuclear genome size in orchids, a plant group with both whole-genome and progressively partial endoreplication. Cytometry 87A: 958–966. [DOI] [PubMed] [Google Scholar]
- Underwood CJ, Henderson IR, Martienssen RA. 2017. Genetic and epigenetic variation of transposable elements in Arabidopsis. Current Opinion in Plant Biology 36: 135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu GT, Cao HX, Reiss B, Schubert I. 2017. Deletion-bias in DNA double-strand break repair differentially contributes to plant genome shrinkage. New Phytologist 214: 1712–1721. [DOI] [PubMed] [Google Scholar]
- Waterworth WM, Drury GE, Bray CM, West CE. 2011. Repairing breaks in the plant genome: the importance of keeping it together. New Phytologist 192: 805–822. [DOI] [PubMed] [Google Scholar]
- Willing E-M, Rawat V, Mandáková T, et al. 2015. Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nature Plants 1: 14023. [DOI] [PubMed] [Google Scholar]
- Zhang H, Dawe RK. 2012. Total centromere size and genome size are strongly correlated in ten grass species. Chromosome Research 20: 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.