Abstract
The present study aimed to investigate whether microRNA (miR)-490-3p can regulate MAPK1 expression, increase proliferation of esophageal squamous cell carcinoma (ESCC) and reduce ESCC cell apoptosis. The Cancer Genome Atlas (TCGA) database was used to explore the functional role of miR-490-3p in ESCC. The expression of miR-490-3p in ESCC tissues and adjacent tissues of patients with ESCC were detected by reverse transcription-quantitative PCR. The effect of miR-490-3p on ESCC cell proliferation and apoptosis were detected by cell counting kit-8 and clone formation assay, and flow cytometry, respectively. The dual luciferase reporter assay was used for detect the regulatory association between miR-490-3p and MAPK1. The TCGA dataset demonstrated that miR-490-3p expression was reduced in ESCC tissues compared with normal tissue. The expression of miR-490-3p was also lower in ESCC tissues compared with adjacent tissues. The expression of miR-490-3p in patients with stage III and IV ESCC were significantly lower than those in stage I and II. In patients with tumor >3 cm, miR-490-3p expression was lower than in patients with tumor <3 cm. Gene set enrichment analysis demonstrated that miR-490-3p may essentially regulate cell apoptosis. In addition, miR-490-3p depletion in TE1 and ECA109 cell lines promoted cell proliferation and inhibited cell apoptosis. The results from dual luciferase reporter assay demonstrated that miR-490-3p may be able to degrade MAPK1. Furthermore, MAPK1 overexpression in TE1 and ECA109 cells partially reversed the effects of miR-490-3p on cell proliferation and apoptosis. Low expression of miR-490-3p may therefore promote the proliferation and inhibit the apoptosis of ESCC cells by regulating MAPK1.
Keywords: microRNA-490-3p, esophageal squamous cell carcinoma, cell proliferation, apoptosis, mitogen-activated protein kinase 1
Introduction
Esophageal squamous cell carcinoma (ESCC) is a common tumor of the digestive tract. The incidence and mortality of ESCC varies among countries, and China has a high ESCC risk (1,2). Thanks to the development of genetic and molecular biology, pathogenesis of ESCC is now better understood. Particularly, the association between microRNAs (miRs) and cancer has been investigated. Recent studies have demonstrated that abnormal expression of miRs is detected in the serum of patients with ESCC (3–5). These findings may provide new direction for further study of the progression of ESCC and to determine novel potential therapeutic targets.
miRs are single-stranded non-coding small RNAs that are typically 18–25 nucleotides in length. They mainly bind at a post-transcriptional level to the target mRNAs, which are subsequently degraded and not translated into protein. miRs can precisely regulate certain biological processes, including cell proliferation, apoptosis, migration and immune responses (6,7). It has been reported that dysregulated miRs might have a role in of tumors development (8). Notably, miR-490-3p has been reported to be associated with the incidence of various types of cancer (1,9–16). In the present study, analysis of miRs expression in ESCC tissues and adjacent tissues from The Cancer Genome Atlas (TCGA) database demonstrated that miR-490-3p was significantly lower in ESCC tissues; however, the role of miR-490-3p in ESCC remains unknown.
Mitogen-activated protein kinases (MAPKs) are a family of serine/threonine kinases that can be activated by various signals (17). Their functions have remained highly evolutionarily conserved (18). MAPK1, also known as ERK2, is a ~42 kD protein and is mediates one of the most important signaling pathways in vivo, with a crucial role in cell growth, differentiation, proliferation, survival and in the inflammatory response (19). A previous study reported that the MAPK1 signaling pathway is crucial in most cells and is regulated at numerous levels. It can respond to external physiological and pathological stimuli, and to extracellular stimuli of the nucleus (17,20,21).
The present study aimed to investigate the effect of miR-490-3p on the proliferation and apoptosis of the ESCC TE1 and ECA109 cell lines, and to determine whether miRNA-490-3p exerts its biological function through regulation of MAPK1 expression.
Materials and methods
Data acquisition and analysis
The clinical data and the corresponding genome-wide expression profile (TCGA-ESCA) using RNA-sequencing of 185 patients with ESCC were obtained from TCGA database (https://cancergenome.nih.gov). A total of 100 cases were subsequently randomly selected for further analysis. The TCGA data was normalization by the MD Anderson Cancer Center. Two-way hierarchical clustering of 88 tumor tissues and 12 adjacent normal mucosa with the 2,114 differentially expressed RNAs were analyzed using Euclidean distance and average linkage clustering. P<0.05 and |log fold change|>1 were defined as cut-off values.
Gene enrichment analysis
Gene Set Enrichment Analysis (GSEA) version 2.2.3 software (http://software.broadinstitute.org/gsea/index.jsp; Broad Institute, Inc.) was used for data analysis. According to the expression level of miR-490-3p, all cases were divided into the miR-490-3p high expression group and low expression group, using the aforementioned cut off values. In the present study, the dataset of C2.CP.KEGG.v6.0 was obtained from the MSigDB database of the GSEA website. The enrichment analysis was performed according to the default weighted enrichment statistics method, and the number of random combinations was set to 1,000 times.
Patients and sample collection
New ESCC tissues and paracarcinoma tissues were obtained from 40 patients, including 18 women and 22 men, aged from 48–69 years old, diagnosed with ESCC and received surgical treatment between July 2016 and September 2017 at The Affiliated Tumor Hospital of Xinjiang Medical University. Patients had not received any treatment prior to surgery and had no family history of ESCC. Patients were diagnosed with ESCC by pathological analysis of the tumors. All patients voluntarily participated in the study and provided signed informed consent. The present study was approved by The Affiliated Tumor Hospital of Xinjiang Medical University Ethics Committee. After radical esophagectomy, tumor tissues tissue did not contain any necrotic area, and adjacent normal tissues that were resected at >5 cm from cancer tissue, were stored in liquid nitrogen.
Cell culture
The poorly differentiated ESCC cell line TE1, the highly differentiated ESCC cell line ECA109 and the control cell line HEEC were purchased from Shanghai American Type Culture Collection. Cells were cultured in Dulbecco's modified eagle medium containing 10% FBS, 1% penicillin and 1% streptomycin and placed at 37°C in a humidified incubator containing 5% CO2 (Gibco; Thermo Fisher Scientific, Inc.). After 48 h incubation, culture medium was replaced every other day. Once cells had reached 80–90% confluence, they were harvested with 0.25% trypsin and passaged at a ratio of 1:2 or 1:3.
Cell transfection
Cells in the logarithmic growth phase were selected for seeding into 24-well plates at 1×105 cells/well. Transfection was performed using the Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). When the cell density reached 70–80%, they were transfected with miR-490-3p mimics (20 µM), miR-490-3p inhibitors (20 µM), pcDNA-MAPK1 (5 µg) and the relative negative controls (Shanghai GenePharma Co., Ltd.). The sequences were as follows: miR-490-3p mimics forward, 5′-CAACCUGGAGGACUCCAUGCCG-3′ and reverse, 5′-AGACCGUCGAUUGGGCCAGUUG-3′; miR-490-3p inhibitors forward, 5′-UUUAGCUGGUACCGACUGUACG-3′ and reverse, 5′-AUUCGGUAGUUUCAGGGCAUAG-3′; negative controls forward 5′-ACCUUCUCAGGCUUGACGUAGA-3′ and reverse, 5′-CCGUAUUAUCUGCCAAGUACGU-3′. At 48 h after transfection, cells were collected for further experiments.
Tissues and cellular RNA extraction
1×106 Cells or 50 mg tissues were collected and lysed by adding 1 ml TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). Cells and tissues were then incubated with 250 µl chloroform, mixed for 30 sec and centrifuged at 10,000 × g at 4°C for 10 min. The aqueous phase was discarded and same volume of pre-cooled isopropanol was added. The precipitate was gently cleaned with 75% ethanol after centrifugation at 10,000 × g for 10 min at 4°C. Diethyl pyrocarbonate (20 µl) water was used to dissolve RNAs. The RNA concentration was measured using a spectrophotometer and stored at −80°C.
Reverse transcription-quantitative PCR (RT-qPCR)
RT was performed using the PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol. RT was performed on ice and produced cDNAs. qPCR was performed according to the miScript SYBR Green PCR kit instructions (Takara Bio, Inc.) for miR-490-3p, MAPK and GAPDH. The PCR amplification conditions were as follows: Pre-denaturation at 94°C for 5 min, followed by 94°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min and 30 sec for 40 cycles.
The primers were designed as follows: miR-490-3p, forward 5′-CGGCGGTCAACCTGGAGGACTCC-3′, reverse 5′-CCAGTGCAGGGTCCGAGGTAT-3′; MAPK1, forward 5′-CGACGCGTCGTTGTAATAAAGCCTCCAG-3′, reverse 5′-GACTAGTCGTTTTCATTTCAATCGTAG-3′; GAPDH, forward 5′-AGCCACATCGCTCAGACAC-3′, reverse 5′-GCCCAATACGACCAAATCC-3′; and U6, forward 5′-CTCGCTTCGGCAGCAGCACATATA-3′, and reverse 5′-AAATATGGAACGCTTCACGA-3′. The 2−ΔΔCq method was used to determine the relative expression levels (22). The level of mRNA expression was normalized to GAPDH, while miRNA expression levels were normalized to U6.
Cell counting kit-8 (CCK-8) assay
At 48 h after transfection, TE1 and ECA109 cells were plated in 96-well plates (1×104 cells/100 µl/well). Five replicates were set for each sample. Cells were cultured at 37°C and 5% CO2 for 0, 24, 48 72 and 96 h. CCK-8 reagent (10 µl; Dojindo Molecular Technologies, Inc.) was then added to cells and incubated for 2 h at 37°C. Absorbance was measured at 450 nm using a microplate reader.
Plate cloning experiment
TE1 and ECA109 cells in the logarithmic growth phase were harvested, and the cell suspension concentration was adjusted to 1×104/ml. Cells were seeded in a 6-well plate at the density of 2,000 cell/well. Following 3–4 days culture at 37°C, cells were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet for 30 min at 25°C. The numbers of clones per well were counted and imaged.
Flow cytometry
Cells were harvested with trypsin and suspended with 100 µl 1X Annexin V binding buffer (AmyJet Scientific, Inc.) at a minimum density of 1×105 cells/ml. Annexin V-FITC was used for cell labeling. Annexin V (5 µl) and propidium iodide (1 µl) were added to the cell suspension, mixed, and incubated at room temperature in the dark for 15 min. 1X Annexin V binding buffer (400 µl) was then added, and cell apoptosis was measured on a flow cytometer. Each sample was analyzed in triplicate and the experiment was repeated three times.
Bioinformatics
The target genes for miR-490-3p were determined through bioinformatics prior to performing functional analysis. The basic information of base sequence, chromosome location and species conservativeness of miR-490-3p was obtained by using miR Base (http://www.mirbase.org/index.shtml). The target genes of miR-490-3p were predicted and the intersection was taken as the gene set for further analysis by four methods, including TargetScan (http://www.targetscan.org/), MicroRanda (http://www.microrna.org/), RNA hybrid (version 2.1.2) and DIANAmT from MicroWalk (umm.uni-heidelberg.de/apps/zmf/mirwalk/) integrated database. Cytoscape software (version 3.7.1; http://cytoscape.org/) and its plug-in Bingo was applied for function enrichment analysis.
Dual luciferase reporter assay
The 3′untranslated region (3′UTR) sequence of MAPK1 was downloaded from the NCBI website (https://www.ncbi.nlm.nih.gov/) to construct the wild-type MAPK1 (MAPK1 WT 3′UTR) and the mutant (MAPK1 MUT 3′UTR) pGL3-plasmids (Hanbio Biotechnology Co., Ltd.). pRL-TK Renilla plasmid (Promega Corporation) was used as the control luciferase for normalization. Cells were plated in 96-well plates at 1×104 cells/well and co-transfected with 50 pmol/l miR-490-3p mimics or negative controls with 80 ng MAPK1 WT 3′UTR or MAPK1 MUT 3′UTR plasmid using Lipofectamine™ 2000. After 48 h, Dual Luciferase® reporter assay system (Promega Corporation) was used to detect fluorescence intensity.
Western blot analysis
Cells were lysed by radioimmunoprecipitation assay buffer (Yeasen), sonicated, centrifuged at 5,000 × g for 10 min at 4°C after which the supernatant was collected. After adding bromophenol blue (9.5 to 0.5 ml), supernatant was boiled for 10 min and stored at −20°C. Proteins (10 µg) were separated by 12% SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Roche Diagnostics). Membranes were then blocked using 5% skimmed milk at 25°C for 1 h. Membranes were incubated with specific primary antibodies at 4°C overnight, which included MAPK1 (1:500; cat. no. ab241580; Abcam) and GAPDH (1:500; cat. no. ab8245; Abcam). Then the membranes were incubated with secondary antibody goat anti-rabbit IgG H&L (1:1,000; cat. no. ab7090; Abcam) the next day at 25°C for 2 h. Protein bands were visualized following a 3-min incubation with enhanced chemiluminescence reagent (Thermo Fisher Scientific, Inc.).
Statistical analysis
SPSS 19.0 (IBM Corp.) was used for statistical analysis. χ2 test was used for classification data and Student's t-test was used for data comparison between two groups. Comparison between multiple groups was performed using One-way ANOVA test followed by the least significant difference post hoc test. Data were expressed as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-490-3p expression level is lower in ESCC tissues from TCGA database
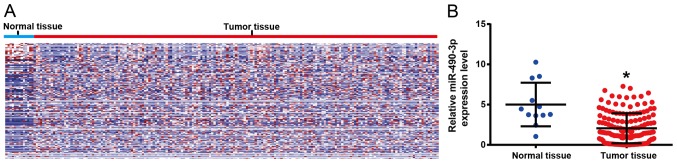
The differentially expressed miRs in ESCC tissues and adjacent tissues from the TCGA database were analyzed. Two-way hierarchical clustering of 988 tumor tissues and 12 adjacent normal mucosa with the 2,114 differentially expressed RNAs were analyzed using Euclidean distance and average linkage clustering (Fig. 1A). P<0.05 and |log fold change|>1 were defined as cut-off values. The results demonstrated that miR-490-3p was significantly lower in ESCC tissues compared with adjacent tissues (Fig. 1B) and chosen for further analysis, as it was the most significant.
Figure 1.
TCGA database demonstrated that miR-490-3p was low in ESCC tissues. (A) miRs differentially expressed in ESCC and paracancerous tissues from the TCGA database. Each row represents an individual gene and each column represents an individual sample. Color gradient from blue to red indicates expression levels from low to high on a normalized value. (B) miR-490-3p was lowly expressed in ESCC tissue. Normal tissue: n=12, ESCC tissue: n=988. *P<0.05 vs. normal tissue. TCGA, The Cancer Genome Atlas; ESCC, esophageal squamous cell carcinoma; miR, microRNA.
Expression of miR-490-3p is reduced in ESCC
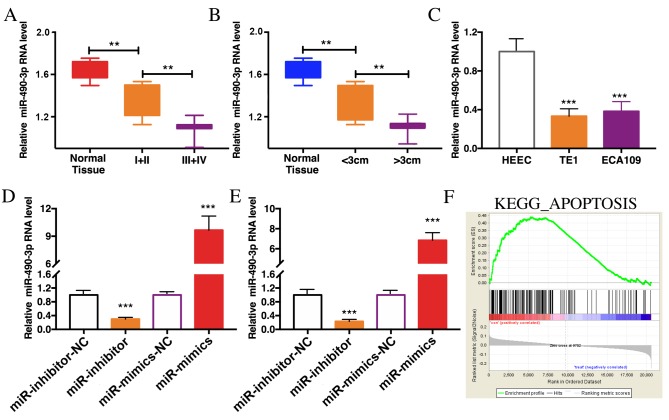
The expression of miR-490-3p in 40 cases of ESCC tissues and paracancerous tissues were determined by RT-qPCR. The results demonstrated that miR-490-3p was expression was lower in ESCC tissues than in paracancerous tissues. In advanced ESCC tissues with tumors >3 cm, miR-490-3p expression level was even lower (Fig. 2A and B). In addition, miR-490-3p expression level was significantly reduced in TE1 and ECA109 cells compared with HEEC cells (Fig. 2C). To further investigate the role of miR-490-3p in ESCC, cells were transfected with miR-490-3p mimics or miR-490-3p inhibitors to overexpress or knockdown miR-490-3p, respectively. The transfection efficiency was verified in TE1 and ECA109 cells (Fig. 2D and E). In addition, GSEA analysis reported that miR-490-3p was associated with in apoptosis regulation in vivo (Fig. 2F).
Figure 2.
miR-490-3p was lowly expressed in ESCC tissue. (A) miR-490-3p was lowly expressed in ESCC tissues, particularly in advanced esophageal tissues. (B) miR-490-3p was lowly expressed in ESCC tissues, particularly in tissues with tumors >3 cm. (C) miR-490-3p was lowly expressed in ESCC cell lines. Effects of miR-490-3p overexpression and depletion in (D) TE1 and (E) ECA109 cells on miR-490-3p expression level. (F) Gene Set Enrichment Analysis demonstrated that miR-490-3p essentially regulated cell apoptosis. **P<0.01, ***P<0.001. ESCC, esophageal squamous cell carcinoma; miR, microRNA; NC, negative control; KEGG, Kyoto Encyclopedia of Genes and Genomes.
miR-490-3p induces apoptosis and suppresses proliferation in ESCC cells
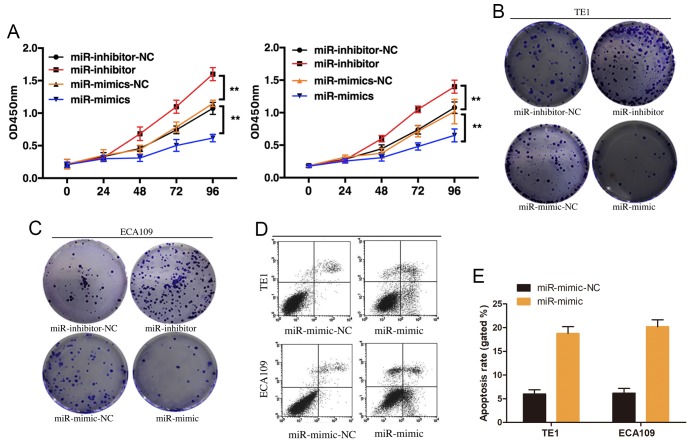
The effect of miR-490-3p on ESCC proliferation was investigated by overexpressing or knocking down miR-490-3p in TE1 and ECA109 cell lines. CCK-8 results demonstrated that miR-490-3p overexpression significantly inhibited ESCC cell proliferation, whereas miR-490-3p knockdown promoted ESCC cell proliferation (Fig. 3A). These results were confirmed by the plate cloning formation experiment (Fig. 3B and C). The effect of miR-490-3p on ESCC cell apoptosis was examined by flow cytometry. The results demonstrated that miR-490-3p overexpression induced ESCC cell apoptosis (Fig. 3D and E). These results suggested that miR-490-3p may induce ESCC cell apoptosis and suppress ESCC cell proliferation.
Figure 3.
miR-490-3p induced ESCC cell apoptosis and inhibited ESCC cell proliferation. (A) miR-490-3p overexpression and depletion in TE1 and ECA109 cell lines significantly inhibited and promoted cell proliferation, respectively. Plate cloning experiments using (B) TE1 and (C) ECA109 cells. (D) Flow cytometry analysis of cell apoptosis demonstrated that miR-490-3p overexpression induced ESCC cell apoptosis. (E) The quantification of flow cytometry analysis demonstrated that miR-490-3p overexpression induced ESCC cell apoptosis. **P<0.01. ESCC, esophageal squamous cell carcinoma; OD, optical density; miR, microRNA; NC, negative control.
miR-490-3p may stimulate MAPK1 degradation
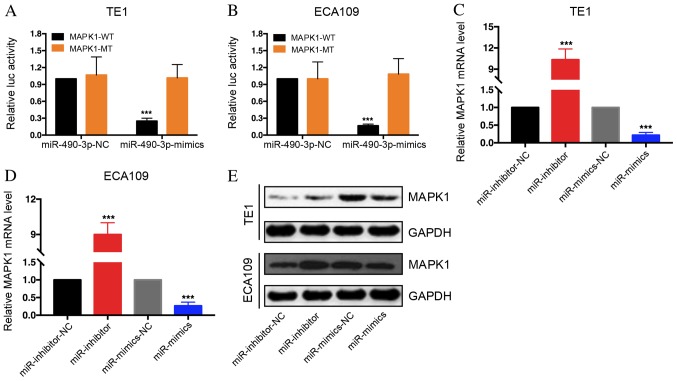
The target genes for miR-490-3p were determined through bioinformatics prior to performing functional analysis. The basic information of base sequence, chromosome location and species conservativeness of miR-490-3p was obtained by using miR Base (http://www.mirbase.org/index.shtml). The target genes of miR-490-3p were predicted and the intersection was taken as the gene set for further analysis by four methods, including TargetScan, MicroRanda, RNA hybrid and DIANAmT from MicroWalk (umm.uni-heidelberg.de/apps/zmf/mirwalk/) integrated database. Cytoscape software (version 3.7.1; http://cytoscape.org/) and its plug-in Bingo was applied for function enrichment analysis. As a result, it was revealed that miR-490-3p may be able to target MAPK1 degradation. Furthermore, luciferase activity of MAPK1-WT 3′UTR group was decreased following miR-490-3p mimics transfection of TE1 and ECA109 cells, whereas no difference was observed in the MAPK1-MUT 3′UTR group (Fig. 4A and B). These results suggested that MAPK1 may bind to miR-490-3p. Subsequently, MAPK1 mRNA expression level was determined following TE1 and ECA109 cell transfection with miR-490-3p mimics and miR-490-3p inhibitors. The results demonstrated that MAPK1 expression level was decreased in the mimics group but increased in the inhibitors group (Fig. 4C and D). Western blot analysis revealed that MAPK1 expression was decreased in miR-490-3p-overexpressing cells; however, MAPK1 expression was increased in miR-490-3p-depleted cells (Fig. 4E). These results indicated that miR-490-3p may regulate MAPK1 expression.
Figure 4.
miR-490-3p overexpression may degrade MAPK1. Dual luciferase reporter assay using (A) TE1 and (B) ECA109 cells demonstrated that miR-490-3p may bind to the 3′UTR of MAPK1. miR-490-3p overexpression significantly downregulated MAPK1 expression level in (C) TE1 and (D) ECA109 cells. miR-490-3p depletion significantly upregulated MAPK1 expression level. (E) miR-490-3p overexpression induced MAPK1 protein downregulation in ESCC cell lines. miR-490-3p depletion induced MAPK1 protein upregulation in ESCC cell lines. ***P<0.001. ESCC, esophageal squamous cell carcinoma; Luc, luciferase; MAPK1, mitogen-activated protein kinase 1; WT, wild-type; MT, mutant; miR, microRNA; NC, negative control.
MAPK overexpression reverses the effect of miR-490-3p on TE1 cell apoptosis
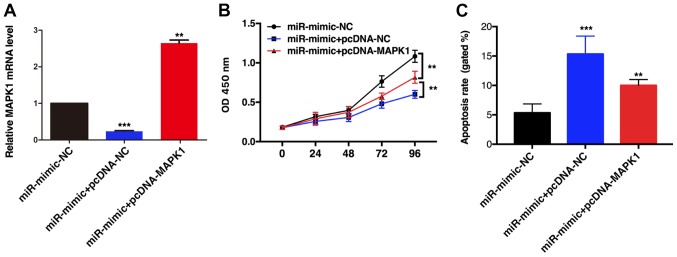
miR-490-3p overexpression significantly inhibited TE1 cell proliferation; however, when MAPK1 and miR-490-3p were simultaneously overexpressed, cell proliferation was increased, although it was still lower than in the control group (Fig. 5A and B). In addition, MAPK1 overexpression partially reversed the miR-490-3p-induced TE1 cell apoptosis (Fig. 5C). These results indicated that miR-490-3p may inhibit TE1 cell proliferation and promote ESCC cell apoptosis by inhibiting MAPK1 expression.
Figure 5.
MAPK1 overexpression reversed miR-490-3p-induced apoptosis on TE1 cells. (A) MAPK1 relative mRNA level in different groups. (B) Following miR-490-3p overexpression, cell proliferation was significantly reduced. MAPK1 overexpression enhanced cell proliferation. (C) Following miR-490-3p overexpression, cell apoptosis was increased. MAPK1 overexpression significantly reduced cells apoptosis. **P<0.01, ***P<0.001. MAPK1, mitogen-activated protein kinase 1; miR, microRNA; NC, negative control; OD, optical density.
Discussion
miRs are small, non-coding RNAs that regulate the expression of their corresponding proteins mainly at a post-transcriptional level. Numerous studies have reported that abnormally expressed miRs in tumors are associated with proto-oncogenes or tumor suppressor genes expression, and with patient sensitivity to chemotherapy and radiotherapy (5–7,23). The determination of tumor-specific miRs and their downstream target genes is therefore crucial to understand the role of miRs in tumor growth, and to develop novel targeted therapeutic strategies. Previous studies reported that miR-490-3p can act as a tumor suppressor gene and regulate various molecular signaling pathways involved in tumor proliferation, migration and invasion (13,15). For instance, miR-490-3p is downregulated in human cancerous gastric tissues, and miR-490-3p silencing can increase SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 expression and promote gastric cancer (11,24). Chen et al (10) reported that miR-490-3p was downregulated in human epithelial ovarian cancer compared with paracancerous tissues. In addition, miR-490-3p is lowly expressed in metastatic ovarian epithelial cancer. Furthermore, miR-490-3p can inhibit tumor cell proliferation by directly downregulating CDK1 expression (25). miR-490-3p was also decreased in osteosarcoma tissues, which promoted proliferation, inhibited apoptosis and cycle progression of osteosarcoma cells by targeting the high mobility group AT-hook 2 gene (14). However, the functional role of miR-490-3p in ESCC pathogenesis has been rarely reported.
The present study aimed to explore the role of miR-490-3p in ESCC pathogenesis and to explore its underlying mechanism. The results demonstrated that miR-490-3p expression was significantly decreased in ESCC tissues and in ESCC cell lines. These results suggested that miR-490-3p may be associated with the biological function of tumor cells. Based on previous studies TE1 and ECA109 cell lines were selected for miR-490-3p functional studies (26,27). The results demonstrated that miR-490-3p depletion increased the proliferation and decreased the apoptosis of TE1 and ECA109 cell lines.
In order to elucidate the underlying mechanism of miR-490-3p at regulating tumor cell biological function, one downstream target of miR-490-3p, MAPK1, was selected. The process of tumor development is closely associated with tumor cell proliferation, differentiation, apoptosis and metastasis (28). The molecular mechanism involves many changes in signaling pathways (28). The MAPK pathway is a crucial signaling pathway that communicates signal from a receptor at the cell surface into the cell nucleus (20). The MAPK pathway can therefore affect numerous biological processes, including cell proliferation, differentiation and apoptosis by directly affecting the transcription and regulation of the associated genes. Bioinformatics analyses demonstrated that MAPK1 was a target gene of miR-490-3p. Furthermore, a luciferase reporter gene assay revealed that miR-490-3p could directly bind to the MAPK1 3′UTR. In addition, miR-490-3p overexpression or depletion induced a decrease or increase in MAPK1 mRNA level and protein expression, respectively. MAPK1 stimulation could also restore the effect of miR-490-3p on proliferation and apoptosis of ESCC cells, which suggested that miR-490-3p may affect ESCC cell proliferation and apoptosis by targeting MAPK1.
In conclusion, the present study demonstrated that miR-490-3p expression was decreased in ESCC tissues. Furthermore, miR-490-3p depletion may promote the proliferation and inhibit the cell apoptosis of ESCC. In addition, MAPK1 may be considered as a target gene of miR-490-3p. This study provided novel insights into the role of miR-490-3p in the pathogenesis of ESCC and may facilitate the development of specific diagnostics or treatments.
Acknowledgements
Not applicable.
Funding
This study was supported by the Xinjiang Autonomous Region Natural Fund (grant. no. 2016D01C356).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BZ and AS designed the study and performed the experiments. BZ, MY and YL collected the data and MY and YL analyzed the data. BZ and AS wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the ethics committee of The Third Clinical Medical College (Affiliated Tumor Hospital) of Xinjiang Medical University. Signed written informed consent was obtained from all participants before the study.
Patient consent for publication
Patients or their guardians provided written informed consent for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mao A. Interventional therapy of Esophageal Cancer. Gastrointest Tumors. 2016;3:59–68. doi: 10.1159/000447512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaib WL, Nammour JP, Gill H, Mody M, Saba NF. The future prospects of immune therapy in gastric and esophageal adenocarcinoma. J Clin Med. 2016;5(pii):E100. doi: 10.3390/jcm5110100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang K, Wu X, Wang J, Lopez J, Zhou W, Yang L, Wang SE, Raz DJ, Kim JY. Circulating miRNA profile in esophageal adenocarcinoma. Am J Cancer Res. 2016;6:2713–2721. [PMC free article] [PubMed] [Google Scholar]
- 4.Malhotra A, Sharma U, Puhan S, Chandra Bandari N, Kharab A, Arifa PP, Thakur L, Prakash H, Vasquez KM, Jain A. Stabilization of miRNAs in esophageal cancer contributes to radioresistance and limits efficacy of therapy. Biochimie. 2019;156:148–157. doi: 10.1016/j.biochi.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jamali L, Tofigh R, Tutunchi S, Panahi G, Borhani F, Akhavan S, Nourmohammadi P, Ghaderian SMH, Rasouli M, Mirzaei H. Circulating microRNAs as diagnostic and therapeutic biomarkers in gastric and esophageal cancer. J Cell Physiol. 2018;233:8538–8550. doi: 10.1002/jcp.26850. [DOI] [PubMed] [Google Scholar]
- 6.Shenouda SK, Alahari SK. MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhou HQ, Chen QC, Qiu ZT, Tan WL, Mo CQ, Gao SW. Integrative microRNA-mRNA and protein-protein interaction analysis in pancreatic neuroendocrine tumors. Eur Rev Med Pharmacol Sci. 2016;20:2842–2852. [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Xuan Y, Yang H, Zhao L, Lau WB, Lau B, Ren N, Hu Y, Yi T, Zhao X, Zhou S, Wei Y. MicroRNAs in colorectal cancer: Small molecules with big functions. Cancer Lett. 2015;360:89–105. doi: 10.1016/j.canlet.2014.11.051. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Chen X, Xiu YL, Sun KX, Zhao Y. MicroRNA-490-3P targets CDK1 and inhibits ovarian epithelial carcinoma tumorigenesis and progression. Cancer Lett. 2015;362:122–130. doi: 10.1016/j.canlet.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 11.Shen J, Xiao Z, Wu WK, Wang MH, To KF, Chen Y, Yang W, Li MS, Shin VY, Tong JH, et al. Epigenetic silencing of miR-490-3p reactivates the chromatin remodeler SMARCD1 to promote Helicobacter pylori-induced gastric carcinogenesis. Cancer Res. 2015;75:754–765. doi: 10.1158/0008-5472.CAN-14-1301. [DOI] [PubMed] [Google Scholar]
- 12.Li W, Guo F, Wang P, Hong S, Zhang C. MiR-221/222 confers radioresistance in glioblastoma cells through activating Akt independent of PTEN status. Curr Mol Med. 2014;14:185–195. doi: 10.2174/1566524013666131203103147. [DOI] [PubMed] [Google Scholar]
- 13.Zhang LY, Liu M, Li X, Tang H. MiR-490-3p modulates cell growth and epithelial to mesenchymal transition of hepatocellular carcinoma cells by targeting endoplasmic reticulum-Golgi intermediate compartment protein 3 (ERGIC3) J Biol Chem. 2013;288:4035–4047. doi: 10.1074/jbc.M112.410506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, Xu G, Liu H, Li T. MicroRNA-490-3p regulates cell proliferation and apoptosis by targeting HMGA2 in osteosarcoma. FEBS Lett. 2015;589:3148–3153. doi: 10.1016/j.febslet.2015.08.034. [DOI] [PubMed] [Google Scholar]
- 15.Tian J, Xu YY, Li L, Hao Q. MiR-490-3p sensitizes ovarian cancer cells to cisplatin by directly targeting ABCC2. Am J Transl Res. 2017;9:1127–1138. [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, Xu X, Xu X, Hu Z, Wu J, Zhu Y, Chen H, Mao Y, Lin Y, Luo J, et al. MicroRNA-490-5p inhibits proliferation of bladder cancer by targeting c-Fos. Biochem Biophys Res Commun. 2013;441:976–981. doi: 10.1016/j.bbrc.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Rezatabar S, Karimian A, Rameshknia V, Parsian H, Majidinia M, Kopi TA, Bishayee A, Sadeghinia A, Yousefi M, Monirialamdari M, Yousefi B. RAS/MAPK signaling functions in oxidative stress, DNA damage response and cancer progression. J Cell Physiol. Feb 27; doi: 10.1002/jcp.28334. doi: 10.1002/jcp.28334 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 18.Krysan PJ, Colcombet J. Cellular complexity in MAPK signaling in plants: Questions and emerging tools to answer them. Front Plant Sci. 2018;9:1674. doi: 10.3389/fpls.2018.01674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatterjee N, Das S, Bose D, Banerjee S, Jha T, Das Saha K. Lipid from infective L. Donovani regulates acute myeloid cell growth via mitochondria dependent MAPK pathway. PLoS One. 2015;10:e120509. doi: 10.1371/journal.pone.0120509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen SX, Zhao F, Huang XJ. MAPK signaling pathway and erectile dysfunction. Zhonghua Nan Ke Xue. 2018;24:442–446. (In Chinese) [PubMed] [Google Scholar]
- 21.Furukawa T. Impacts of activation of the mitogen-activated protein kinase pathway in pancreatic cancer. Front Oncol. 2015;5:23. doi: 10.3389/fonc.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Hummel R, Sie C, Watson DI, Wang T, Ansar A, Michael MZ, Van der Hoek M, Haier J, Hussey DJ. MicroRNA signatures in chemotherapy resistant esophageal cancer cell lines. World J Gastroenterol. 2014;20:14904–14912. doi: 10.3748/wjg.v20.i40.14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah AA, Leidinger P, Backes C, Keller A, Karpinski P, Sasiadek MM, Blin N, Meese E. A set of specific miRNAs is connected with murine and human gastric cancer. Genes Chromosomes Cancer. 2013;52:237–249. doi: 10.1002/gcc.22024. [DOI] [PubMed] [Google Scholar]
- 25.Wang LL, Sun KX, Wu DD, Xiu YL, Chen X, Chen S, Zong ZH, Sang XB, Liu Y, Zhao Y. DLEU1 contributes to ovarian carcinoma tumourigenesis and development by interacting with miR-490-3p and altering CDK1 expression. J Cell Mol Med. 2017;21:3055–3065. doi: 10.1111/jcmm.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Pan X, Hu Z. MiR-502 mediates esophageal cancer cell TE1 proliferation by promoting AKT phosphorylation. Biochem Biophys Res Commun. 2018;501:119–123. doi: 10.1016/j.bbrc.2018.04.188. [DOI] [PubMed] [Google Scholar]
- 27.Liang N, Song X, Xie J, Xu D, Liu F, Yu X, Tian Y, Liu Z, Qiao L, Zhang J. Effect of galectin-3 on the behavior of Eca109 human esophageal cancer cells. Mol Med Rep. 2015;11:896–902. doi: 10.3892/mmr.2014.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim EK, Choi EJ. Compromised MAPK signaling in human diseases: An update. Arch Toxicol. 2015;89:867–882. doi: 10.1007/s00204-015-1472-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.