Abstract
The regulatory roles of circular RNAs (circRNAs) in cancer are attracting increasing attention. The aim of the present study was to explore the roles of circRNAs in pancreatic ductal adenocarcinoma (PDAC) using microarray data. The circRNA and microRNA (miRNA) microarray data were downloaded from Gene Expression Omnibus. A total of 256 differentially expressed circRNAs were obtained by analyzing the circRNA microarray data from 26 pairs of PDAC and adjacent normal tissues. Differentially expressed miRNAs were analyzed using a dataset of 6 PDAC tissues and 5 non-neoplastic pancreas samples (GSE43796); 20 differentially expressed miRNAs were detected. circRNA/miRNA interactions were predicted between differentially expressed circRNAs and miRNAs using miRanda and RNAhybrid algorithms and 51 circRNA/miRNA interactions were obtained. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using gene symbols of differentially expressed circRNAs demonstrated that 41 circRNAs were enriched in 17 pathways. Subnetworks that were associated with apoptosis or proliferation were extracted from the 17 pathways and a new network was constructed using Cytoscape software, which identified that mitogen-activated protein kinase, PI3K/AKT and WNT/β-catenin signaling pathways may be associated with PDAC development. In conclusion, 256 differentially expressed circRNAs and 20 differentially expressed miRNAs were identified in PDAC tissues compared with normal tissues; the circRNA/miRNA interactions and the networks of KEGG pathways provided a global view of the function of these differentially expressed circRNAs and miRNAs.
Keywords: circular RNA, microRNA, pancreatic cancer, pathway analysis
Introduction
Pancreatic cancer is one of the top five causes of cancer-associated mortality with a fatality rate of 95%; the majority of patients with pancreatic cancer have a delayed diagnosis, as early diagnosis is difficult (1). Recent data showed that pancreatic cancer has the lowest 5-year relative survival rate (9%) among all cancer types (2). Pancreatic ductal adenocarcinoma (PDAC) accounts for a majority of pancreatic cancer cases and is an aggressive and difficult malignancy to treat (3). Despite advancing knowledge of the mechanisms of carcinogenicity and improvement in diagnosis and management, the prognosis remains poor for patients with PDAC (3). Therefore, new mechanisms and biomarkers are needed to improve the prognosis and detection of PDAC in patients.
MicroRNAs (miRNAs) are a class of small non-coding RNAs (ncRNAs), 18–25 nucleotides long, that negatively control gene expression at the mRNA and protein level (4). Accumulating evidence has suggested that miRNAs are dysregulated in various types of cancers, including PDAC (5). As the absence of symptoms in early disease results in a late diagnosis of PDAC, numerous miRNAs, such as miR-21, miR-155, miR-196 and miR-210, have been identified as biomarkers for PDAC (5). Understanding the mechanisms contributing to miRNA dysregulation may help to improve PDAC diagnosis and treatment.
Another class of ncRNAs are circular RNAs (circRNAs), which function as miRNA sponges, thus serving as competitive inhibitors and suppressing the ability of the miRNA to bind its mRNA targets (6,7). Although circRNAs were discovered in RNA viruses in the 1970s, these molecules were considered as molecular flukes or products of aberrant RNA splicing (7). Recent evidence indicates that circRNAs may serve important roles in cancers, including bladder cancer (8), esophageal squamous cell carcinoma (9), breast cancer (10), basal cell carcinoma (11), cutaneous squamous cell carcinoma (12), colorectal cancer (13) and PDAC (14). In the present study, microarray data downloaded from Gene Expression Omnibus (GEO) were used to explore the roles of circRNAs and miRNAs in PDAC and to provide novel insights into PDAC biology.
Materials and methods
Microarray data
Raw gene expression profile datasets GSE69362, GSE79634 and GSE43796 were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo). The datasets GSE69362 and GSE79634 were analyzed on the GPL19978 Agilent-069978 Arraystar Human CircRNA microarray V1 platform, which analyzes 5,396 human circRNAs at the same time. The dataset GSE43796 was analyzed on the GPL15159 Agilent-031181 Unrestricted Human miRNA V16.0 Microarray platform, which can detect 1,205 mature human miRNAs.
The GSE69362 dataset accessed the expression of circRNAs in six pairs of PDAC and adjacent normal tissues. The GSE79634 dataset accessed the expression of circRNAs in 20 pairs of samples. GSE43796 detected the expression of miRNAs in solid-pseudopapillary neoplasm of pancreas (n=14), ductal adenocarcinoma (n=6), neuroendocrine tumor (n=6) and non-neoplastic pancreas (n=5). The six PDAC tissues and the five non-neoplastic pancreases from GSE43796 were included in the present study.
Data analysis
Differentially expressed genes were identified using the limma package in the R software (version 3.3.2; http://www.r-project.org) (15). Raw data was read and background was corrected and normalized. Probes that were ≥10% brighter compared with the negative controls on at least one-half of arrays were averaged for each gene. The array weights were estimated and used in the linear model analysis. Paired or unpaired Student's t-test was used to identify differentially expressed genes between tumor tissues and normal tissues. The Benjamini-Hochberg method was used to correct for multiple comparisons (16). Differentially expressed genes between tumor tissues and normal tissues were identified, using fold-change >2 and adjusted P<0.05 as selection criteria.
miRNA prediction
As certain sequences of circRNAs in circBase may differ from the mature sequence of circRNAs (17), sequences of the circRNAs were extracted using circPrimer (version 1.2; http://www.bioinf.com.cn) (18), which was developed by our laboratory. As exon-derived circRNAs have been demonstrated to serve as miRNA sponges, the 256 differentially expressed circRNAs were annotated using circPrimer and only 214 exon-derived circRNAs were used for further analysis.
The sequences of the 214 circRNAs and 20 differentially expressed miRNAs were saved as FASTA format. circRNA/miRNA interactions were predicted using circMir (version 1.0; http://www.bioinf.com.cn), which used the miRanda (19) and RNAhybrid (20) algorithms to predict miRNAs that bind to spliced junctions of circRNAs. As the prediction algorithm often suffers from high false positive rates, only the circRNA/miRNA interactions predicted by the two independent algorithms were used. Cytoscape software version 3.1.1 (http://cytoscape.org) was utilized to construct a possible functional network of the differentially expressed circRNAs and miRNAs.
Pathway analysis of circRNA gene symbols
circRNA host genes were submitted to the web-based tool DAVID (http://david.abcc.ncifcrf.gov) to perform a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The following default thresholds were used: Count=2 and EASE (a modified Fisher's Exact test P-value)=0.1. The enriched pathways and gene symbols were imported into Cytoscape software version 3.1.1 to construct a network.
Results
Overview of the datasets
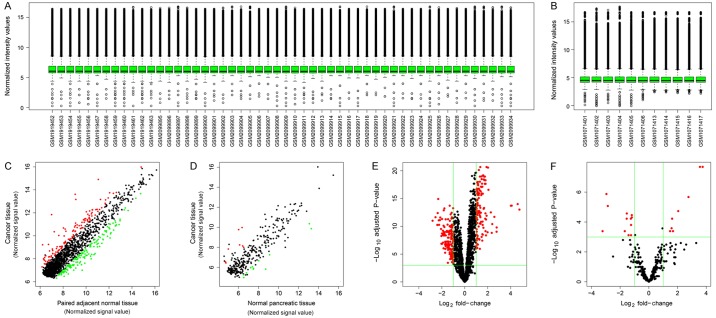
The distribution of normalized signal intensities of the datasets indicated that microarray quality for circRNA and miRNA was good (Fig. 1A and B, respectively). The expression levels of the majority of genes were correlated between PDAC and normal tissues, although there were a number of differentially expressed circRNAs and miRNAs (Fig. 1C and D, respectively). Volcano plots were generated to visualize the relationship of fold-change and statistical significance for circRNAs and miRNAs (Fig. 1E and F, respectively).
Figure 1.
Overview of the microarray datasets. (A) Box plots of the distributions of normalized signal intensities of the circRNA datasets. (B) Box plots for the distributions of normalized signal intensities of the miRNA datasets. (C) Scatter plots of the circRNA microarray signal values of between PDAC and adjacent normal tissues. Red dots represent upregulated circRNAs compared with normal tissues; green dots represent downregulated circRNAs compared with normal tissues; black dots represent equally expressed circRNAs. (D) Scatter plots of the miRNA microarray signal values between PDAC and normal tissues. Red dots represent upregulated miRNAs compared with normal tissues; green dots represent downregulated miRNAs compared with normal tissues; black dots represent equally expressed miRNAs. (E) Volcano plot of circRNA datasets. Red dots represent differentially expressed circRNAs. (F) Volcano plot of miRNA datasets. Red dots represent differentially expressed miRNAs. circRNA, circular RNA; miRNA, microRNA; PDAC, pancreatic ductal adenocarcinoma.
Identification of differentially expressed genes
Compared with adjacent normal tissues, there were 256 differentially expressed circRNAs in PDAC tissues, including 115 upregulated circRNAs and 141 downregulated circRNAs. The top 40 differentially expressed circRNAs in PDAC are presented in Table I, and the full list is presented in Table SI. Eight upregulated miRNAs and 12 downregulated miRNAs were identified in PDAC (Table II).
Table I.
Top 40 differentially expressed circRNAs in pancreatic ductal adenocarcinoma.
| A, Upregulated circRNAs | ||||
|---|---|---|---|---|
| circRNA | Host gene | Fold change | P-value | Adjusted P-valuea |
| hsa_circ_0000977 | NOL10 | 27.155 | 1.498×10−7 | 2.223×10−6 |
| hsa_circ_0006220 | TADA2A | 24.448 | 3.806×10−8 | 8.269×10−7 |
| hsa_circ_0001666 | FAM120B | 17.041 | 5.381×10−8 | 1.096×10−6 |
| hsa_circ_0043278 | TADA2A | 16.234 | 5.970×10−8 | 1.110×10−6 |
| hsa_circ_0078297 | MTHFD1L | 6.870 | 9.875×10−6 | 5.566×10−5 |
| hsa_circ_0013912 | POLR3C | 6.834 | 2.484×10−5 | 1.198×10−4 |
| hsa_circ_0003600 | SOX13 | 5.984 | 8.372×10−7 | 8.265×10−6 |
| hsa_circ_0044436 | KAT7 | 5.272 | 4.677×10−6 | 3.019×10−5 |
| hsa_circ_0018909 | VDAC2 | 5.108 | 3.209×10−5 | 1.463×10−4 |
| hsa_circ_0029634 | ZMYM2 | 4.829 | 2.919×10−7 | 3.679×10−6 |
| hsa_circ_0001907 | LOC100507412 | 4.182 | 1.461×10−9 | 6.333×10−8 |
| hsa_circ_0066147 | SFMBT1 | 4.079 | 2.067×10−12 | 1.165×10−9 |
| hsa_circ_0092314 | RANBP1 | 3.967 | 2.826×10−5 | 1.317×10−4 |
| hsa_circ_0050102 | PGPEP1 | 3.741 | 6.531×10−7 | 6.735×10−6 |
| hsa_circ_0006117 | PTPRA | 3.722 | 1.245×10−12 | 1.053×10−9 |
| hsa_circ_0000912 | FCHO1 | 3.613 | 1.603×10−6 | 1.327×10−5 |
| hsa_circ_0080210 | GRB10 | 3.607 | 5.627×10−5 | 2.310×10−4 |
| hsa_circ_0030292 | FAM124A | 3.515 | 3.366×10−10 | 2.159×10−8 |
| hsa_circ_0082452 | EXOC4 | 3.503 | 1.274×10−10 | 1.134×10−8 |
| hsa_circ_0005105 | SEC24A | 3.484 | 3.878×10−7 | 4.401×10−6 |
| B, Downregulated circRNAs | ||||
| circRNA | Host gene | Fold change | P-value | Adjusted P-valuea |
| hsa_circ_0000518 | RPPH1 | 6.986 | 2.148×10−6 | 1.649×10−5 |
| hsa_circ_0005556 | NBAS | 5.853 | 9.288×10−7 | 8.823×10−6 |
| hsa_circ_0005918 | FCHSD2 | 5.249 | 5.710×10−6 | 3.537×10−5 |
| hsa_circ_0081188 | SLC25A13 | 5.129 | 3.563×10−7 | 4.156×10−6 |
| hsa_circ_0013587 | LRIG2 | 4.969 | 1.231×10−8 | 3.304×10−7 |
| hsa_circ_0006110 | USP34 | 4.813 | 1.952×10−6 | 1.536×10−5 |
| hsa_circ_0005273 | PTK2 | 4.716 | 5.497×10−6 | 3.455×10−5 |
| hsa_circ_0003930 | GGNBP2 | 4.478 | 1.848×10−6 | 1.474×10−5 |
| hsa_circ_0000511 | RPPH1 | 4.398 | 9.923×10−7 | 9.271×10−6 |
| hsa_circ_0000517 | RPPH1 | 4.193 | 1.349×10−5 | 7.288×10−5 |
| hsa_circ_0005394 | ZC3H7A | 4.102 | 6.545×10−5 | 2.594×10−4 |
| hsa_circ_0092367 | SNORD116-14 | 4.099 | 3.443×10−4 | 1.036×10−3 |
| hsa_circ_0000520 | RPPH1 | 3.981 | 3.628×10−4 | 1.082×10−3 |
| hsa_circ_0003831 | PPP2R5C | 3.950 | 1.410×10−6 | 1.210×10−5 |
| hsa_circ_0006944 | GTF2I | 3.746 | 5.175×10−5 | 2.172×10−4 |
| hsa_circ_0061749 | BRWD1 | 3.745 | 1.933×10−5 | 9.955×10−5 |
| hsa_circ_0000958 | PPP1R12C | 3.704 | 3.575×10−5 | 1.603×10−4 |
| hsa_circ_0000662 | AXIN1 | 3.700 | 2.699×10−4 | 8.420×10−4 |
| hsa_circ_0079385 | ZDHHC4 | 3.605 | 9.123×10−4 | 2.374×10−3 |
| hsa_circ_0001288 | SETD2 | 3.600 | 6.598×10−6 | 4.043×10−5 |
The P-value was adjusted using the Benjamini-Hochberg method. circRNA; circular RNA.
Table II.
Differentially expressed miRNAs in pancreatic ductal adenocarcinoma.
| A, Upregulated miRNAs | |||
|---|---|---|---|
| miRNA | Fold change | P-value | Adjusted P-valuea |
| hsa-miR-210-3p | 13.480 | 2.649×10−6 | 4.584×10−4 |
| hsa-miR-135b-5p | 11.830 | 2.289×10−6 | 4.584×10−4 |
| hsa-miR-203a-3p | 6.798 | 3.989×10−5 | 3.450×10−3 |
| hsa-miR-222-3p | 4.139 | 1.787×10−4 | 8.834×10−3 |
| hsa-miR-196b-5p | 3.216 | 1.923×10−3 | 3.379×10−2 |
| hsa-miR-200a-5p | 3.080 | 5.337×10−4 | 1.467×10−2 |
| hsa-miR-221-3p | 3.003 | 1.204×10−3 | 2.777×10−2 |
| hsa-miR-181b-5p | 2.791 | 1.953×10−3 | 3.379×10−2 |
| B, Downregulated miRNAs | |||
| miRNA | Fold change | P-value | Adjusted P-valuea |
| hsa-miR-148a-3p | 9.347 | 1.815×10−3 | 3.379×10−2 |
| hsa-miR-144-3p | 7.768 | 2.443×10−5 | 2.818×10−3 |
| hsa-miR-451a | 7.255 | 1.090×10−4 | 6.285×10−3 |
| hsa-miR-376c-3p | 3.198 | 1.902×10−3 | 3.379×10−2 |
| hsa-miR-204-5p | 2.916 | 4.908×10−4 | 1.467×10−2 |
| hsa-miR-154-5p | 2.897 | 2.396×10−4 | 1.036×10−2 |
| hsa-miR-381-3p | 2.665 | 2.865×10−3 | 4.425×10−2 |
| hsa-miR-654-3p | 2.382 | 5.511×10−4 | 1.467×10−2 |
| hsa-miR-377-3p | 2.365 | 2.942×10−3 | 4.425×10−2 |
| hsa-miR-136-3p | 2.359 | 9.045×10−4 | 2.235×10−2 |
| hsa-miR-557 | 2.275 | 3.002×10−4 | 1.154×10−2 |
| hsa-miR-487b-3p | 2.271 | 3.903×10−4 | 1.350×10−2 |
The P-value was adjusted using the Benjamini-Hochberg method. circRNA; circular RNA.
Prediction of circRNA/miRNA interaction
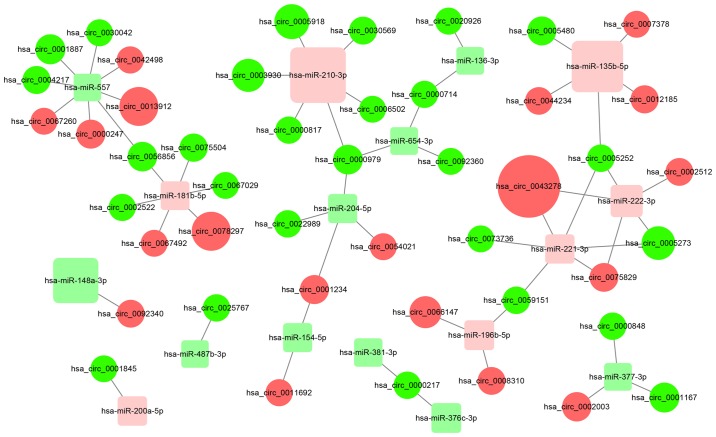
circRNAs may upregulate the expression of target genes of miRNAs by sequestering the miRNAs. In the present study, a total of 51 circRNA/miRNA interactions were identified and a possible functional network of the differentially expressed circRNAs and miRNAs was generated (Fig. 2). The detailed results including miRNA binding sites are presented in Table SII.
Figure 2.
Networks of circRNA/miRNA interactions predicted using miRanda and RNAhybrid software. Red circles represent upregulated circRNAs; green circles represent downregulated circRNAs; pink squares represent upregulated miRNAs; light green squares represent downregulated miRNAs. The size of the circles and squares represents the fold-change of the genes. circRNA, circular RNA; miRNA, microRNA.
KEGG pathway analysis
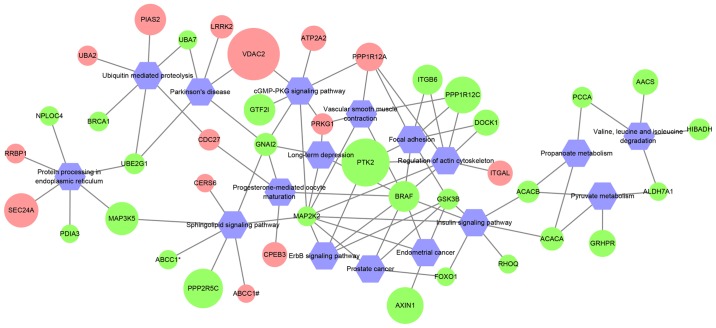
Previous studies have shown that circRNAs modulate the expression of their host genes (6,7). Thus, KEGG pathway enrichment analysis was performed using host genes of differentially expressed circRNAs. A total of 41 host genes were enriched in 17 pathways (Fig. 3; Table SIII). B-Raf proto-oncogene, serine/threonine kinase (BRAF) and mitogen-activated protein kinase kinase 2 (MAP2K2) interacted with the highest number of pathways, which suggested that these two genes may serve important roles in PDAC.
Figure 3.
Network of KEGG pathways and circRNA host gene symbols. Red circles represent upregulated circRNA host genes; green circles represent downregulated circRNA host genes; the size of the circles represents the fold-change of the circRNAs. As both hsa_circ_0000676 and hsa_circ_0000679 are derived from ABCC1, they are indicated as ABCC1# and ABCC1*, respectively. circRNA, circular RNA; KEGG, Kyoto Encyclopedia of Genes and Genomes.
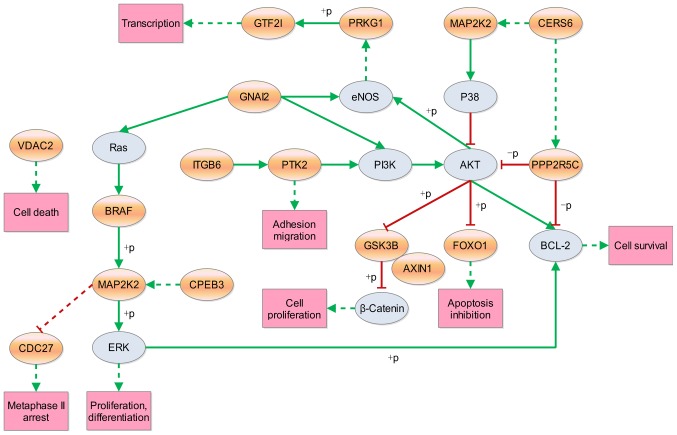
Since apoptosis and proliferation are associated with cancer development and progression, the subnetworks of these pathways were extracted from the 17 enriched KEGG pathways. Subsequently, a new network was constructed with these extracted subnetworks, which included 16 host genes that potentially affect cell survival directly or indirectly (Fig. 4). Thus, target circRNAs were obtained for future studies.
Figure 4.
Pathways associated with apoptosis or proliferation. Orange ellipses represent the host gene symbols for differentially expressed circRNAs and gray ellipses represent the downstream genes regulated by parental genes of the circRNAs in the KEGG pathways; green solid lines represent a direct activation effect; green dotted lines represent an indirect activation effect; red solid lines represent a direct inhibition effect; red dotted lines represent an indirect inhibition effect. circRNA, circular RNA; KEGG, Kyoto Encyclopedia of Genes and Genomes; +p, phosphorylation; -p, dephosphorylation.
Discussion
Two former studies from one research group have profiled the expression of circRNAs in PDAC (14,21). However, only six pairs of PDAC and adjacent normal tissues were profiled and the sample size was relatively small. In the present study, comprehensive analysis with 26 samples from GEO was performed to explore the putative roles of circRNAs in PDAC and to provide novel insights into PDAC biology. The regulatory roles of circRNAs in cancer are attracting increasing attention. Li et al (14) have identified a number of dysregulated circRNAs in PDAC. Another study further investigated the expression level of one dysregulated circRNA termed circ-LDLRAD3, which was upregulated in pancreatic cancer cell lines as well as tumors and plasma from patients with pancreatic cancer, and circ-LDLRAD3 was demonstrated to be associated with invasion and metastasis of pancreatic cancer (22). circRNA exerts its regulatory roles through sequestering cancer-related miRNAs (13,23–26). Therefore, differentially expressed circRNAs and miRNAs in PDAC were used to predict circRNA/miRNA interactions. circRNAs may upregulate the expression of target genes of miRNAs by sequestering the miRNAs. In addition, if circRNAs and their host genes share the same miRNA binding sites, the circRNAs may upregulate the expression of their host genes. In the present study, a total of 51 circRNA/miRNA interactions were identified using 256 differentially expressed circRNAs and 20 differentially expressed miRNAs and a network of the circRNA/miRNA interactions was produced to illustrate the relationships of these genes. The network contained 41 circRNA nodes, 14 miRNA nodes and 51 edges. The number of connections indicates gene involvement in various pathways in PDAC; therefore, the nodes connecting multiple genes may be worth further investigation using experimental methods.
circRNAs are encoded in exons and/or introns of their parental genes (27). Previous studies have shown that circRNAs modulate the expression of their parental genes (6,7,28,29). To explore the potential roles of circRNAs, the predominant pathways of the gene symbols of the differentially expressed circRNAs were identified by a pathway-mapping tool. BRAF and MAP2K2 interacted with the highest number of pathways. BRAF and MAP2K2 are two important members of MAPK signaling pathway, which is associated with PDAC (30,31). The results of the present study indicated that the majority of the inspected genes achieved their effects through the PI3K/AKT signaling pathway. Pancreatic acinar-to-ductal metaplasia is an initiating event that is induced through the PI3K/AKT signaling pathway and can progress to PDAC (32). Another pathway involving cell proliferation is the Wnt/β-catenin signaling pathway. Sox15 exerts its tumor-suppressive effects in pancreatic cancer by suppressing the Wnt/β-catenin signaling pathway (33). Recent data has suggested that the WNT/β-catenin signaling pathway upregulated the expression of cellular communication network factor 1 and consequently enhanced pancreatic cancer development and malignancy (34). In the present study, the aim was to identify circRNA-associated pathways that are involved in the development of PDAC.
As well as acting as miRNA sponges, circRNAs may exert their function through protein binding (35), protein coding (36,37), modulating transcriptional activity of RNA polymerase II (38) and competing with linear splicing (39). Cellular localization has been linked with physiological function and functional mechanism of circRNAs (40,41). Most circRNAs comprise of exonic sequences and are located in the cytoplasm, whereas a small number of intron-derived circRNAs are located in the nucleus (42). Therefore, it is necessary to confirm the cellular localization of a circRNA before studying its roles.
The potential limitations of the present study need to be considered when interpreting the results. Firstly, the results were not verified in clinical samples; differentially expressed circRNAs and miRNAs may be verified using reverse transcription-quantitative PCR in larger clinical samples. Secondly, the circRNA/miRNA interactions were predicted using a bioinformatics method and should be confirmed by the luciferase reporter assay. Thirdly, host gene symbols of circRNAs were used to carry out the KEGG pathway enrichment analysis; whether these circRNAs exert an effect in the enriched pathways should be confirmed using experimental methods.
In conclusion, the potential roles of circRNAs in PDAC were explored using bioinformatics. In total, 256 differentially expressed circRNAs and 20 differentially expressed miRNAs were identified in PDAC tissues compared with normal tissues. The roles of circRNAs in PDAC were explored by prediction of circRNA/miRNA interactions and KEGG pathway enrichment analysis. The circRNA-associated pathways presented in present study could help uncover the effect of circRNAs on PDAC development. Further study confirming the effects of these circRNAs on PDAC development are required in order to improve PDAC diagnosis and therapy.
Supplementary Material
Acknowledgements
Not applicable
Funding
This study was funded by The National Natural Science Foundation of China (grant no. 81602551) and the young talents program of Jiangsu Cancer Hospital (grant no. 2017YQL-10).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
QZ and JYW interpreted the data and wrote the paper. SLZ, SYZ and SJY collected the data and conducted the analysis. SLZ designed this study and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wachsmann MB, Pop LM, Vitetta ES. Pancreatic ductal adenocarcinoma: A review of immunologic aspects. J Investig Med. 2012;60:643–663. doi: 10.2310/JIM.0b013e31824a4d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Stark A, Eibl G. Pancreatic ductal adenocarcinoma. https://www.pancreapedia.org/reviews/pancreatic-ductal-adenocarcinoma. [May 23;2015 ];Pancreapedia: Exocrine Pancreas Knowledge Base, Version 1.0. 2015 [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Hernandez YG, Lucas AL. MicroRNA in pancreatic ductal adenocarcinoma and its precursor lesions. World J Gastrointest Oncol. 2016;8:18–29. doi: 10.4251/wjgo.v8.i1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, Xu D, Lin HK, Gong Z. Circular RNAs in cancer: Novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472–480. [PMC free article] [PubMed] [Google Scholar]
- 7.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, Xu Y, Hu J, Dong G, Xu PL, Yin R. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nair AA, Niu N, Tang X, Thompson KJ, Wang L, Kocher JP, Subramanian S, Kalari KR. Circular RNAs and their associations with breast cancer subtypes. Oncotarget. 2016;7:80967–80979. doi: 10.18632/oncotarget.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sand M, Bechara FG, Sand D, Gambichler T, Hahn SA, Bromba M, Stockfleth E, Hessam S. Circular RNA expression in basal cell carcinoma. Epigenomics. 2016;8:619–632. doi: 10.2217/epi-2015-0019. [DOI] [PubMed] [Google Scholar]
- 12.Sand M, Bechara FG, Gambichler T, Sand D, Bromba M, Hahn SA, Stockfleth E, Hessam S. Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2016;83:210–218. doi: 10.1016/j.jdermsci.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, Liang L. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Hao X, Wang H, Liu Z, He Y, Pu M, Zhang H, Yu H, Duan J, Qu S. Circular RNA expression profile of pancreatic ductal adenocarcinoma revealed by microarray. Cell Physiol Biochem. 2016;40:1334–1344. doi: 10.1159/000453186. [DOI] [PubMed] [Google Scholar]
- 15.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110 doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong S, Wang J, Zhang Q, Xu H, Feng J. CircPrimer: A software for annotating circRNAs and determining the specificity of circRNA primers. BMC Bioinformatics. 2018;19:292. doi: 10.1186/s12859-018-2304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34 (Web Server Issue) 2006:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu S, Song W, Yang X, Wang J, Zhang R, Zhang Z, Zhang H, Li H. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genom Data. 2015;5:385–387. doi: 10.1016/j.gdata.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X, Wang S, Wang GX, Sun SY. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23:8345–8354. doi: 10.3748/wjg.v23.i47.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nan A, Chen L, Zhang N, Liu Z, Yang T, Wang Z, Yang C, Jiang Y. A novel regulatory network among LncRpa, CircRar1, MiR-671 and apoptotic genes promotes lead-induced neuronal cell apoptosis. Arch Toxicol. 2017;91:1671–1684. doi: 10.1007/s00204-016-1837-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 27.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144. doi: 10.1016/j.canlet.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 30.Papademetrio DL, Lompardía SL, Simunovich T, Costantino S, Mihalez CY, Cavaliere V, Álvarez É. Inhibition of survival pathways MAPK and NF-κB triggers apoptosis in pancreatic ductal adenocarcinoma cells via suppression of autophagy. Target Oncol. 2016;11:183–195. doi: 10.1007/s11523-015-0388-3. [DOI] [PubMed] [Google Scholar]
- 31.Herner A, Sauliunaite D, Michalski CW, Erkan M, De Oliveira T, Abiatari I, Kong B, Esposito I, Friess H, Kleeff J. Glutamate increases pancreatic cancer cell invasion and migration via AMPA receptor activation and Kras-MAPK signaling. Int J Cancer. 2011;129:2349–2359. doi: 10.1002/ijc.25898. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Bai X, Zhang Q, Wen L, Su W, Fu Q, Sun X, Lou Y, Yang J, Zhang J, et al. The hepatitis B virus X protein promotes pancreatic cancer through modulation of the PI3K/AKT signaling pathway. Cancer Lett. 2016;380:98–105. doi: 10.1016/j.canlet.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 33.Thu KL, Radulovich N, Becker-Santos DD, Pikor LA, Pusic A, Lockwood WW, Lam WL, Tsao MS. SOX15 is a candidate tumor suppressor in pancreatic cancer with a potential role in Wnt/β-catenin signaling. Oncogene. 2014;33:279–288. doi: 10.1038/onc.2012.595. [DOI] [PubMed] [Google Scholar]
- 34.Sano M, Driscoll DR, DeJesus-Monge WE, Quattrochi B, Appleman VA, Ou J, Zhu LJ, Yoshida N, Yamazaki S, Takayama T, et al. Activation of WNT/β-catenin signaling enhances pancreatic cancer development and the malignant potential Via Up-regulation of Cyr61. Neoplasia. 2016:785–794. doi: 10.1016/j.neo.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. Circ-ZNF609 Is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 39.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Shan G. Circular RNAs remain peculiarly unclear in biogenesis and function. Sci China Life Sci. 2015;58:616–618. doi: 10.1007/s11427-015-4855-y. [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Huang C, Wang X, Shan G. Circular RNAs in eukaryotic cells. Curr Genomics. 2015;16:312–318. doi: 10.2174/1389202916666150707161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S, Zhao Y. Circular RNAs: Characteristics, function, and role in human cancer. Histol Histopathol. 2018;33:887–893. doi: 10.14670/HH-11-969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.