Abstract
Rapidly predicting an accurate three dimensional geometry of a molecule is a crucial task for cheminformatics and across a wide range of molecular modeling. Consequently, developing a fast, accurate, and open implementation of structure prediction is necessary for reproducible cheminformatics research. We introduce a fragment-based coordinate generation implementation for Open Babel, a widely-used open source toolkit for cheminformatics. The new implementation improves speed and stereochemical accuracy, while retaining or improving accuracy of bond lengths, bond angles, and dihedral torsions. Input molecules are broken into fragments by cutting at rotatable bonds. The coordinates of fragments are set according to a fragment library, prepared from open crystallographic databases. Since the coordinates of multiple atoms are decided at once, coordinate prediction is accelerated over the previous rules-based implementation in Open Babel, as well as the widely-used distance geometry methods in RDKit. This new implementation will be beneficial for a wide range of applications, including computational property prediction in polymers, molecular materials and drug design.
Electronic supplementary material
The online version of this article (10.1186/s13321-019-0372-5) contains supplementary material, which is available to authorized users.
Keywords: Coordinate generation, Fragments, Molecular geometry
Introduction
Accurate prediction of the three-dimensional structure of a molecule is critical to a wide range of cheminformatics and molecular modeling tasks, since electrostatic, intermolecular, and other conformation-driven properties depend on the interatomic distances. There is an increasing interest in the “inverse design” of molecules [1] with optimal or near-optimal properties. For example, generative neural networks [2–4] and genetic algorithms [5–7] create molecules with desirable target properties. Moreover, many computational chemistry simulations, including molecular dynamics and quantum chemistry require full three-dimensional structures to run.
Consequently, there have been many proposed methods for three-dimensional coordinate generation, including rule-based, [8, 9] fragment-based [10, 11], and distance geometry embedding methods [12–18]. There are a few free or open source packages capable of coordinatte generation, including BALL [19], FROG [20, 21], RDKit [22], and Open Babel [23]. The latter, which is highly popular, has used a rule-based coordinate builder with a small set of ring fragments, followed by force field minimization. Fragment-based approaches [10] have reported increased accuracy and speed over Open Babel, since no force field minimization is required.
In this work, we discuss an open source implementation of a new fragment-based approach for coordinate generation in Open Babel, with improved accuracy and performance. We compare the stereochemical accuracy with the previous implementation and the open source RDKit distance geometry method, as well as speed and geometric accuracy, measured by heavy-atom root mean square displacement with experimental crystal structures (RMSD), bond distance, bond angle, and torsional/dihedral angle errors. RDKit is chosen as a baseline because it is widely used and a benchmark paper [24] describes it as “competitive with the commercial algorithms”. We also discuss molecules with large geometric or stereochemical errors and future work to improve both geometric and stereochemical accuracy while retaining fast performance.
Implementation
The new implementation, using a fragment database, required generation of a suitable fragment library, as well as code in Open Babel to perform the new coordinate generation method.
The new fragment-based coordinate generation requires several steps: (1) break the input molecule into fragments, (2) look up fragments from the library, and (3) generate a 3D structure by stitching fragments together. Figure 1 shows an overview of the method. Since fragments include multiple atoms, placement of fragments improves both speed and accuracy over the rule-based method, in which the position of heavy atoms were determined one-by-one.
Fig. 1.
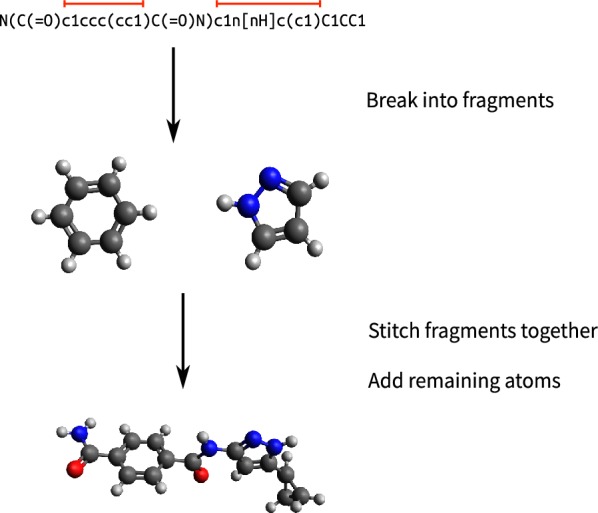
An overview of fragment-based coordinate generation. An input molecule is given without 3D information (e.g. SMILES). The molecule is broken into fragments, which are retrieved from the fragment database. The coordinates of all atoms in a matched fragment are determined in one step, accelerating overall speed of coordinate generation. Fragments and non-fragment atoms are stitched together to generate the final structure
All figures are produced by Avogadro version 1.2 from the corresponding SDF files [25].
Generating the fragment library
Before fragment-based coordinate generation can be implemented, a library of known fragments must be created. For this implementation, any molecular substructure which does not have rotatable bonds is considered a fragment. Thus, each molecule is divided into fragments by cutting at all rotatable bonds, including both rings and large non-rotatable functional groups. Small fragments (less than 5 atoms) are not currently stored into the library.
In generating the library of known fragment geometries, we collected 3D structure information from the Crystallography Open Database (COD) [26], the Platinum Dataset [24], and Ligand Expo [27]. We stored only fragments with at least 5 atoms that occurred at least 3 times in the superset of these repositories, creating a total of 5,779 fragments. For each fragment the canonical SMILES was stored—ensuring that only unique substructures were retained. When the same fragment was encountered multiple times, only the first conformation found in the database was stored. Future work will focus on including averaged consensus geometries from similar fragment conformers (e.g., chair vs. twist-boat cyclohexane).
In addition to this main fragment library, the pre-existing Open Babel database of generic ring fragments was retained. This includes 1000 of the most common ring fragments from analysis of the NCI Open Database [28] and ZINC [29], as well as ring templates from atoms in size, stored as generic SMARTS patterns [30, 31]. This additional library is intended to ensure other ring fragments not explicitly covered in the larger fragment database have approximate matches (e.g., if the stereochemistry or elemental composition differs slightly). Using this auxiliary database is discussed below.
Breaking down fragments
Coordinate prediction starts by breaking the query molecule into multiple fragments of non-rotatable bonds. For each fragment, the canonical SMILES of each fragment is determined.
Fragment search
Using the canonical SMILES of a fragment, the coordinates of all atoms in a fragment are retrieved from the flat-file database. To improve performance, an index file is used to determine the file offset of the particular coordinates. The speedup provided by the index is discussed below.
If the exact canonical SMILES is not found in the fragment database, the fragment is tested against the SMARTS patterns of general ring fragments in the auxiliary database. If a fragment is not found in both databases, the atoms are handled by the rules-based atom-by-atom builder. In the set of 4548 Platinum compounds, there were 9741 fragments which have at least five atoms, and 7852 (80.6%) of fragments are found in the COD-based rigid fragment database, 1,887 (19.4%) are partially found in general ring database, and only two fragments are not found in either database.
Stitching fragments
Building up the entire geometry requires connecting fragments and atom-by-atom rules-based coordinate generation. A working molecule is prepared in the beginning of this process. It only includes atoms of the input molecule, i.e. all bonds are removed. Atoms are iterated by depth-first search order of original molecule. Any atom that has already been determined is skipped from further processing. Otherwise, it is checked for a fragment match. All atoms not matching fragments are connected one-by-one to the working molecule by the previous Open Babel rules-based builder code.
For each fragment match, the geometry of the match is retrieved from the database, and translated to connect to the neighboring atom in the working molecule. The bond vector between the existing atom and the new fragment is determined based on the perceived hybridization of the atom (e.g., sp, sp2, sp3), the covalent radii of the two elements, and the bond order (i.e., single, double, triple, aromatic).
Results and discussion
We evaluated the performance of our method on the Platinum dataset, including 4548 organic ligands from the Protein Data Bank [32]. For testing, we used 4432 fragments from the open crystallographic database (COD) to avoid overlap of “training” and test sets. If fragments were drawn from test set molecules, RMSD and other geometric errors would reduce unfairly as fragments know the “answer” of the prediction. For comparison, we considered the most recent release of Open Babel (v. 2.4.1), RDKit (Release 2018.09.1) with the ETKDG method [33] and this new implementation. Each molecule was supplied to the programs as the corresponding SMILES string.
Across the entire set of molecules, we considered the time to generate coordinates and heavy-atom root mean square deviation (RMSD) between the generated and reference molecule. As the RMSD is highly susceptible to differences in conformers, we also considered the mean bond length error, mean bond angle error, and dihedral angle error. Finally, we tested the “success” of retaining the stereochemistry of the original SMILES. All experiments are conducted as single-core processes on a Thinkpad X1 Carbon laptop with Core i7-7500U (2.70GHz), 16GB RAM and Ubuntu 16.04 running on Docker (18.09.1).
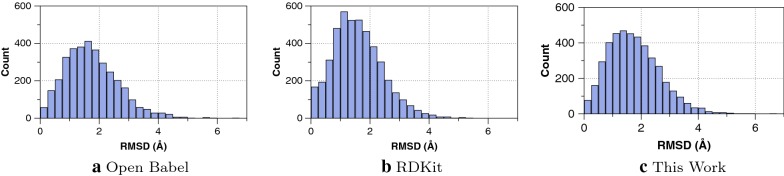
As compiled in Table 1 the new implementation improves the stereochemical success rate from 76.3% to 93.9% while dramatically decreasing the time required by almost a factor of two (93.7 s to 54.8 s). The geometric accuracy increases very slightly, in part due to overall decreases in bond and dihedral/torsion errors. The differences between methods in RMSD, bond errors, angles and torsions are all statistically significant through analysis of variance (ANOVA) with p-values less than . Illustrated in Fig. 2, the distribution of RMSD across the entire set of 4548 ligands is fairly similar between the original Open Babel implementation, RDKit ETKDG and the new implementation. The relatively high RMSD distribution is largely due to differences in conformations between the stochastic single pose considered here and an ensemble of diverse conformers needed to find low-RMSD geometries.[35–37]
Table 1.
Comparison between implementations
| Software | Time (s) | RMSD (Å) | Bond (Å) | Angle () | Torsion () | TFD | Success (%) |
|---|---|---|---|---|---|---|---|
| Open Babel | 93.7 | 1.75 | 0.055 | 2.40 | 48.8 | 0.27 | 76.3 |
| RDKit (ETKDG) | 274.6 | 1.59 | 0.060 | 2.87 | 43.9 | 0.21 | 99.5 |
| This work | 54.8 | 1.75 | 0.049 | 2.49 | 44.1 | 0.27 | 93.9 |
The performance on 4548 molecules in the Platinum dataset is shown. Time column shows the total time to process all molecules in second. RMSD column shows mean RMSD. Bond, Angle, Torsion columns show mean error of each. TFD column shows mean of the torsion fingerprint deviation [34]. Success indicates the percent of predicted molecules whose InChIKey match that of the original molecule. RMSD and mean error are calculated over successful molecules
Fig. 2.
Histograms of heavy-atom RMSD (Å) for a the original Open Babel rule-based coordinate generation, b RDKit distance geometry method, and c current work
In addition to the mean torsion angle error between the generated geometries and the experimental geometries, we computed the torsion fingerprint deviation (TFD) [34] using RDKit, as an established metric for comparison of torsional errors. The metric ignores hydrogen atoms and minimizes effects of dihedral angles with multiple symmetric atoms. Both the mean torsion angle errors and TFD metrics in Table 1 indicate evident differences in dihedral angles between generated geometries and experimental—the main cause for the relatively high observed RMSD.
The fragment-based method is much faster than other methods because it can determine the coordinates of many atoms at once from the database. Compared to ETKDG, bond length errors and angle errors are generally better (e.g., 0.049 Å vs. 0.060 Å, respectively and 2.49 vs. 2.87, respectively). On the other hand, RMSD and torsion errors are slightly worse than ETKDG, possibly because the current implementation does not consider torsion angle explicitly. Some stereo errors remain, likely because of issues with poor layout of some non-fragment bonds, resulting in incorrect stereochemistry.
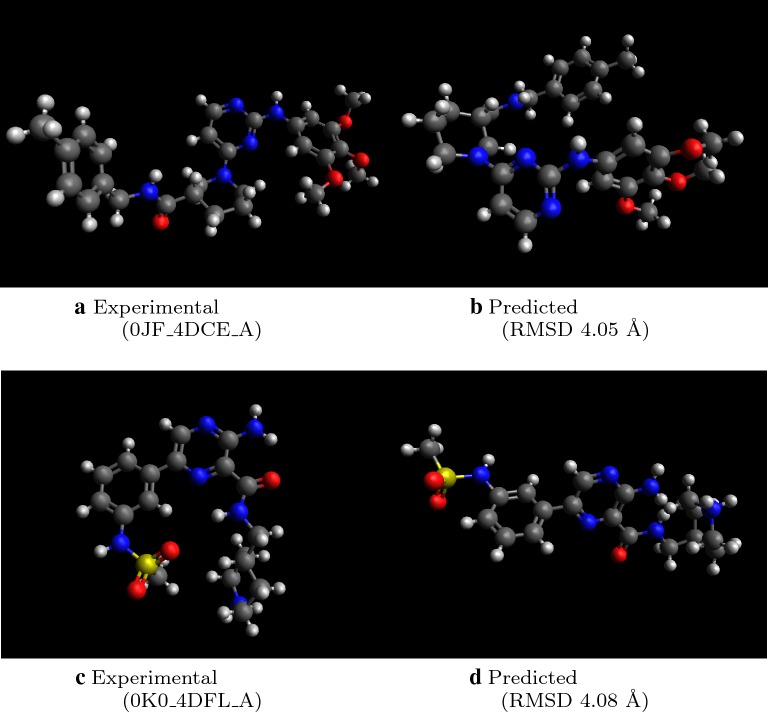
Overall, the new implementation is a notable improvement. For example, Fig. 3 indicates two example molecules with very low RMSD to the experimental structure. The processing time is much faster than both released versions of Open Babel and RDKit.
Fig. 3.
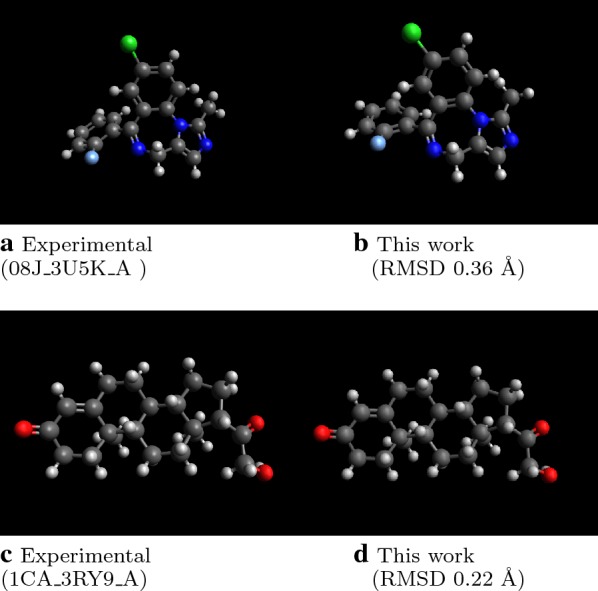
Examples of molecules with low RMSD. Across the molecules in the Platinum set the heavy-atom RMSD between predicted structures and experiment 5.2% falls below 0.5 Å, and 20.5% falls below 1.0 Å, without further force field optimization
While errors in dihedral/torsion angles exist, the purpose of this study is not to find the conformer that best matches experiment by generating various conformers, but rapidly generating initial geometries for further processing. Some evaluation papers (e.g. [24, 36]) report better RMSD for RDKit or Confab. This is because they generate multiple, geometrically diverse conformers and to find the best RMSD. Such conformer generation is recommended subsequent to creating an initial three-dimensional geometry if desired.[36, 38–45]
Analysis of problem molecules
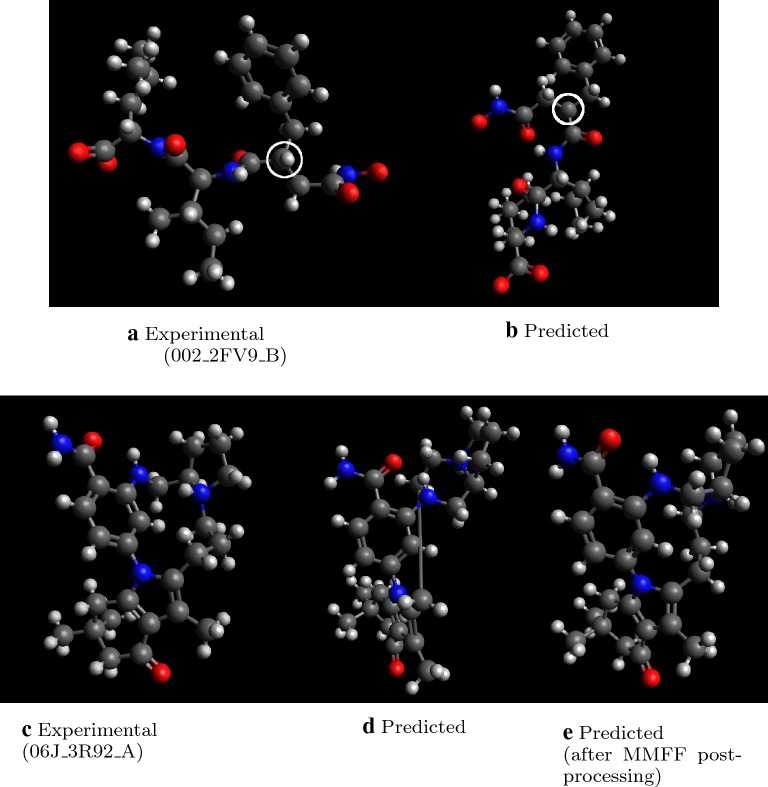
We find that 9.6% of molecules have RMSD above 3.5 Å or incorrect stereochemistry, compared to 26.8% for the original Open Babel implementation, and only 2.8% for RDKit ETKDG. Figure 4 shows two examples of predicted molecule with high RMSD. In both cases, the main differences between experimental and predicted structures come from inter-fragment dihedral angles. In the bottom example, Fig. 4c, d the predicted conformation is more extended than in experiment. While additional post-processing with conformational searches help to minimize such differences, further work to find patterns of inter-fragment dihedral angle preferences will also improve initial predictions.
Fig. 4.
Examples of molecules with high RMSD > 4.0 Å. Note that differences in dihedral angles produce apparently different geometries as judged by RMSD
Beyond poor placement of fragments and choice of dihedral angles, some molecules exhibit incorrect stereochemistry after coordinate generation. Figure 5 shows two examples of molecule with stereochemistry errors. In the first case, an incorrect geometry at the circled carbon yields a difference in stereochemistry after hydrogen atom placement.
Fig. 5.
Examples of molecules with incorrect stereochemistry. Note that the bottom case indicates a case with a long ring closure bond in a macrocycle
In the second case, an extended ring-closure bond occurs in a macrocycle. This structure is generated by stitching small ring fragments together, and they are placed one by one i.e. without considering overall structure. As a result, a long ring-closure bond is inevitable to make ends meet. Post-processing by MMFF optimization (discussed later) reduces these strange connections, but stereochemistry can become scrambled in the process.
As noted above, distance geometry methods such as RDKit ETKDG [33] and stochastic proximity embedding [18] provide greater stereochemistry success rates. The implementation could potentially be improved by using distance geometry to create fragment geometries for species not in the database. Other techniques to augment generation of unusual ring systems and macrocycles should also be explored [46, 47].
Effects of implementation and post-processing
In order to accelerate the fragment search, we prepared an index file, which stores only the canonical SMILES of fragments and the corresponding position in fragment geometry database. When a match occurs, the file offset of the query fragment is retrieved and the database can be read directly to that position without requiring parsing or searching the entire database. While the latter is currently only 744 kb in size, as indicated in Table 2 the use of this index file improves performance by a factor of (i.e., 140.9 s without the index to 54.8 s with the index).
Table 2.
Effect of implementation
| Difference from final | Time (s) | RMSD (Å) | Bond (Å) | Angle () | Torsion () | TFD | Success (%) |
|---|---|---|---|---|---|---|---|
| No index | 140.9 | 1.78 | 0.056 | 2.77 | 45.5 | 0.30 | 92.8 |
| No generic rings | 20.6 | 2.01 | 0.103 | 4.75 | 51.5 | 0.51 | 89.4 |
| Match generic rings before fragments | 56.7 | 1.78 | 0.061 | 2.76 | 46.9 | 0.31 | 92.7 |
| Final implementation | 54.8 | 1.75 | 0.049 | 2.49 | 44.1 | 0.27 | 93.9 |
The performance on 4548 molecules in the Platinum dataset is shown. Time column shows the total time to process all molecules in second. RMSD column shows mean RMSD in Å. Bond, Angle, Torsion columns show mean error of each. TFD column shows mean of the torsion fingerprint deviation [34]. Success indicates the percent of predicted molecules whose InChIKey match that of the original molecule. RMSD and mean error are calculated over successful molecules
Beyond the use of the index file, as described above, we also used an auxiliary database of general ring fragments to improve database hit rate. As indicated in Table 2, we tried using the generic fragments before and after placing fragments. Using the generic rings results in better RMSD, in exchange for somewhat longer prediction time (e.g., 20.6 s vs. 54.8 s). More importantly, the generic fragments reduce bond and angle errors and increase stereochemical success. By searching ring fragments before rigid fragments, the prediction accuracy slightly improved but speed and success rate deteriorated. The final implementation places generic ring fragments after exact matching with rigid fragments.
We also evaluated the effect of post-processing after coordinate generation. The default –gen3d option for Open Babel performs coordinate generation followed by MMFF94 [48–52] geometry optimization and conformer searching, increasing processing time in favor of producing fewer poor geometries (i.e., incorrectly extended ring closure bonds). As illustrated in Table 3, we find that both methods improve RMSD, bond and angle errors and somewhat increase stereochemical accuracy (to 94.0%) at the cost of increased processing time.
Table 3.
Effect of post-processing with MMFF94
| Speed | Time (s) | RMSD (Å) | Bond (Å) | Angle () | Torsion () | TFD | Success (%) |
|---|---|---|---|---|---|---|---|
| Fastest (No MMFF) | 54.8 | 1.75 | 0.049 | 2.49 | 44.1 | 0.27 | 93.9 |
| Fast (100 MMFF) | 372.7 | 1.72 | 0.049 | 2.90 | 44.1 | 0.26 | 93.6 |
| Med (200 MMFF + conf. search) | 732.1 | 1.60 | 0.048 | 2.52 | 43.0 | 0.23 | 93.9 |
The performance on 4548 molecules in the Platinum dataset is shown. Time column shows the total time to process all molecules in second. RMSD column shows mean RMSD in Å. Bond, Angle, Torsion columns show mean error of each. TFD column shows mean of the torsion fingerprint deviation [34]. Success indicates the percent of predicted molecules whose InChIKey match that of the original molecule. RMSD and mean error are calculated over successful molecules
The current default Open Babel conformer search is a weighted stochastic rotor search, changing the likelihood of different dihedral angles on the basis of evaluated MMFF94 energies. That is, during the Monte Carlo search, a high-energy conformation will lead to lower likelihood of the associated dihedral angles being chosen in subsequent iterations.
We would generally suggest that users should use both force field optimization and conformer search to reduce bond, angle and RMSD errors, since the resulting processing time is still an average of 0.16s per compound with a single core process on a laptop. This is set as the default option in Open Babel, although users can opt for “fastest” processing if other optimization or conformer search methods are desired.
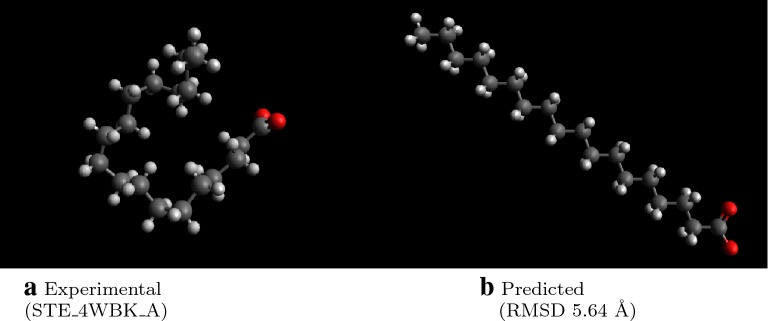
Despite the geometry optimization and low-energy conformer search, the RMSD remains fairly high. In some cases, this occurs because the lowest energy conformation is not necessarily the same as that in the experimental crystal structure [53]. For example in Fig. 6, the generated structure reflects an extended alkyl chain, the low energy conformation, but the experimental structure is folded. Generating geometrically diverse conformers is one method to find geometries matching such experimental pose. As stated above, performing a thorough conformer search is recommended to find either low-energy structures for modeling or diverse geometries to match experimental conformations found in crystal structures.
Fig. 6.
Examples of molecules with high RMSD due to differing dihedral angles. Note that while the experimental conformer reflects a folded alkyl chain, the predicted geometry favors the low-energy extended chain
Finally, we considered use of a larger fragment database by extracting fragments from all of the COD, Ligand Expo and the Platinum set itself. This larger fragments database includes 5,779 fragments, an additional 1,347 fragments more than the COD-only database, discussed above. Results are compiled in Additional file 1: Table S1. In 9,741 fragments (≥ 5 atoms), 9,004 (92.4%) were found in the rigid fragment database. The number of matches increased from 7,852 (80.6%) in case of using the COD-only database. This increase of matches reduced execution time (54.8 s to 28.1 s, respectively), since more fragments could be placed before searching general ring fragments. However, since almost all fragments were found in either of database even in case of COD-only database, the larger fragment data set did not contribute to increased accuracy.
Conclusion
We developed a highly efficient open source coordinate prediction method based on a fragment library. The new implementation improves both speed and stereochemical accuracy over the previous rules-based Open Babel implementation. We find that remaining issues often result from missing fragments, resulting in extended ring-closure bonds and in incorrect dihedral angles. Adding explicit rules to handle rare ring fragments and macrocycles will improve these issues. In some cases, molecules with large RMSD to experimental geometries reflect a difference between the low-energy conformer generated by this implementation and the experimental pose, a known problem [53, 54].
We note that increasing the size of the fragment database decreases the generation time, since more atoms can be placed in one step. As an open source, open data method, we anticipate further improvements in the performance of the implementation over time. For example, drawing from sources such as PubChemQC [55, 56] and ZINC [29] will enable incorporating an increasing number of structurally diverse fragments.
Finally, we note that future work, blending this fragment-based method with distance geometry methods (e.g., ETKDG) used by RDKit will combine the speed of fragment-based placement with improved geometric and stereochemical accuracy. As noted above, fragment methods face challenges on less common, macrocyclic, or systems with overlapping fragments. On the other hand, distance-geometry methods face challenges in producing planar aromatic ring systems, which can be found in fragment databases. By combining both methods, we anticipate improved performance across multiple metrics.
Availability and requirements
Project name: Open Babel
Project home page: http://openbabel.org/
Operating system(s): Platform independent
Programming languages: C++, Python, Ruby, Java, C#
Other requirements: Modern C++ compiler
License: GNU GPL v2.
Additional file
Additional file 1. Histgrams of bond length errors, angle errors, torsion angle errors, torsion fingerprint deviation. Plot of relationship between RMSD and molecular weight, and execution time and molecular weight.
Acknowledgements and funding
The authors appreciate helpful comments and discussion from Noel O’Boyle and Greg Landrum. GRH thanks the National Science Foundation (CHE-1800435) for support. N.Y. thanks the Google Summer of Code 2018 program for support.
Authors' contributions
NY implemented the method and wrote paper. GH supervised the whole project. Both authors read and approved the final manuscript.
Availability of data and materials
All the data, including timing logs, SDF files of all 3D coordinates from Open Babel v2.4.1, RDKit, and this implementation, plus evaluation code is available at https://github.com/n-yoshikawa/ob-fragment-generation. The code and fragment database have been successfully merged into the Open Babel development “master” codebase, available at https://github.com/openbabel/openbabel, intended to be part of the future 3.0 release.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Naruki Yoshikawa, Email: naruki-yoshikawa@g.ecc.u-tokyo.ac.jp.
Geoffrey R. Hutchison, Email: geoffh@pitt.edu
References
- 1.Sanchez-Lengeling B, Aspuru-Guzik A. Inverse molecular design using machine learning: generative models for matter engineering. Science. 2018;361(6400):360–365. doi: 10.1126/science.aat2663. [DOI] [PubMed] [Google Scholar]
- 2.Gómez-Bombarelli R, Wei JN, Duvenaud D, Hernández-Lobato JM, Sánchez-Lengeling B, Sheberla D, Aguilera-Iparraguirre J, Hirzel TD, Adams RP, Aspuru-Guzik A. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent Sci. 2018;4(2):268–276. doi: 10.1021/acscentsci.7b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segler MH, Kogej T, Tyrchan C, Waller MP. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent Sci. 2017;4(1):120–131. doi: 10.1021/acscentsci.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olivecrona M, Blaschke T, Engkvist O, Chen H. Molecular de-novo design through deep reinforcement learning. J Cheminform. 2017;9(1):48. doi: 10.1186/s13321-017-0235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshikawa N, Terayama K, Sumita M, Homma T, Oono K, Tsuda K. Population-based de novo molecule generation, using grammatical evolution. Chem Lett. 2018;47(11):1431–1434. doi: 10.1246/cl.180665. [DOI] [Google Scholar]
- 6.Kanal IY, Owens SG, Bechtel JS, Hutchison G. Efficient computational screening of organic polymer photovoltaics. J Phys Chem Lett. 2013;4(10):1613–1623. doi: 10.1021/jz400215j. [DOI] [PubMed] [Google Scholar]
- 7.O’Boyle NM, Campbell CM, Hutchison G. Computational design and selection of optimal organic photovoltaic materials. J Phys Chem C. 2011;115(32):16200–16210. doi: 10.1021/jp202765c. [DOI] [Google Scholar]
- 8.Sadowski J, Gasteiger J. From atoms and bonds to three-dimensional atomic coordinates: automatic model builders. Chem Rev. 1993;93(7):2567–2581. doi: 10.1021/cr00023a012. [DOI] [Google Scholar]
- 9.Gasteiger J, Rudolph C, Sadowski J. Automatic generation of 3d-atomic coordinates for organic molecules. Tetrahedron Comput Methodol. 1990;3(6):537–547. doi: 10.1016/0898-5529(90)90156-3. [DOI] [Google Scholar]
- 10.Andronico A, Randall A, Benz RW, Baldi P. Data-driven high-throughput prediction of the 3-D structure of small molecules: review and progress. J Chem Inf Model. 2011;51(4):760–776. doi: 10.1021/ci100223t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kothiwale S, Mendenhall JL, Meiler J. Bcl: Conf: small molecule conformational sampling using a knowledge based rotamer library. J Cheminf. 2015;7(1):47. doi: 10.1186/s13321-015-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crippen GM, Smellie AS, Peng JW. Use of augmented Lagrangians in the calculation of molecular conformations by distance geometry. J Chem Inf Comput Sci. 1988;28(3):125–128. doi: 10.1021/ci00059a001. [DOI] [PubMed] [Google Scholar]
- 13.Havel TF, Crippen GM, Kuntz ID, Blaney JM. The combinatorial distance geometry method for the calculation of molecular conformation. II. Sample problems and computational statistics. J Theor Biol. 1983;104(3):383–400. doi: 10.1016/0022-5193(83)90113-3. [DOI] [PubMed] [Google Scholar]
- 14.Havel TF, Kuntz ID, Crippen GM. The combinatorial distance geometry method for the calculation of molecular conformation. I. A new approach to an old problem. J Theor Biol. 1983;104(3):359–381. doi: 10.1016/0022-5193(83)90112-1. [DOI] [PubMed] [Google Scholar]
- 15.Blaney JM, Dixon JS. Distance geometry in molecular modeling. In: Lipkowitz KB, Boyd DB, editors. Reviews in computational chemistry. New York: Wiley; 2007. pp. 299–335. [Google Scholar]
- 16.Spellmeyer DC, Wong AK, Bower MJ, Blaney JM. Conformational analysis using distance geometry methods. J Mol Graph Modell. 2003;15(1):18–36. doi: 10.1016/S1093-3263(97)00014-4. [DOI] [PubMed] [Google Scholar]
- 17.Agrafiotis DK, Xu H. A self-organizing principle for learning nonlinear manifolds. Proc Natl Acad Sci USA. 2002;99(25):15869–15872. doi: 10.1073/pnas.242424399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agrafiotis DK, Xu H, Zhu F, Bandyopadhyay D, Liu P. Stochastic proximity embedding: methods and applications. Mol Inf. 2010;29(11):758–770. doi: 10.1002/minf.201000134. [DOI] [PubMed] [Google Scholar]
- 19.Hildebrandt A, Dehof AK, Rurainski A, Bertsch A, Schumann M, Toussaint NC, Moll A, Stöckel D, Nickels S, Mueller SC, et al. Ball-biochemical algorithms library 1.3. BMC Bioinf. 2010;11(1):531. doi: 10.1186/1471-2105-11-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leite TB, Gomes D, Miteva MA, Chomilier J, Villoutreix BO, Tufféry P. Frog: a free online drug 3d conformation generator. Nucleic acids Res. 2007;35(suppl–2):568–572. doi: 10.1093/nar/gkm289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miteva MA, et al. Frog2: Efficient 3D conformation ensemble generator for small compounds. Nucleic acids Res. 2010;38(Suppl–2):622–677. doi: 10.1093/nar/gkq325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.RDKit: Open-source cheminformatics. http://www.rdkit.org. Accessed 1 Mar 2019
- 23.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open babel: an open chemical toolbox. J Cheminf. 2011;3(1):33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedrich N-O, de Bruyn Kops C, Flachsenberg F, Sommer K, Rarey M, Kirchmair J. Benchmarking commercial conformer ensemble generators. J Chem Inf Model. 2017;57(11):2719–2728. doi: 10.1021/acs.jcim.7b00505. [DOI] [PubMed] [Google Scholar]
- 25.Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminf. 2012;4(1):17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gražulis S, Daškevič A, Merkys A, Chateigner D, Lutterotti L, Quirós M, Serebryanaya NR, Moeck P, Downs RT, Le Bail A. Crystallography open database (cod): an open-access collection of crystal structures and platform for world-wide collaboration. Nucleic Acids Res. 2012;40(D1):420–427. doi: 10.1093/nar/gkr900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Z, Chen L, Maddula H, Akcan O, Oughtred R, Berman HM, Westbrook J. Ligand depot: a data warehouse for ligands bound to macromolecules. Bioinformatics. 2004;20(13):2153–2155. doi: 10.1093/bioinformatics/bth214. [DOI] [PubMed] [Google Scholar]
- 28.NCI Open Database. https://cactus.nci.nih.gov/download/nci/. Accessed 1 Mar 2019
- 29.Irwin JJ, Shoichet BK. Zinc—a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45(1):177–182. doi: 10.1021/ci049714+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sayle R (1997) 1st-class smarts patterns. In: EuroMUG 97
- 31.Skillman G, Kuntz T (1998) Recursive smarts for synthetic chemists. In: EuroMUG 98
- 32.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riniker S, Landrum GA. Better informed distance geometry: using what we know to improve conformation generation. J Chem Inf Model. 2015;55(12):2562–2574. doi: 10.1021/acs.jcim.5b00654. [DOI] [PubMed] [Google Scholar]
- 34.Schulz-Gasch T, Schärfer C, Guba W, Rarey M. Tfd: torsion fingerprints as a new measure to compare small molecule conformations. J Chem Inf Model. 2012;52(6):1499–1512. doi: 10.1021/ci2002318. [DOI] [PubMed] [Google Scholar]
- 35.Friedrich N-O, Meyder A, Bruyn Kops C, Sommer K, Flachsenberg F, Rarey M, Kirchmair J. High-quality dataset of protein-bound ligand conformations and its application to benchmarking conformer ensemble generators. J Chem Inf Model. 2017;57:529–539. doi: 10.1021/acs.jcim.6b00613. [DOI] [PubMed] [Google Scholar]
- 36.Ebejer J-P, Morris GM, Deane CM. Freely available conformer generation methods: how good are they? J Chem Inf Model. 2012;52(5):1146–1158. doi: 10.1021/ci2004658. [DOI] [PubMed] [Google Scholar]
- 37.O’Boyle NM, Vandermeersch T, Flynn CJ, Maguire AR, Hutchison G. Confab—systematic generation of diverse low-energy conformers. J Cheminf. 2011;3(1):8. doi: 10.1186/1758-2946-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan L, Hutchison GR, Morris GM. Bayesian optimization for conformer generation. J Cheminf. 2019;11(1):32. doi: 10.1186/s13321-019-0354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iuzzolino L, Reilly AM, McCabe P, Price SL. Use of crystal structure informatics for defining the conformational space needed for predicting crystal structures of pharmaceutical molecules. J Chem Theory Comput. 2017;13(10):5163–5171. doi: 10.1021/acs.jctc.7b00623. [DOI] [PubMed] [Google Scholar]
- 40.Gunby NR, Masters SL, Crittenden DL. Embracing chemical and structural diversity with UCONGA: a universal conformer generation and analysis program. J Mol Graph Modell. 2017;77:286–294. doi: 10.1016/j.jmgm.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 41.Hawkins PCD. Conformation generation: the state of the art. J Chem Inf Model. 2017;57(8):1747–1756. doi: 10.1021/acs.jcim.7b00221. [DOI] [PubMed] [Google Scholar]
- 42.Gürsoy O, Smieško M. Searching for bioactive conformations of drug-like ligands with current force fields: how good are we? J Cheminform. 2017;9(1):29. doi: 10.1186/s13321-017-0216-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cleves AE, Jain AN. ForceGen 3D structure and conformer generation: from small lead-like molecules to macrocyclic drugs. J Comput Aided Mol Des. 2017;31(5):419–439. doi: 10.1007/s10822-017-0015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kothiwale S, Mendenhall JL, Meiler J. BCL: C onf : small molecule conformational sampling using a knowledge based rotamer library. J Cheminf. 2015;7(1):47. doi: 10.1186/s13321-015-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S, Bolton EE, Bryant SH. PubChem3D: conformer ensemble accuracy. J Cheminf. 2013;5(1):1. doi: 10.1186/1758-2946-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner V, Jantz L, Briem H, Sommer K, Rarey M, Christ CD. Computational macrocyclization: from de novo macrocycle generation to binding affinity estimation. ChemMedChem. 2017;12(22):1866–1872. doi: 10.1002/cmdc.201700478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedrich N-O, Flachsenberg F, Meyder A, Sommer K, Kirchmair J, Rarey M. Conformator: a novel method for the generation of conformer ensembles. J Chem Inf Model. 2018;59(2):731–742. doi: 10.1021/acs.jcim.8b00704. [DOI] [PubMed] [Google Scholar]
- 48.Halgren TA. Merck molecular force field. i. basis, form, scope, parameterization, and performance of mmff94. J Comput Chem. 1996;17(5–6):490–519. doi: 10.1002/(SICI)1096-987X(199604)17:5/6<490::AID-JCC1>3.0.CO;2-P. [DOI] [Google Scholar]
- 49.Halgren TA. Merck molecular force field. II. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions. J Comput Chem. 1996;17(5–6):520–552. doi: 10.1002/(SICI)1096-987X(199604)17:5/6<520::AID-JCC2>3.0.CO;2-W. [DOI] [Google Scholar]
- 50.Halgren TA. Merck molecular force field. III. Molecular geometries and vibrational frequencies for MMFF94. J Comput Chem. 1996;17(5–6):553–586. doi: 10.1002/(SICI)1096-987X(199604)17:5/6<553::AID-JCC3>3.0.CO;2-T. [DOI] [Google Scholar]
- 51.Halgren TA, Nachbar RB. Merck molecular force field. IV. Conformational energies and geometries for MMFF94. J Comput Chem. 1996;17(5–6):587–615. [Google Scholar]
- 52.Halgren TA. Merck molecular force field. V. Extension of MMFF94 using experimental data, additional computational data, and empirical rules. J Comput Chem. 1996;17(5–6):616–641. doi: 10.1002/(SICI)1096-987X(199604)17:5/6<616::AID-JCC5>3.0.CO;2-X. [DOI] [Google Scholar]
- 53.Peach ML, Cachau RE, Nicklaus MC. Conformational energy range of ligands in protein crystal structures: the difficult quest for accurate understanding. J Mol Recogn. 2017;30(8):2618. doi: 10.1002/jmr.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sitzmann M, Weidlich IE, Filippov IV, Liao C, Peach ML, Ihlenfeldt W-D, Karki RG, Borodina YV, Cachau RE, Nicklaus MC. PDB ligand conformational energies calculated quantum-mechanically. J Chem Inf Model. 2012;52(3):739–756. doi: 10.1021/ci200595n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakata M (2015) The pubchemqc project: A large chemical database from the first principle calculations. In: AIP conference proceedings, vol. 1702, p. 090058. AIP Publishing
- 56.Nakata M, Shimazaki T. Pubchemqc project: a large-scale first-principles electronic structure database for data-driven chemistry. J Chem Inf Model. 2017;57(6):1300–1308. doi: 10.1021/acs.jcim.7b00083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Histgrams of bond length errors, angle errors, torsion angle errors, torsion fingerprint deviation. Plot of relationship between RMSD and molecular weight, and execution time and molecular weight.
Data Availability Statement
All the data, including timing logs, SDF files of all 3D coordinates from Open Babel v2.4.1, RDKit, and this implementation, plus evaluation code is available at https://github.com/n-yoshikawa/ob-fragment-generation. The code and fragment database have been successfully merged into the Open Babel development “master” codebase, available at https://github.com/openbabel/openbabel, intended to be part of the future 3.0 release.