Abstract
The aim of the present study was to examine the expression and clinical significance of long non-coding RNA (lncRNA)-CCDC26 in patients with acute myeloid leukemia (AML), and to investigate the potential functions of CCDC26. The Gene Expression Omnibus database and reverse transcription-quantitative polymerase chain reaction analysis were used to detect the expression levels of CCDC26 in patients with AML and healthy volunteers. Clinical data for 93 patients with AML were collected to analyze the clinical significance of CCDC26. Weighted gene co-expression network analysis (WGCNA), a protein-protein interaction (PPI) network, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were used to examine the functions of CCDC26. The expression levels of CCDC26 in the initially diagnosed and relapsed patients with AML were significantly upregulated compared with the control group. The upregulated expression level of CCDC26 in patients with AML was significantly associated with age, anemia, risk stratification and remission. Furthermore, patients with a high CCDC26 expression level had a poorer overall survival (P=0.0105). In addition, the area under the curve (AUC)1year and AUC2year of CCDC26 for overall survival were 0.722 and 0.686, respectively. WGCNA, PPI network and KEGG pathway analysis revealed that CCDC26 was involved in the regulation of a number of biological processes. lncRNA-CCDC26 may serve as a novel biomarker for monitoring the progression and predicting the clinical outcome of AML.
Keywords: biomarker, long non-coding RNA-CCDC26, acute myeloid leukemia, prognosis, weighted gene co-expression network analysis
Introduction
Acute myeloid leukemia (AML) is a heterogeneous malignant clonal disease originating from hematopoietic stem or myeloid progenitor cells (1). AML is the most common type of leukemia in adults, with an incidence rate of 3.7 per 100,000 worldwide, as it progresses rapidly, with the natural course of the disease being only 11–20 weeks (2). Despite recent advances in the diagnosis, treatment and prognosis of AML, the overall survival rate remains <50% (3). Therefore, further studies are required, in order to examine the underlying mechanisms of AML pathogenesis and prognosis, as well as the search for novel treatments. Following extensive research on the pathogenesis of AML over the past few years, abnormal epigenetic modifications have been reported as an important mechanism for the occurrence and development of leukemia (4,5). Long non-coding RNA (lncRNA), a recently discovered epigenetic modification mechanism, serves a critical role in cell function and gene regulation (6,7).
lncRNAs are longer than 200 nucleotides, and are a members of the non-protein-coding RNA family. Numerous studies have demonstrated that lncRNA serves an important role in the pathogenesis and progression of AML (8,9). lncRNA-CCDC26 is located at chromosomal locus 8q24 and is associated with low-grade glioma, chronic myeloid leukemia-derived K562 cells and pediatric AML (10–13). Furthermore, previous studies have reported that CCDC26, also referred to as retinoid modification (RAM), is upregulated in AML cell lines (HL60 and THP-1), while CCDC26 regulates the differentiation and apoptosis of the acute monocytic leukemia cell line PLB985, which is induced by retinoic acid (12,14).
These results have indicated that CCDC26 is associated with AML; however, the expression level and the clinical significance of lncRNA-CCDC26 in adult patients with AML remains unclear. The aim of the present study was therefore to examine the expression of CCDC26 in clinical bone marrow samples of AML and AML cell lines, and evaluate its role in the progression and poor prognosis of AML.
Materials and methods
Patient samples
Bone marrow samples from 93 patients who were diagnosed with AML, based on the World Health Organization Morphology, Immunology, Cytogenetics Molecular biology classification criteria (15), were collected at the Guangzhou First People's Hospital (Guangzhou, China) between November 2014 and March 2018. All participants provided their written informed consent to participate in this study. The age of the patients ranged from 11 to 81 years, with an average of 40 years, including 51 females and 42 males. Among the 93 patients, 45 were at the stage of initial diagnosis of AML, 7 were relapses, 24 post-chemotherapy and 17 post-hematopoietic stem cell transplantation (HSCT). The inclusion criteria were as follows: i) Blasts ≥20% of bone marrow nucleated cells (ANC); ii) blasts <20% of ANC, however with t(15;17), t(8;21) or inv (16)/t(16;16); and iii) clearly diagnosed AML. The exclusion criteria were as follows: i) Age <18 years; and ii) incomplete follow-up information. Mononuclear cells (MNCs) were isolated from bone marrow samples with Ficoll (GE Healthcare Life Sciences, Little Chalfont, UK) and stored at −80°C with TRIzol® reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA) for as long as required. MNCs isolated from the bone marrow included monocytes, lymphocytes, hematopoietic stem cells, and hematopoietic progenitor cells. Bone marrow samples from ten anonymized healthy volunteers were collected between March 2017 and August 2017 and were included as the control samples. The age ranged from 23 to 45 years, with an average of 31 years, and included 5 females and 5 males. The present study was approved by the Ethics Committee of Guangzhou First People's Hospital (Guangzhou, China).
The clinical characteristics of the patients are listed in Table I. Clinical data were followed up until May 1, 2018. The median follow-up time for surviving patients was 388 days (range, 122–1,275 days).
Table I.
Association between CCDC26 expression and clinicopathological factors of acute myeloid leukemia (n=45).
| CCDC26 expression | CCDC26 expression | ||||
|---|---|---|---|---|---|
| Characteristics | High n=36 (%) | Low n=9 (%) | P-valuea | Mean ± standard deviation | P-valueb |
| Sex | 0.117 | 0.412 | |||
| Male | 21 (58.3) | 2 (22.2) | 7.942±8.551 | ||
| Female | 15 (41.7) | 7 (77.8) | 6.039±6.701 | ||
| Age (years) | 0.654 | 0.031c | |||
| ≥60 | 9 (25.0) | 1 (11.1) | 13.667±10.455 | ||
| <60 | 27 (75.0) | 8 (88.9) | 5.111±5.528 | ||
| Anemia | 0.514 | 0.026c | |||
| Yes | 30 (83.3) | 6 (66.7) | 7.815±8.277 | ||
| No | 6 (16.7) | 3 (33.3) | 3.799±3.126 | ||
| Fever | 0.573 | 0.697 | |||
| Yes | 26 (72.2) | 5 (55.6) | 6.707±7.796 | ||
| No | 10 (27.8) | 4 (44.4) | 7.687±7.644 | ||
| Hemorrhage | 0.204 | 0.854 | |||
| Yes | 17 (47.2) | 7 (77.8) | 6.811±7.889 | ||
| No | 19 (52.8) | 2 (22.2) | 7.242±7.612 | ||
| Extramedullary infiltration | 0.106 | 0.538 | |||
| Yes | 25 (69.4) | 3 (33.3) | 6.455±7.076 | ||
| No | 11 (30.6) | 6 (66.7) | 7.929±8.722 | ||
| WBC (×109/l) | 1.000 | 0.893 | |||
| ≥40 | 10 (27.8) | 2 (22.2) | 7.271±8.126 | ||
| <40 | 26 (72.2) | 7 (77.8) | 6.918±7.634 | ||
| LDH (U/l) | 1.000 | 0.654 | |||
| >240 | 30 (83.3) | 7 (77.8) | 6.77±7.368 | ||
| ≤240 | 6 (16.7) | 2 (22.2) | 8.133±9.454 | ||
| Risk stratification | 0.002d | 0.003d | |||
| Poor/Intermediate | 23 (63.9) | 0 (0.0) | 9.886±8.176 | ||
| Favorable | 13 (36.1) | 9 (100) | 3.419±5.248 | ||
| Remission | 0.226 | 0.030c | |||
| PR or NR | 9 (25.0) | 0 (0.0) | 11.926±8.395 | ||
| CR | 27 (75.0) | 9 (100) | 5.783±7.085 | ||
χ2 test
Student's t-test.
P<0.05
P<0.01. WBC, white blood cell; LDH, lactate dehydrogenase; PR, partial remission; NR, no remission; CR, complete remission.
Risk stratification was based on the cytogenetic and molecular anomaly background of the National Comprehensive Cancer Network Guidelines for the treatment of acute myeloid leukemia (16). Since only 3 patients at initial-diagnosis stage were of intermediate risk, risk stratification was performed by classifying patients into poor/intermediate and favorable risk groups.
Public data from the gene expression omnibus (GEO) database
Evaluation of the CCDC26 expression level was performed using the publicly available GEO database (https://www.ncbi.nlm.nih.gov/geo/). GSE85030 was downloaded from the report of Lei et al (17), which contained 6 AML and 2 normal control samples.
Extraction of bone marrow MNCs
Bone marrow samples were superimposed on the surface of 5 ml Ficoll (1.077 g/ml; GE Healthcare Life Sciences), and horizontal centrifugation at 670 × g, at 25°C for 20 min was subsequently performed in the centrifuge (Centrifuge 5810; Eppendorf, Hamburg, Germany). Following centrifugation, the liquid was divided into three layers, with a narrow white cloudy layer mainly composed of MNCs at the interface between the upper and middle layers. The cloudy layer was aspirated into another centrifuge tube, and the MNCs were washed twice with RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.). The proportion of MNCs was estimated on May-Grünwald-Giemsa-stained cytocentrifugate preparations using light microscopy at ×400 magnification. A two-step process was performed for staining, firstly 50% May-Grünwald at 25°C for 3–5 min, secondly 10% buffered Giemsa solution for 10–30 min then running water for 1–3 min. The MNCs selected for analysis contained a minimum of 90% blasts following separation. Pellets of 2–10,000,000 MNCs were stored in TRIzol® reagent and immediately frozen at −80°C.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from MNCs using TRIzol reagent, according to the manufacturer's protocol. An RT kit (Promega Corporation, Madison, WI, USA) was used to reverse transcribe total RNA to cDNA, following the manufacturer's protocol. The reverse transcription method was a 2-step process, with a total 20 µl. Firstly, RNA 500 ng, 0.5 µl oligo (dT) (0.5 µg/reaction), 0.5 µl random primer (0.5 µg/reaction) and RNase free double distilled water (ddH2O), up to 5 µl, was added together and incubated at 70°C for 5 min, and subsequently rapidly cooled on ice for 5 min. Secondly, 4.0 µl GoScriptTM 5X reaction buffer, 1.7 µl MgCl2 (final concentration 2.0 mM), 1.0 µl 0.5 mM dNTPs, 0.3 µl ribonuclease inhibitor (20 U), 1.0 µl reverse transcriptase, and ddH2O to a total of 15 µl was subsequently added. After mixing, samples were incubated at 42°C for 60 min and inactivated at 70°C for 15 min. RT-qPCR was carried out with SYBR Green Master Mix (Promega Corporation). The qPCR was performed at a total 10 µl and included the following: 5 µl GoTaq 2X Master Mix, 0.5 µl forward primer (10 µM), 0.5 µl reverse primer (10 µM), 2 µl cDNA (diluted 10-fold) and 2 µl ddH2O. The following thermocycling conditions were used for the qPCR: Intital denaturation at 95°C for 10 min; 40 cycles of 95°C 15 sec and 60°C for 1 min. Primer sequences were as follows: 18SrRNA, 5′-CGGCGGCTTTGGTGACTCTAGA-3′ forward and 5′-CCTGCTGCCTTCCTTGGATGTG-3′ reverse; CCDC26, 5′-CCTTGTACAGTGTTGCCTCAGC-3′ forward and 5′-GCAGTCTTCGGCATTCTCCCA-3′ reverse. The results were normalized to the expression of 18SrRNA and are presented as a fold change (2−ΔΔCq) (18). Each experiment was repeated in triplicate.
Screening for differentially expressed mRNAs
The differentially expressed mRNAs between AML and normal control samples in GSE85030 were screened using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/). The criteria for screening were adjusted P<0.05 and |log2FC|>1.
Weighted gene co-expression network analysis (WGCNA)
WGCNA, which can identify clusters (modules) of highly correlated genes, is a systematic biology method used to describe correlation patterns among genes in microarray samples (19). Compared to Pearson's correlation coefficient, WGCNA uses the soft threshold to provide more extensive and accurate correlations between genes. The R package WGCNA (19) was used to construct a weighted correlation network between lncRNA-CCDC26 and differentially expressed mRNAs. The module colors under the dendrogram represent the module assignment determined by the Dynamic Tree Cut. The k-core score was used to determine the core genes of the co-expression networks. The lncRNA-co-expressed mRNA cluster was obtained with a soft threshold of 0.85 for further functional analysis of lncRNA-CCDC26.
Pathway analysis and protein-protein interaction (PPI) network establishment
The WGCNA-calculated lncRNA-CCDC26-co-expressed mRNAs were subjected to pathway analysis and PPI network establishment. The Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov/) tool was used for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The PPI network of CCDC26-co-expressed mRNAs was established using Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; http://string-db.org/). The highly important nodes are represented by the central nodes in the network; nodes in the network represent genes, and edges represent interactions between nodes.
Statistical analysis
All statistical analysis was conducted using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA), GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) and R software 3.5.1 (https://www.r-project.org/). Student's t-test and χ2 tests were performed to compare the significance of differences between two groups, as appropriate. One-way ANOVA test followed by the Bonferroni test was used for multiple comparisons. Overall survival rates were calculated via the Kaplan-Meier method and the log-rank test was used for comparison. Receiver operating characteristic (ROC) curves were constructed using the survival ROC package (20) in R software. The data are presented as mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of CCDC26 is upregulated in AML
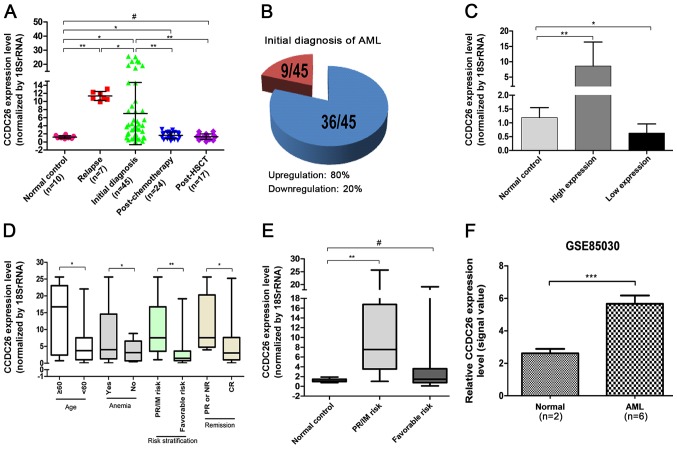
The relative expression level of CCDC26 was detected in 93 AML bone marrow samples by RT-qPCR and normalized to 18rRNA. As indicated in Fig. 1A and B, CCDC26 expression was significantly upregulated in 80% of patients with an initial AML diagnosis (P=0.018) and in patients who had relapsed (P=0.001). At the same time, the expression of CCDC26 in patients with AML relapses was significantly higher compared with patients with an initial AML diagnosis (P=0.041). The level of CCDC26 expression was distinctly decreased in patients with AML following chemotherapy (P=0.001) and HSCT (P=0.002) compared with the patients with an initial AML diagnosis. In addition, compared with the normal control, the expression of CCDC26 in patients with AML following chemotherapy remained high (P=0.045), however there was no significant difference in patients with AML following transplantation (P=0.725). These results indicated that the relative expression of CCDC26 in patients with AML was upregulated, but returned to normal levels following effective treatment. As indicated in Fig. 1C, the absolute value of CCDC26 for upregulation in AML was 7.415 compared with the normal control (P=0.006), while the absolute value of CCDC26 for downregulation in AML was 0.562 compared with the normal control (P=0.039).
Figure 1.
CCDC26 is upregulated in AML. (A) CCDC26 expression was analyzed at different treatment stages of AML and in healthy donors. (B) Pie chart indicating the proportion of AML samples with up-(blue) and downregulated (red) expression of CCDC26. (C) High and low expression level of CCDC26 in patients with AML. (D) Expression of CCDC26 according to clinical parameters of AML. (E) CCDC26 expression in different groups of AML risk stratification. (F) The expression level of CCDC26 was analyzed in the publicly available Gene Expression Omnibus dataset GSE85030. Error bars, mean ± standard errors of the mean. *P<0.05, **P<0.01, ***P<0.001; #P>0.05. AML, acute myeloid leukemia; PR/IM risk, poor/intermediate risk; PR, partial remission; NR, no remission; CR, complete remission; HSCT, hematopoietic stem cells transplantation.
This result was confirmed in the publicly available GEO expression dataset GSE85030. The relative expression level of CCDC26 in AML was higher compared with that in the normal control (Fig. 1F).
CCDC26 upregulation is associated with age, anemia, risk stratification and remission
The association between CCDC26 expression and clinical parameters of patients with an initial AML diagnosis was investigated. Higher CCDC26 expression was observed in patients with old age (≥60 years), anemia, poor/intermediate risk and partial remission or no remission, while the expression was lower in patients at a young age (<60 years), without anemia, favorable risk and complete remission (CR), (P=0.031, 0.026, 0.003 and 0.03, respectively) (Fig. 1D and Table I). No association was observed between the expression level of CCDC26 and other clinical factors, including sex, fever, hemorrhage, white blood cells (WBC), lactate dehydrogenase (LDH) or extramedullary infiltration. Compared with the normal control, the expression of CCDC26 in patients with AML with poor/intermediate risk was significantly increased (P=0.002), however there was no significant difference in the expression of CCDC26 in patients with AML with favorable-risk stratification (P>0.05; Fig. 1E).
Association between clinicopathological factors and CCDC26 expression, analyzed using the χ2 test
The expression level of CCDC26 was divided into two groups: High (above normal control) and low (under normal control). The χ2 test was used to evaluate differences in clinical parameters between the two groups. As presented in Table I, the expression of CCDC26 is associated with risk stratification (P=0.002). In the present study, no significant difference was observed in the association between CCDC26 expression and other clinical parameters, including sex, age, anemia, fever, hemorrhage, WBC, LDH, extramedullary infiltration and remission.
CCDC26 upregulation is associated with poor prognosis in patients with AML
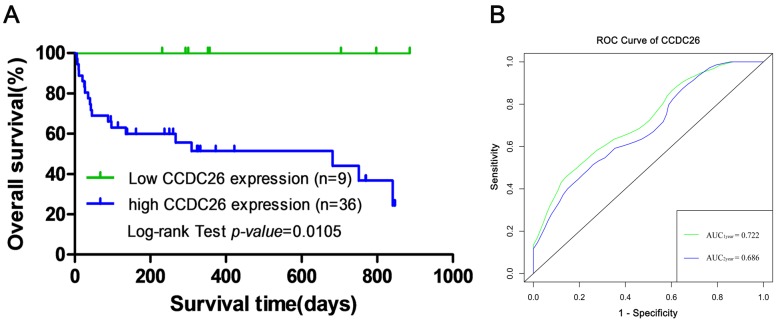
To examine the association between CCDC26 expression levels and the prognosis of patients with AML, Kaplan-Meier survival analysis and the log-rank test were performed to evaluate the association between CCDC26 expression and overall survival. Kaplan-Meier survival analysis indicated a significant difference in overall survival between the CCDC26 high- and low-expression groups in 45 patients with an initial AML diagnosis (log-rank test; χ2=6.550; P=0.0105; Fig. 2A). The ROC curve was used to evaluate the predictive ability of CCDC26. As presented in Fig. 2B, the AUC1year and AUC2year of CCDC26 according to the ROC curve were 0.722 and 0.686, respectively, which indicated that the expression level of CCDC26 can be used to predict overall survival in AML.
Figure 2.
CCDC26 expression is associated with prognosis. (A) Kaplan-Meier survival curves of patients with acute myeloid leukemia were stratified by CCDC26 expression levels. (B) Predictive ability of CCDC26 for overall survival rate. ROC, receiver operating characteristic; AUC, area under the curve.
Co-expression module establishment
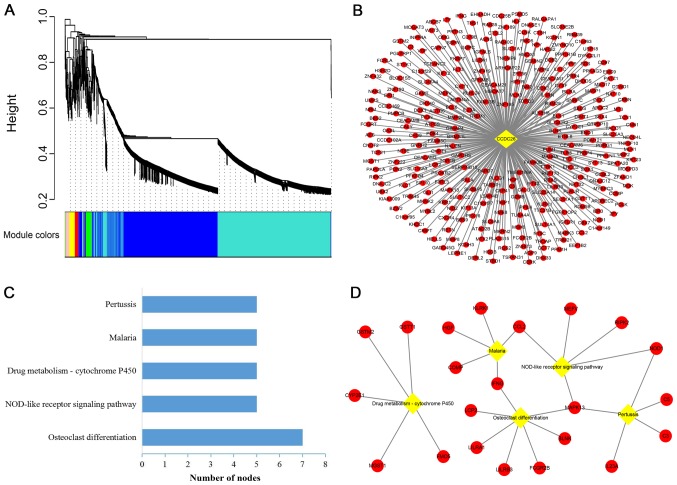
WGCNA is a systematic biology method used to find clusters (modules) of highly correlated genes and examine the potential biological patterns. CCDC26 and a total of 5,020 differentially expressed mRNAs in GSE85030 were involved in the construction of a weighted gene co-expression network. As indicated in Fig. 3A, the cluster dendrogram contained nine co-expression modules (gray, red, green, yellow, black, pink, brown, blue and turquoise). The co-expression network of CCDC26 was in the turquoise module, which contained 289 mRNAs (Fig. 3B).
Figure 3.
Weighted gene co-expression network and pathway analysis of CCDC26 in GSE85030. (A) Cluster dendrogram built based on the dissimilarity of topological overlap, with nine module colors. The module colors under the dendrogram represent the module assignment determined by the Dynamic Tree Cut. Height represents hierarchies. (B) A co-expression network of CCDC26 and 289 mRNAs was built in the turquoise module. (C) KEGG pathway analysis of co-expressed genes of CCDC26. The bar plot indicates the number of enriched genes in the significant pathways (P<0.05). (D) Interaction and overlap of significant pathways. Diamonds represent significant pathways, and circles represent the mRNAs in the pathway. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Pathway analysis and PPI network establishment of co-expressed genes
To predict the potential biological processes of CCDC26 in AML, KEGG was used to enrich and analyze 289 CCDC26-co-expressed genes in the DAVID database. According to the KEGG terms, CCDC26 was enriched in five important pathways, including ‘osteoclast differentiation’, ‘NOD-like receptor signaling pathway’, ‘drug metabolism-cytochrome P450’, ‘malaria’ and ‘pertussis’ (Fig. 3C). ‘Osteoclast differentiation’ and ‘NOD-like receptor signaling pathway’ were considered to serve the most important roles in the network, since the other pathways strongly depended on their linkages (Fig. 3D).
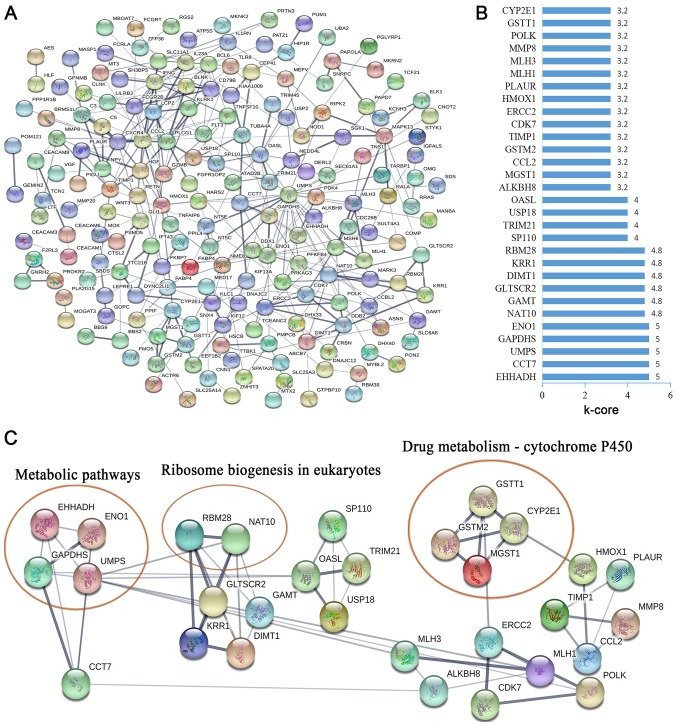
To further investigate the function of CCDC26-co-expressed genes at the protein level, the STRING database was used to screen for functional genes, thus providing a visual annotated network revealing the structural and functional properties of the proteins. The PPI network was composed of 187 nodes and 345 edges (Fig. 4A). The k-core was used to determine the core genes of the PPI network, and the 30 highest k-core genes were contained within four core subnetworks (Fig. 4B). The four core subnetworks were enriched in three pathways, including ‘metabolic pathways’, ‘ribosome biogenesis in eukaryotes’ and ‘drug metabolism-cytochrome P450’ (Fig. 4C).
Figure 4.
PPI networks constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins database. (A) PPI networks of co-expressed genes of CCDC26 were constructed based on the confidence scores of experimental and computational interaction. (B) The 30 top k-core genes in the network. (C) The 30 top k-core genes involved in four core subnetworks were enriched in three pathways. Ellipses represent the signaling pathway. PPI, protein-protein interaction.
Discussion
AML is a highly malignant tumor of the hematopoietic system with substantial heterogeneity in cytogenetics and molecular genetics (21). Over the past few years, with the rapid development of molecular biology, researchers have discovered that chromosomal abnormalities, and gene fusions and mutations serve an important role in the clinical evaluation, prognosis and treatment of patients with AML. However, these molecular abnormalities are not yet comprehensively used as clinical prognostic molecular markers and therapeutic targets (22,23). Therefore, further studies are crucial for identifying novel AML molecular markers and therapeutic targets for guiding treatment or prognostic analysis.
Recently, the aberrant expression of lncRNAs has emerged as an important factor in a number of biological processes (e.g. cell proliferation and apoptosis) and human diseases, particularly in cancer (17,24–26). Studies have reported that lncRNA-CCDC26 not only serves an important role in the development of low-grade glioma, but is also significantly increased in the chronic myeloid leukemia cell line K562, and pediatric AML and AML cell lines HL60 and THP-1 (10–13). In addition, CCDC26, also known as RAM, regulates the differentiation and apoptosis of the AML cell line PLB985, which is induced by retinoic acid (12,14).
In the present study, it was reported that the level of lncRNA-CCDC26 in AML was significantly higher compared with that in healthy individuals. The high expression of CCDC26 in patients with AML was distinctly associated with a number of clinical parameters, including age, anemia, risk stratification and remission. Furthermore, the high CCDC26 expression in AML was associated with poor prognosis. These results indicated that high CCDC26 expression was associated with AML oncogenesis and may serve as a prognostic indicator for AML.
Despite the significant progress reported in the treatment of AML, only 50–60% of elderly patients achieve CR following chemotherapy, and the 5-year overall survival rate following intensive induction chemotherapy is 15–30% (27,28). The 5-year overall survival rate of patients with AML with high-risk factors has been reported to be only 13% (29). However, allogeneic (allo)-HSCT is an effective treatment following chemotherapy remission (30). A study reported that patients aged 40–60 years, who achieved allo-HSCT remission, had a higher 5-year overall survival rate compared with those who only achieved chemotherapy remission (57 and 40%, respectively) (30). Chemotherapy cannot completely eliminate quiescent leukemia cells and minimal residual disease was still present (31,32). In the present study, although the expression of CCDC26 could not fully return to normal levels, it significantly decreased compared with that in patients with an initial diagnosis of AML. However, during the transplantation process, pretreatment with high-dose chemotherapy can eliminate a number of quiescent leukemia cells in the bone marrow. Studies have also reported that donor lymphocytes can play a graft-vs.-leukemia role (33–35). In the present study, the expression of CCDC26 following transplantation was lower compared with that in patients with an initial AML diagnosis, and returned to normal levels. These data indicated that CCDC26 could have a clinical therapeutic effect.
A large number of clinical trials and retrospective studies have reported that the 5-year survival rates for patients with AML with poor-risk stratification are 2–14% (36–39). In the present study, the expression level of CCDC26 in patients with AML with poor/intermediate-risk stratification were higher compared with that in healthy individuals, while the expression in patients with AML with favorable-risk stratification was at a normal level. Furthermore, the prognosis of patients with AML with relapsed or refractory AML is poor, and the 3-year overall survival is estimated at <10% (40,41). It was indicated that the expression of CCDC26 in relapsed patients was significantly higher compared with that in patients with an initial AML diagnosis. These findings suggested that CCDC26 can serve as an indicator of risk stratification in AML.
In order to further investigate the functions of CCDC26, co-expression and PPI networks were constructed in the present study, and KEGG pathway analysis was performed. The analysis revealed that CCDC26 was involved in the regulation of a number of pathways, including ‘osteoclast differentiation’, ‘NOD-like receptor signaling pathway’, ‘metabolic pathways’, ‘ribosome biogenesis in eukaryotes’ and ‘drug metabolism-cytochrome P450’, which served an important role in the pathogenesis and progression of AML.
In conclusion, CCDC26 was indicated to be upregulated in patients with AML, and this upregulation was associated with the progression of AML and poor prognosis. These results suggested that lncRNA-CCDC26 may serve as a novel biomarker for monitoring the progression and predicting the clinical outcome of patients with AML. However, further studies are required in order to determine additional biological characteristics of CCDC26 and their potential involvement in the underlying mechanisms of AML in larger cohort studies.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81500126), the Medical Science and Technology of Research Project of Guangdong (grant no. 2013B021800065) and the Natural Science Foundation of Guangdong Province, China (grant no. 2015A030313727) awarded to CW.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
CW and SW designed the study. CC and PW performed the experiments, CC, PW, WM, YZ, WZ, TD, MZ and XC analyzed the data. CC, PW, CW and SW wrote the manuscript. All authors reviewed the manuscript and approved the final version.
Ethics approval and consent to participate
All research conducted was in line with generally accepted ethical principles and was approved by the Research Ethics Committee of Guangzhou First People's Hospital (Guangzhou, China). The collection and analysis of AML samples was performed in accordance with the principles outlined in the Declaration of Helsinki. All participants provided their written informed consent to participate in this study. The consent procedure was approved by the aforementioned ethics committee. Personal information for the samples involved in the study was anonymized.
Patient consent for publication
Written informed consent was provided by each patient.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 2.Deschler B, Lübbert M. Acute myeloid leukemia: Epidemiology and etiology. Cancer. 2006;107:2099–2107. doi: 10.1002/cncr.22233. [DOI] [PubMed] [Google Scholar]
- 3.Ferrara F. Unanswered questions in acute myeloid leukaemia. Lancet Oncol. 2004;5:443–450. doi: 10.1016/S1470-2045(04)01512-8. [DOI] [PubMed] [Google Scholar]
- 4.Ntziachristos P, Mullenders J, Trimarchi T, Aifantis I. Mechanisms of epigenetic regulation of leukemia onset and progression. Adv Immunol. 2013;117:1–38. doi: 10.1016/B978-0-12-410524-9.00001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woods BA, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Immunol Rev. 2015;263:22–35. doi: 10.1111/imr.12246. [DOI] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Han BW, Chen YQ. Potential pathological and functional links between long noncoding RNAs and hematopoiesis. Sci Signal. 2013;6:re5. doi: 10.1126/scisignal.2004099. [DOI] [PubMed] [Google Scholar]
- 8.Mer AS, Lindberg J, Nilsson C, Klevebring D, Wang M, Grönberg H, Lehmann S, Rantalainen M. Expression levels of long non-coding RNAs are prognostic for AML outcome. J Hematol Oncol. 2018;11:52. doi: 10.1186/s13045-018-0596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen ZH, Wang WT, Huang W, Fang K, Sun YM, Liu SR, Luo XQ, Chen YQ. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017;24:212–224. doi: 10.1038/cdd.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Jin T, Zhang J, Lou H, Yang B, Li Y, Chen C, Zhang Y. Polymorphisms of TREH, IL4R and CCDC26 genes associated with risk of glioma. Cancer Epidemiol. 2012;36:283–287. doi: 10.1016/j.canep.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Wang S, Hui Y, Li X, Jia Q. Silencing of lncRNA CCDC26 restrains the growth and migration of glioma cells in vitro and in vivo via targeting miR-203. Oncol Res. 2018;26:1143–1154. doi: 10.3727/096504017X14965095236521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano T, Yoshikawa R, Harada H, Harada Y, Ishida A, Yamazaki T. Long noncoding RNA, CCDC26, controls myeloid leukemia cell growth through regulation of KIT expression. Mol Cancer. 2015;14:90. doi: 10.1186/s12943-015-0364-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radtke I, Mullighan CG, Ishii M, Su X, Cheng J, Ma J, Ganti R, Cai Z, Goorha S, Pounds SB, et al. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci USA. 2009;106:12944–12949. doi: 10.1073/pnas.0903142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin W, Rossin A, Clifford JL, Gronemeyer H. Co-resistance to retinoic acid and TRAIL by insertion mutagenesis into RAM. Oncogene. 2006;25:3735–3744. doi: 10.1038/sj.onc.1209410. [DOI] [PubMed] [Google Scholar]
- 15.Swerdlow SH, Campo E, Harris NL, et al. Lyon: IARC; 2008. WHO classifcation of tumours of haematopoietic and lymphoid tissues. [Google Scholar]
- 16.O'Donnell MR, Tallman MS, Abboud CN, Altman JK, Appelbaum FR, Arber DA, Bhatt V, Bixby D, Blum W, Coutre SE, et al. Acute myeloid Leukemia, Version 3.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15:926–957. doi: 10.6004/jnccn.2017.0116. [DOI] [PubMed] [Google Scholar]
- 17.Lei L, Xia S, Liu D, Li X, Feng J, Zhu Y, Hu J, Xia L, Guo L, Chen F, et al. Genome-wide characterization of lncRNAs in acute myeloid leukemia. Brief Bioinform. 2018;19:627–635. doi: 10.1093/bib/bbx007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341X.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 21.Salerno L, Romeo G, Modica MN, Amata E, Sorrenti V, Barbagallo I, Pittalà V. Heme oxygenase-1: A new druggable target in the management of chronic and acute myeloid leukemia. Eur J Med Chem. 2017;142:163–178. doi: 10.1016/j.ejmech.2017.07.031. [DOI] [PubMed] [Google Scholar]
- 22.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, Beverloo HB, Moorhouse MJ, van der Spek PJ, Löwenberg B, Delwel R. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 23.Hussaini MO, Mirza AS, Komrokji R, Lancet J, Padron E, Song J. Genetic landscape of acute myeloid leukemia interrogated by Next-generation Sequencing: A large cancer center experience. Cancer Genomics Proteomics. 2018;15:121–126. doi: 10.21873/cgp.20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Wang W, Cao L, Li Z, Wang X. Long Non-coding RNA CCAT1 Acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells. 2016;39:330–336. doi: 10.14348/molcells.2016.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernando TR, Contreras JR, Zampini M, Rodriguez-Malave NI, Alberti MO, Anguiano J, Tran TM, Palanichamy JK, Gajeton J, Ung NM, et al. The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia. Mol Cancer. 2017;16:126. doi: 10.1186/s12943-017-0692-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaidzik VI, Teleanu V, Papaemmanuil E, Weber D, Paschka P, Hahn J, Wallrabenstein T, Kolbinger B, Köhne CH, Horst HA, et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia. 2016;30:2160–2168. doi: 10.1038/leu.2016.207. [DOI] [PubMed] [Google Scholar]
- 27.Burnett AK, Milligan D, Goldstone A, Prentice A, McMullin MF, Dennis M, Sellwood E, Pallis M, Russell N, Hills RK, et al. The impact of dose escalation and resistance modulation in older patients with acute myeloid leukaemia and high risk myelodysplastic syndrome: The results of the LRF AML14 trial. Br J Haematol. 2009;145:318–332. doi: 10.1111/j.1365-2141.2009.07604.x. [DOI] [PubMed] [Google Scholar]
- 28.Löwenberg B, Ossenkoppele GJ, van Putten W, Schouten HC, Graux C, Ferrant A, Sonneveld P, Maertens J, Jongen-Lavrencic M, von Lilienfeld-Toal M, et al. High-dose daunorubicin in older patients with acute myeloid leukemia. N Engl J Med. 2009;361:1235–1248. doi: 10.1056/NEJMoa0901409. [DOI] [PubMed] [Google Scholar]
- 29.Versluis J, Hazenberg CLE, Passweg JR, van Putten WL, Maertens J, Biemond BJ, Theobald M, Graux C, Kuball J, Schouten HC, et al. Post-remission treatment with allogeneic stem cell transplantation in patients aged 60 years and older with acute myeloid leukaemia: A time-dependent analysis. Lancet Haematol. 2015;2:e427–e436. doi: 10.1016/S2352-3026(15)00148-9. [DOI] [PubMed] [Google Scholar]
- 30.Cornelissen JJ, Versluis J, Passweg JR, van Putten WL, Manz MG, Maertens J, Beverloo HB, Valk PJ, van Marwijk Kooy M, Wijermans PW, et al. Comparative therapeutic value of post remission approaches in patients with acute myeloid leukemia aged 40–60 years. Leukemia. 2015;29:1041–1050. doi: 10.1038/leu.2014.332. [DOI] [PubMed] [Google Scholar]
- 31.Hira VVV, Van Noorden CJF, Carraway HE, Maciejewski JP, Molenaar RJ. Novel therapeutic strategies to target leukemic cells that hijack compartmentalized continuous hematopoietic stem cell niches. Biochim Biophys Acta Rev Cancer. 2017;1868:183–198. doi: 10.1016/j.bbcan.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Behbehani GK, Samusik N, Bjornson ZB, Fantl WJ, Medeiros BC, Nolan GP. Mass cytometric functional profiling of acute myeloid leukemia defines Cell-cycle and immunophenotypic properties that correlate with known responses to therapy. Cancer Discov. 2015;5:988–1003. doi: 10.1158/2159-8290.CD-15-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolb HJ, Schattenberg A, Goldman JM, Hertenstein B, Jacobsen N, Arcese W, Ljungman P, Ferrant A, Verdonck L, Niederwieser D, et al. Graft-versus-leukemia effect of donor lymphocyte transfusions in marrow grafted patients. Blood. 1995;86:2041–2050. [PubMed] [Google Scholar]
- 34.Orti G, Barba P, Fox L, Salamero O, Bosch F, Valcarcel D. Donor lymphocyte infusions in AML and MDS: Enhancing the graft-versus-leukemia effect. Exp Hematol. 2017;48:1–11. doi: 10.1016/j.exphem.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 35.Dickinson AM, Norden J, Li S, Hromadnikova I, Schmid C, Schmetzer H, Jochem-Kolb H. Graft-versus-Leukemia effect following hematopoietic stem cell transplantation for leukemia. Front Immunol. 2017;8:496. doi: 10.3389/fimmu.2017.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from cancer and leukemia group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 37.Slovak ML, Kopecky KJ, Cassileth PA, Harrington DH, Theil KS, Mohamed A, Paietta E, Willman CL, Head DR, Rowe JM, et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: A southwest oncology Group/Eastern cooperative oncology group study. Blood. 2000;96:4075–4083. [PubMed] [Google Scholar]
- 38.Grimwade D, Walker H, Harrison G, Oliver F, Chatters S, Harrison CJ, Wheatley K, Burnett AK, Goldstone AH, Medical Research Council Adult Leukemia Working Party The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): Analysis of 1065 patients entered into the United Kingdom medical research council AML11 trial. Blood. 2001;98:1312–1320. doi: 10.1182/blood.V98.5.1312. [DOI] [PubMed] [Google Scholar]
- 39.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, Goldstone A. The importance of diagnostic cytogenetics on outcome in AML: Analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- 40.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 41.Rowe JM, Tallman MS. How I treat acute myeloid leukemia. Blood. 2010;116:3147–3156. doi: 10.1182/blood-2010-05-260117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.