Abstract
Pancreatic cancer is a deadly disease, the deadliest of all the solid tumors; the 5-year survival rate of patients with this disease is ~8%. Previously, high-throughput sequencing has led to the discovery of novel small non-coding RNAs, also known as transfer RNA-derived fragments (tRFs). Studies have suggested that tRFs may be novel biomarkers for certain diseases. However, the expression of tRFs in pancreatic cancer has yet to be characterized. In the present study, the expression levels of tRFs observed in clinical pancreatic cancer samples were analyzed, quantitative PCR (qPCR) was performed to validate the tRFs expression levels and bioinformatics predictions were analyzed. The results revealed that the pancreatic cancer samples screened out a total of 48 tRFs and transfer RNA halves (tiRNAs). There were four tRFs and tiRNAs selected for qPCR validation; the findings were consistent with the sequencing results. Bioinformatic predictions revealed that AS-tDR-000064 was predicted to have 2,450 target genes; AS-tDR-000069 was predicted 445 target genes; AS-tDR-000102 was predicted 746 target genes; and AS-tDR-001391 was predicted 216 target genes. Gene Ontology (GO) analyses demonstrated that the target genes of AS-tDR-000064 were mostly enriched in ‘the regulation of cellular processes’ (Biological Process), ‘the synapses’ (Cellular Component) and ‘enzyme binding’ (Molecular Function). The target genes of AS-tDR-000069 were mostly enriched in ‘signaling’ (Biological Process), ‘the plasma membrane’ (Cellular Component) and ‘phosphatidylinositol 3-kinase (PI3K) binding’(Molecular Function), the target genes of AS-tDR-000102 were mostly enriched in ‘axon development’ (Biological Process), ‘the synapse’ (Cellular Component) and ‘sequence-specific DNA binding’ (Molecular Function) and the target genes of AS-tDR-001391 were mostly enriched in ‘the neuromuscular processes’ (Biological Process), the neurons’ (Cellular Component) and ‘PDZ domain binding’ (Molecular Function). The Kyoto Encyclopedia of Genes and Genomes pathway analyses revealed that the target genes of AS-tDR-000064 were mostly enriched in ‘the Ras signaling pathway’, the target genes of AS-tDR-000069 were mostly enriched in ‘the cancer pathways’, the target genes of AS-tDR-000102 were mostly enriched in ‘axon guidance’ and the target genes of AS-tDR-001391 were mostly enriched in ‘the PI3K/protein kinase-B signaling pathway’.
Keywords: transfer RNA-derived fragments, transfer RNA halves, high-throughput sequencing, quantitative reverse-transcription PCR, bioinformatics prediction, Gene Ontology analysis, Kyoto Encyclopedia of Genes and Genomes pathway analysis
Introduction
Pancreatic cancer is a deadly disease, the deadliest of all the solid tumors; almost all patients diagnosed with pancreatic cancer will eventually succumb to the disease (1). This disease has a low survival rate at just 5 years, which has only increased by 3% over the last 20 years, from 5% in 1997 to 8% in 2017 in the United States (2,3). However, the incidence of pancreatic cancer has increased by 49% within the USA alone over the past 20 years (2,3). By the year 2030, pancreatic cancer is predicted to be the second leading cause of cancer-associated mortality; thus, it is crucial for the public and for physicians to start investing time in pancreatic cancer research for the purposes of improving the current situation and future treatment options (4). This type of cancer has a possible curative treatment in surgical resection. However, only 15–20% of patients with this disease will be able to undergo surgical resection, as most are diagnosed in the late stages of disease when surgery is no longer feasible (5). Thus, researching new diagnostic biomarkers to identify pancreatic cancer at an earlier stage is currently an area of great interest.
Previously, high-throughput sequencing led to the discovery of novel small non-coding RNAs known as transfer RNA (tRNA)-derived fragments (tRFs) (6–8). tRFs cover all the domains of life (archaea, bacteria and eukarya) (9–11). They were thought to be random products of tRNA, but research has revealed that they are their own biological entities (9,10). Humans produce tRFs, consisting of dicer or angiogenin, including yeast with endonucleases, such as ribonuclease T2 (1). Transfer RNA (tRNA)-derived fragments (tRFs) comprise two groups, sized 30 to 35 nucleotides (nt), named tRNA halves (tiRNA), including those that are stress-induced and those with a full-length tRNA anticodon loop cleaved by endonucleases, which generates the tiRNAs. Another group are approximately 20 nt in length, and are referred to as tRFs, which can be separated from the tRNAs to form a 5′- and a 3′-sequence and stemming out of the 5′- and the 3′-site of a mature tRNA (7,12,13). The precursor tRNAs generate the 5′-lead and 3′-tail sequences that are also classified as tRFs (8,14,15).
There have been reports of tRFs in an acquired metabolic disorder (16), breast cancer (17), developmental disorders (18), lung (19) and colorectal cancer (20). This demonstrates that tRFs may serve a significant role in tumorigenesis. The mechanisms of tRFs are diverse. tRFs regulate gene expression post-translation; with the main factor being displaced for eukaryotic initiation factor 4G translation, tRFs directly inhibit protein synthesis (21–23). A 3′-tRF from lymphoma B-cells suppresses proliferation and regulates responses to DNA damage in a similar way to microRNAs (24). tRFs were indicated to compete for binding sites against the binding protein Y-BOX binding protein 1 (YBX1), which may suppress cell proliferation and invasion through the stabilization of oncogenic transcripts. Thus, tRFs counteract the protein YBX1 and suppress tumor progress (17).
In summary, the aforementioned studies demonstrate that tRFs play important roles in tumorigenesis. The studies also suggest that tRFs may be novel biomarkers of diseases, yet the expression profile of tRFs in pancreatic cancer remains unknown. Thus, in the present study, tRF expression levels in clinical samples of pancreatic cancer were analyzed using tRF and tiRNA sequencing. Subsequently, the expression levels of selected tRFs and tiRNAs were analyzed using quantitative PCR (qPCR), and bioinformatics predictions were analyzed.
Materials and methods
Sample collection
The three patients with pancreatic cancer involved in the present study were recruited in June 2017 at the Affiliated Changzhou No. 2 People's Hospital of Nanjing Medical University (Jiangsu, China). All patients were female and 51–69 years of age, with an average age of 61.7 years. Three pairs of pancreatic cancer and tissues adjacent to the tumor sites at a distance of 2 cm were selected for tRFs and tiRNAs sequencing. The paired pancreatic cancer and adjacent normal tissue samples were obtained from the postoperative specimens; the adjacent normal tissue samples were used as the control group for the tRFs and tiRNAs sequencing.
The staging of pancreatic cancer is dependent on a Tumor-Node-Metastasis (TNM) system that was jointly designed by the American Joint Committee on Cancer and the Union for International Cancer Control (25). The patients' clinical stages were T2N1M0, T2N2M0 and T2N0M0 (Table I). The paired pancreatic cancer samples, along with associated normal tissues, were obtained from post-operative specimens and immediately preserved in −80°C liquid nitrogen. Each patient provided a written informed consent form, and the Ethics Committee of the Affiliated Changzhou No. 2 People's Hospital of Nanjing Medical University (Changzhou, China) approved the study protocol.
Table I.
Patient characteristics.
| Number | Age, years | Sex | Tumor size, cm | Tumor stage | Pathological description |
|---|---|---|---|---|---|
| 1 | 65 | Female | 2.5×2.5×3.0 | T2N1M0 | Middle-differentiated adenocarcinoma invaded nerve, metastasis in 2/7 lymph nodes |
| 2 | 51 | Female | 2.5×2.5×2.5 | T2N2M0 | Middle-differentiated adenocarcinoma invaded nerve and Vater, metastasis in 5/11 lymph nodes |
| 3 | 69 | Female | 3.5×3.5×3.0 | T2N0M0 | Middle, low-differentiated adenocarcinoma invaded duodenal wall, no lymph node metastasis |
T, tumor; N, node; M, metastasis.
tRFs and tiRNAs sequencing
The pancreatic cancer and adjacent normal tissue samples were used to extract total RNA. The amount of purified total RNA was 1–2 µg. Prior to the tRF and tiRNA sequencing, the quantity of each RNA was measured in a Nanodrop instrument (Thermo Fisher Scientific, Inc.). The integrity of each RNA was checked using agarose gel (2%) electrophoresis (2 µg per lane). tRFs and tiRNAs have heavy RNA modifications that interfere with the small RNA-sequencing (RNA-seq) library construction (26). Prior to tRF and tiRNA sequencing, the following pretreatments were performed: 3′-aminoacyl (positively charged) deacetylation to 3′-OH for 3′adaptor ligation, 2′,3′-cyclic phosphate removal to 3′-OH for 3′adaptor ligation, 5′-OH phosphorylation to 5′-P for 5′adaptor ligation, and m1A and m3C demethylation for efficient RT. Sequencing libraries were size-selected (50-bp single-read at 10 Mb reads) for the RNA biotypes to be sequenced using an automated gel cutter (Thermo Fisher Scientific, Inc.). These libraries were qualified and quantified using an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc.). For standard small RNA sequencing using the Illumina NextSeq instrument (Illumina, Inc.), the sequencing type was 50-bp single-read at 10 Mb reads. Comprehensive data and Student's paired t-test statistical analysis were performed using the Arraystar tRFs and tiRNA-seq data analysis package (version 4.1; DNASTAR). When comparing two groups of profile differences (such as disease vs. control), the normalized tag number of tRNAs annotated in the Genomic tRNA Database (http://gtrnadb.ucsc.edu) was used, including the tag count of each sample, the ‘fold change’ (i.e., the ratio of the group averages) between the groups for each tRF/tiRNA was computed. The statistical significance of the difference was estimated using a paired Student's t-test. tRFs/tiRNAs with fold change ≥2 and P≤0.05 were selected as significantly differentially expressed (DE) tRFs/tiRNAs. The analysis outputs and the differentially expressed genes can be filtered and ranked according to fold change, P-value, etc., using the Data and Sort & Filter functions in Microsoft Excel (any version; Microsoft Corporation).
Reverse transcription-quantitative (RT-q)-PCR
In the present study, four tRFs and tiRNAs were selected for PCR validation. Total RNA was extracted from the pancreatic cancer samples and the adjacent normal tissue samples using cetyltrimethylammonium bromide buffer (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA was reverse transcribed using the rtStar™ First-Strand cDNA Synthesis kit (3′ and 5′adaptor; catalog no. AS-FS-003; Arraystar). The temperature conditions for RT were 37°C for 15 min followed by 85°C for 5 sec. tRFs and tiRNAs exhibit heavy RNA modifications that interfere with PCR (26); therefore, prior to PCR, the following pretreatments were performed: 3′-aminoacyl (positively charged) deacetylation to 3′-OH for 3′adaptor ligation, 2′,3′-cyclic phosphate removal to 3′-OH for 3′adaptor ligation, 5′-OH phosphorylation to 5′-P for 5′adaptor ligation, and m1A and m3C demethylation for efficient RT. Following this, a 3′adaptor ligation was performed, connecting the 5′adaptor and RT ensued. The primers for qPCR were designed by KangChen Biotech Co., Ltd. (Table II). Subsequently, qPCR was performed using the ViiA 7 real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with FAM fluorophore (Ai Mei Jie Technology Co. Ltd.). The thermocycling conditions were as follows: 95°C for 5 sec, followed by 30 cycles of 60°C for 30 sec and dissociation at 95°C for 1 min and 55°C 1 min. The 2−ΔΔCq method (27) was used to analyze the PCR data, and U6 was used as a reference gene. A Mann-Whitney U test was used to determine the statistical significance of the qPCR expression values. P<0.05 was considered to indicate a statistically significant difference. Statistical analyses were performed using GraphPad Prism (version 6.0; GraphPad Software, Inc.).
Table II.
tRFs and tiRNAs selected for validation via quantitative PCR.
| Gene | Primer sequence | Product length, bp |
|---|---|---|
| U6 | F: 5′-GCTTCGGCAGCACATATACTAAAAT-3′ | 89 |
| R: 5′-CGCTTCACGAATTTGCGTGTCAT-3′ | ||
| AS-tDR-000064 | F: 5′-CAGTCCGACGATCATCCCAC-3′ | 45 |
| R: 5′-TGCTCTTCCGATCTTGGTGG-3′ | ||
| AS-tDR-000069 | F: 5′-AGTTCTACAGTCCGACGATCTCTC-3′ | 49 |
| R: 5′-CTCTTCCGATCTTGGAGGTTC-3′ | ||
| AS-tDR-000102 | F: 5′-CTACAGTCCGACGATCTCCCC-3′ | 45 |
| R: 5′-TCTTCCGATCTTGGTGGAGATG-3′ | ||
| AS-tDR-001391 | F: 5′-GGCTCGTTGGTCTAGGGGTATG-3′ | 52 |
| R: 5′-ACGTGTGCTCTTCCGATCTGAA-3′ |
tRFs, transfer RNA-derived fragments; tiRNAs, tRNA halves.
Predicted targets of various tRFs and tiRNAs expressions
miRanda (http://www.microrna.org/microrna/home.do) and TargetScan (http://www.targetscan.org/) were used to predict targets of various tRFs and tiRNAs expressions. miRanda and TargetScan assign binding scores of tRFs and target genes; a higher score indicates stronger binding. The 35 target genes with the highest binding scores were selected as the candidate target genes.
Functional DE-tRF and tiRNA enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID version 6.8; http://david.ncifcrf.gov/) is a powerful tool that assists in exploiting the functions of the genes of interest. tRFs and tiRNAs with fold change ≥2 and P≤0.05 were selected as significantly DE. As tRFs and tiRNAs can act on target genes to downregulate their expression, to identify the possible biological pathways or functions that could be regulated by the DE-tRFs and tiRNAs, the target genes were analyzed using DAVID. Target genes were screened for significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp/) pathways and Gene Ontology (GO; http://geneontology.org), assessing the biological processes, molecular functions and cellular components. The GO project offers a controlled vocabulary for describing genes and the attributes of gene products within all organisms. There are three domains covered by GO: Cellular components, biological processes and molecular functions. The Bioconductor's (https://www.bioconductor.org) top GO function was used for the Fisher's exact test when locating whether additional overlap existed between the GO annotation list and the DE list, compared with the expected amount. The top GO indicated a P-value that demonstrated the importance of the GO enrichment term for DE genes. Therefore, the lower the P-value, the greater the significance of the GO term (P≤0.05 was suggested). The pathway analysis model was used for the functional gene mapping analysis of the KEGG pathway. The P-value (Fisher's P-value, EASE-score or hypergeometric P-value) indicated the importance of the pathway that was associated with specific conditions; the lower the P-value, the more important the pathway. The KEGG and GO selection criteria had P-values of <0.05.
Results
tRF and tiRNA sequencing
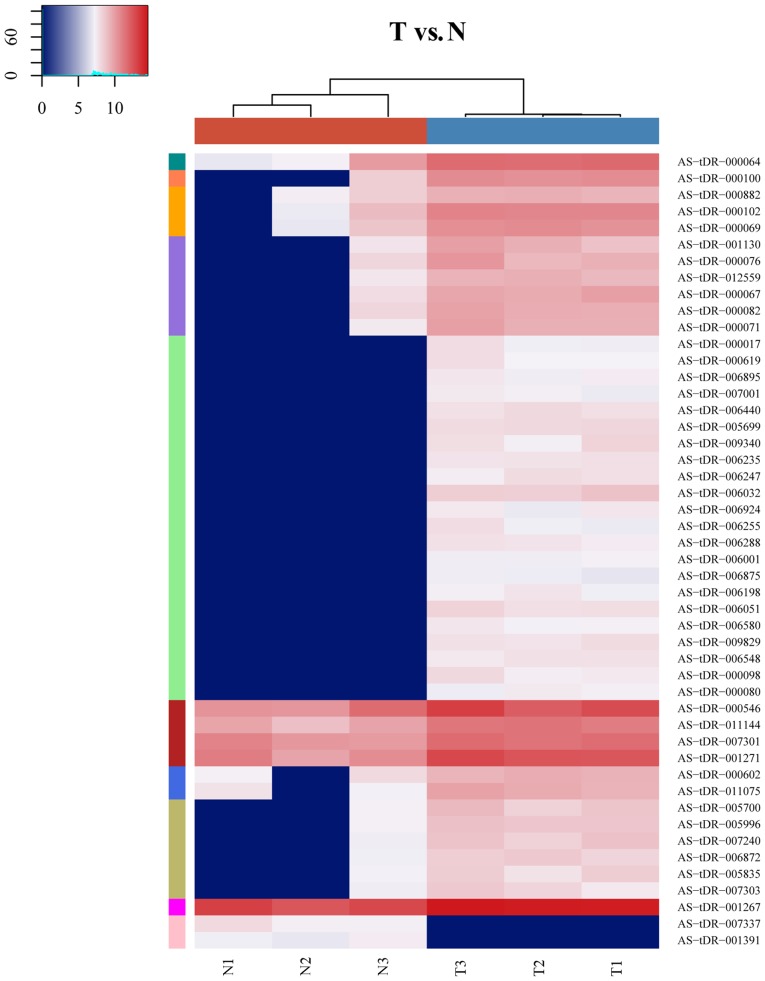
A total of 48 DE-tRFs and tiRNAs were screened from the pancreatic cancer samples and adjacent normal tissue samples, including 46 upregulated tRFs and tiRNAs and 2 downregulated tRFs and tiRNAs (Table III). A heat map of tRFs and tiRNAs is presented in Fig. 1.
Table III.
The 48 various tRF and tiRNA expressed within the pancreatic cancer samples.
| A, Upregulated tDRs | ||||
|---|---|---|---|---|
| tDRs_ID | tRNA | tDR type | Fold change, T/N | P-value |
| AS-tDR-000546 | Glu-CTC-1-1 | tRF-5 | 3.515352854 | 0.028505313 |
| AS-tDR-011144 | Glu-TTC-2-1 | tiRNA-5 | 3.316925679 | 0.024712693 |
| AS-tDR-007301 | Glu-TTC-2-1 | tiRNA-5 | 2.218473266 | 0.030904399 |
| AS-tDR-001267 | Glu-TTC-2-1 | tiRNA-5 | 2.919735986 | 0.011554406 |
| AS-tDR-000882 | Lys-CTT-1-1 | tRF-5 | 3.900728682 | 0.027078399 |
| AS-tDR-001130 | Val-CAC-1-1 | tRF-5 | 10.18152502 | 0.023608439 |
| AS-tDR-005700 | Val-CAC-1-1 | tRF-3 | 7.86901301 | 0.008481533 |
| AS-tDR-000076 | Val-CAC-1-1 | tRF-3 | 8.846767284 | 0.025715623 |
| AS-tDR-000602 | Glu-TTC-1-1 | tRF-5 | 4.750337157 | 0.047733824 |
| AS-tDR-011075 | Glu-TTC-1-1 | tiRNA-5 | 6.476649011 | 0.033682034 |
| AS-tDR-001271 | Glu-TTC-1-1 | tiRNA-5 | 4.07538365 | 0.028804625 |
| AS-tDR-000017 | Gly-CCC-2-1 | tRF-5 | 172.3909573 | 0.037686709 |
| AS-tDR-005996 | Leu-AAG-1-1 | tRF-3 | 8.00593945 | 0.008751401 |
| AS-tDR-007240 | Leu-AAG-1-1 | tRF-3 | 8.343736727 | 0.017853307 |
| AS-tDR-000064 | Leu-AAG-1-1 | tRF-3 | 7.269608569 | 0.012542769 |
| AS-tDR-012559 | Leu-AAG-1-1 | tRF-3 | 9.5247251 | 0.017505317 |
| AS-tDR-006872 | Leu-AAG-1-1 | tRF-3 | 7.046389294 | 0.030169694 |
| AS-tDR-005835 | Val-AAC-2-1 | tRF-3 | 6.171351562 | 0.034156521 |
| AS-tDR-000619 | Glu-TTC-4-1 | tRF-5 | 184.2695023 | 0.02810437 |
| AS-tDR-007303 | Gly-GCC-4-1 | tRF-5 | 6.198056479 | 0.014263475 |
| AS-tDR-006895 | Ala-CGC-1-1 | tRF-3 | 168.4545482 | 0.009518626 |
| AS-tDR-000102 | Ala-CGC-1-1 | tRF-3 | 8.371135493 | 0.00433924 |
| AS-tDR-007001 | Gln-CTG-1-1 | tRF-3 | 156.4192978 | 0.008799826 |
| AS-tDR-000069 | Gln-CTG-1-1 | tRF-3 | 8.166729416 | 0.008389211 |
| AS-tDR-006440 | Gln-CTG-1-1 | tRF-3 | 237.5981942 | 0.003454101 |
| AS-tDR-005699 | Val-AAC-chr6-84 | tRF-3 | 266.3540372 | 0.001098637 |
| AS-tDR-009340 | Val-AAC-chr6-84 | tRF-3 | 231.4538129 | 0.027702861 |
| AS-tDR-000100 | Ala-AGC-2-1 | tRF-3 | 12.7381447 | 0.004182236 |
| AS-tDR-006235 | Ala-AGC-2-1 | tRF-3 | 216.309212 | 0.001144849 |
| AS-tDR-006247 | Ala-AGC-2-1 | tRF-3 | 217.0466322 | 0.013086257 |
| AS-tDR-006032 | Cys-GCA-11-1 | tRF-3 | 369.7327691 | 0.009656002 |
| AS-tDR-006924 | Cys-GCA-11-1 | tRF-3 | 168.3240454 | 0.018648999 |
| AS-tDR-000067 | Cys-GCA-11-1 | tRF-3 | 10.73455381 | 0.022127251 |
| AS-tDR-006255 | Val-CAC-3-1 | tRF-3 | 171.1292469 | 0.043234111 |
| AS-tDR-006288 | Val-CAC-3-1 | tRF-3 | 200.0867071 | 0.005066061 |
| AS-tDR-006001 | Leu-CAG-1-1 | tRF-3 | 144.0176019 | 0.00182195 |
| AS-tDR-000082 | Leu-CAG-1-1 | tRF-3 | 8.426255616 | 0.001442425 |
| AS-tDR-006875 | Leu-CAG-1-1 | tRF-3 | 129.3419781 | 0.002049951 |
| AS-tDR-006198 | Leu-CAG-1-1 | tRF-3 | 170.0118027 | 0.011517777 |
| AS-tDR-006051 | Gln-TTG-1-1 | tRF-3 | 255.8832937 | 0.008529051 |
| AS-tDR-000071 | Gln-TTG-1-1 | tRF-3 | 13.1479288 | 0.002794513 |
| AS-tDR-006580 | Asp-GTC-2-1 | i-tRF | 165.7441719 | 0.007588423 |
| AS-tDR-009829 | pre-Leu-AAG-1-2 | tRF-1 | 226.3986073 | 0.002930597 |
| AS-tDR-006548 | Val-TAC-1-1 | tRF-3 | 208.6565351 | 0.003334287 |
| AS-tDR-000098 | Ala-AGC-1-1 | tRF-3 | 207.9900163 | 0.023503917 |
| AS-tDR-000080 | Leu-TAG-2-1 | tRF-3 | 158.4247545 | 0.00803969 |
| B, Downregulated tDRs | ||||
| tDRs_ID | tRNA | tDR type | Fold change, T/N | P-value |
| AS-tDR-007337 | pre-Pro-TGG-3-2 | tRF-1 | −196.1316037 | 0.028326535 |
| AS-tDR-001391 | Pro-CGG-1-1 | tiRNA-5 | −146.1942975 | 0.012735921 |
tDR, tRNA-derived small RNA; tRF, transfer RNA-derived fragment; tiRNA, tRNA halve; T/N, tumor/normal tissue.
Figure 1.
Heat map indicating the expression levels of various tRFs and tiRNAs. Each square represents a gene, and its color represents the amount of expression of the gene. The higher the amount of expression, the darker the color (red is upregulated, green is downregulated). On the heatmap, dark green, dark orange, light orange, purple, green, blue, olive, magenta, pink and dark red represent low, intermediate to high tRFs and tiRNAs expression, respectively. The colors in the hatmap represent relative tRFs and tiRNAs expression as indicated in the color key. tRFs, transfer RNA-derived fragments; tiRNAs, tRNA halves; T, tumor tissue; N, normal tissue.
Reverse transcription-quantitative (RT-q)-PCR
In the present study, four DE-tRFs and tiRNAs (AS-tDR-000064, AS-tDR-000069, AS-tDR-000102 and AS-tDR-001391), which exhibited abundant expression and were significantly different between the pancreatic cancer samples and adjacent normal tissue samples were selected for qPCR validation; the results demonstrated that AS-tDR-000064, AS-tDR-000069 and AS-tDR-000102 were insignificantly upregulated to in the pancreatic cancer samples compared with adjacent normal tissue samples, and AS-tDR-001391 was insignificantly downregulated (Table IV), which was comparable with the sequencing results presented in Table III. However, the P-values did not indicate significant differences due to the small sample size.
Table IV.
Quantitative PCR results of tRFs and tiRNAs selected for validation.
| tDRs_ID | tRNA | tDR type | Fold change, T/N | P-value |
|---|---|---|---|---|
| AS-tDR-000064 | Leu-AAG-1-1 | tRF-3 | 1.45 | 0.54775 |
| AS-tDR-000069 | Gln-CTG-1-1 | tRF-3 | 1.20 | 0.80352 |
| AS-tDR-000102 | Ala-CGC-1-1 | tRF-3 | 1.36 | 0.51009 |
| AS-tDR-001391 | Pro-CGG-1-1 | tiRNA-5 | 0.24 | 0.07624 |
tRFs, transfer RNA-derived fragments; tiRNAs, tRNA halves; T/N, tumor/normal tissue.
Predicted target genes of various tRFs and tiRNAs and functional DE-tRF and tiRNA enrichment analysis
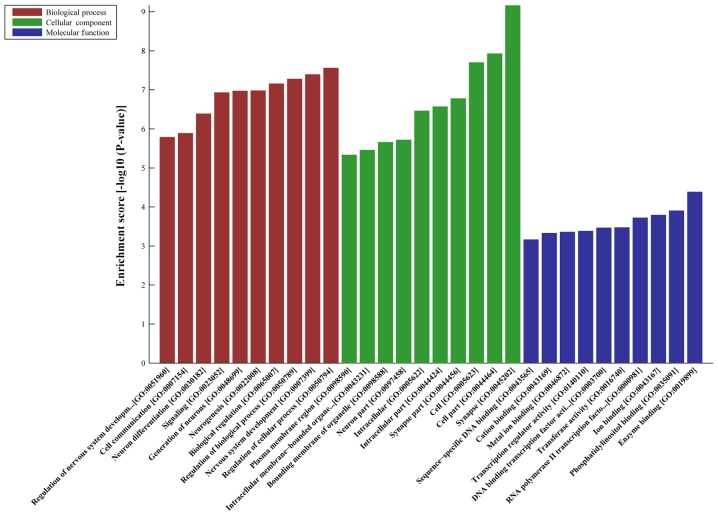
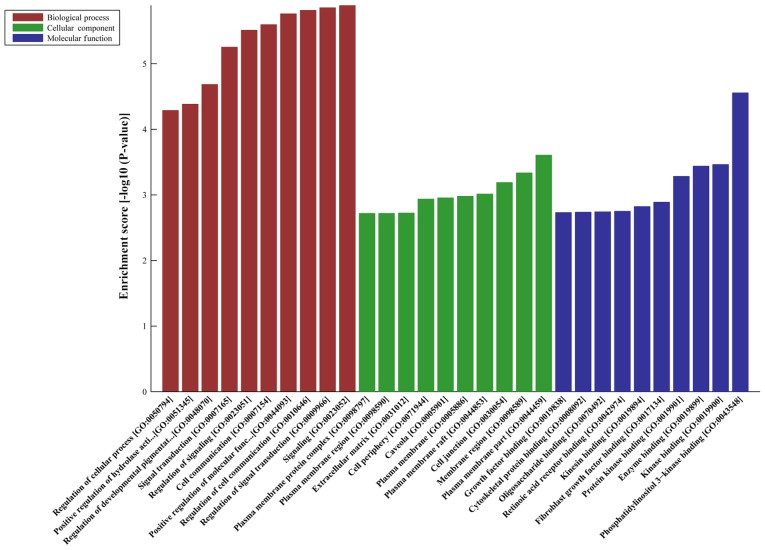
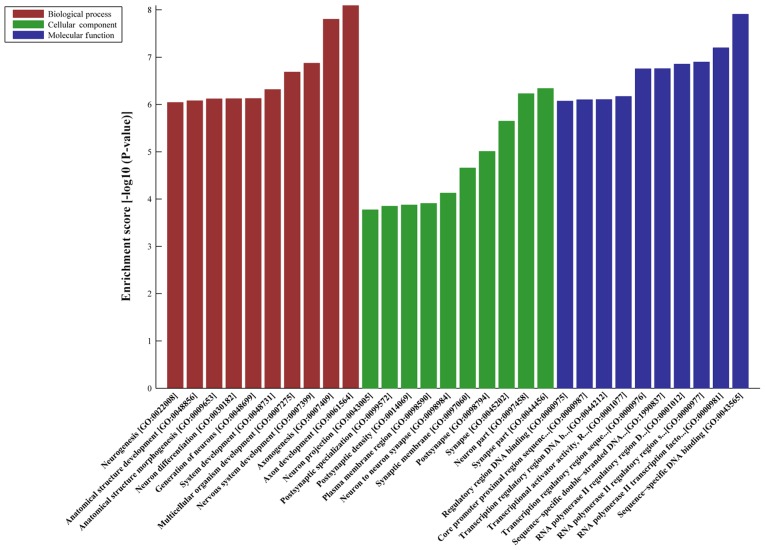
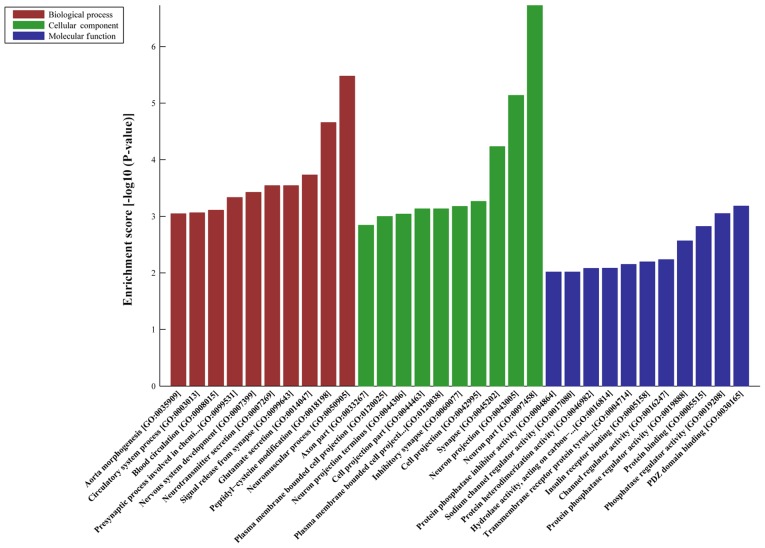
DE-AS-tDR-000064 was predicted to have 2,450 target genes by miRanda and TargetScan; DE-AS-tDR-000069 was predicted 445 target genes; DE-AS-tDR-000102 was predicted 746 target genes; and DE-AS-tDR-001391 was predicted 216 target genes. GO analyses revealed that the target genes of AS-tDR-000064 were mostly enriched in ‘Regulation of cellular process’ (Biological Process), ‘Synapse’ (Cellular Component) and ‘Enzyme binding’ (Molecular Function) (Fig. 2). The target genes of AS-tDR-000069 were mostly enriched in ‘Signaling’ (Biological Process), ‘Plasma membrane part’ (Cellular Component) and ‘Phosphatidylinositol 3-kinase binding’ (Molecular Function) (Fig. 3). The target genes of AS-tDR-000102 were mostly enriched in ‘Axon development’ (Biological Process), ‘Synapse part’ (Cellular Component) and ‘Sequence-specific DNA binding’ (Molecular Function) (Fig. 4). The target genes of AS-tDR-001391 were mostly enriched in ‘Neuromuscular process’ (Biological Process), ‘Neuron part’ (Cellular Component) and ‘PDZ domain binding’ (Molecular Function) (Fig. 5).
Figure 2.
GO analysis of AS-tDR-000064. The target genes of AS-tDR-000064 were most enriched in ‘Regulation of cellular process’ (Biological Process), ‘Synapse’ (Cellular Component) and ‘Enzyme binding’ (Molecular Function). GO, gene ontology.
Figure 3.
GO analysis of AS-tDR-000069. The target genes of AS-tDR-000069 were most enriched in ‘Signaling’ (Biological Process), ‘Plasma membrane part’ (Cellular Component) and ‘Phosphatidylinositol 3-kinase binding’ (Molecular Function). GO, gene ontology.
Figure 4.
GO analysis of AS-tDR-000102. The target genes of AS-tDR-000102 were mostly enriched in ‘Axon development’ (Biological Process), ‘Synapse part’ (Cellular Component) and ‘Sequence-specific DNA binding’ (Molecular Function). GO, gene ontology.
Figure 5.
GO analysis of AS-tDR-001391. The target genes of AS-tDR-001391 were mostly enriched in ‘Neuromuscular process’ (Biological Process), ‘Neuron part’ (Cellular Component) and ‘PDZ domain binding’ (Molecular Function). GO, gene ontology.
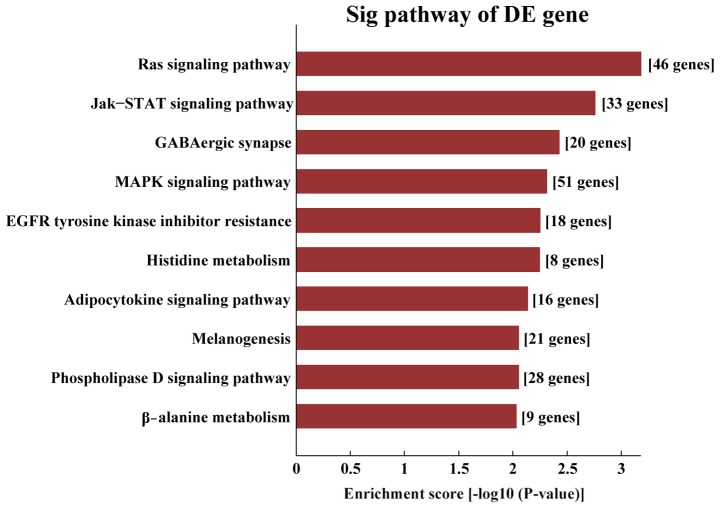
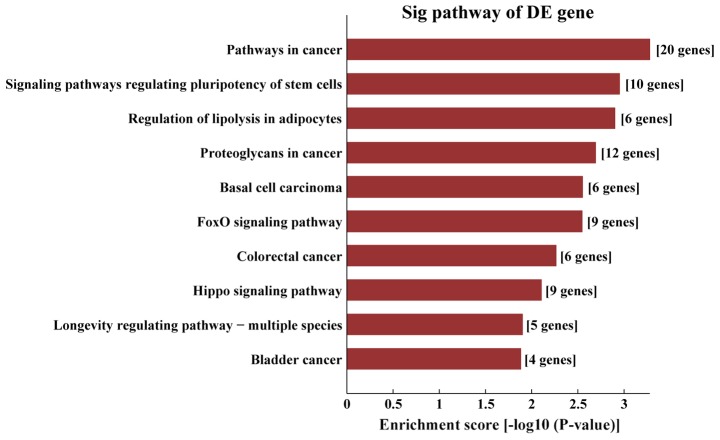
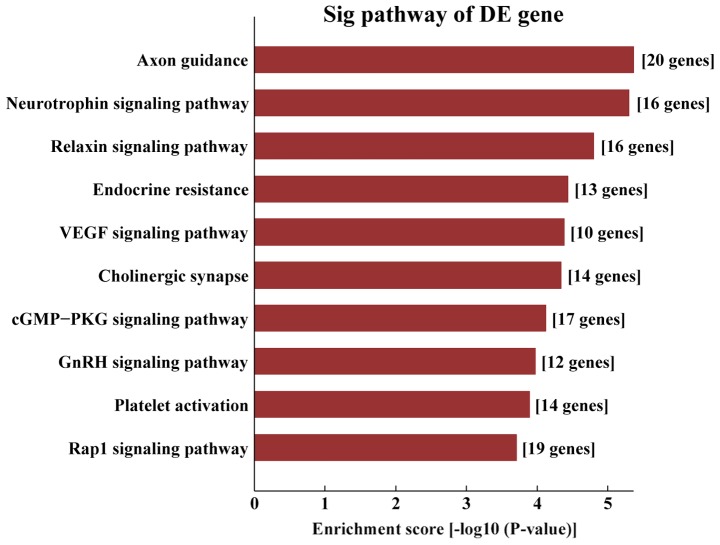
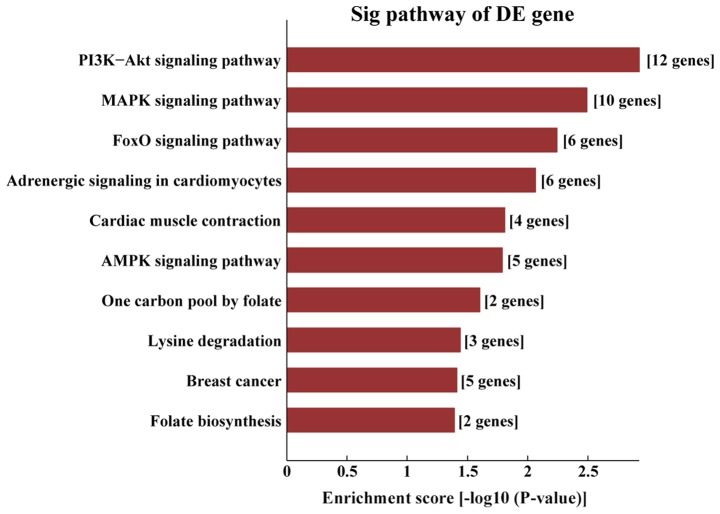
KEGG pathway analysis demonstrated that the target genes of AS-tDR-000064 were mostly enriched in the ‘Ras signaling pathway’ (Fig. 6), the target genes of AS-tDR-000069 were mostly enriched in ‘the cancer pathways’ (Fig. 7), the target genes of AS-tDR-000102 were mostly enriched in ‘axon guidance’ (Fig. 8) and the target genes of AS-tDR-001391 were mostly enriched in ‘the PI3K/protein kinase-B (Akt) signaling pathway’ (Fig. 9).
Figure 6.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of AS-tDR-000064. The target genes of AS-tDR-000064 were most enriched in the ‘Ras signaling pathway’.
Figure 7.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of AS-tDR-000069. The target genes of AS-tDR-000069 were most enriched in ‘the cancer pathways’.
Figure 8.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of AS-tDR-000102. The target genes of AS-tDR-000102 were most enriched in ‘axon guidance’.
Figure 9.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of AS-tDR-001391. The target genes of AS-tDR-001391 were most enriched in ‘the PI3K/protein kinase-B (Akt) signaling pathway’.
Discussion
The technological advances in sequence technology, along with the increase in sncRNA research, have resulted in the discovery of a number of new small RNAs, including tRFs and tiRNAs. Initially, tRFs and tiRNAs were described as a large group of species, and information regarding their functions within cells has been increasing. While studies have described tRFs and tiRNAs expression in human cell lines, the actual retention within human tissue remains unknown (8,17,28,29).
In the present study, an Illumina NextSeq instrument was used to analyze standard small RNA sequences. Using the criteria of log2FC≥2 and P-value<0.05 with the Arraystar tRFs and tiRNA-seq data analysis package (version 4.1; DNASTAR), a total of 48 DE-tRFs and tiRNAs were screened from the pancreatic cancer samples and the adjacent normal tissue samples, including 46 upregulated tRFs and tiRNAs and two downregulated tRF and tiRNA. Of these, four tRFs and tiRNAs were selected for further validation by qPCR, and the results demonstrated that AS-tDR-000064, AS-tDR-000069 and AS-tDR-000102 were upregulated in the pancreatic cancer samples, though not significantly, compared with adjacent normal tissue samples, and AS-tDR-001391 was downregulated, though not significantly, which was consistent with the sequencing data, with AS-tDR-001391 expression levels being the most different between the pancreatic cancer samples and the adjacent normal tissue samples. MiRanda and TargetScan were used to predict targets of differentially expressed tRFs and tiRNAs. The target genes were analyzed using DAVID to screen for significantly enriched KEGG pathways and Gene Ontology biological processes, molecular functions and cellular components.
It should be noted that the present study had limitations. First, although the regulatory roles of AS-tDR-000064, AS-tDR-000069, AS-tDR-000102 and AS-tDR-001391, which are associated with tumors, have been demonstrated by sequencing, their expression levels were not validated further, and thus, the expression and role of systemic tumor-associated pathways in functional experiments were not demonstrated. Secondly, as only three patients were included in the present study, additional consideration was not given to the different stages of the patients with pancreatic cancer recruited for tRFs and tiRNAs sequencing. Finally, the results from the present study are based only on tRFs and tiRNAs sequencing analysis and bioinformatics predictions, and further verification is required. These results form the foundation of a primary study and can be used to an extent in future research, particularly regarding selecting the direction of future studies on pancreatic cancer.
In the present study, after the sequencing results were obtained, further qPCR verification was performed in the samples. tRFs and tiRNAs (AS-tDR-000064, AS-tDR-000069, AS-tDR-000102 and AS-tDR-001391) with high expression and a large statistical difference between the cancer samples and adjacent normal tissue samples based on sequencing were selected for verification. Whether the upregulation or downregulation of these 4 tRFs and tiRNAs are associated with the generation and/or progression of human pancreatic cancer requires further investigation in a large clinical cohort. In addition, the results of the present study are based bioinformatics analysis alone. Previous studies have revealed tRFs and tiRNAs to be present in acquired metabolic disorders (16), breast cancer (17), lung cancer (19), colorectal cancer (20), B-cell lymphoma (24), stress injuries (29), neurodegenerative diseases (30) and prostate cancer (31), among others, but, to the best of our knowledge, tRFs and tiRNAs in pancreatic cancer have not been reported.
The importance of the assessed tRFs and tiRNAs in the enrichment of cancer-associated pathways was demonstrated in the present study, including ‘the Ras signaling pathway’, ‘cancer pathways’, ‘axon guidance’ and ‘the PI3K/Akt signaling pathway’, which suggests potential uses of AS-tDR-000064, AS-tDR-000069, AS-tDR-000102 and AS-tDR-001391 as diagnostic and therapeutic biomarkers for pancreatic cancer. However, to confirm these results, further experiments are required. The expression levels of tRF-1391 in clinical samples of pancreatic cancer and pancreatic cancer cell lines will be validated as the next step in its potential as a tumor marker.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Foundation for Changzhou's High-level Health Talent Cultivation (grant no. 2016CZBJ007), the Social Development Foundation of Science and Technology of Jiangsu (grant no. BE2016658), the Changzhou Sci&Tech Program (grant no. CE20165020) and Project of Changzhou Medical Innovation Team (grant no. CCX201807).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
XQ and CZ designed the study. The experiment was performed by LJ. XQ and CZ provided some of the samples and experiment methods. LJ wrote the paper. CZ reviewed and edited the manuscript. All the authors reviewed and approved the revised manuscript.
Ethics approval and consent to participate
The Affiliated Changzhou No. 2 People's Hospital of Nanjing Medical University (Changzhou, China) approved the current study. All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, Neale RE, Tempero M, Tuveson DA, Hruban RH, Neoptolemos JP. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 2.Parker SL, Tong T, Bolden S, Wingo PA. Cancer statistics, 1997. CA Cancer J Clin. 1997;47:5–27. doi: 10.3322/canjclin.47.1.5. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 5.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–1049. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 6.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson DM, Parker AR. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Martens-Uzunova ES, Olvedy M, Jenster G. Beyond microRNA-novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013;340:201. doi: 10.1016/j.canlet.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 11.Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. 2009;185:43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haussecker D, Huang YA, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar P, Mudunuri SB, Anaya J, Dutta A. tRFdb: A database for transfer RNA fragments. Nucleic Acids Res. 2015;43:D141–D145. doi: 10.1093/nar/gku1138. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 17.Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao Y, Sun Q, Liu X, Wang P, Wu R, Ma Z. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des. 2017;90:730–738. doi: 10.1111/cbdd.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang B, Yang H, Cheng X, Wang D, Fu S, Shen W, Zhang Q, Zhang L, Xue Z, Li Y, et al. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res. 2017;77:3194–3206. doi: 10.1158/0008-5472.CAN-16-3146. [DOI] [PubMed] [Google Scholar]
- 21.Gebetsberger J, Zywicki M, Künzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909. doi: 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S, Sun L, Kragler F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009;150:378–387. doi: 10.1104/pp.108.134767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is downregulated in B cell lymphoma. Proc Natl Acad Sci USA. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edge SB, Compton CC. The American Joint Committee on Cancer the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 26.Durdevic Z, Schaefer M. tRNA modifications: Necessary for correct tRNA-derived fragments during the recovery from stress? Bioessays. 2013;35:323–327. doi: 10.1002/bies.201200158. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Møller S, Trapman J, Bangma CH, Litman T, Visakorpi T, Jenster G. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2012;31:978–991. doi: 10.1038/onc.2011.304. [DOI] [PubMed] [Google Scholar]
- 29.Mishima E, Inoue C, Saigusa D, Inoue R, Ito K, Suzuki Y, Jinno D, Tsukui Y, Akamatsu Y, Araki M, et al. Conformational change in transfer RNA is an early indicator of acute cellular damage. J Am Soc Nephrol. 2014;25:2316–2326. doi: 10.1681/ASN.2013091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 31.Olvedy M, Scaravilli M, Hoogstrate Y, Visakorpi T, Jenster G, Martens-Uzunova ES. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget. 2016;7:24766–24777. doi: 10.18632/oncotarget.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.