Abstract
The effect of long non-coding RNA (lncRNA) THOR on proliferation and migration of colon cancer cells was investigated. Lentiviral vector expressing lncRNA THOR shRNA was used to establish colon cancer SW620 lncRNA THOR knockdown cell line (experimental group), and at the same time, a control vector cell line (control group) was established by empty vector virus. Proliferation ability of the two groups was analyzed by CCK8 and EdU methods. Migration ability of the cells was analyzed by Transwell method. Xenograft tumor method was used to analyze the in vivo proliferation ability of the two groups of cells. mRNA levels of lncRNA THOR target genes were analyzed by reverse transcription-quantitative PCR (RT-qPCR). Compared with control cells, the cell proliferation ability of the experimental group was significantly decreased (P<0.05). Compared with the control group, the cell migration ability of the experimental group was significantly decreased (P<0.05). The tumor growth rate of the experimental group in the mice was significantly lower than that of the control group (P<0.05). Compared with the control group, mRNA levels of lncRNA THOR target genes IGF2BP1, SOX9 and c-myc in the experimental group were significantly downregulated (P<0.05). The results indicated that lncRNA THOR knockdown can significantly downregulate the expression of genes involved in tumor proliferation and migration, promote tumor cell proliferation and migration, indicating that lncRNA THOR plays an important role in colon cancer.
Keywords: lncRNA THOR, colon cancer, proliferation, migration
Introduction
Colon cancer is one of the common types of nausea and digestive tract tumors, which has become a disease that seriously affects human health (1). The molecular mechanism of colon cancer development and progression is still unclear. Long non-coding RNAs (lncRNAs) are a subgroup of non-coding RNAs composed of more than 200 nucleotides, playing an important role in many biological processes such as dose-compensation effect, epigenetic regulation, cell cycle and cell differentiation regulation (2). Previously, it was believed that lncRNA genes had no biological function and were therefore called junk DNA. Recent studies have found that lncRNAs are abnormally expressed in many tumors such as lung cancer, glioma, liver cancer and breast cancer (3–6). lncRNA THOR is a conserved lncRNA identified in testicular tissue in 2017 (7). It was reported that lncRNA THOR can significantly promote the proliferation of osteosarcoma cells and renal cancer cells (8,9). In addition, it was found that lncRNA THOR could enhance the stem cell characteristics of nasopharyngeal carcinoma cells, thereby increasing the resistance of nasopharyngeal carcinoma cells to chemotherapy (10). Early high-throughput sequencing revealed that the expression of lncRNA THOR in colon cancer tissues was significantly higher than that in adjacent normal tissues. However, effect of lncRNA THOR on the biological behavior of colon cancer cells has not been reported. This study analyzed the effect of lncRNA THOR on proliferation and migration of colon cancer cells.
Materials and methods
General information
Thirty female Balb/c nude mice weighing 20 g were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. The mice (6 per cage) were kept in SPF animal room at 25°C with a 12 h light/12 h dark cycle and free access to food and water and humidity was 50%. Colon cancer cell line SW620 was purchased from ATCC. CCK8 cell proliferation assay kit and BeyoClick™ EdU-488 cell proliferation assay kit were purchased from Biyuntian Biotechnology Co., Ltd. Transwell kit was purchased from Corning. MTT assay kit and RNA extraction kit were purchased from Sigma-Aldrich; Merck KGaA. RNA extraction kit (RC101), reverse transcription kit (R323-01) and universal high-sensitivity dye quantitative PCR detection kit (Q431-02) were purchased from Nanjing Nuoweizan Biological Co., Ltd. Control shRNA lentivirus, and Lenti-THOR-shRNA lentivirus were obtained from Shandong Weizhen Biological Co., Ltd. The study was approved by the Ethics Committee of Jinan Central Hospital Affiliated to Shandong University (Jinan, China).
lncRNA THOR knockdown cell line
Colon cancer cell line SW620 [SW-620] (ATCC® CCL-227™) was cultured in DMEM medium (10% FBS) twice to restore cell status. After digestion with 0.25% trypsin, the cell concentration was adjusted to 1×105 cells/ml and inoculated in 12-well plate with 1 ml per well. After 12 h of cell adherence, the control shRNA lentivirus and Lenti-THOR-shRNA lentivirus were added respectively, and the cell-to-virus titer ratio was 1:100. Culture medium was replaced by fresh medium at 24 h after transfection, followed by cell culture for additional 4 h. Then cells were treated with 1 µg/ml puromycin for 24 h, and viable cells were observed under the microscope (ZEISS) to observe GFP fluorescence. When GFP in all cells is expressed, stable cell line is successfully constructed. Lenti-THOR-shRNA stable expression cell line was used as the experimental group, and control shRNA stable expression cell line was used as the control group.
Preparation of a reverse transcription-quantitative PCR (RT-qPCR) system. The same number of living cells were collected both from the control and experimental groups. Total RNA was extracted using an RNA extraction kit, and then the RNA was reverse-transcribed into cDNA using a reverse transcription kit to serve as the template for RT-qPCR. The temperature protocol for reverse transcription was 37°C for 30 min, 85°C for 15 sec and 4°C for storage. cDNA template was diluted 1:100 before use. PCR reaction systems included: 2X SYBR-Green premix 10 µl, template 8.8 µl, upstream primers 0.6 µl and downstream primers 0.6 µl. PCR reaction conditions were: 95°C for 30 sec, followed by 40 cycles of 95°C for 10 sec and 60°C for 30 sec. Sequences of primers used in PCR reactions are shown in Table I. β-actin was the reference gene. The method of quantification was 2−ΔΔCq (11).
Table I.
Sequences of primers used in PCR reactions.
| Genes | Forward primers (5′-3′) | Reverse primers (5′-3′) |
|---|---|---|
| lncRNA THOR | ACAATCGAGCAAGGCAGTGA | TGGCCAAGACCTGCTGTTAG |
| β-actin | GGCCCAGAATGCAGTTCGCCTT | AATGGCACCCTGCTCACGCA |
| c-myc | CCCTCCACTCGGAAGGACTA | GCTGGTGCATTTTCGGTTGT |
| SOX9 | GCTGCGAAGTGGAAACCATC | CCTCCTTCTGCACACATTTGAA |
| IGF2BP1 | CAGGAGATGGTGCAGGTGTTTATC C | GTTTGCCATAGATTCTTCCCTGAGC |
Cell proliferation analysis (CCK8 and EDU methods)
Control and experimental cells were trypsinized at a cell density of 5×105 cells/ml, and seeded in 96-well plates with 100 µl per well. Cells were cultured for 6 h at 37°C in a 5% CO2 incubator. After that, 10 µl of CCK solution was added 12, 24, 36, 48 and 72 h later. After that, cells were cultivated for additional 3 h. Finally, OD values at 450 nm were measured using a microplate reader (BioTek) to reflect cell proliferation.
Cells of the control and experimental groups were digested and inoculated into a 12-well plate. After the cells were completely adhered, the EdU was diluted to 50 µM with a cell culture medium, and 100 µl was added to each well. Cells were incubated for 2 h in a 37°C and 5% CO2 incubator. After that, cells were washed three times with PBS, and then fixed with 4% paraformaldehyde at room temperature for 20 min. After washing three times with PBS, nuclei were stained with DAPI. After washing three times with PBS, cells were observed under a microscope.
Cell migration analysis (Transwell method)
The control and experimental cells were trypsinized to prepare a single cell suspension, and the cell density was adjusted to 5×105 cells/ml, and inoculated into the Transwell upper chamber, 100 µl per well. The lower chamber was filled with fresh cell culture medium. Cells were incubated in a 37°C and 5% CO2 incubator for 24 h, and were fixed with paraformaldehyde. Then the upper chamber membrane was fixed with 1% crystal violet (MXB Biotechnologies) at room temperature for 5 min. Migrating cells were observed under an optical microscope, and 20 visual fields were selected for both the experimental and control groups to calculate the average number of migrating cells.
Xenograft tumor
After cells of the control and experimental groups were trypsinized, cell density was adjusted to 1×108/ml. Thirty Balb/c nude mice were randomly divided into the control and experimental groups. Tumor cells (100 µl) were injected into the fat pad of each mouse, and mice were raised in SPF-level animal house. Tumor formation and growth in the mice were observed. Tumor length (L) and width (W) were measured. Tumor volume was calculated by the following formula: V = 1/2 × L × W2.
Statistical analysis
All data were analyzed by SPSS 17.0 statistical software (SPSS, Inc.). Measurement data were expressed as mean ± standard deviation and compared by t-test. P<0.05 indicated a difference with statistical significance.
Results
Comparison of lncRNA THOR levels in the control and experimental groups
The lncRNA THOR knockdown stable cell line was established by lentiviral-mediated THOR-shRNA. The expression level of lncRNA THOR in the control group (1.21±0.21) was significantly higher than that in the experimental group (0.28±0.10), and the difference was statistically significant (P<0.05) (Fig. 1).
Figure 1.
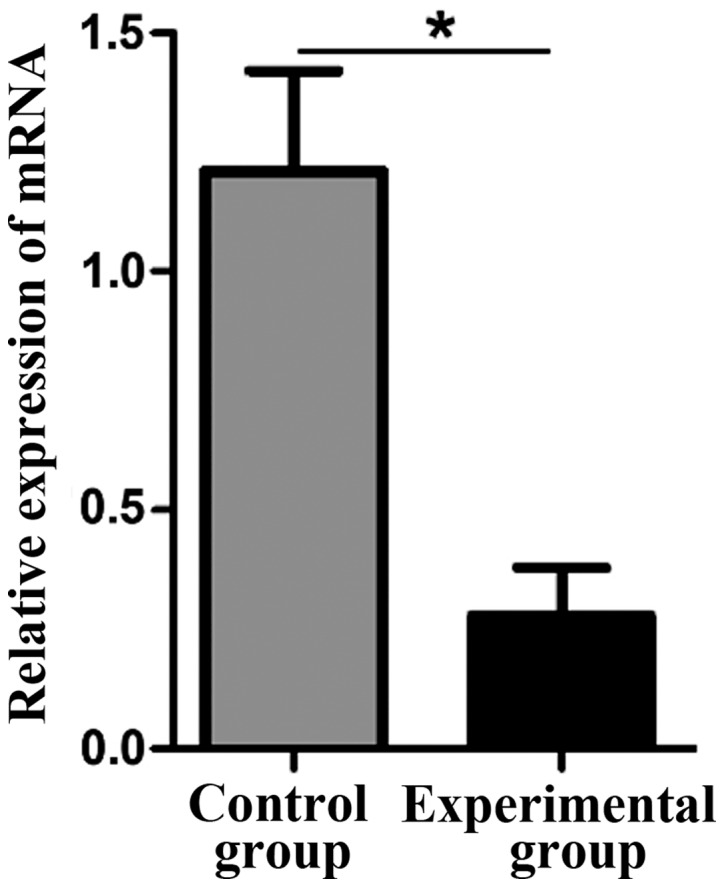
Comparison of lncRNA THOR levels between the control and experimental groups. *P<0.05 vs. the control group. lncRNA, long non-coding RNA.
Effect of lncRNA THOR on cell proliferation
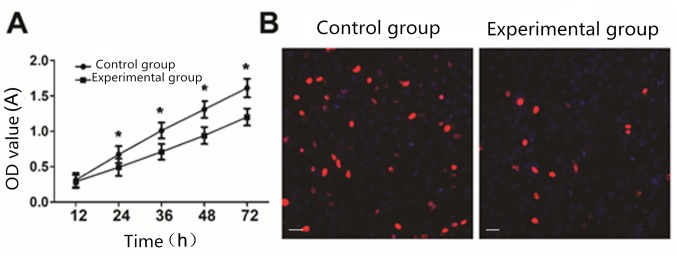
CCK8 analysis showed that compared with the control cells, cell proliferation ability of the experimental group was significantly reduced (P<0.05) (Fig. 2A). EdU staining analysis showed that the cell proliferation ability of the experimental group was significantly decreased compared with that of the control group (P<0.05) (Fig. 2B).
Figure 2.
Effect of lncRNA THOR on the proliferation of colon cancer cells. (A) Cell proliferation between the control shRNA group and Lenti-THOR-shRNA experimental group measured using CCK-8 assay. *P<0.05 vs. the control group. (B) Fluorescent micrographs of EdU staining (red) between the control shRNA group and Lenti-THOR-shRNA experimental group. Cell enlargement, ×100. Scale bar, 20 µm. lncRNA, long non-coding RNA.
Effect of lncRNA THOR on cell migration
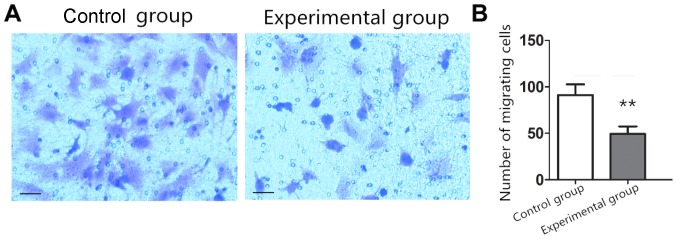
Compared with the number of migrating cells in the control group (91.23±11.44), the number of migrating cells in the experimental group (49.32±8.14) decreased significantly (P<0.05) (Fig. 3).
Figure 3.
Effect of lncRNA THOR on cell migration. (A) Cell migration between shRNA control group and Lenti-THOR-shRNA experimental group analyzed by Transwell. Cell enlargement, ×200. Scale bar, 20 µm. (B) Quantitative analysis of the average number of migrating cells in Fig. 2A. **P<0.01 vs. the control group. lncRNA, long non-coding RNA.
Effect of lncRNA THOR on tumor growth
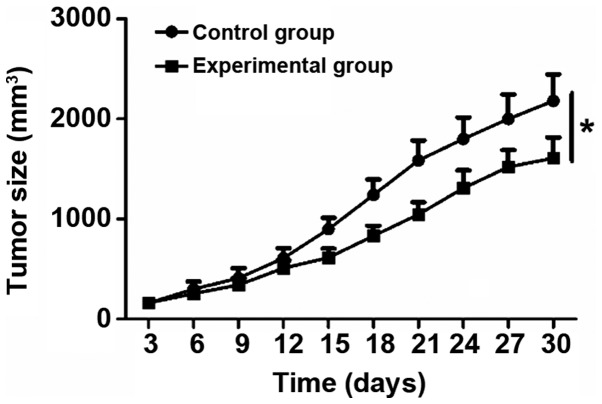
Control and experimental group cells were inoculated into nude mice, and cell proliferation rate in vivo was analyzed. Compared with the control cells, growth rate of tumors in the experimental group was significantly reduced (P<0.05) (Fig. 4).
Figure 4.
Effect of lncRNA THOR on tumor growth by in vivo tumor formation experiment. *P<0.05 vs. the control group. lncRNA, long non-coding RNA.
Effect of lncRNA THOR knockdown on its target genes
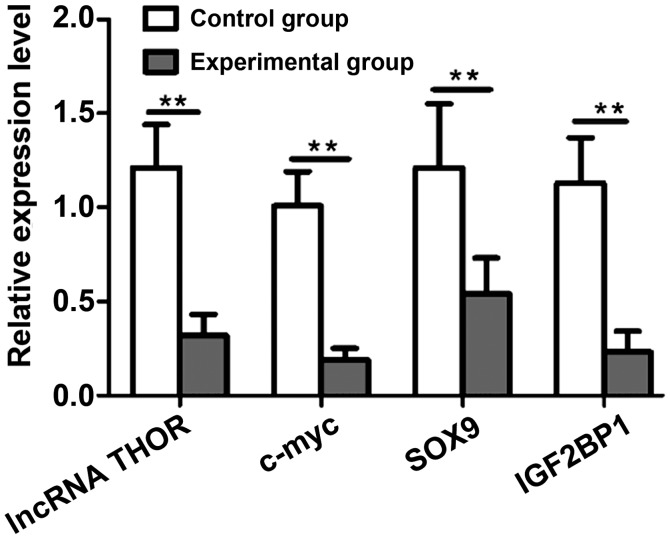
Effect of lncRNA THOR knockdown on its target genes at mRNA level was further analyzed by RT-qPCR. Compared with the control group, the mRNA levels of IGF2BP1, SOX9 and c-myc in the experimental group were significantly reduced (P<0.05) (Fig. 5).
Figure 5.
Effect of lncRNA THOR knockdown on the relative expression level of target gene mRNA by RT-qPCR. **P<0.01 vs. the control group. lncRNA, long non-coding RNA.
Discussion
Colon cancer, as the third most common type of malignant tumor in the world, seriously affects human health (12). Despite the efforts made in cancer diagnosis, many cancer patients are still diagnosed at advanced stages. Therefore, novel early diagnosis biomarkers are needed (13). Studies have shown that lncRNAs participate in many biological processes by regulating gene expression (14). It has been reported that lncRNA CCTA1, ATB and HOTAIR participate in the regulation of cell behavior of colon cancer by mediating the downstream signaling pathways (15,16). Because of the important role of lncRNA in tumorigenesis, lncRNAs have been used as a novel tumor marker and tumor therapeutic target. This study analyzed the role of lncRNA THOR in the pathogenesis of colon cancer.
At cellular level, lncRNA THOR knockdown led to significantly inhibited proliferation and migration of colon cancer SW620 cells. At animal level, lncRNA THOR knockdown mediated the significantly inhibited growth of tumors in mice. It indicated that lncRNA THOR plays an important role in the development of colon cancer.
Sox9 plays an important role in early embryonic development, cell fate determination and differentiation of tissues and organs, sex determination, occurrence and development of nervous system and cartilage (17). The high expression of Sox9 is related to the size of the tumors, TNM stage, lymph node metastasis and differentiation of colorectal cancer patients (18). lncRNA THOR maintains the stability and activity of IGF2BP1 through conservative interaction with IGF2BP1 mRNA (7). The results showed that knockdown of lncRNA THOR resulted in significant downregulation of the target genes SOX9 and IGF2BP1 mRNA, suggesting that lncRNA THOR has important significance in stabilizing SOX9 and IGF2BP1 mRNA. C-myc, as an oncogene, plays an important role in tumorigenesis (19). This study also found that knockdown of lncRNA THOR in colon cancer cells resulted in a significant decrease in c-myc mRNA level, which further demonstrated that lncRNA THOR, was involved in the occurrence and progress of colon cancer genes.
Collectively, lncRNA THOR, as an oncogene, promotes the proliferation and migration of colon cancer cells, which can be used as a therapeutic target for colon cancer treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YL and XY drafted the paper and performed PCR. XY and LW were responsible for CCK8 assay and Transwell method. All the authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Jinan Central Hospital Affiliated to Shandong University (Jinan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Yang W, Chen Z, Chen J, Meng Y, Feng B, Sun L, Dou L, Li J, Cui Q, et al. Long noncoding RNA lncSHGL recruits hnRNPA1 to suppress hepatic gluconeogenesis and lipogenesis. Diabetes. 2018;67:581–593. doi: 10.1111/1753-0407.12632. [DOI] [PubMed] [Google Scholar]
- 3.Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. lncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018;9:722. doi: 10.1038/s41419-018-0759-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mondal T, Juvvuna PK, Kirkeby A, Mitra S, Kosalai ST, Traxler L, Hertwig F, Wernig-Zorc S, Miranda C, Deland L, et al. Sense-antisense lncRNA pair encoded by locus 6p22.3 determines neuroblastoma susceptibility via the USP36-CHD7-SOX9 regulatory axis. Cancer Cell. 2018;33:417–434.e7. doi: 10.1016/j.ccell.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 5.Liao Y, Cheng S, Xiang J, Luo C. lncRNA CCHE1 increased proliferation, metastasis and invasion of non-small lung cancer cells and predicted poor survival in non-small lung cancer patients. Eur Rev Med Pharmacol Sci. 2018;22:1686–1692. doi: 10.26355/eurrev_201803_14581. [DOI] [PubMed] [Google Scholar]
- 6.Niknafs YS, Han S, Ma T, Speers C, Zhang C, Wilder-Romans K, Iyer MK, Pitchiaya S, Malik R, Hosono Y, et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat Commun. 2016;7:12791. doi: 10.1038/ncomms12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosono Y, Niknafs YS, Prensner JR, Iyer MK, Dhanasekaran SM, Mehra R, Pitchiaya S, Tien J, Escara-Wilke J, Poliakov A, et al. Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell. 2017;171:1559–1572.e1520. doi: 10.1016/j.cell.2017.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Chen M, Xu Y, Chen X, Zhou P, Zhao X, Pang F, Liang W. Long non-coding RNA THOR promotes human osteosarcoma cell growth in vitro and in vivo. Biochem Biophys Res Commun. 2018;499:913–919. doi: 10.1016/j.bbrc.2018.04.019. [DOI] [PubMed] [Google Scholar]
- 9.Ye XT, Huang H, Huang WP, Hu WL. lncRNA THOR promotes human renal cell carcinoma cell growth. Biochem Biophys Res Commun. 2018;501:661–667. doi: 10.1016/j.bbrc.2018.05.040. [DOI] [PubMed] [Google Scholar]
- 10.Gao L, Cheng XL, Cao H. lncRNA THOR attenuates cisplatin sensitivity of nasopharyngeal carcinoma cells via enhancing cells stemness. Biochimie. 2018;152:63–72. doi: 10.1016/j.biochi.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Shahbazian H, Nasuri Y, Hosseini SM, Arvandi S, Razzaghi S. A report of the frequency of colorectal carcinoma and involved lymph nodes in South-West Iran. Indian J Med Paediatr Oncol. 2016;37:38–41. doi: 10.4103/0971-5851.177014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Xie H, Gao Q, Zhan H, Xiao H, Zou Y, Zhang F, Liu Y, Li J. Colon cancer associated transcripts in human cancers. Biomed Pharmacother. 2017;94:531–540. doi: 10.1016/j.biopha.2017.07.073. [DOI] [PubMed] [Google Scholar]
- 14.Kim C, Kang D, Lee EK, Lee JS. Long noncoding RNAs and RNA-binding proteins in oxidative stress, cellular senescence, and age-related diseases. Oxid Med Cell Longev. 2017;2017:2062384. doi: 10.1155/2017/2062384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng H, Wang JM, Li M, Tang R, Tang K, Su Y, Hou Y, Zhang J. Long non-coding RNAs: New biomarkers for prognosis and diagnosis of colon cancer. Tumour Biol. 2017;39:1010428317706332. doi: 10.1177/1010428317706332. [DOI] [PubMed] [Google Scholar]
- 16.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 18.Dong C, Wilhelm D, Koopman P. Sox genes and cancer. Cytogenet Genome Res. 2004;105:442–447. doi: 10.1159/000078217. [DOI] [PubMed] [Google Scholar]
- 19.Weidensdorfer D, Stöhr N, Baude A, Lederer M, Köhn M, Schierhorn A, Buchmeier S, Wahle E, Hüttelmaier S. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA. 2009;15:104–115. doi: 10.1261/rna.1175909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.