Abstract
Gastric cancer (GC) is one of the most common types of malignant cancer and is associated with poor prognosis. Although the prognosis of patients with GC is associated with grade, stage and lymph node metastases, these traditional clinical features are inadequate to predict the outcome of GC. Therefore, there has been an increased focus on identifying novel molecular biomarkers for early diagnosis and prognosis, in order to improve outcomes in GC. In the present study, an integrative analysis of microRNA (miRNA) expression profiles, mRNA expression profiles and clinical characteristics was performed in a large cohort of patients with GC in order to identify an integrative prognostic model for improving postoperative risk classification. An integrative mRNA/miRNA signature (IMMIS), comprised of three miRNAs and one mRNA, was identified from a large number of differentially expressed miRNAs and mRNAs using univariate and multivariate Cox regression analysis. The prognostic value of the IMMIS was validated in the discovery cohort, testing cohort and The Cancer Genome Atlas (TCGA) cohort. The present results suggested that the identified signature had a reliable predictive performance and could classify the patients into high- and low-risk groups with significantly different overall survival times. In the discovery cohort, the hazard ratio (HR) was 2.805 with a 95% CI=1.722–4.567 (P<0.001). The median overall survival time as 1.49 vs. 3.85 years. In the testing cohort, the HR was 1.625 with a 95% CI=1.004–2.638 (P=0.039) and the median overall survival time was 2.17 vs. 4.62 years. In the TCGA cohort, the HR was 2.139 with a 95% CI=1.519–3.012 (P<0.001) and the median overall survival time was 1.53 vs. 4.62 years. The IMMIS constituted a reliable independent prognostic factor compared with clinical covariates, including age, sex, grade and stage, as indicated by multivariate and stratified analyses. Furthermore, comparative analysis revealed that the predictive value of the IMMIS was superior to the mRNA-based signature alone. The present results suggested the potential value of the IMMIS as a promising novel biomarker for improving the clinical management of patients with GC.
Keywords: gastric cancer, mRNA expression profiles, microRNA expression profiles, signature, prognosis
Introduction
Gastric cancer (GC), additionally referred to as stomach cancer, is the one of most common types of digestive cancer worldwide and is the third leading cause of cancer-related death (1). China is one of the high-risk areas for digestive cancer, and GC is ranked the second most common cancer behind lung cancer in China (2). Notably, an estimated 477.7 individuals per 100,000 newly diagnosed cases and 158.7 per 100,000 mortalities were reported in China in 2015 (2). Gastric adenocarcinomas are the most common histological type of GC, which account for ~90% of GC cases (3). Although the survival rate of patients with GC has improved over the past years, the prognosis remains unsatisfactory, with a 5-year survival rate of 10% for patients with localized gastric adenocarcinoma and 50% for patients with early resectable gastric adenocarcinoma (4). Although the prognosis of patients with GC is associated with grade, stage and lymph node metastases, these traditional clinical features are inadequate to predict the outcome of GC. Therefore, there is an increasing focus on identifying novel molecular biomarkers for early diagnosis and prognosis to improve outcomes in patients with GC.
With the development of high throughput sequencing technologies for transcriptional analysis, including microarray and RNA sequencing (-Seq), researchers have increased their efforts in trying to identify molecular biomarkers for predicting the outcome of patients with GC at the transcriptional levels. Several mRNA or micro RNA (miRNA) expression signatures have been proposed to predict the survival of patients with GC. For example, Chen et al (5) developed a three-mRNA survival prediction model using cDNA microarray data from 18 pairs of cancerous and noncancerous gastric tissues. A previous study performed by Cho et al (6) constructed a six-gene expression-based prognostic risk score to predict the likelihood of relapse after curative resection. For miRNAs, Ding et al (7) identified an eight-miRNA signature for predicting the overall survival in 380 patients with gastric adenocarcinoma. However, previous studies on other types of cancer have shown that, notwithstanding the importance of individual RNA, intrinsic multi-RNA-based expression signatures have greater prognostic value (8–10). However, to the best of our knowledge, no integrated studies concerning their prognostic significance have been reported yet.
The aim of the present study was to develop and assess the predictive value of an integrated multi-gene signature by analyzing mRNA and miRNA expression profiles, and clinical information in a large cohort of patients with GC.
Materials and methods
Patient datasets
The Cancer Genome Atlas (TCGA) database (cancergenome.nih.gov/) was used to identify the following data: i) Normalized level-3 RNA-Seq data of 375 GC samples and 32 normal tissue samples obtained with HTSeq (version 0.6.1; cancergenome.nih.gov/); ii) normalized level-3 miRNA-Seq data of 436 GC samples; and iii) 41 normal tissue samples obtained with Illumina HiSeq (Illumina, Inc.). For all datasets, corresponding clinical data were collected and analyzed. The Ensembl gene ID of protein-coding genes was retrieved from the HUGO Gene Nomenclature Committee database (www.genenames.org/). Cross-referencing of the Ensembl gene ID with tumor barcodes was performed. Furthermore, data of patients with incomplete expression data and follow-up information were removed from the analysis. A total of 18,528 mRNAs in 407 samples, 1,573 miRNAs in 477 patients and 438 samples with survival information were retained for further analysis in the present study.
Development of an integrative mRNA/miRNA signature (IMMIS)
Differentially expressed mRNAs and miRNAs in GC and normal tissue samples were detected using the DEGseq2 package (11). An adjusted P<0.05 was used with Benjamini-Hochberg (B-H) correction and a |log2 (fold change)| >1 applied. Subsequently, univariate Cox proportional hazards analyses were used to identify prognostic mRNAs and miRNAs from the differentially expressed mRNAs and miRNAs. Finally, an IMMIS was constructed as the linear combination of expression values of each prognostic mRNA and miRNA, which was weighted by their estimated regression coefficients in the multivariate Cox regression analysis (12–18).
Statistical analysis
The differences in overall survival time between the high- and low-risk groups were assessed using Kaplan-Meier survival plots and log-rank tests. Univariate and multivariate Cox regression analyses for overall survival were performed on the individual clinical variables with and without the IMMIS in each cohort. Hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated. All statistical analyses were performed using R-version3.5.2 (http://www.R-project.org/) (19).
Results
Identification of differentially expressed mRNAs and miRNAs between GC samples and normal tissue samples
By comparing the expression profiles of mRNAs and miRNAs between GC samples and normal tissue samples, a total of 4,221 mRNAs and 201 miRNAs were identified to be differentially expressed in GC samples and normal tissue samples according to the DEGseq2 package method with |log2 (fold change)| >1 and an adjusted P<0.05 after B-H adjustment. These profiles were used for subsequent survival analyses. Among the differentially expressed autophagy-associated mRNAs and miRNAs, 2,055 mRNAs and 138 miRNAs were upregulated, and 2,166 mRNAs and 63 miRNAs were downregulated in GC samples compared with normal tissue samples.
Identification of independent prognostic mRNAs and miRNAs
In order to identify and validate an IMMIS, samples, corresponding clinical data, and mRNA and miRNA expression profiles were initially cross-referenced with tumor barcodes. A total of 361 samples were retained for subsequent survival analyses. All 361 samples with survival information were randomly divided into two equal patient cohorts: i) The discovery cohort (n=181); and ii) testing cohort (n=180). The detailed clinical information of the discovery cohort and testing cohort are provided in Table I. Statistical analysis indicated that the clinical characteristics were similar in the discovery and testing cohorts (Table I). The association between the expression level of differentially expressed mRNAs and miRNAs with the overall survival of patients with GC using the univariate Cox analyses was then examined. A total of 12 RNAs (including 11 mRNAs and 1 miRNA; FDR<0.05) were observed to be significantly associated with overall survival, and were considered candidate prognostic RNAs. All 12 prognostic RNAs were identified as risk factors with positive coefficients (Table II), and high expression of these RNAs was associated with a poor outcome.
Table I.
Clinical features of the patients with gastric cancer in the discovery cohort, testing cohort and the entire TCGA cohort.
| Clinicopathological characteristic | Discovery cohort, n=181 | Testing cohort, n=180 | Entire TCGA cohort, n=361 | P-value | High risk group, n=180 | Low risk group, n=181 | P-value |
|---|---|---|---|---|---|---|---|
| Age | 0.424a | 0.492a | |||||
| <68 | 87 | 94 | 181 | 95 | 86 | ||
| ≥68 | 92 | 82 | 174 | 84 | 90 | ||
| Sex | 0.713a | 0.773a | |||||
| Male | 117 | 112 | 229 | 116 | 113 | ||
| Female | 64 | 68 | 132 | 64 | 68 | ||
| Stage | 0.128a | 0.133a | |||||
| I | 29 | 20 | 49 | 27 | 22 | ||
| II | 58 | 50 | 108 | 52 | 56 | ||
| III | 62 | 82 | 144 | 80 | 64 | ||
| IV | 21 | 16 | 37 | 13 | 24 | ||
| Unknown | 11 | 12 | 23 | 8 | 15 | ||
| Grade | 0.489b | 0.404b | |||||
| G1 | 2 | 6 | 8 | 3 | 5 | ||
| G2 | 64 | 67 | 131 | 60 | 71 | ||
| G3 | 109 | 104 | 213 | 112 | 101 | ||
| Unknown | 6 | 3 | 9 | 5 | 4 | ||
| Status | 0.942a | <0.001a | |||||
| Alive | 106 | 111 | 217 | 87 | 130 | ||
| Deceased | 75 | 69 | 144 | 93 | 51 | ||
| Therapy | 0.916a | 0.193a | |||||
| Chemotherapy | 67 | 72 | 139 | 63 | 76 | ||
| Ancillary | 8 | 7 | 15 | 10 | 5 | ||
| Unknown | 106 | 112 | 207 | 107 | 100 |
χ2-test
Fisher's exact test. TCGA, The Cancer Genome Atlas.
Table II.
Information of mRNAs and microRNAs in the integrative signature.
| Ensembl ID | Gene symbol | Genomic location | P-value | Hazard ratio | Coefficient |
|---|---|---|---|---|---|
| ENSG00000146090 | RASGEF1C | Chr 5: 179,527,795-179,636,153(−) | P<0.001 | 2.248 | 0.810 |
| ENSG00000120129 | DUSP1 | Chr 5: 172,195,093-172,198,198(−) | P<0.001 | 1.004 | 0.004 |
| ENSG00000157927 | RADIL | Chr 7: 4,836,686-4,923,350(−) | P<0.001 | 2.358 | 0.858 |
| ENSG00000134917 | ADAMTS8 | Chr 11: 130,274,820-130,298,888(−) | P<0.001 | 1.107 | 0.102 |
| ENSG00000144476 | ACKR3 | Chr 2: 237,476,430-237,491,001(+) | P<0.001 | 1.061 | 0.059 |
| ENSG00000130005 | GAMT | Chr 19: 1,397,091-1,401,569(−) | P<0.001 | 1.041 | 0.040 |
| ENSG00000183287 | CCBE1 | Chr 18: 57,098,172-57,364,612(−) | P<0.001 | 1.496 | 0.403 |
| ENSG00000106366 | SERPINE1 | Chr 7: 100,770,370-100,782,547(+) | P<0.001 | 1.010 | 0.010 |
| ENSG00000163673 | DCLK3 | Chr 3: 36,753,913-36,781,352(−) | P<0.001 | 7.497 | 2.014 |
| ENSG00000163530 | DPPA2 | Chr 3: 109,012,635-109,035,364(−) | P<0.001 | 1.756 | 0.563 |
| ENSG00000029559 | IBSP | Chr 4: 88,720,733-88,733,074(+) | P<0.001 | 1.184 | 0.168 |
| hsa-mir-184 | miR-184 | Chr 15: 79,209,788-79,209,871(+) | P<0.001 | 1.010 | 0.010 |
RASGEF1C, RasGEF domain family member 1C; DUSP1, dual specificity phosphatase 1; RADIL, Rap associating with DIL domain; ADAMTS8, ADAM metallopeptidase with thrombospondin type 1 motif 8; ACKR3, atypical chemokine receptor 3; GAMT, guanidinoacetate N-methyltransferase; CCBE1, collagen and calcium binding EGF domains 1; SERPINE1, serpin family E member 1; DCLK3, doublecortin like kinase 3; DPPA2, developmental pluripotency associated 2; IBSP, integrin binding sialoprotein; miR-, microRNA.
Establishment of an IMMIS for survival prediction in the discovery cohort
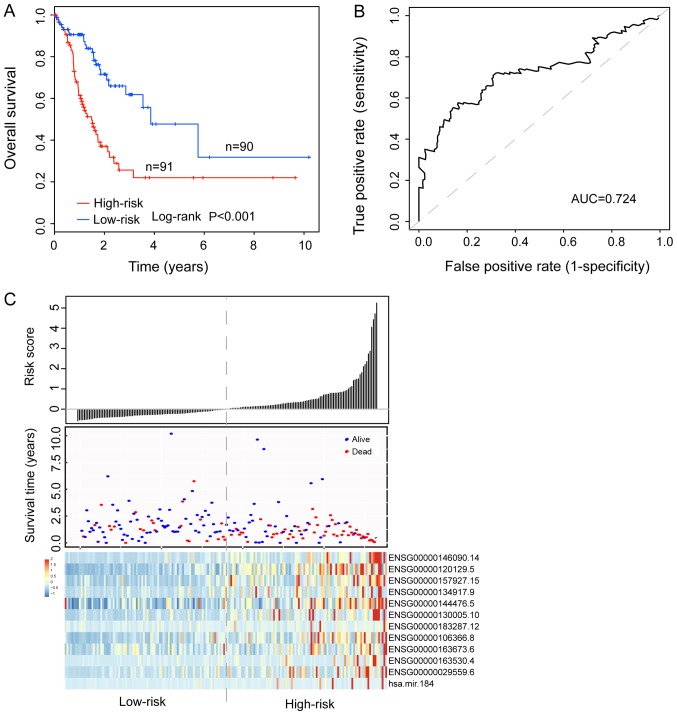
To construct an IMMIS for survival prediction, the 12 RNAs were fitted into a multivariate Cox regression model in the discovery cohort. Notably, the IMMIS was constructed using the determined expression of 12 prognostic RNAs and multivariate Cox regression coefficient as follows: Risk score=(0.5677× RasGEF domain family member 1C) + [0.0035× dual specificity phosphatase 1 (DUSP1)] + (0.0807× Rap associating with DIL domain) + [0.0281× ADAM metallopeptidase with thrombospondin type 1 motif 8 (ADAMTS8)] + [(−0.0095× atypical chemokine receptor 3 (ACKR3)] + (0.0096× guanidinoacetate N-methyltransferase) + [(−0.1882× collagen and calcium binding EGF domains 1 (CCBE1)] + (0.0024× serpin family E member 1) + (0.4322× doublecortin like kinase 3) + (0.6609× developmental pluripotency associated 2) + (0.1356× integrin binding sialoprotein) + (0.0110× miRNA-184). With the identified IMMIS, 181 patients with GC from the discovery cohort were divided into a high-risk group (n=91) and a low-risk group (n=90) according to the median risk score value (0.646), which was considered the cut-off value. Survival analysis showed that patients in the high-risk group had significantly shorter overall survival compared with those in the low-risk group, and median survival was 1.49 vs. 3.85 years, respectively. (P<0.001; Fig. 1A). The result of univariate Cox regression analysis indicated that the HR of the high-risk group compared with the low-risk group regarding overall survival was 2.805 (P<0.001; 95% CI=1.722–4.567; Table III). The 3- and 5-year overall survival rates of patients in the high-risk group were 25.7 and 22%, respectively, which were significantly lower compared with those of patients in the low-risk group. Specifically, he 3- and 5- year overall survival rates of patients in the low-risk group were 61.8 and 47.7%, respectively. Notably, the area under the curve for the IMMIS prognostic model was 0.724 for the 3-year overall survival rate (Fig. 1B). In addition, the distribution of risk scores, the survival status of patients and expression patterns of the 12 prognostic RNAs in the IMMIS are presented in Fig. 1C.
Figure 1.
Development of an IMMIS in the discovery cohort. (A) Kaplan-Meier survival curves of overall survival between high- and low-risk groups. Overall survival was significantly increased in the low-risk group compared with the high-risk group. (B) Time-dependent receiver operating characteristic curves at 3 years of overall survival. The AUC for the IMMIS prognostic model was 0.724 for the 3-year overall survival rate. (C) Distribution of risk scores, survival status of patients and expression patterns of the 12 prognostic RNAs in the IMMIS. IMMIS, integrative mRNA/microRNA signature; AUC, area under the curve.
Table III.
Univariate and multivariate Cox regression analysis of the IMMIS and other clinicopathological variables. Discovery cohort, n=181
| Variables | Univariate model | Multivariate model | |||||
|---|---|---|---|---|---|---|---|
| Characteristics | Comparison groups | Hazard ratio | 95% Confidence interval | P-value | Hazard Ratio | 95% Confidence interval | P-value |
| IMMIS | High- Vs. Low-risk | 2.805 | 1.722–4.567 | <0.001 | 2.613 | 1.560–4.375 | <0.001 |
| Age | <68 vs. ≥68 years | 1.032 | 1.008–1.056 | 0.007 | 1.039 | 1.011–1.067 | 0.005 |
| Sex | Male vs. female | 1.025 | 0.634–1.658 | 0.920 | 1.007 | 0.601–1.688 | 0.979 |
| Stage | I and II vs. III and IV | 1.549 | 0.950–2.527 | 0.079 | 1.868 | 1.096–3.181 | 0.022 |
| Grade | G1 vs. G2 and G3 | 0.987 | 0.612–1.593 | 0.957 | 0.937 | 0.552–1.591 | 0.811 |
IMMIS, integrative mRNA/microRNA signature; TCGA, The Cancer Genome Atlas.
Validation of the IMMIS for survival prediction in the testing cohort and entire TCGA cohort
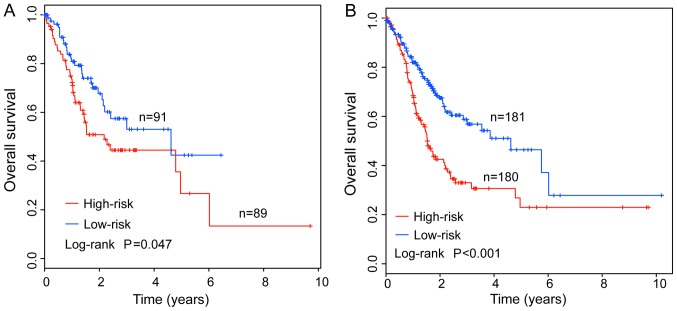
To confirm the predictive value of the IMMIS for survival prediction, the predictive ability of the IMMIS in the testing cohort and entire TCGA cohort was analyzed. With the same risk score model and cut-off derived from the discovery cohort, the IMMIS classified 89 and 91 patients of the testing cohort into high- and low-risk groups, respectively. Consistent with the findings in the discovery cohort, the overall survival time of the high-risk group patients was significantly shorter than that of the patients in the low-risk group. Specifically, the median survival time was 2.17 and. 4.62 years in patients in the high- and low-risk groups, respectively. (P=0.047; Fig. 2A). The 3- and 5-year overall survival rates of patients in the high-risk group were 44.5 and 26.7%, whereas the corresponding rates were 53 and 42.4% in the low-risk group, respectively. In the entire TCGA cohort, 361 patients were classified in either the high-risk (n=180) and low-risk (n=181) groups. Notably, overall survival was significantly different between the high- years and low-risk groups according to the IMMIS and the cut-off value derived from the discovery cohort. The median survival was 1.53 and 4.62 years in the high- and low-risk groups, respectively. (P<0.001; Fig. 2B). The 3- and 5-year overall survival rates of patients in the high-risk group were 35.6 and 26.1%, whereas the corresponding rates were 57.6 and 45.3% in the low-risk group, respectively. The HR of the high-risk group compared with the low-risk group regarding overall survival was 1.625 (P=0.039, 95% CI=1.004–2.638) in the testing cohort and 2.139 (P<0.001, 95% CI=1.519–3.012) in the entire TCGA cohort (Table IV).
Figure 2.
Validation of an integrative mRNA/microRNA signature. Kaplan-Meier survival curves of overall survival between high- and low-risk groups in the (A) testing cohort and (B) entire The Cancer Genome Atlas cohort.
Table IV.
Univariate and multivariate Cox regression analysis of the IMMIS and other clinicopathological variables.
| A, Testing cohort, n=180 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Variables | Univariate model | Multivariate model | ||||||
| Characteristics | Comparison groups | Hazard ratio | IMMIS | High- vs. Low-risk | Hazard ratio | IMMIS | High- vs. Low-risk | |
| IMMIS | High vs. Low | 1.625 | 1.004–2.638 | 0.039 | 1.269 | 1.001–2.112 | 0.049 | |
| Age | <68 vs. ≥68 years | 1.016 | 0.992–0.402 | 0.191 | 1.026 | 1.001–1.052 | 0.045 | |
| Sex | Male vs. female | 1.607 | 0.955–2.704 | 0.074 | 1.696 | 0.971–2.962 | 0.063 | |
| Stage | I and II vs. III and IV | 2.157 | 1.259–3.694 | 0.005 | 1.804 | 1.033–3.150 | 0.038 | |
| Grade | G1 vs. G2 and G3 | 1.757 | 1.057–2.921 | 0.030 | 1.851 | 1.050–3.265 | 0.033 | |
| B, Entire TCGA cohort, n=361 | ||||||||
| Variables | Univariate model | Multivariate model | ||||||
| Characteristics | Comparison groups | Hazard ratio | IMMIS | High- vs. Low-risk | Hazard ratio | IMMIS | High- vs. Low-risk | |
| IMMIS | High vs. Low | 2.139 | 1.519–3.012 | <0.001 | 1.844 | 1.288–2.638 | 0.001 | |
| Age, n=355 | <68 vs. ≥68 years | 1.024 | 1.007–1.041 | 0.004 | 1.029 | 1.011–1.047 | 0.001 | |
| Sex | Male vs. female | 1.282 | 0.901–1.825 | 0.167 | 1.269 | 0.876–1.840 | 0.208 | |
| Stage, n=338 | I and II vs. III and IV | 1.751 | 1.228–1.866 | 0.002 | 1.723 | 1.195–2.485 | 0.004 | |
| Grade | G1 vs. G2 and G3 | 1.319 | 0.932–1.866 | 0.118 | 1.250 | 0.862–1.814 | 0.239 | |
IMMIS, integrative mRNA/microRNA signature; TCGA, The Cancer Genome Atlas.
Independence of the IMMIS from other clinicopathological factors
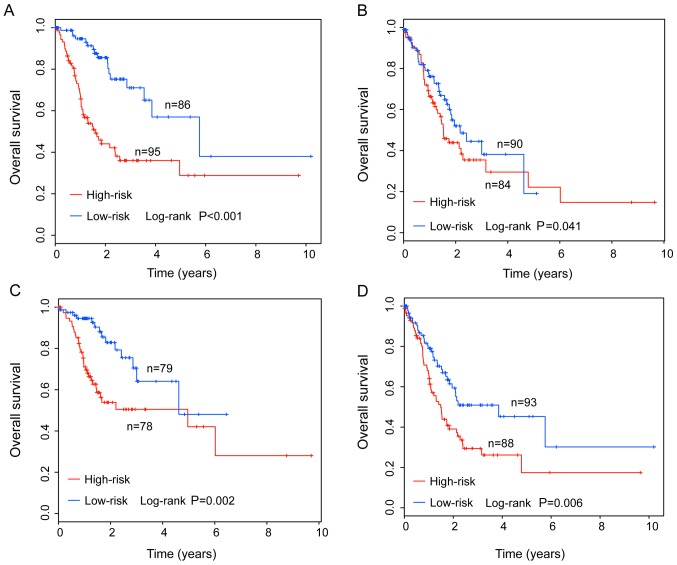
To examine whether the predictive value of the IMMIS was independent of other clinicopathological factors associated with patients with GC, multivariate Cox regression analysis was used to compare the performance of the IMMIS and other clinicopathological factors, including age, sex, stage and grade. The results from the multivariate Cox regression analysis indicated that the IMMIS, stage and age were significantly associated with overall survival of patients with GC (Table III). Therefore, stratification analysis on age and stage to determine whether the IMMIS was independent of age and stage was also performed. First, 355 patients (excluding 6 patients without age data) were classified into a younger stratum (n=181) and an elder stratum (n=174) according to the median age (68 years). Results of stratification analysis for age revealed that, within each age stratum, the IMMIS could further subdivide the patients into the high- and low-risk groups with significantly different overall survival. The median survival was 1.61 and 5.75 years in the high- and low-risk groups, respectively (P<0.001), for the younger group. The median survival was 1.53 and 2.17 years in the high- and low-risk groups, respectively, (P=0.041) for the elder group (Fig. 3A and B). Subsequently, 338 patients (excluding 23 patients without stage data) were classified into an early-stage stratum (stage I/II; n=157) and an advanced-stage stratum (stage III/IV; n=181) according to the GC stage (20). Results of stratification analysis based on stage indicated that, within each stage stratum, the IMMIS could further subdivide the patients into the high- and low-risk groups with significantly different overall survival. The median survival was 4.62 and 4.96 years in the high- and low-risk groups, respectively (P=0.002), for the early-stage group, and 1.49 and 3.85 years in the high- and low-risk groups, respectively (P=0.006) for the advanced-stage group (Fig. 3C and D). Multivariate and stratification analysis demonstrated that the survival prediction ability of the IMMIS was independent of other clinicopathological factors associated with survival prediction in patients with GC.
Figure 3.
Stratified analysis by age and stage. Kaplan-Meier survival curves of overall survival rates between high- and low-risk groups in (A) younger, <68 years, and (B) older, ≥68 years, patients. Kaplan-Meier survival curves of overall survival between high- and low-risk groups in (C) early-stage, I/II, and (D) advanced-stage, III/IV, patients.
Performance comparison of IMMIS with mRNA-based signature
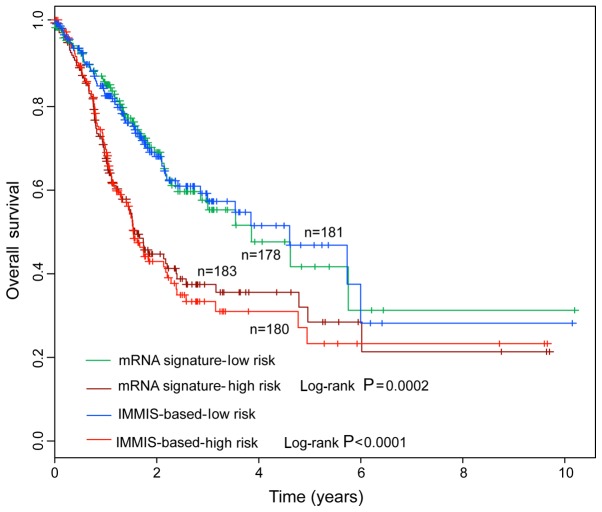
The predictive value of the IMMIS compared with an mRNA-based signature was also evaluated. A total of 11 mRNAs of the IMMIS were used to construct an mRNA-based signature. When applied to the entire TCGA cohort, the IMMIS and mRNA-based signatures reliably classified the patients into high-risk group and low-risk group with significantly different overall survival (Fig. 4). However, Kaplan-Meier survival curves revealed that patients in the high-risk group predicted by the IMMIS had a poorer prognosis compared with those in the high-risk group predicted by the mRNA-based signature, and patients in low-risk group predicted by the IMMIS had an improved prognosis compared with those in the low-risk group predicted by the mRNA-based signature (Fig. 4). The present results suggested that the IMMIS may have improved prognosis prediction ability compared with the mRNA-based signature.
Figure 4.
Comparison of the prognostic value of the IMMIS and mRNA-based signature for survival prediction. IMMIS, integrative mRNA/microRNA signature.
Discussion
The Tumor-Node-Metastasis (TNM) classification of malignant tumors is widely used for the staging and risk stratification of patients with GC (20). However, patients with similar clinical features and those who belong to the same TNM stage may have different clinical outcomes (20). Advances in molecular biology and particularly in the omics sciences have recently demonstrated the complex heterogeneity of GC characterized by genetic and epigenetic changes (21). The findings imply the potential of molecular aberrations as alternative biomarkers for aiding TNM staging and improving prognosis predictions of patients with GC (13,14). Accumulating evidence has revealed that the dysregulated expression of mRNAs and miRNAs has critical roles in the development and progression of GC (20–29), highlighting the applications of miRNAs and mRNAs as molecular biomarkers for predicting the prognoses of patients with GC in clinical practice. Although increasing efforts have been made to discover novel mRNA- or miRNA-based expression signatures for improving prognosis prediction, the predictive signatures previously developed mainly consider a single type of RNA. Several studies have investigated and revealed the prospect of the combination of multiple RNA types for risk stratification in several types of cancer, including colon cancer, breast cancer, glioblastoma multiforme and ovarian cancer (8,9,30). However, it is still unknown whether multi-RNA-based signature could substantially increase the prognostic value in GC.
In the present study, an integrative analysis of mRNA expression profiles, miRNA expression profiles and clinical information in a large cohort of patients with GC was performed. After dividing the entire TCGA cohort into a discovery cohort and a testing cohort, a novel multi-RNA-based signature, consisting of 11 mRNAs and one miRNA, was identified. The predictive value of the identified signature was successfully validated in the testing cohort, which reflects the reproducibility of the integrative signature. Moreover, the integrative signature could act as an independent factor for survival prediction in patients with GC. Notably, comparative analysis revealed that the predictive value of the IMMIS was superior compared with an mRNA-based signature alone. Therefore, the present results suggested that the IMMIS may be a promising signature to effectively identify patients with GC who are at high risk of mortality and guide individualized therapy choices.
The majority of genes included in the integrative signature have been experimentally demonstrated to be associated with GC. For example, DUSP1, a member of the threonine-tyrosine dual-specificity phosphatase family, is involved in cellular proliferation, differentiation and apoptosis, and is additionally associated with tumor carcinogenesis progression (31). Teng et al (32) demonstrated that DUSP1 can induce apatinib resistance by activating the mitogen-activated protein kinase-signaling pathway in patients with GC. The expression status of ADAMTS8 has been investigated in GC, and the expression levels are significantly increased in patients with GC (33). In addition, ADAMTS8 is associated with grade and tumor size (34). Kim et al (35) investigated the expression of ACKR3 and its clinical relevance in GC, and found that ACKR3 expression is associated with aggressive behavior and poor prognosis in GC. A recent study has suggested that CCBE1 may have an important role in the progression of gastrointestinal stromal tumors by enhancing angiogenesis and mediating resistance to imatinib (36). Notably, miRNA-184 has been reported to inhibit cell proliferation and invasion, and functions as a potential oncogene in several types of cancer (37,38). A recent study has indicated that overexpression of miRNA-184 is associated with poor outcome in GC (7).
In conclusion, in the present study, a novel IMMIS was constructed, which could effectively stratify patients into low- and high-risk groups of mortality. With further prospective study, the IMMIS may add more information to the current TNM staging system and could improve prognosis prediction. However, there were some limitations in the present study. Firstly, the integrative signature was only based on available TCGA datasets, and no other independent patient datasets were tested. Secondly, the biological functions of the mRNAs and miRNA incorporated in the integrated signature require further experimental studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YYW conceived and designed the experiments. XY, FMZ, ZWG and WYK performed the experiments and analyzed the data. YYW wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Only publicly available datasets were used and ethical approval was not received in the present study, as it was not necessary.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Hu B, El Hajj N, Sittler S, Lammert N, Barnes R, Meloni-Ehrig A. Gastric cancer: Classification, histology and application of molecular pathology. J Gastrointest Oncol. 2012;3:251–261. doi: 10.3978/j.issn.2078-6891.2012.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orditura M, Galizia G, Sforza V, Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J, Savastano B, Mabilia A, et al. Treatment of gastric cancer. World J Gastroenterol. 2014;20:1635–1649. doi: 10.3748/wjg.v20.i7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CN, Lin JJ, Chen JJ, Lee PH, Yang CY, Kuo ML, Chang KJ, Hsieh FJ. Gene expression profile predicts patient survival of gastric cancer after surgical resection. J Clin Oncol. 2005;23:7286–7295. doi: 10.1200/JCO.2004.00.2253. [DOI] [PubMed] [Google Scholar]
- 6.Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17:1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding B, Gao X, Li H, Liu L, Hao X. A novel microRNA signature predicts survival in stomach adenocarcinoma. Oncotarget. 2017;8:28144–28153. doi: 10.18632/oncotarget.15961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong Y, Wang R, Peng L, You W, Wei J, Zhang S, Wu X, Guo J, Xu J, Lv Z, Fu Z. An integrated lncRNA, microRNA and mRNA signature to improve prognosis prediction of colorectal cancer. Oncotarget. 2017;8:85463–85478. doi: 10.18632/oncotarget.20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volinia S, Croce CM. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc Natl Acad Sci USA. 2013;110:7413–7417. doi: 10.1073/pnas.1304977110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bing Z, Tian J, Zhang J, Li X, Wang X, Yang K. An integrative model of miRNA and mRNA expression signature for patients of breast invasive carcinoma with radiotherapy prognosis. Cancer Biother Radiopharm. 2016;31:253–260. doi: 10.1089/cbr.2016.2059. [DOI] [PubMed] [Google Scholar]
- 11.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J, Chen X, Wang Z, Guo M, Shi H, Wang X, Cheng L, Zhou M. A potential prognostic long non-coding RNA signature to predict metastasis-free survival of breast cancer patients. Sci Rep. 2015;5:16553. doi: 10.1038/srep16553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun J, Cheng L, Shi H, Zhang Z, Zhao H, Wang Z, Zhou M. A potential panel of six-long non-coding RNA signature to improve survival prediction of diffuse large-B-cell lymphoma. Sci Rep. 2016;6:27842. doi: 10.1038/srep27842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou M, Guo M, He D, Wang X, Cui Y, Yang H, Hao D, Sun J. A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J Transl Med. 2015;13:231. doi: 10.1186/s12967-015-0556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou M, Xu W, Yue X, Zhao H, Wang Z, Shi H, Cheng L, Sun J. Relapse-related long non-coding RNA signature to improve prognosis prediction of lung adenocarcinoma. Oncotarget. 2016;7:29720–29738. doi: 10.18632/oncotarget.8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou M, Zhang Z, Zhao H, Bao S, Sun J. A novel lncRNA-focus expression signature for survival prediction in endometrial carcinoma. BMC Cancer. 2018;18:39. doi: 10.1186/s12885-017-3983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou M, Zhang Z, Zhao H, Bao S, Cheng L, Sun J. An immune-related six-lncRNA signature to improve prognosis prediction of glioblastoma multiforme. Mol Neurobiol. 2018;55:3684–3697. doi: 10.1007/s12035-017-0572-9. [DOI] [PubMed] [Google Scholar]
- 18.Zhou M, Zhong L, Xu W, Sun Y, Zhang Z, Zhao H, Yang L, Sun J. Discovery of potential prognostic long non-coding RNA biomarkers for predicting the risk of tumor recurrence of breast cancer patients. Sci Rep. 2016;6:31038. doi: 10.1038/srep31038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Core Team. R, corp-author. R Foundation for Statistical Computing; Vienna: 2012. A language and environment for statistical computing. [Google Scholar]
- 20.Liu JY, Peng CW, Yang XJ, Huang CQ, Li Y. The prognosis role of AJCC/UICC 8th edition staging system in gastric cancer, a retrospective analysis. Am J Transl Res. 2018;10:292. [PMC free article] [PubMed] [Google Scholar]
- 21.Chia NY, Tan P. Molecular classification of gastric cancer. Ann Oncol. 2016;27:763–769. doi: 10.1093/annonc/mdw040. [DOI] [PubMed] [Google Scholar]
- 22.Jiang HB, Yang TJ, Lu P, Ma YJ. Gene expression profiling of gastric cancer. Eur Rev Med Pharmacol Sci. 2014;18:2109–2115. [PubMed] [Google Scholar]
- 23.Marimuthu A, Jacob HK, Jakharia A, Subbannayya Y, Keerthikumar S, Kashyap MK, Goel R, Balakrishnan L, Dwivedi S, Pathare S, et al. Gene expression profiling of gastric cancer. J Proteomics Bioinform. 2011;4:74–82. doi: 10.4172/jpb.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeno A, Takemasa I, Doki Y, Yamasaki M, Miyata H, Takiguchi S, Fujiwara Y, Matsubara K, Monden M. Integrative approach for differentially overexpressed genes in gastric cancer by combining large-scale gene expression profiling and network analysis. Br J Cancer. 2008;99:1307–1315. doi: 10.1038/sj.bjc.6604682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishimoto T, Baba H, Izumi D, Sugihara H, Kurashige J, Iwatsuki M, Tan P. Current perspectives toward the identification of key players in gastric cancer microRNA dysregulation. Int J Cancer. 2016;138:1337–1349. doi: 10.1002/ijc.29627. [DOI] [PubMed] [Google Scholar]
- 26.Pan HW, Li SC, Tsai KW. MicroRNA dysregulation in gastric cancer. Curr Pharm Des. 2013;19:1273–1284. doi: 10.2174/1381612811319070011. [DOI] [PubMed] [Google Scholar]
- 27.Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J, Sung JJ. MicroRNA dysregulation in gastric cancer: A new player enters the game. Oncogene. 2010;29:5761–5771. doi: 10.1038/onc.2010.352. [DOI] [PubMed] [Google Scholar]
- 28.Zhou M, Hu L, Zhang Z, Wu N, Sun J, Su J. Recurrence-associated long non-coding RNA signature for determining the risk of recurrence in patients with colon cancer. Mol Ther Nucleic Acids. 2018;12:518–529. doi: 10.1016/j.omtn.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou M, Zhao H, Wang X, Sun J, Su J. Analysis of long noncoding RNAs highlights region-specific altered expression patterns and diagnostic roles in Alzheimer's disease. Brief Bioinform: 2019;20:598–608. doi: 10.1093/bib/bby021. [DOI] [PubMed] [Google Scholar]
- 30.Guo L, Peng Y, Meng Y, Liu Y, Yang S, Jin H, Li Q. Expression profiles analysis reveals an integrated miRNA-lncRNA signature to predict survival in ovarian cancer patients with wild-type BRCA1/2. Oncotarget. 2017;8:68483–68492. doi: 10.18632/oncotarget.19590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen J, Zhang Y, Yu H, Shen B, Liang Y, Jin R, Liu X, Shi L, Cai X. Role of DUSP1/MKP1 in tumorigenesis, tumor progression and therapy. Cancer Med. 2016;5:2061–2068. doi: 10.1002/cam4.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teng F, Xu Z, Chen J, Zheng G, Zheng G, Lv H, Wang Y, Wang L, Cheng X. DUSP1 induces apatinib resistance by activating the MAPK pathway in gastric cancer. Oncol Rep. 2018;40:1203–1222. doi: 10.3892/or.2018.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou M, Zhao H, Xu W, Bao S, Cheng L, Sun J. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol Cancer. 2017;16:16. doi: 10.1186/s12943-017-0580-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilic MO, Aynekin B, Kara A, Icen D, Demircan K. Differentially regulated ADAMTS1, 8, and 18 in gastric adenocarcinoma. Bratisl Lek Listy. 2017;118:71–76. doi: 10.4149/BLL_2017_014. [DOI] [PubMed] [Google Scholar]
- 35.Kim N, Baek SW, Ryu H, Choi YS, Song IC, Yun HJ, Jo DY, Kim SY, Lee HJ. Atypical chemokine receptor ACKR3 expression is associated with aggressive behavior and poor prognosis in gastric cancer. Cancer Res. 2016;76(Suppl 14):3947. [Google Scholar]
- 36.Tian GA, Zhu CC, Zhang XX, Zhu L, Yang XM, Jiang SH, Li RK, Tu L, Wang Y, Zhuang C, et al. CCBE1 promotes GIST development through enhancing angiogenesis and mediating resistance to imatinib. Sci Rep. 2016;6:31071. doi: 10.1038/srep31071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng Z, Wang HZ, Li X, Wu Z, Han Y, Li Y, Chen G, Xie X, Huang Y, Du Z, Zhou Y. MicroRNA-184 inhibits cell proliferation and invasion, and specifically targets TNFAIP2 in Glioma. J Exp Clin Cancer Res. 2015;34:27. doi: 10.1186/s13046-015-0142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu G, Liu J, Wu Z, Wu X, Yao X. MicroRNA-184 inhibits cell proliferation and metastasis in human colorectal cancer by directly targeting IGF-1R. Oncol Lett. 2017;14:3215–3222. doi: 10.3892/ol.2017.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.