Abstract
Non-small-cell lung cancer (NSCLC) is the most common cause of cancer-associated mortality worldwide. MicroRNAs (miRs) are a class of small non-coding RNAs that are commonly dysregulated in human cancer. The aim of the current study was to evaluate the effect of miR-296-3p on the cell migration and invasion of NSCLC. Pairs of tumor tissues and para-cancerous tissues (n=50) were collected from patients with NSCLC, and the expression of miR-296-3p was analyzed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Additionally, tumor cell viability, migration and invasion were examined in vitro using Cell Counting Kit-8, wound healing and Matrigel assays, respectively. Furthermore, potential targets of miR-296-3p were screened for using TargetScan and validated using a dual-luciferase reporter assay. The expression levels of phosphoinositide-3-kinase (PI3K), AKT serine/threonine kinase (AKT), mammalian target of rapamycin (mTOR), matrix metallopeptidase 2 (MMP2) and SRY-box 4 (SOX4) were detected by RT-qPCR and western blot analysis. The data indicated that miR-296-3p was downregulated in tumor tissues compared with adjacent normal tissues. Overexpression of miR-296-3p inhibited NSCLC cell viability, migration and invasion in vitro. Furthermore, apurinic/apyrimidinic endodeoxyribonuclease 1 (APEX1) was identified as a direct target of miR-296-3p. APEX1 expression was upregulated in tumor tissues compared with para-cancerous tissues, and the mRNA and protein expression levels of APEX1 were decreased following transfection of NSCLC cells with miR-296-3p mimics compared with control cells. Additional investigations revealed that miR-296-3p was involved in regulating the PI3K/AKT/mTOR signaling pathway, and miR-296-3p mimics decreased the mRNA and protein expression levels of MMP2 and SOX4. In summary, the findings demonstrated that miR-296-3p may function as a tumor suppressor, and inhibits the migration and invasion of NSCLC cells by targeting APEX1. miR-296-3p is therefore a potential therapeutic molecular modulator of NSCLC.
Keywords: non-small-cell lung cancer, microRNA-296-3p, apurinic/apyrimidinic endodeoxyribonuclease 1, migration, invasion
Introduction
Non-small-cell lung cancer (NSCLC) is a heterogeneous class of lung cancer, and in 2015 ranked as the most common cause of cancer-associated mortality worldwide (1). The majority of patients with NSCLC are diagnosed during the metastatic period, and the median survival time following diagnosis is 1 year (2). Although surgical techniques and biological treatments have been developed, the overall 5-year survival rate is 18% (3). In addition, patients diagnosed at the advanced stage are not suitable for surgery (3). Thus, the identification of novel targeted agents may produce effective methods for treating patients with advanced NSCLC (4). However, the underlying molecular mechanisms of NSCLC remain poorly defined.
MicroRNAs (miRNAs/miRs) are a class of endogenous, non-coding RNAs, which are 18–25 nucleotides in length and can regulate gene expression by inhibiting translation and/or degradation of mRNA (5). The regulation of gene expression by miRNAs is mediated via base pairing with 3′-untranslated regions (3′-UTRs) in mRNA molecules (6,7). miRNAs are commonly dysregulated in cancer, which affects tumorigenesis processes and the expression of specific genes (7). miR-296 is located at the chromosome 20q13.32 genomic locus. It functions as a tumor suppressor in cervical, pancreatic and colorectal cancer by regulating specific targets (8–10). miR-296-3p is derived from the 3′ arm of mature miR-296, and miR-296-5p is derived from the 5′ arm (11). In NSCLC, miR-296-5p has been demonstrated to suppress tumor progression by targeting polo like kinase 1 (12). In addition, miR-296-3p has been reported to inhibit NSCLC cell proliferation, promote cell apoptosis, and enhance resistance to cisplatin and paclitaxel (13). However, to the best of our knowledge, whether miR-296-3p is involved in the migration and invasion of NSCLC has not been investigated.
The aim of the current study was to investigate the biological and functional role of miR-296-3p in NSCLC. The effects of miR-296-3p on NSCLC cell migration and invasion in vitro were investigated, which indicated that miR-296-3p may exhibit tumor-suppressive functions. Understanding the molecular mechanism of miR-296-3p may provide novel therapeutic targets for the treatment of NSCLC.
Materials and methods
Clinical tissue specimens
NSCLC tissues and adjacent normal tissues (the distance between tumor tissues and normal tissues was 5 cm) were collected from 50 patients with NSCLC, including 29 male and 21 female patients, with a median age of 59.92 years (range, 45–75 years) that received surgery at Xi'an High-Tech Hospital (Xi'an, China) and Shaanxi Provincial People's Hospital (Xi'an, China) between September 2015 and December 2016. All specimens were immediately frozen in liquid nitrogen and stored at −80°C prior to further use. The present study was approved by the Ethics Committees of Xi'an High-Tech Hospital and Shaanxi Provincial People's Hospital. Written informed consent was obtained from all patients.
Cell culture
A549 cells were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in RPMI-1640 medium (GE Healthcare Life Sciences) with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 1% (w/v) penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.). The cells were maintained at 37°C with 5% CO2.
Cell transfection
miR-296-3p mimics and corresponding negative control (NC) vectors were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). A549 cells were transfected with 30 nM miR-296-3p mimics (5′-GAGGGUUGGGUGGAGGCUCUCC-3′) and miR-NC (5′-UUCUCCGAACGUGUCACGU-3′) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Subsequently, RNA was reverse transcribed to cDNA using the PrimeScript™ RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). RT conditions were as follows: 37°C for 15 min and 85°C for 5 sec. RT-qPCR was performed using SYBR® Premix Ex Taq™ kit (Takara Biotechnology Co., Ltd.) using an ABI 7500 real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR reaction conditions were as follows: 95°C for 10 min, followed by 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec (40 cycles). U6 was used as the internal control for miR-296-3p, and GAPDH was used as the internal control for apurinic/apyrimidinic endodeoxyribonuclease 1 (APEX1), phosphoinositide-3-kinase (PI3K), AKT serine/threonine kinase (AKT), mammalian target of rapamycin (mTOR), matrix metallopeptidase 2 (MMP2) and SRY-box 4 (SOX4). All samples were normalized to internal controls and the fold change was calculated using the 2−ΔΔCq method (14). The sequences of the primers used were as follows: miR-296-3p forward, 5′-GAGGGTTGGGTGGAGGCTCTCC-3′; U6 forward, 5′-ATGACACGCAAATTCGTGAAGC-3′; the reverse primers of miR-296-3p and U6 used universal primers. APEX1 forward, 5′-TGAAGCCTTTCGCAAGTTCCT-3′ and reverse, 5′-TGAGGTCTCCACACAGCACAA-3′; PI3K forward, 5′-CATCACTTCCTCCTGCTCTAT-3′ and reverse, 5′-CAGTTGTTGGCAATCTTCTTC-3′; AKT forward, 5′-GGACAACCGCCATCCAGACT-3′ and reverse, 5′-GCCAGGGACACCTCCATCTC-3′; mTOR forward, 5′-ATTTGATCAGGTGTGCCAGT-3′ and reverse, 5′-GCTTAGGACATGGTTCATGG-3′; MMP2 forward, 5′-GACCTTGACCAGAACACCATCG-3′ and reverse, 5′-GCTGTATTCCCGACCGTTGAAC-3′; SOX4 forward, 5′-GTGAGCGAGATGATCTCGGG-3′ and reverse, 5′-CAGGTTGGAGATGCTGGACTC-3′; and GAPDH forward, 5′-TGCCAAATATGATGACATCAAGAA-3′ and reverse 5′-GGAGTGGGTGTCGCTGTTG-3′.
Western blot analysis
Proteins were extracted from tissues and cells using radioimmunoprecipitation assay lysis buffer, and the concentration of each sample was measured using a bicinchoninic acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). The proteins (30 µg) were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% not-fat milk in 0.05% TBS-Tween-20 (TBST) for 1 h at room temperature. The membranes were incubated with primary antibodies against APEX1 (cat. no. ab137708; 1:2,000), PI3K (cat. no. Ab191606; 1:1,000), phospho (p)-PI3K (cat. no. ab192651; 1:1,000), AKT (cat. no. Ab179463; 1:10,000), p-AKT (cat. no. ab131443; 1:1,000), mTOR (cat. no. Ab2732; 1:2,000), p-mTOR (p-S2448; cat. no. ab109268; 1:1,000), MMP2 (cat. no. ab92536; 1:1,000), SOX4 (cat. no. ab80261; 1:500) and GAPDH (cat. no. ab181602; 1:1,000) overnight at 4°C. All primary antibodies were purchased from Abcam (Cambridge, MA, USA). Subsequently, the membranes were washed in TBST and then incubated with secondary antibody (goat anti-rabbit IgG horseradish peroxidase-conjugated antibody; cat. no. ab205718; 1:2,000; Abcam) at room temperature for 1 h. Protein expression signals were detected using enhanced chemiluminescence reagents (GE Healthcare, Chicago, IL, USA) with GAPDH used as the loading control. Data was analyzed by Image-Pro Plus 6.0 software (Media Cybernetics, Inc.).
Cell viability assay
Cell viability was analyzed using the Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Cells (4×103 cells/well) were seeded into 96-well plates and cultured at 37°C with 5% CO2 for 0, 12, 24 and 48 h. CCK-8 reagent (10 µl) was added into each well. Subsequently, cells were cultured at 37°C for 2 h. The optical density value was measured at a wavelength of 450 nm using a microplate reader.
Wound healing assay
Cells (4×105) were seeded in 6-well plates. When confluence reached >90%, a 10-µl pipette tip was used to scratch two parallel wounds in the cell monolayer. The cell debris was washed twice with PBS and cells were incubated at 37°C with 5% CO2 in serum-free RPMI-1640 medium. Following 24 h of incubation at 37°C, images of the cells were captured using a light microscope (magnification, ×200) and the width of the wound was calculated using a standard caliper.
Matrigel assays
A549 cells (2×105 cells/ml) were seeded in the upper chamber of Transwell filled with serum-free RPIM-1640 medium inserts coated with 100 µl Matrigel (BD Biosciences, San Jose, CA, USA) at 37°C for 4 h. The bottom chamber was filled with RPMI-1640 medium containing 10% FBS. Following incubation for 24 h, the cells that had invaded into the lower chambers were fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet for 15 min at room temperature. Cells were imaged and quantified from 5 randomly-selected fields using a light microscope (magnification, ×200).
Prediction of target genes and dual-luciferase reporter assay
The potential targets of miR-296-3p were predicted using the TargetScan online tool (targetscan.org/). The DNA regions coding wild-type (WT) APEX1 3′-untranslated region (3′-UTR; miR-296-3p binding sites) and a mutant sequence (miR-296-3p binding site deletion) were inserted into a pmirGLO vector (Promega Corporation). miR-296-3p mimics and miR-NC were co-transfected into A549 cells with WT and mutant vectors using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After transfection for 48 h, firefly luciferase activity was normalized to Renilla luciferase activity using the dual-luciferase reporter assay system (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol.
Statistical analysis
All experiments were repeated three times. All data are analyzed by GraphPad Prism 7 software (GraphPad, Inc.) presented as the mean ± standard deviation. Student's t-test was used to analyze two groups, except for the comparison between tumor tissues and normal tissues where a paired Student's t-test was used. One-way analysis of variance followed by the Newman-Keuls test was used to compare the differences among multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulated expression of miR-296-3p in NSCLC tissues
In order to determine the role of miR-296-3p in NSCLC, 50 pairs of tumor tissues and adjacent normal tissues were collected from patients with NSCLC. miR-296-3p expression was detected by RT-qPCR, revealing that miR-296-3p expression was significantly lower in tumor tissues compared with corresponding normal tissues (P<0.01; Fig. 1).
Figure 1.
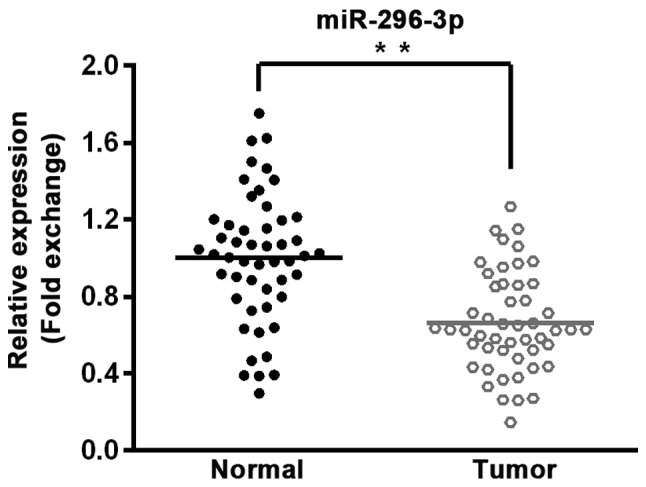
Expression of miR-296-3p is significantly reduced in non-small-cell lung cancer tissues compared with para-cancerous tissues. Reverse transcription-quantitative polymerase chain reaction was used to determine the expression levels in tumor tissues and adjacent normal tissues. n=50. **P<0.01. miR, microRNA.
miR-296-3p inhibits NSCLC cell viability, migration and invasion in vitro
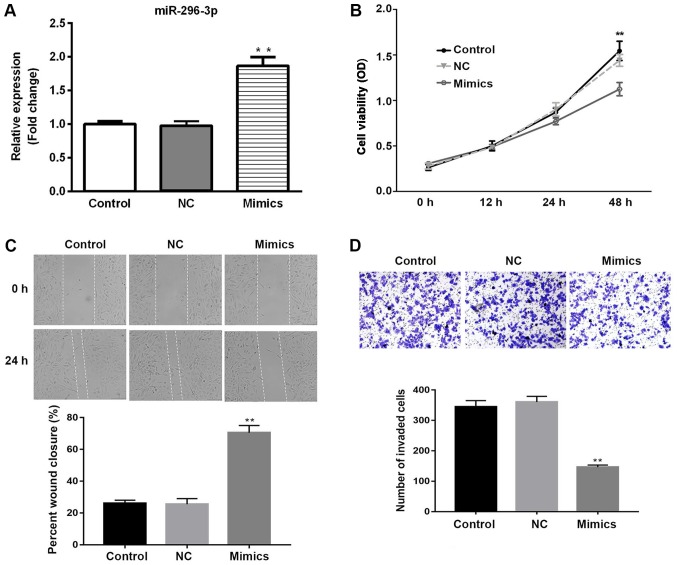
Transfection of miR-296-3p mimics into NSCLC cells was used to investigate the functional role of miR-296-3p. Initially, the transfection efficiency of miR-296-3p was determined by RT-qPCR. miR-296-3p expression was significantly upregulated in A549 cells transfected with miR-296-3p mimic compared with the miR-NC group (P<0.01; Fig. 2A). The effects of miR-296-3p overexpression on cell viability, migration and invasion were determined by CCK-8, wound healing and Matrigel assays, respectively. Cell viability was significantly downregulated in the miR-296-3p mimics group compared with the miR-NC-transfected cells at 48 h (P<0.01), with similar viability rates in the miR-NC and untransfected control group (Fig. 2B). As presented in Fig. 2C, miR-296-3p mimic inhibited the migration of cells at 24 h, compared with the miR-NC group (P<0.01). Similarly, overexpression of miR-296-3p significantly reduced the number of invaded cells, suggesting that miR-296-3p suppressed cell invasion in vitro (P<0.01; Fig. 2D). These findings indicate that overexpression of miR-296-3p exhibits tumor suppressive effects via reduced cell viability, migration and invasion.
Figure 2.
Overexpression of miR-296-3p inhibits non-small-cell lung cancer cell viability, migration and invasion. (A) A549 cells were transfected with miR-296-3p mimics and miR-NC. The transfection efficiency was determined by reverse transcription-quantitative polymerase chain reaction. (B) Cell viability was analyzed by the Cell Counting Kit-8 assay. (C) Wound healing assay was used to measure cell migration (magnification, ×200). (D) Matrigel assay was used to evaluate cell invasion (magnification, ×200). Data are presented as the mean ± standard deviation. **P<0.01 vs. NC. miR, microRNA; NC, negative control; OD, optical density.
APEX1 is a potential target of miR-296-3p
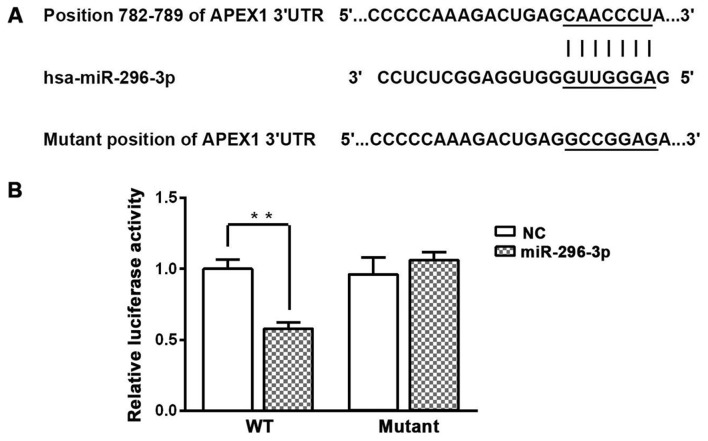
To determine how miR-296-3p mediates the aforementioned effects in NSCLC cells, bioinformatics analysis was used to predict the potential targets of miR-296-3p. The results indicated that miR-296-3p directly binds to position 782–789 of the APEX1 mRNA 3′-UTR (Fig. 3A). A dual-luciferase reporter assay was used to validate this prediction. Overexpression of miR-296-3p significantly decreased luciferase activity in A549 cells transfected with WT APEX1 3′-UTR compared with the miR-NC group (P<0.01; Fig. 3B). However, overexpression of miR-296-3p did not effect the luciferase activity in A549 cells transfected with the mutant APEX1 3′-UTR.
Figure 3.
APEX1 is a direct target of miR-296-3p. (A) WT sequence of the putative miR-296-3p binding site in the APEX1 3′-UTR was predicted by bioinformatics analysis. (B) Dual-luciferase reporter assay was performed following transfection of A549 cells with plasmids containing WT or mutant APEX1 3′-UTR. Data are presented as the mean ± standard deviation. **P<0.01. APEX1, apurinic/apyrimidinic endodeoxyribonuclease 1; miR, microRNA; 3′-UTR, 3′-untranslated region; NC, negative control; WT, wild-type.
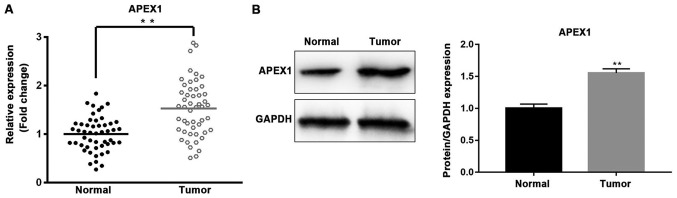
Furthermore, the expression of APEX1 was analyzed in NSCLC tissues and cells. The mRNA and protein expression levels of APEX1 were significantly higher in tumor tissues compared with normal tissues (P<0.01; Fig. 4A and B). The expression levels of APEX1 mRNA and protein were significantly downregulated in A549 cells transfected with miR-296-3p mimics compared with the miR-NC group (P<0.05; Fig. 5A, I and J).
Figure 4.
Expression of APEX1 is upregulated in non-small-cell lung cancer tissues compared with para-cancerous tissues. (A) The mRNA expression level of APEX1 in tumor tissues and adjacent normal tissues was assessed by reverse transcription-quantitative polymerase chain reaction. n=50. (B) Western blot analysis measured the protein expression level of APEX1 in tumor tissues and corresponding normal tissues. GAPDH was used as the loading control. **P<0.01 vs. Normal. APEX1, apurinic/apyrimidinic endodeoxyribonuclease 1.
Figure 5.
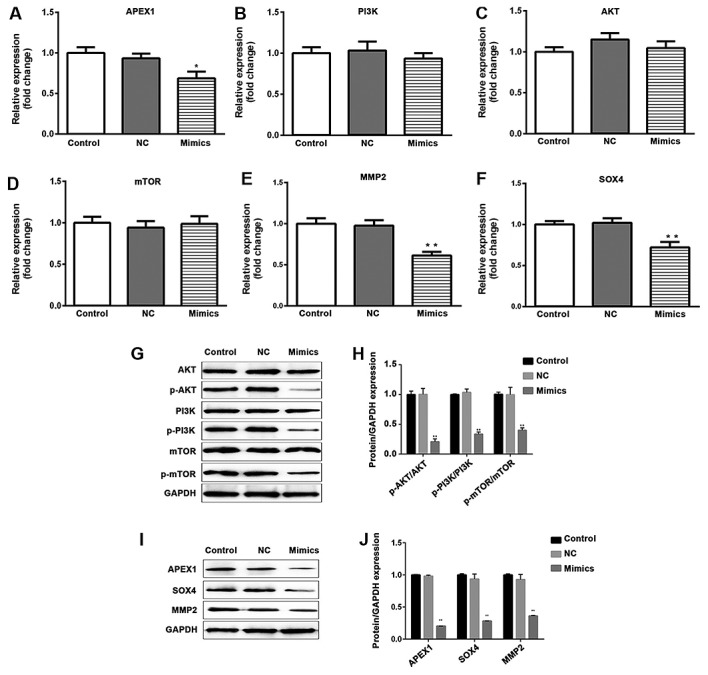
Overexpression of miR-296-3p does not affect the mRNA and protein expression levels of PI3K, AKT and mTOR; however, it does reduce the phosphorylation of PI3K, AKT and mTOR. mRNA and protein levels of APEX1, MMP2 and SOX4 were decreased following transfection of A549 cells with miR-296-3p mimics. The mRNA expression levels of (A) APEX1, (B) PI3K, (C) AKT, (D) mTOR, (E) MMP2 and (F) SOX4 were detected using reverse transcription-quantitative polymerase chain reaction. (G) Protein levels of total PI3K, p-PI3K, total AKT, p-AKT, total mTOR and p-mTOR were examined using western blot analysis. GAPDH was used as a normalization control. (H) Quantification of the ratios of p-AKT/AKT, p-PI3K/PI3K and p-mTOR/mTOR. (I) Protein levels of APEX1, MMP2 and SOX4 were measured by western blot analysis. (J) The fold change of proteins/GAPDH according to the APEX1, MMP2 and SOX4 protein levels. Data are presented as the mean ± standard deviation. *P<0.05, **P<0.01 vs. NC. miR, microRNA; PI3K, phosphoinositide-3-kinase; AKT, AKT serine/threonine kinase; mTOR, mammalian target of rapamycin; MMP2, matrix metallopeptidase 2; SOX4, SRY-box 4; p-, phosphorylated-; NC, negative control.
miR-296-3p regulates the PI3K/AKT/mTOR signaling pathway and the expression of MMP2 and SOX4
In order to investigate how miR-296-3p regulates the signaling associated with cellular processes, RT-qPCR was used to evaluate the mRNA expression levels of PI3K, AKT, mTOR, MMP2 and SOX4. As presented in Fig. 5B-F, overexpression of miR-296-3p did not induce a change in PI3K, AKT and mTOR expression at the mRNA level (P>0.05); however, it did significantly decrease the mRNA and protein levels of MMP2 and SOX4 (P<0.01), compared with the miR-NC group (Fig. 5E, F, I and J). The protein expression levels of total PI3K, AKT and mTOR were not affected by miR-296-3p; however, the phosphorylation levels of PI3K, AKT and mTOR were significantly reduced in A549 cells transfected with miR-296-3p mimic (P<0.01; Fig. 5G and H).
Discussion
In the present study, the molecular mechanism of miR-296-3p in NSCLC was investigated, and the results revealed that miR-296-3p acted as a tumor suppressor in NSCLC. miR-296-3p was demonstrated to inhibit NSCLC cell viability, migration and invasion in vitro, and was revealed to directly target APEX1 mRNA.
Previous studies have examined the expression and role of miR-296-3p in human cancer. For example, miR-296-3p was demonstrated to be upregulated in prostate cancer, which increased tumor cell resistance to natural killer cells by targeting intercellular adhesion molecule-1 (15). By contrast, miR-296-3p has been reported to suppress tumor cell proliferation, migration and invasion in several types of malignancy. In choroidal malignant melanoma, overexpression of miR-296-3p suppressed tumor cell proliferation, migration and invasion (16). Additionally, miR-296-3p was downregulated in glioblastoma cells and decreased cell proliferation and invasion by inhibition of potassium voltage-gated channel subfamily H member 1 (17). The current study revealed that miR-296-3p expression was lower in NSCLC tissues compared with para-cancerous tissues, which was consistent with a previous study by Luo et al (13). Furthermore, overexpression of miR-296-3p inhibited tumor cell viability, migration and invasion in vitro. These results indicate that miR-296-3p exerts antitumor effects in NSCLC. However, the underlying molecular mechanism of how miR-296-3p mediates these effects in NSCLC remained unclear.
Bioinformatics analysis was used to predict the potential mRNA targets of miR-296-3p. The results indicated that miR-296-3p may bind to the 3′-UTR of APEX1 at nucleotides 782–789. A dual-luciferase reporter assay revealed that APEX1 was a direct target of miR-142-3p. APEX1 is a DNA repair enzyme that recognizes and cleaves apurinic/apyrimidinic sites in damaged DNA (18). APEX1 is commonly upregulated in human cancer, including in prostate cancer (19), osteosarcoma (20) and human melanoma (21). Furthermore, high expression of APEX1 is associated with unfavorable prognosis in breast cancer and osteosarcoma (20,22). APEX1 is involved in regulating biological behavior in human cancer. For example, APEX1 has been demonstrated to promote colon cancer tumorigenicity and progression in vitro and in vivo (23). Furthermore, overexpression of APEX1 is an independent predictor of osteosarcoma local recurrence and metastasis (24). Additionally, APEX1 was established as a prognostic indicator in NSCLC, and cytoplasmic expression of APEX1 was associated with poor survival rates (25). However, the expression and functional roles of APEX1 in NSCLC have remained unclear; therefore, the expression of APEX1 was detected in the present study. The results demonstrated that APEX1 was significantly upregulated in NSCLC tissues at the mRNA and protein levels compared with para-cancerous tissues, which was consistent with previous studies in other cancer types, such as prostate cancer, osteosarcoma and human melanoma (19–21). Additionally, the expression of APEX1 was significantly downregulated by transfection with miR-296-3p mimic in NSCLC cells; this suggested that overexpression of miR-296-3p may inhibit NSCLC cell migration and invasion by targeting APEX1.
The PI3K signaling pathway is commonly activated in human cancer (26). AKT and mTOR are major effectors that act downstream of PI3K (27). PI3K is activated by various extracellular cytokines via auto-phosphorylation of cell surface receptors (27). Once activated, PI3K can phosphorylate downstream kinases, including AKT, and activated AKT phosphorylates other downstream targets to regulate numerous cellular processes (28–30). The PI3K/AKT/mTOR signaling pathway is an important mediator of tumor cell proliferation, apoptosis, migration, invasion, metabolism and angiogenesis (28,31). MMP2 and SOX4 are also involved in regulating cell migration and invasion (32,33). To the best of our knowledge, no previous studies have investigated the association between APEX1, the PI3K/AKT/mTOR pathway, and MMP2 and SOX4. In the current study, overexpression of miR-296-3p reduced the phosphorylation of PI3K, AKT and mTOR, but did not affect their mRNA expression level. These results suggest that miR-296-3p may have an inhibitory effect on the PI3K/AKT/mTOR signaling pathway, although this action potentially occurs at the translation/post-translation level, rather than by affecting mRNA stability. Additionally, miR-296-3p reduced the mRNA and protein levels of MMP2 and SOX4. These findings suggest that miR-296-3p may inactivate the PI3K/AKT/mTOR signaling pathway and suppress the expression of MMP2 and SOX4 by targeting APEX1, resulting in reduced cell migration and invasion.
In conclusion, the present study demonstrated that miR-296-3p was downregulated in NSCLC tissues compared with para-cancerous tissues. Additionally, miR-296-3p inhibited NSCLC cell migration and invasion, reduced PI3K/AKT/mTOR signaling, and reduced the expression of MMP2 and SOX4, potentially by targeting APEX1. APEX1 was validated as a novel target of miR-296-3p, and these insights may be useful for identifying novel therapeutic targets for the clinical treatment of patients with NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data genarated or analyzed during the present study are included in this published article.
Authors' contributions
LW and YZ contributed to study design. LW and RC performed experiments and data analysis. LW was a major contributor in writing the manuscript. All author have read and approved the final manuscript.
Ethics approval and consent to participate
The human sample collection was approved by the Ethics Committees of Xi'an High-Tech Hospital and Shaanxi Provincial People's Hospital. All patients provided written informed consent.
Patient consent for publication
Patients provided consent for the publication of the present study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, Petrella F, Spaggiari L, Rosell R. Non-small-cell lung cancer. Nat Rev Dis Primers. 2015;1:15009. doi: 10.1038/nrdp.2015.9. [DOI] [PubMed] [Google Scholar]
- 2.Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382:720–731. doi: 10.1016/S0140-6736(13)61715-8. [DOI] [PubMed] [Google Scholar]
- 3.Skjefstad K, Johannessen C, Grindstad T, Kilvaer T, Paulsen EE, Pedersen M, Donnem T, Andersen S, Bremnes R, Richardsen E, et al. A gender specific improved survival related to stromal miR-143 and miR-145 expression in non-small cell lung cancer. Sci Rep. 2018;8:8549. doi: 10.1038/s41598-018-26864-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Custodio A, Méndez M, Provencio M. Targeted therapies for advanced non-small-cell lung cancer: Current status and future implications. Cancer Treat Rev. 2012;38:36–53. doi: 10.1016/j.ctrv.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 6.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lv L, Wang X. MicroRNA-296 targets specificity protein 1 to suppress cell proliferation and invasion in cervical cancer. Oncol Res. 2018;26:775–783. doi: 10.3727/096504017X15132494420120. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Li H, Li J, Shi B, Chen F. MicroRNA-296 targets AKT2 in pancreatic cancer and functions as a potential tumor suppressor. Mol Med Rep. 2017;16:466–472. doi: 10.3892/mmr.2017.6602. [DOI] [PubMed] [Google Scholar]
- 10.He Z, Yu L, Luo S, Li M, Li J, Li Q, Sun Y, Wang C. miR-296 inhibits the metastasis and epithelial-mesenchymal transition of colorectal cancer by targeting S100A4. BMC Cancer. 2017;17:140. doi: 10.1186/s12885-017-3121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Ouyang XP, Jiang T, Zheng XL, He PP, Zhao GJ. MicroRNA-296: A promising target in the pathogenesis of atherosclerosis? Mol Med. 2018;24:12. doi: 10.1186/s10020-018-0012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu C, Li S, Chen T, Hu H, Ding C, Xu Z, Chen J, Liu Z, Lei Z, Zhang HT, et al. MiR-296-5p suppresses cell viability by directly targeting PLK1 in non-small cell lung cancer. Oncol Rep. 2016;35:497–503. doi: 10.3892/or.2015.4392. [DOI] [PubMed] [Google Scholar]
- 13.Luo W, Lin Y, Meng S, Guo Y, Zhang J, Zhang W. miRNA-296-3p modulates chemosensitivity of lung cancer cells by targeting CX3CR1. Am J Transl Res. 2016;8:1848–1856. [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Chen Q, Yan J, Wang Y, Zhu C, Chen C, Zhao X, Xu M, Sun Q, Deng R, et al. MiRNA-296-3p-ICAM-1 axis promotes metastasis of prostate cancer by possible enhancing survival of natural killer cell-resistant circulating tumour cells. Cell Death Dis. 2013;4:e928. doi: 10.1038/cddis.2013.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Hu Y, Cui J, Zhou Y, Chen L. Coordinated targeting of MMP-2/MMP-9 by miR-296-3p/FOXCUT exerts tumor-suppressing effects in choroidal malignant melanoma. Mol Cell Biochem. 2018;445:25–33. doi: 10.1007/s11010-017-3248-x. [DOI] [PubMed] [Google Scholar]
- 17.Ba Y, Liao H, Liu T, Zeng X, Xiao F, Luo L, Guo H, Guo L. MiR-296-3p regulates cell growth and multi-drug resistance of human glioblastoma by targeting ether-à-go-go (EAG1) Eur J Cancer. 2013;49:710–724. doi: 10.1016/j.ejca.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Liu G, Xia L, Zhou Q, Xiong J, Xian J, Du M, Zhang L, Liao L, Su X, et al. A polymorphism in the DNA repair domain of APEX1 is associated with the radiation-induced pneumonitis risk among lung cancer patients after radiotherapy. Br J Radiol. 2014;87:20140093. doi: 10.1259/bjr.20140093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelley MR, Cheng L, Foster R, Tritt R, Jiang J, Broshears J, Koch M. Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clin Cancer Res. 2001;7:824–830. [PubMed] [Google Scholar]
- 20.Wang D, Luo M, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: Enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol Cancer Ther. 2004;3:679–686. [PubMed] [Google Scholar]
- 21.Yang S, Irani K, Heffron SE, Jurnak F, Meyskens FL., Jr Alterations in the expression of the apurinic/apyrimidinic endonuclease-1/redox factor-1 (APE/Ref-1) in human melanoma and identification of the therapeutic potential of resveratrol as an APE/Ref-1 inhibitor. Mol Cancer Ther. 2005;4:1923–1935. doi: 10.1158/1535-7163.MCT-05-0229. [DOI] [PubMed] [Google Scholar]
- 22.Woo J, Park H, Sung SH, Moon BI, Suh H, Lim W. Prognostic value of human apurinic/apyrimidinic endonuclease 1 (APE1) expression in breast cancer. PLoS One. 2014;9:e99528. doi: 10.1371/journal.pone.0099528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim MH, Kim HB, Yoon SP, Lim SC, Cha MJ, Jeon YJ, Park SG, Chang IY, You HJ. Colon cancer progression is driven by APEX1-mediated upregulation of Jagged. J Clin Invest: 2013;(pii):65521. doi: 10.1172/JCI65521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Yang D, Cogdell D, Du X, Li H, Pang Y, Sun Y, Hu L, Sun B, Trent J, et al. APEX1 gene amplification and its protein overexpression in osteosarcoma: Correlation with recurrence, metastasis, and survival. Technol Cancer Res Treat. 2010;9:161–169. doi: 10.1177/153303461000900205. [DOI] [PubMed] [Google Scholar]
- 25.Puglisi F, Aprile G, Minisini AM, Barbone F, Cataldi P, Tell G, Kelley MR, Damante G, Beltrami CA, Di Loreto C. Prognostic significance of Ape1/ref-1 subcellular localization in non-small cell lung carcinomas. Anticancer Res. 2001;21:4041–4049. [PubMed] [Google Scholar]
- 26.Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: Its functions and alterations in human cancer. Apoptosis. 2004;9:667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- 27.Engelman JA. Targeting PI3K signalling in cancer: Opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 28.Lv X, Li CY, Han P, Xu XY. MicroRNA-520a-3p inhibits cell growth and metastasis of non-small cell lung cancer through PI3K/AKT/mTOR signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:2321–2327. doi: 10.26355/eurrev_201804_14822. [DOI] [PubMed] [Google Scholar]
- 29.Tsurutani J, Fukuoka J, Tsurutani H, Shih JH, Hewitt SM, Travis WD, Jen J, Dennis PA. Evaluation of two phosphorylation sites improves the prognostic significance of Akt activation in non-small-cell lung cancer tumors. J Clin Oncol. 2006;24:306–314. doi: 10.1200/JCO.2005.02.4133. [DOI] [PubMed] [Google Scholar]
- 30.Testa JR, Bellacosa A. AKT plays a central role in tumorigenesis. Proc Natl Acad Sci USA. 2001;98:10983–10985. doi: 10.1073/pnas.211430998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruhn MA, Pearson RB, Hannan RD, Sheppard KE. AKT-independent PI3-K signaling in cancer-emerging role for SGK3. Cancer Manag Res. 2013;5:281–292. doi: 10.2147/CMAR.S35178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang XX, Cheng Q, Zhang SN, Qian HY, Wu JX, Tian H, Pei DS, Zheng JN. PAK5-Egr1-MMP2 signaling controls the migration and invasion in breast cancer cell. Tumour Biol. 2013;34:2721–2729. doi: 10.1007/s13277-013-0824-x. [DOI] [PubMed] [Google Scholar]
- 33.Du Q, Liu J, Zhang X, Zhang X, Zhu H, Wei M, Wang S. Propofol inhibits proliferation, migration, and invasion but promotes apoptosis by regulation of Sox4 in endometrial cancer cells. Braz J Med Bio Res. 2018;51:e6803. doi: 10.1590/1414-431X20176803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data genarated or analyzed during the present study are included in this published article.