Abstract
The survival rate of patients with pancreatic cancer is between 3 and 5%. The neddylation pathway is overactive in multiple cancer types and is associated with poor prognosis. In recent years, the neddylation process has become a popular research target for the development of novel cancer therapies. However, the activation level of the pathway, and whether its targeting sensitizes pancreatic cancer cells to cisplatin treatment is currently unclear. In the present study, using reverse transcription-quantitative PCR and western blot analyses, the neddylation pathway was observed to be overactivated at the protein, but not the mRNA level. In addition, by analyzing The Cancer Genome Atlas data, it was demonstrated that high expression levels of NEDD8 activating enzyme E1 subunit 1 were observed to be a predictor of poor prognosis for patients with pancreatic cancer. Cisplatin enhanced the cytotoxic effects of MLN4924 both in vitro and in vivo according to Cell Counting kit-8 assays and an in vivo tumor model. Further mechanistic studies, including western blotting and immunohistochemistry assays, revealed that combined MLN4924 and cisplatin treatment induced higher levels of DNA damage by increasing the accumulation of well-defined cullin-ring ligase substrates, such as chromatin licensing and DNA replication factor 1, origin recognition complex subunit 1, p21, p27 and phosphorylated IκBα. The results of the present study support the clinical use of combined neddylation inhibitor and cisplatin treatment, which may improve the survival of, and impart other benefits for patients with pancreatic cancer.
Keywords: neddylation, MLN4924, DNA damage, pancreatic cancer
Introduction
Pancreatic cancer is the seventh leading cause of cancer-associated mortality in males and females worldwide, with high incidence rates in both sexes (1,2). Curative therapy comprises surgical resection; however, the five-year survival rate for patients remains <5% (3). In addition, the current first-line chemotherapeutic treatment, gemcitabine, has not improved the low overall survival rate (3). Therefore, there is an urgent requirement for novel therapeutic targets and strategies for the treatment of patients with pancreatic cancer.
Neddylation is a post-translational modification of proteins (4). Neural precursor cell-expressed, developmentally downregulated 8 (NEDD8) is a ubiquitin-like protein that activates the largest member of the ubiquitin E3 ligase family, cullin ring ligase (CRL) (5). Substrate-bound NEDD8 is unable to degrade proteins; however, it is able to modulate their conformation, stability, localization and function (6). The neddylation pathway has been observed to be overactivated in multiple cancer types, including liver (7), lung (8) and colon cancer (9). These previous studies suggested that neddylation serves an important role in cancer. MLN4924 (also known as pevonedistat) is a specific neddylation inhibitor that binds irreversibly to NEDD8 activating enzyme E1 subunit 1 (NAE1) (10). MLN4924 has shown promising efficacy against a variety of cancers, including liver cancer (11), lung cancer (8), cholangiocarcinoma (12) and glioblastoma (13). In addition, MLN4924 has been demonstrated to enhance the effects of autophagy inhibitors in liver cancer (14), as well as sensitizing pancreatic cells to radiotherapy by promoting DNA damage (15). Notably, MLN4924 was revealed to enhance the therapeutic effects of gemcitabine in pancreatic cancer by increasing cancer cell apoptosis (16). However, its role in modulating neddylation and its efficacy in combination regimens require further study.
In the present study, the effect of combined MLN4924 and cisplatin treatment in pancreatic cancer was investigated, and the expression profile of neddylation pathway members was examined. The results demonstrated that molecules involved in the neddylation pathway were overexpressed at the protein, but not the mRNA level, and only NAE1 overexpression was a predictor of poor prognosis in patients with pancreatic cancer. Combined MLN4924 and cisplatin treatment exhibited a marked therapeutic effect on pancreatic cancer cells both in vivo and in vitro. These mechanistic studies revealed that combined therapy suppressed pancreatic cancer cell growth by promoting DNA damage.
Materials and methods
Cell lines, culture conditions and reagents
The human pancreatic cancer cell lines BXPC3 and SW1990 (purchased from the Cell Bank of Typical Culture Preservation Commission of the Chinese Academy of Sciences) were cultured in DMEM (HyClone; GE Healthcare Life Sciences) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin solution at 37°C (5% CO2). MLN4924 and cisplatin were purchased from MedChemExpress. For in vitro studies, an MLN4924 stock solution (50 mM) was prepared in DMSO, aliquoted and stored at −20°C until use at a final concentration of 0.3 µM. For in vivo studies, MLN4924 was dissolved in 15% 2-hydroxypropyl-β-cyclodextrin (HPBCD); the solution was freshly prepared each week and stored in the dark at 4°C prior to use at 20 mg/kg. Cisplatin was used at a concentration of 0.25 µM for in vitro, and 3 mg/kg for in vivo studies.
Patients and specimens
The pancreatic cancer tissue samples used in the present study were derived from patients that underwent primary curative tumor resection at the Department of Hepatobiliary Surgery (Yongchuan Hospital, Chongqing, China) between October 2015 and February 2016. A total of 12 paired tumor and normal adjacent samples, from nine males and three females with an average age of 46 years, were used for western blotting analysis, and 42 paired tumor and normal adjacent tissues (including the 12 paired samples used for western blotting analysis), from 32 males and 10 females with an average age of 51 years, were used for reverse transcription-quantitative PCR (RT-qPCR) analysis. Patients had not received any radio- or chemotherapeutic treatment prior to surgery. Ethical approval was obtained from the Ethics Committee of Yongchuan Hospital of Chongqing Medical University (Chongqing, China) and written informed consent for tissue collection and the use of patient data for scientific purposes was obtained from all participants.
Immunohistochemical tissue staining
Subcutaneous tumor tissues from mice were immunohistochemically stained with anti-cleaved (c)-caspase3 (1:100; cat. no. 9664S, Cell Signaling Technology, Inc.) and anti-Ki-67 (1:100; cat. no. 9449S, Cell Signaling Technology, Inc.) primary antibodies. Tissue sections (5-µm) were formalin-fixed in 10% formaldehyde at 20°C for 24 h, paraffin-embedded, dehydrated and subjected to peroxidase blocking using an immunohistochemistry kit (cat. no. GK500705; Dako; Agilent Technologies, Inc.) for 5 min at room temperature according to the manufacturer's protocol. Primary antibodies were added and the sections were incubated at room temperature for 30 min. The sections were incubated for another 30 min at room temperature with the no. 2 solution of the EnVision Detection kit (cat. no. GK500705; Dako; Agilent Technologies, Inc.). Subsequently, the sections were incubated in prepared 3,3′-diaminobenzidine from the kit at room temperature for 10 min. The slides were then counterstained with hematoxylin (cat. no. S3309; Dako; Agilent Technologies, Inc.) for 30 sec at room temperature. The stained slides were observed under a light microscope (Leica DMI3000 M; magnification, ×40) and images were captured. The percentage of Ki-67-positive cells was calculated using ImageJ software (version 1.50i; National Institutes of Health).
The Cancer Genome Atlas (TGCA) database
TCGA (https://cancergenome.nih.gov/) database was used for the purposes of this study. The dataset was analyzed using UALCAN (www.ualcan.path.uab.edu).
Cell viability assessment
BXPC3 and SW1990 cells were seeded in triplicate into 96-well plates at a density of 1×103 cells/well. The cells were subsequently treated with DMSO, MLN4924, cisplatin or MLN4924 + cisplatin at the indicated doses (0.3 µM MLN; 0.5 µM cisplatin) for 48 h. Cell viability was assessed using the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) and the ATPlite™ kit (PerkinElmer, Inc.) according to the manufacturer's instructions.
Clonogenic assay
BXPC3 and SW1990 cells were trypsinized and seeded in triplicate in 6-well plates (300 cells/well). Following overnight culture at 37°C (5% CO2), the cells were incubated with DMSO (dilution, 1:1,000), MLN4924, cisplatin or MLN4924 + cisplatin (0.3 µM MLN; 0.5 µM cisplatin) for 12 days. Colonies were fixed with 4% paraformaldehyde at room temperature for 30 min and stained with 0.4% crystal violet solution in methanol for 30 min at room temperature, images were captured using a camera, and those with >30 cells were counted by three independent investigators. Representative results of 3 independent experiments are presented.
RT-qPCR analysis
Total RNA was isolated from cells and tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions, and all samples were then treated with RNase-free DNase (Ambion; Thermo Fisher Scientific, Inc.). The RT reaction was performed using 2 µg total RNA/sample with the PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol. PCR amplification was performed using the Power SYBR® Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) and the ABI 7900 thermocycler (Applied Biosystems; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. The thermocycling conditions were as follows: 95°C for 2 min; followed by 40 cycles of 95°C for 10 sec, 60°C for 30 sec and 72°C for 30 sec; dissociation curve (95°C for 15 sec, 60°C for 15 sec and 95°C for 15 sec). The primer sequences were as follows: Human NAE1, forward, 5′-GCTGTTGTCATACTTCTC-3′, and reverse, 5′-TCGTGGACATAATCATCT-3′; human ubiquitin like modifier activating enzyme 3 (UBA3), forward, 5′-TGGTGTTGGTGCTTGTAA-3′, and reverse, 5′-GTTGTATGCTTCCTCCTCTAA-3′; NEDD8-conjugating enzyme Ubc12 (UBC12), forward, 5′-CAGAAGAAGGAGGAGGAGTC-3′, and reverse, 5′-GAAGTTGAGGAGGTCGTCTG-3′; β-actin, forward, 5′-CATGTACGTTGCTATCCAGGC-3′, and reverse, 5′-CTCCTTAATGTCACGCACGAT-3′. The data were quantified using the 2−ΔΔCq method (17) and the results are presented as relative expression levels.
Western blot analysis
Cell and tissue lysates were prepared using RIPA buffer (Beyotime Institute of Biotechnology), and protein concentrations were determined using an enhanced bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology). Protein samples (50 µg/lane) were separated using SDS-PAGE on a 12% gel, then proteins were transferred to PVDF membranes, blocked with 5% BSA (cat. no. ST023; Beyotime Institute of Biotechnology) in TBS with Tween-20 (dilution, 1:1,000) for 2 h at room temperature, and probed using primary antibodies against histone γ-H2AX (1:1,000; cat. no. 9718; Cell Signaling Technology, Inc.), chromatin licensing and DNA replication factor 1 (CDT1; 1:1,000; cat. no. 8064S; Cell Signaling Technology, Inc.), origin recognition complex subunit 1 (ORC1; 1:1,000; cat. no. 4731S; Cell Signaling Technology, Inc.), phosphorylated (p)-IκBα (S32; 1:1,000; cat. no. D155066-0100; Sangon Biotech Co., Ltd.), NAE1 (1:1,000; cat. no. ab187142; Abcam), UBA3 (1:1,000; cat. no. ab124728; Abcam), UBC12 (1:1,000; cat. no. ab109507; Abcam), p21 (1:1,000; cat. no. ab109520; Abcam), p27 (1:1,000; cat. no. ab32034; Abcam), GAPDH (1:2,000; cat. no. ab181602, Abcam), p-checkpoint kinase (CHK)1 (1:1,000; cat. no. ab58567; Abcam), p-CHK2 (1:1,000; cat. no. ab32148; Abcam), c-poly (ADP-ribose) polymerase 1 (PARP; 1:1,000; cat. no. ab32561; Abcam), c-caspase-3 (1:1,000; cat. no. 9664; Cell Signaling Technology, Inc.), CHK1 (1:1,000; cat. no. ab40866; Abcam), CHK2 (1:1,000; cat. no. ab47433; Abcam), H2AX (1:1,000; cat. no. ab11175; Abcam), IκBα (1:1,000; cat. no. ab32518; Abcam) and cullin 1 (1:1,000; cat. no. sc-135874; Santa Cruz Biotechnology, Inc.) at 4°C overnight. Subsequently, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:1,000; cat. nos. 7077, 7074 and 7076; Cell Signaling Technology, Inc.) at room temperature for 2 h. All membranes were imaged using the LAS400 system (GE Healthcare Life Sciences) and an enhanced chemiluminescent kit (cat. no. 32106; Thermo Fisher Scientific, Inc.). The results were quantified using ImageJ software (version 1.50i; National Institutes of Health) with GAPDH as the loading control. All the bands presented together possessed the same loading control.
Assessment of the antitumor effects of MLN4924 and cisplatin in vivo
Male athymic nude mice (n=32; weight, 17–19 g; age, 5 weeks) were purchased from the Shanghai Experimental Animal Center. The housing conditions were as follows: Temperature, 18–29°C; daily temperature difference, ≤3°C; relative humidity, 40–70%; fresh air ventilation, 10 times/h; airflow speed, ≤0.18 m/sec; pressure difference, 25 Pa; cleanliness level, 10,000; ammonia concentration l5 mg/m3; noise, ≤60 dB; illumination, 150–300 Lux; light/dark cycle, 12 h/12 h; and free access to food and water. BXPC3 cells were trypsinized, resuspended in PBS and subcutaneously injected into the groins of the mice (5×106 cells/injection). At 1 week post-injection, the tumor-bearing mice were randomly divided into 4 groups (n=8 mice/group). When the tumors had reached an average size of 250 mm3, mice were treated by subcutaneous injection with 15% HPBCD, MLN4924 (20 mg/kg), cisplatin (2.5 mg/kg) or MLN4924 + cisplatin twice a week for 4 weeks (the largest tumor volume was 1,854 mm3, the longest tumor length was 15.94 mm). Tumor size was measured weekly using vernier calipers and tumor volume was calculated using the following formula: Volume=π/6 (length × width × height) (18). At the end of the study, the mice were sacrificed using CO2 followed by cervical dislocation (the flow rate of CO2 was 20% of the volume of the chamber/min), and tumor tissues and blood were collected. Tumor tissues were fixed with 10% formaldehyde at 20°C for 24 h. Paraffin-embedded tissues were sectioned (5-µm thick) for immunohistochemical analysis of c-caspase-3 and Ki-67. Blood samples were extracted from the tail vein and centrifuged at 500 × g for 10 min at room temperature. Serum samples were collected for the biochemical assays of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels using ELISA kits (cat. nos. MBS016898 and MBS775731; MyBioSource, Inc.) according to the manufacturer's protocols and the Modular DDP Analyzer (Roche Applied Science) according to the manufacturer's instructions. Ethical approval was obtained from the Ethics Committee of Yongchuan Hospital of Chongqing Medical University, and all procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Statistical analysis
The results from three independent experiments are presented as the mean ± standard deviation. Paired Student's t-tests were used for the comparison of parameters between two groups, and multiple comparisons were performed using one-way analysis of variance followed by the Student-Newman-Keuls post hoc test. SPSS software (version 17.0; SPSS, Inc.) was used for statistical analyses.
Results
Neddylation is overactivated in pancreatic cancer, and high NAE1 expression is an indicator of poor patient prognosis
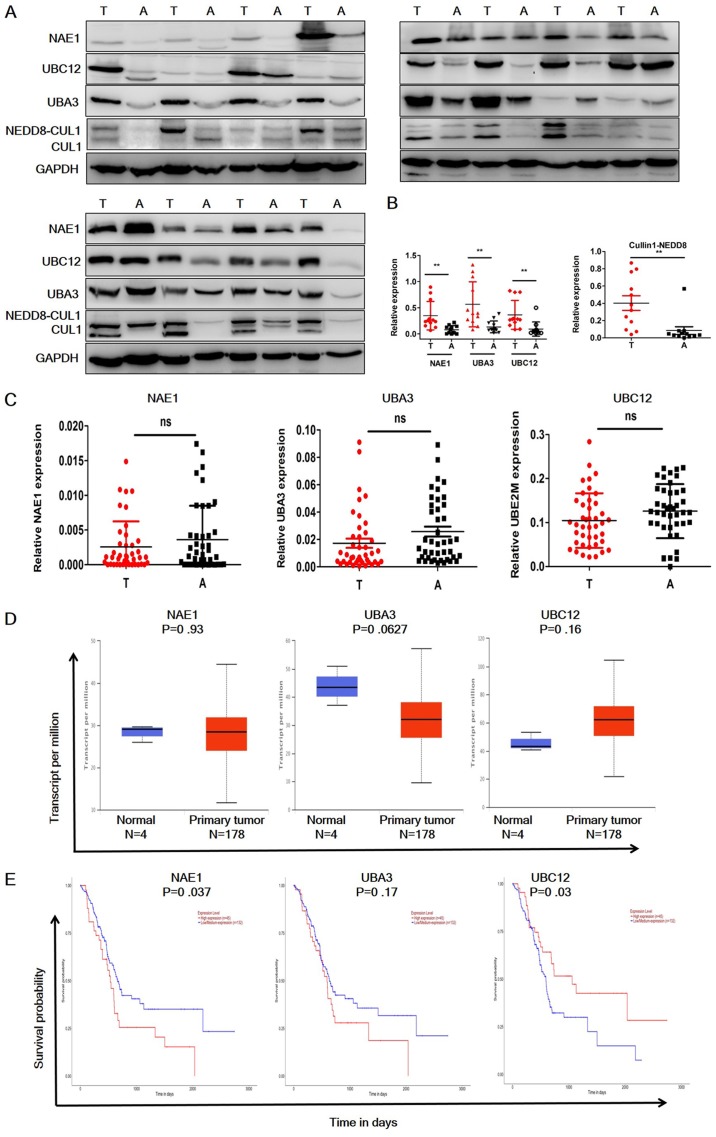
To evaluate the status of the neddylation pathway in pancreatic cancer, the protein expression levels of NAE1, UBA3 (subunits of E1), UBE12 (E2) and cullin 1 in 12 paired-pancreatic cancer and adjacent normal tissues were analyzed (Fig. 1A and B). NAE1, UBA3 and UBE12 were significantly overexpressed in pancreatic cancer samples, and neddylated cullin 1 expression was significantly higher in tumor tissues compared with the adjacent normal tissues. RT-qPCR analysis was then performed to investigate the mRNA expression levels of these molecules (Fig. 1C). In contrast to the protein expression levels, the RT-qPCR results demonstrated that NAE1, UBA3 and UBE12 mRNA expression levels were not significantly elevated in tumor tissues. These data were further confirmed by consulting TCGA datasets (Fig. 1D). UBC12 expression appeared to be elevated in the TCGA dataset; however, this was not identified as statistically significant. Notably, the TCGA datasets indicated that NAE1 overexpression was a predictor of poor prognosis in patients with pancreatic cancer, whereas high expression levels of UBA3 were not a predictor of poor prognosis (Fig. 1E), and low UBC12 expression predicted a poor prognosis (Fig. 1E). These data indicated that neddylation pathway members are only overexpressed at the protein level in pancreatic cancer, and high NAE1 and low UBC12 expression may be indicators of poor patient prognosis.
Figure 1.
Expression profile of neddylation pathway members in pancreatic cancer tissues. (A) Western blot analysis of the expression levels of neddylation pathway molecules in pancreatic cancer and adjacent normal tissues (n=12). (B) Quantification of western blot results. A total of 12 cancer and normal adjacent tissue pairs were analyzed. (C) Reverse transcription-quantitative PCR analysis of the mRNA expression levels of neddylation pathway molecules in pancreatic cancer and adjacent normal tissues. (D) Relative mRNA expression levels of neddylation pathway molecules in pancreatic cancer and adjacent normal tissues in The Cancer Genome Atlas dataset. (E) Kaplan-Meier plots showing the survival of patients with pancreatic cancer, grouped by high and low mRNA expression levels. The results are presented as the mean ± standard deviation. **P<0.01. T, tumor; A, adjacent; NAE-1, NEDD8 activating enzyme E1 subunit; UBA3, ubiquitin like modifier activating enzyme 3; UBC12, NEDD8-conjugating enzyme Ubc12; NEDD8, Neural precursor cell-expressed, developmentally downregulated 8; CULl, cullin 1.
MLN4924 enhances the therapeutic effect of cisplatin in pancreatic cancer cells in vitro
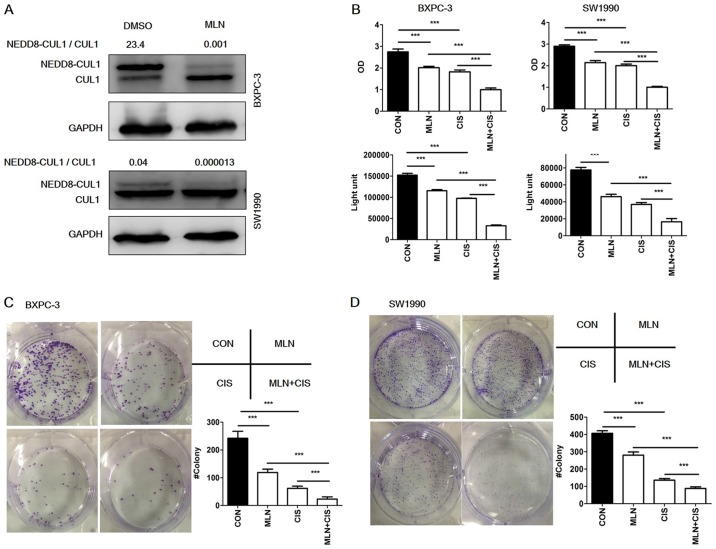
Overactivation of the neddylation pathway in pancreatic cancer indicates that it may present a potential therapeutic target. To evaluate the effects of combined MLN4924 and cisplatin treatment in pancreatic cancer cells, inhibition of neddylation using MLN4924 was first demonstrated (Fig. 2A). To investigate the effect of combined MLN4924 and cisplatin treatment on the proliferation of pancreatic cancer cells, BXPC3 and SW1990 cells were then treated with MLN4924, cisplatin or MLN4934 + cisplatin at the indicated doses (0.3 µM MLN; 0.5 µM cisplatin) for 48 h, and analyzed using ATPlite and CCK-8 cell viability assays. As presented in Fig. 2B, MLN4924 or cisplatin treatment alone significantly suppressed the proliferation of both cell lines, and the combined administration of these agents significantly further suppressed proliferation compared with monotherapy. Cell clonogenic survival analysis revealed comparable results (Fig. 2C and D), indicating that MLN4924 suppresses the proliferation of pancreatic cancer cells in vitro, and that combined MLN4924 + cisplatin treatment demonstrates a greater effect than treatment with each agent alone.
Figure 2.
MLN4924 + cisplatin treatment suppresses pancreatic cancer proliferation in vitro. (A) MLN4924 inhibited the neddylation of CUL1 in pancreatic cancer cells. Cells were treated with MLN4924 for 24 h and analyzed by western blotting. (B) Effects of MLN4924 + cisplatin treatment on the viability of BXPC3 and SW1990 cells in vitro. Cells were treated with DMSO, MLN4924, cisplatin or MLN4924 + cisplatin for 48 h (0.3 µM MLN; 0.5 µM cisplatin) and viability was assessed using ATPlite and Cell Counting Kit-8 assays, respectively. (C and D) Effects of MLN4924 + cisplatin treatment on clonogenic survival. BXPC3 and SW1990 cells were treated with DMSO, MLN4924, cisplatin or MLN4924 + cisplatin for 12 days (0.3 µM MLN; 0.5 µM cisplatin). All experiments were repeated 3 times and the results are presented as the mean ± standard deviation. ***P<0.001. DMSO, dimethyl sulfoxide; CON, control; MLN, MLN4924; CIS, cisplatin; NEDD8, Neural precursor cell-expressed, developmentally downregulated 8; CULl, cullin 1.
MLN4924 enhances the therapeutic effect of cisplatin in pancreatic cancer in vivo
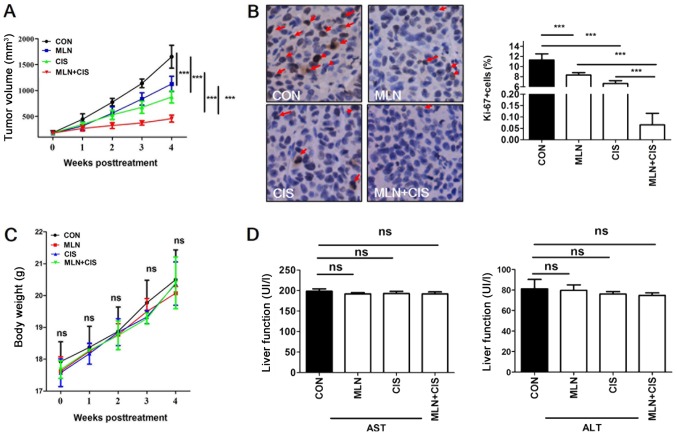
To evaluate the antitumor effects of MLN4924 + cisplatin treatment in vivo, a mouse BXPC3 ×enograft tumor model was established and mice were treated with MLN4924, cisplatin or MLN4924 + cisplatin. Compared with monotherapy, MLN4924 + cisplatin significantly inhibited tumor growth (Fig. 3A). In addition, positive Ki-67 staining was significantly decreased in the MLN4924 + cisplatin group compared with the monotherapy and control groups (Fig. 3B). These results indicate that the proliferation of pancreatic tumor cells in the MLN4924 + cisplatin-treated group was significantly suppressed. During the treatment period, no obvious side effects, such as weight loss or reduced liver function were observed (Fig. 3C and D). These results demonstrated that MLN4924 + cisplatin suppressed pancreatic tumor growth in vivo.
Figure 3.
MLN4924 + cisplatin treatment suppresses pancreatic cancer cell proliferation in vivo. (A) Mouse subcutaneous BXPC3 ×enograft tumor models were treated with MLN4924 + cisplatin twice per week, and tumor size was determined using calipers once per week. (B) Representative immunohistochemical staining analysis images of Ki-67 expression in xenograft tumors (magnification, ×400). Positively stained cells are indicated with arrows and quantified. (C) Weights of the mice were measured twice per week during the treatment period and no obvious changes were observed. (D) Liver function of treated mice was determined by measuring the serum levels of ALT and AST. The results are presented as the mean ± standard deviation. ***P<0.001. CON, control; MLN, MLN4924; CIS, cisplatin; ALT, alanine aminotransferase; AST, aspartate aminotransferase; ns, not significant.
MLN4924 enhances the tumor-suppressive effects of cisplatin by inducing DNA damage and apoptosis
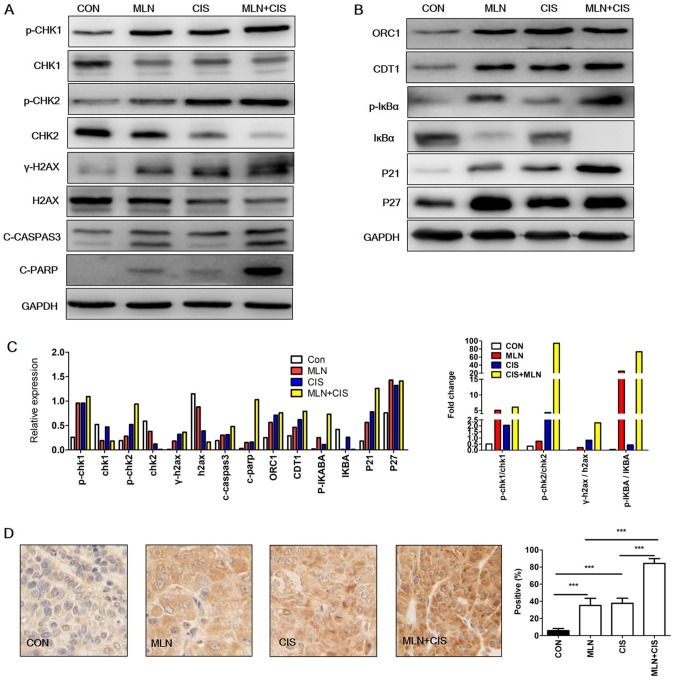
MLN4924 is a promising anticancer agent as it induces the accumulation of CRL substrates that promote DNA damage, cell apoptosis and autophagy (6). Cisplatin also demonstrates anticancer effects via the induction of DNA damage. It was therefore hypothesized that the enhanced anticancer effects of MLN4924 combined with cisplatin may be due to increased levels of DNA damage resulting from the accumulation of CRL substrates. To test this hypothesis, pancreatic cancer cells were treated with MLN4924, cisplatin or MLN4924 + cisplatin. The DNA damage markers p-CHK 1, p-CHK2 and γ-H2AX were subsequently analyzed. As indicated in Fig. 4A and C, p-CHK1, p-CHK2 and γ-H2AX expression levels were markedly increased in the MLN4924 + cisplatin-treated group compared with the monotherapy and control groups. These results indicated that MLN4924 + cisplatin treatment enhanced DNA damage in pancreatic cancer cells.
Figure 4.
Enhanced DNA damage and accumulation of CRL substrates results in cell apoptosis. (A and B) Western blot analysis of BXPC3 cells treated with MLN4924 + cisplatin for 24 h. (C) Quantification of western blotting and the ratio of p-/total protein. Each column represents a single quantification. (D) Representative immunohistochemical staining images of C-caspase-3 expression in xenograft tumors (magnification, ×400). The results are presented as the mean ± standard deviation. ***P<0.001. C-caspase-3, cleaved caspase-3; CRL, cullin ring ligase; CON, control; MLN, MLN4924; CIS, cisplatin; CHK, checkpoint kinase; γ-H2AX, phospho-histone H2AX; C-PARP, cleaved-poly (ADP-ribose) polymerase 1; ORC1, origin recognition complex subunit 1; p-, phosphorylated; CDT1, chromatin licensing and DNA replication factor 1.
In order to investigate the mechanisms underlying increased DNA damage following MLN4924 + cisplatin treatment, the levels of CRL substrates (known to induce DNA damage) were determined. As presented in Fig. 4B and C, accumulation of the DNA replication license proteins CDT1 and ORC1 was observed in the MLN4924 + cisplatin group compared with in the control group. Similarly, a marked increase in the CRL substrates p27, p-IκBα and p21 was observed in the MLN4924 + cisplatin treatment group. These results indicated that the enhanced DNA damage caused by MLN4924 + cisplatin treatment may be due to the increased accumulation of CRL substrates.
It has been reported that DNA damage induces apoptosis (19). Given that MLN4924 + cisplatin enhanced DNA damage in pancreatic cancer cells in the present study, it was hypothesized that this treatment may also lead to increased apoptosis. As shown in Fig. 4A and C, an increase in the expression level of apoptosis markers c-caspase-3 and C-PARP was observed, compared with the monotherapy and the control groups. Similarly, immunohistochemical staining analysis of c-caspase-3 expression in mouse xenograft tumors revealed that the levels of this apoptosis-associated protein were also significantly increased (Fig. 4D). These results indicate that MLN4924 + cisplatin enhanced DNA damage and induced the expression of apoptosis-associated proteins.
Discussion
Pancreatic cancer is a human malignancy associated with high mortality rates. Despite recommended treatment with gemcitabine in the clinic, the 5-year survival rates for patients remain low worldwide (1). Overactivation of the neddylation pathway has been observed in specific malignancies and indicates poor prognosis (6,8,12,13). However, to the best of our knowledge, its mRNA expression in pancreatic cancer had not been previously studied. A previous study demonstrated that neddylation pathway-associated molecules, such as NAE1, UBA3 and UBC12 were overexpressed at the protein level in pancreatic cancer tissues when compared with adjacent normal tissues (8). In addition, the levels of neddylated cullin 1, a known neddylation substrate (8) were observed to be markedly increased in pancreatic cancer tissue samples when compared with adjacent normal tissues. However, the results of the present study demonstrated that no differences in the mRNA expression levels of these molecules were observed in pancreatic cancer tissues compared with adjacent normal tissues. UBC12 appeared to be elevated in the TCGA dataset; however, this was not statistically significant. In addition, by consulting TCGA database, only NAE1 overexpression was demonstrated to be a predictor of poor patient prognosis in pancreatic cancer. This contradicts previous studies involving lung cancer and cholangiocarcinoma (8,12), but is consistent with the results presented by Li et al (16). However, low UBC12 expression predicted a poor prognosis, which was inconsistent with a previous study (8). UBC12 may not only function as an E2 of the neddylation process. The potential mechanism requires further investigation in the future.
Neddylation inhibitors have demonstrated promising anticancer effects in certain malignancies by inducing DNA damage, apoptosis, cell cycle arrest, cell senescence and autophagy (11,20–23). Combined MLN4924 and gemcitabine treatment has been demonstrated to significantly suppress pancreatic cancer growth by inducing apoptosis, via the elevation of NADPH oxidase activator 1 expression in vitro and in vivo (16). In addition, combined MLN4924 and radiotherapy treatment was observed to significantly inhibit pancreatic cancer growth by promoting DNA damage (15). As MLN4924 and cisplatin are both known to induce DNA damage and are used in clinical practice for the treatment of specific malignancies, the effects of combined MLN4924 and cisplatin treatment on pancreatic cancer cells were investigated in the present study. This hypothesis was verified in esophageal squamous cell carcinoma (24). The results demonstrated that MLN4924 + cisplatin treatment significantly suppressed pancreatic cancer growth both in vitro and in vivo, and that combined treatment was well tolerated by the mice.
As MLN4924 + cisplatin therapy induced significant effects on the proliferation of pancreatic cancer cells, mechanistic studies were subsequently performed. The results demonstrated that MLN4924 + cisplatin-mediated suppression of cell proliferation may be related to enhanced DNA damage and increased levels of apoptosis-associated proteins. This was consistent with a previous study involving combined MLN4924 and radiotherapy treatment (15). Additionally, inhibition of CHK1 (which is involved in the DNA damage response) using a specific inhibitor (SCH 900776) was also observed to increase the anticancer activity of MLN4924 in pancreatic cancer cells (25). MLN4924 induces DNA damage by promoting the accumulation of the DNA replication license proteins CDT1 and ORC1, as well as additional CRL substrates (6). Consistent with these observations, a significant accumulation of CDT1, ORC1, p27, p-IκBα and p21 following MLN4924 + cisplatin treatment was observed in pancreatic cancer cells.
The constitutive photomorphogenesis 9 (COP9) signalosome (CSN) complex was revealed to be responsible for the de-neddylation of Cul family proteins (26). CSN complex subunit 5 is a key component of the CSN complex (28), which was found to be overexpressed in a number of cancer types, including pancreatic cancer (27,28). As neddylation and de-neddylation enzymes are both overexpressed in cancer, it was speculated that the neddylation dynamics were tightly regulated. Hua et al (13) demonstrated that the neddylation pathway was overactivated in glioblastoma and that MLN4924 suppressed GBM cell (U251 and A172) growth significantly; however, neddylated cul1 is expressed to a greater degree in U251 cells than A172 cells. In the present study, the levels of neddylated Cul1 varied between BXPC3 and SW1990 cells; however, this did not alter the effects of MLN4924. These results were consistent with another glioblastoma study (13).
In conclusion, the results of the present study indicated that the neddylation pathway is overactivated in pancreatic cancer, and that the expression levels of key molecules within this pathway are upregulated. In addition, MLN4924 + cisplatin treatment significantly suppressed pancreatic cancer cell growth in vitro and in vivo. MLN4924 + cisplatin treatment may suppress pancreatic cancer cell proliferation by promoting DNA damage and increasing the expression of apoptosis-associated proteins via enhanced CDT1, ORC1, p27, p-IκBα and p21 accumulation.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Nature Science Foundation of Chongqing Yongchuan Science and Technology Commission (grant no. Ycstc-2017nc5004).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YZ, YSI and HL contributed to the study design, analysis and interpretation of the data. HL conceived the study. YZ, YSI, QHP, and YGZ performed the experiments, and YZ, YSI and YGZ performed the statistical analyses. HL drafted the manuscript and supervised the study. All of the authors approved the final manuscript.
Ethics approval and consent to participate
Ethical approval was obtained from the Research Ethics Committee of Chongqing Medical University, and written informed consent regarding tissue collection and the use of patient data for scientific purposes was obtained from all recruited patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 4.Rabut G, Peter M. Function and regulation of protein neddylation. ‘Protein modifications: Beyond the usual suspects’ review series. EMBO Rep. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enchev RI, Schulman BA, Peter M. Protein neddylation: Beyond cullin-RING ligases. Nat Rev Mol Cell Biol. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, Morgan MA, Sun Y. Targeting neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid Redox Signal. 2014;21:2383–2400. doi: 10.1089/ars.2013.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, Li L, Yu G, Ying W, Gao Q, Zhang W, Li X, Ding C, Jiang Y, Wei D, et al. The neddylation-cullin 2-RBX1 E3 ligase axis targets tumor suppressor RhoB for degradation in liver cancer. Mol Cell Proteomics. 2015;14:499–509. doi: 10.1074/mcp.M114.045211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Wang M, Yu G, Chen P, Li H, Wei D, Zhu J, Xie L, Jia H, Shi J, et al. Overactivated neddylation pathway as a therapeutic target in lung cancer. J Natl Cancer Inst. 2014;106:dju083. doi: 10.1093/jnci/dju083. [DOI] [PubMed] [Google Scholar]
- 9.Xie P, Zhang M, He S, Lu K, Chen Y, Xing G, Lu Y, Liu P, Li Y, Wang S, et al. The covalent modifier Nedd8 is critical for the activation of Smurf1 ubiquitin ligase in tumorigenesis. Nat Commun. 2014;5:3733. doi: 10.1038/ncomms4733. [DOI] [PubMed] [Google Scholar]
- 10.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 11.Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D, Pan Y, Ding C, Qian J, Wu L, et al. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012;72:3360–3371. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 12.Gao Q, Yu GY, Shi JY, Li LH, Zhang WJ, Wang ZC, Yang LX, Duan M, Zhao H, Wang XY, et al. Neddylation pathway is up-regulated in human intrahepatic cholangiocarcinoma and serves as a potential therapeutic target. Oncotarget. 2014;5:7820–7832. doi: 10.18632/oncotarget.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hua W, Li C, Yang Z, Li L, Jiang Y, Yu G, Zhu W, Liu Z, Duan S, Chu Y, et al. Suppression of glioblastoma by targeting the overactivated protein neddylation pathway. Neuro Oncol. 2015;17:1333–1343. doi: 10.1093/neuonc/nov066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen P, Hu T, Liang Y, Jiang Y, Pan Y, Li C, Zhang P, Wei D, Li P, Jeong LS, et al. Synergistic inhibition of autophagy and neddylation pathways as a novel therapeutic approach for targeting liver cancer. Oncotarget. 2015;6:9002–9017. doi: 10.18632/oncotarget.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei D, Li H, Yu J, Sebolt JT, Zhao L, Lawrence TS, Smith PG, Morgan MA, Sun Y. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Res. 2012;72:282–293. doi: 10.1158/0008-5472.CAN-11-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Zhou W, Li L, Wu J, Liu X, Zhao L, Jia L, Sun Y. Inhibition of neddylation modification sensitizes pancreatic cancer cells to gemcitabine. Neoplasia. 2017;19:509–518. doi: 10.1016/j.neo.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Feldman JP, Goldwasser R, Mark S, Schwartz J, Orion I. A mathematical model for tumor volume evaluation using two-dimensions. J Appl Quant Methods. 2009;4:455–462. [Google Scholar]
- 19.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: Rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116:1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 21.Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 2010;70:10310–10320. doi: 10.1158/0008-5472.CAN-10-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, Xiong X, Jia L, Sun Y. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis. 2012;3:e386. doi: 10.1038/cddis.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blank JL, Liu XJ, Cosmopoulos K, Bouck DC, Garcia K, Bernard H, Tayber O, Hather G, Liu R, Narayanan U, et al. Novel DNA damage checkpoints mediating cell death induced by the NEDD8-activating enzyme inhibitor MLN4924. Cancer Res. 2013;73:225–234. doi: 10.1158/0008-5472.CAN-12-1729. [DOI] [PubMed] [Google Scholar]
- 24.Lin S, Shang Z, Li S, Gao P, Zhang Y, Hou S, Qin P, Dong Z, Hu T, Chen P. Neddylation inhibitor MLN4924 induces G2 cell cycle arrest, DNA damage and sensitizes esophageal squamous cell carcinoma cells to cisplatin. Oncol Lett. 2018;15:2583–2589. doi: 10.3892/ol.2017.7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JA, Song C, Rong Y, Kuang T, Wang D, Xu X, Yuan J, Luo K, Qin B, Nowsheen S, et al. Chk1 inhibitor SCH 900776 enhances the antitumor activity of MLN4924 on pancreatic cancer. Cell Cycle. 2018;17:191–199. doi: 10.1080/15384101.2017.1405194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cope GA, Deshaies RJ. COP9 signalosome: A multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. doi: 10.1016/S0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- 27.Fukumoto A, Ikeda N, Sho M, Tomoda K, Kanehiro H, Hisanaga M, Tsurui Y, Tsutsumi M, Kato JY, Nakajima Y. Prognostic significance of localized p27Kip1 and potential role of Jab1/CSN5 in pancreatic cancer. Oncol Rep. 2004;11:277–284. [PubMed] [Google Scholar]
- 28.Pan Y, Claret FX. Targeting Jab1/CSN5 in nasopharyngeal carcinoma. Cancer Lett. 2012;326:155–160. doi: 10.1016/j.canlet.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.