Abstract
E3 ubiquitin-protein ligase (HUWE1) has previously been identified as a HECT domain-containing ubiquitin ligase (E3) that is involved in several signaling pathways, transcriptional regulation, neural differentiation, DNA damage responses and apoptosis. However, the function of HUWE1 in the various types of cancer remains unclear. A previous study indicated that HUWE1 exhibited different roles depending on the cancer type due to the ubiquitination of various substrates. The objective of the present study was to determine whether HUWE1 can be employed as a prognostic indicator in human cancer. The expression of HUWE1 was examined using the Oncomine database, and gene alterations during carcinogenesis, copy number alterations and mutations of HUWE1 were then analyzed using cBioPortal, which is the International Cancer Genome Consortium and the Tumorscape database. Furthermore, the association between HUWE1 expression and patient survival was evaluated using Kaplan-Meier plotter and the PrognoScan databases. In addition, the present study attempted to establish the functional association between HUWE1 expression and cancer phenotypes, and the results revealed that HUWE1 may serve as a diagnostic marker or therapeutic target for certain types of cancer. HUWE1 may serve an oncogenic role in breast, brain and prostate cancer, while it may serve an anti-oncogenic role in colorectal cancer and certain lung cancers. The function of HUWE1 and its mechanisms require more in-depth and extensive investigation in future studies.
Keywords: HECT domain-containing ubiquitin ligase, cancer, diagnostic marker, therapeutic target
Introduction
Tumorigenesis results from dysregulation of oncogenes and tumor suppressors that influence cellular proliferation, differentiation, senescence or apoptosis (1). Protein degradation is essential for homeostasis and the survival of cells (2). Ubiquitination targets proteins for proteasome-mediated degradation by the covalent attachment of one or more ubiquitin molecules to a lysine residue (2–4). In addition to regulating protein turnover, this post-translational modification contributes to other cellular processes, including the regulation of protein trafficking and subcellular distribution, signal transduction, cell cycle, apoptosis and DNA repair (5–7).
Protein ubiquitination is performed by a trio of enzymes, termed ubiquitin-activating enzyme, ubiquitin-conjugating enzyme and ubiquitin protein ligase (E3). The E3 ligases determine the substrate specificity of the ubiquitination reactions (3,6,8). E3 ubiquitin ligases are a large family of proteins that are classified into three major structurally distinct types: N-end rule E3s; E3s containing the homology to E6AP C-terminus (HECT) domain; and E3s with a Really Interesting New Gene finger domain, including its derivatives, the Plant Homeo-Domain and the U-Box (9). HECT E3s were first reported in 1995, as the first E3s described (2). HECT E3s serve important roles in sporadic and hereditary human diseases, including cancer, cardiovascular (Liddle's syndrome) and neurological (Angelman syndrome) disorders, and in disease-relevant processes, including bone homeostasis, immune responses and retroviral budding (10).
E3 ubiquitin-protein ligase (HUWE1) is a recently-identified 500 kDa HECT domain-containing E3 ligase, that serves a critical role in proteasomal degradation of several proteins, including p53, c-Myc, myeloid cell leukemia sequence 1 (Mcl-1), cell division cycle 6 (Cdc6), histones, N-Myc, Msx-interacting-zinc finger 1(Miz1), DNA topoisomerase 2-binding protein 1, DNA polymerase β, mitofusin 2, histone deacetylase 2 and Ras association domain family member 1 (1,11–21). However, whether any of these existing or other unidentified substrates mediate the major function of HUWE1 in cell proliferation remains unclear. One study demonstrated that silencing HUWE1 increased apoptosis by ubiquitination and degradation of Mcl-1, while another study reported that silencing HUWE1 increased survival by ubiquitination and degradation of p53 in the same cell line (1,6,14). Experiments performed in a HDM2-null genetic background confirmed that ARF-induced stabilization of p53 involved HUWE (22). However, other data could not demonstrate the inhibitory activity of ARF toward HUWE1 (15). Furthermore, HUWE1 was incapable of regulating p53 abundance in response to DNA-damage stress, while other substrates, including Mcl-1 and Cdc6, were ubiquitylated and degraded (16). In addition, the steady-state protein levels of p53 were not increased by depletion of HUWE1 in neuroblastoma cells (6,21). Another study demonstrated that HUWE1 assembled Lysine (Lys) 63-linked polyubiquitin chains on c-Myc and this modification was revealed to be required for gene activation by c-Myc (15). However, other data revealed that HUWE1 ubiquitylates N-Myc via Lys48-mediated linkages and targets it for destruction by the proteasome (21), and post-translational modifications of c-Myc do not appear to be required for its interaction with p300 (23,24).
Previous studies have indicated that HUWE1 may either serve an oncogenic or anti-oncogenic function depending on the type of cancer (14,25,26). In addition, it has been demonstrated that HUWE1 serves important roles in proteasomal degradation of several proteins in various types of cancer (1,11–21). However, the precise role of HUWE1 expression remains controversial due to conflicting evidence. Notably, the HUWE1 gene exhibits an important role in various types of tumor, including lung, breast and colorectal carcinomas (1,8,14,15). Compared with normal tissue the expression levels of HUWE1 are higher in numerous types of primary human tumors, predominantly solid tumor, including lung, breast, colon and prostate carcinoma, as well as glioblastoma (14). By contrast, HUWE1 is undetectable or expressed at very low levels in normal epithelium, in benign polyps, and pancreatic cancer (14,27). These results suggest that HUWE1 may serve an important role depending on the type of cancer.
In order to analyze the expression of HUWE1 in various types of cancer and to investigate the molecular mechanism of HUWE1 in the regulation of tumor development, the present study evaluated HUWE1 expression using the Oncomine database. Gene alterations during carcinogenesis, copy number alterations and mutations of HUWE1 were also examined using cBioPortal, which is the International Cancer Genome Consortium and the Tumorscape database.
Materials and methods
Oncomine database analysis
Expression levels of HUWE1 in cancer vs. normal tissues were analyzed using the ‘cancer vs. normal’ filter in the Oncomine database (https://www.oncomine.org/resource/login.html) (28,29). All data conforming to the criteria of P<0.01, fold-change >2 and a gene rank percentile <10%, were included in the present study (20–33). The advanced analysis criteria were adjusted as follows: P<0.0001, fold-change >2 and gene ranking in the top 10%. A heat map was used to present the expression profile of HUWE1 in various cancer types.
Kaplan-Meier analysis
The Kaplan-Meier plotter database (http://kmplot.com/analysis/) (34) was used to assess the effect of 54,675 genes on survival using 10,461 cancer samples. This included 5,143 breast, 1,816 ovarian, 2,437 lung and 1,065 gastric cancer cases with a mean follow-up of 69, 40, 49 and 33 months, respectively. The primary purpose of the tool is for meta-analysis-based biomarker assessment. The association between the expression levels of HUWE1 and survival rates in breast, gastric, ovarian and lung cancer were analyzed using the Kaplan-Meier plotter. The hazard ratio with a 95% confidence interval and log rank P-value were calculated.
Prognoscan database analysis
The PrognoScan database (http://dna00.bio.kyutech.ac.jp/PrognoScan/) (35) was searched to determine the association between the expression levels of HUWE1 and survival rates in various cancer types. The threshold was adjusted to Cox P<0.05.
Identifying the proteins that interact with HUWE1
The search tool for the retrieval of interacting genes/proteins (STRING v.10) analysis tool (https://string-db.org/) was used to identify interacting proteins, with ‘HUWE1 (Homo sapiens)’ used as the query. Several previously identified partners have been genetically verified and therefore served as the foundation for revealing other protein partners in the network. If any identified proteins were not specific to the HUWE1 network, they were excluded from the gene signature (36).
cBioPortal database analysis
The cBioPortal (http://cbioportal.org) was used to investigate mutations and copy number alterations (CNAs) of the HUWE1 gene and the predicted protein partners in various cancer types. The cBioPortal is a website used for investigating, visualizing and analyzing multidimensional cancer genomics data (37,38). The threshold criteria of studies were dataset ≥100 samples and samples with >20% alteration frequency (37).
Statistical analysis
The Prognoscan and Kaplan-Meier plots were used to generate survival curves. All results were reviewed with a P-value from a log-rank test, and with Oncomine and heat maps. Oncomine reported the statistical significance with a P-value.
Results
HUWE1 transcript expression by cancer type
To investigate the function of HUWE1 in different cancer types, the Oncomine database was used to compare HUWE1 expression levels between tumor and normal tissues. The HUWE1 mRNA expression levels in the tissue of origin were selected and compared using the ‘cancer vs. normal’ filter. For inclusion and further evaluation, data matching the following criteria were selected: P<0.01 and fold-change >2, or P<0.0001 and fold-change >2. Statistical analyses, including P-values, two-tailed Student's t-test and multiple testing corrections, were performed using the Oncomine default algorithms. Compared with normal tissue, HUWE1 was overexpressed in various tumor types, while it exhibited lower expression levels in other tumor types. These results indicated that HUWE1 may serve either an oncogenic or tumor suppressor function depending on the cancer type (Fig. 1). Accordingly, further detailed analyses of HUWE1 were performed.
Figure 1.
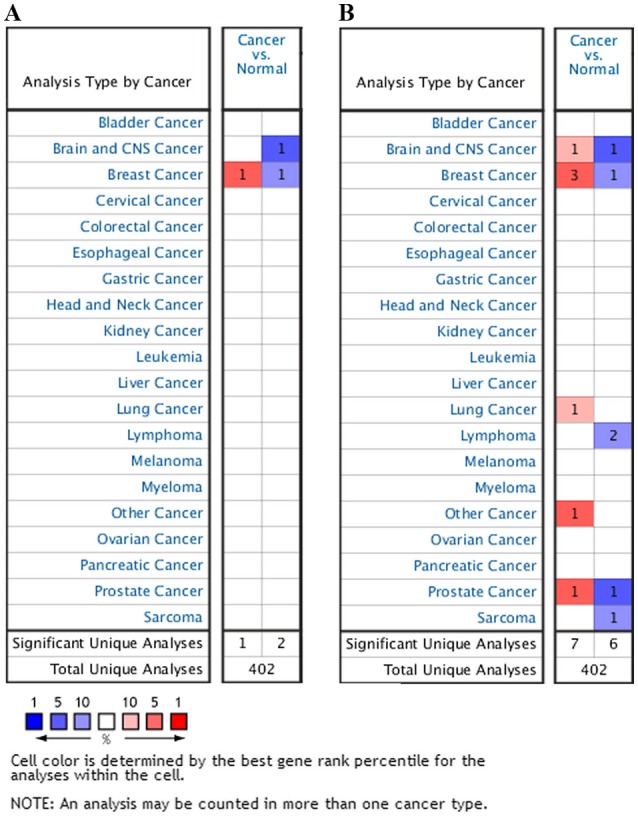
HUWE1 mRNA expression levels in various cancer types compared with normal tissue controls. The left column in red indicates the number of datasets in which the expression level of HUWE1 was upregulated and the right column in blue represents the number of datasets in which the expression level of HUWE1 was downregulated in cancer tissues compared with normal tissues. The search criteria threshold was set at: (A) P<0.0001, fold-change >2 and a gene rank percentile of 10%, and (B) P<0.001, fold-change >2 and a gene rank percentile of 10%. HUWE1, E3 ubiquitin-protein ligase.
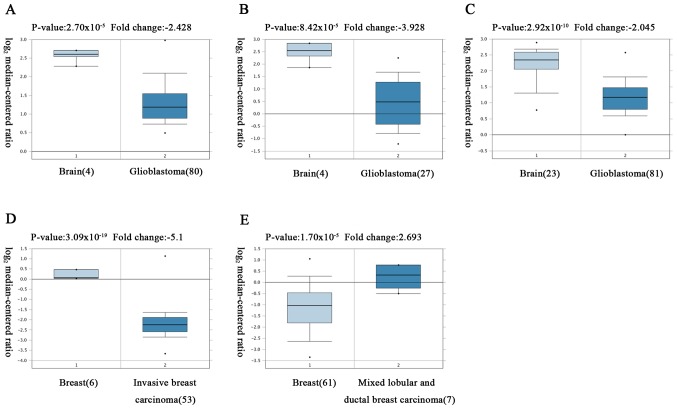
The analysis revealed that HUWE1 was overexpressed in leukemia and lung cancer, but was under expressed in glioblastoma, lymphoma, sarcoma and testicular seminoma tissues compared with normal tissues
However, the expression of HUWE1 in breast and prostate cancers remains controversial (Fig. 2; Table I).
Figure 2.
Analysis of HUWE1 in different cancer types according to the Oncomine database. HUWE1 expression levels in normal (left column) and cancer tissue (right column) were obtained from the Oncomine database. The fold-change of HUWE1 in different types of cancer was identified from the analysis. (A) The fold-change of HUWE1 in glioblastoma compared with normal brain tissues from Murat Brain Statistics dataset. (B) The fold-change of HUWE1 in glioblastoma compared with normal brain tissues from Bredel Brain 2 Statistics dataset. (C) The fold-change of HUWE1 in glioblastoma compared with normal brain tissues from Sun Brain Statistics dataset. (D) The fold-change of HUWE1 in invasive breast carcinoma compared with normal breast tissue from Finak Breast Statistics dataset. (E) The fold-change of HUWE1 in mixed lobular and ductal breast carcinoma compared with normal breast tissue from TCGA Breast Statistics dataset. HUWE1, E3 ubiquitin-protein ligase.
Table I.
Association of HUWE1 expression with survival of patients with cancer.
| Cancer type | n | P-value | HR (95% CI) | Endpoint | Dataset | Probe ID | ln (HR) |
|---|---|---|---|---|---|---|---|
| Bladder cancer | 30 | 3.05×10−2 | 3.42 (1.12–10.42) | Overall survival | GSE5287 | 208598_s_at | 1.2296 |
| Brain cancer | 74 | 3.16×10−2 | 0.23 (0.06–0.88) | Overall survival | GSE4412-GPL96 | 208598_s_at | −1.4783 |
| Breast cancer | 158 | 2.07×10−2 | 3.69 (1.22–11.15) | Overall survival | GSE3143 | 34372_at | 1.3054 |
| Breast cancer | 204 | 2.68×10−2 | 0.47 (0.24–0.92) | Relapse free survival | GSE12276 | 207783_x_at | −0.7499 |
| Breast cancer | 77 | 2.47×10−2 | 0.01 (0.00–0.57) | Relapse free survival | GSE9195 | 207783_x_at | −4.4615 |
| Breast cancer | 77 | 2.07×10−2 | 0.01 (0.00–0.45) | Distant metastasis free survival | GSE9195 | 207783_x_at | −5.2755 |
| Breast cancer | 136 | 3.91×10−3 | 0.26 (0.10–0.65) | Distant metastasis free survival | GSE12093 | 208598_s_at | −1.3632 |
| Breast cancer | 200 | 1.84×10−2 | 1.54 (1.08–2.21) | Distant Metastasis Free survival | GSE11121 | 208599_at | 0.4321 |
| Breast cancer | 117 | 3.52×10−2 | 7.17 (1.15–44.82) | Distant metastasis free survival | E-TABM-158 | 208599_at | 1.9697 |
| Breast cancer | 236 | 1.38×10−2 | 1.57 (1.10–2.24) | Disease specific survival | GSE3494-GPL96 | 214673_s_at | 0.44818 |
| Breast cancer | 236 | 1.25×10−2 | 5.04 (1.42–17.91) | Disease specific survival | GSE3494-GPL96 | 208598_s_at | 1.6168 |
| Breast cancer | 249 | 4.97×10−3 | 4.08 (1.53–10.89) | Disease Free Survival | GSE4922-GPL96 | 208598_s_at | 1.4066 |
| Breast cancer | 198 | 7.58×10−3 | 0.58 (0.39–0.87) | Distant metastasis free survival | GSE7390 | 207783_x_at | −0.5376 |
| Breast cancer | 198 | 5.87×10−3 | 0.57 (0.39–0.85) | Overall survival | GSE7390 | 207783_x_at | −0.5539 |
| Breast cancer | 198 | 2.54×10−2 | 0.63 (0.43–0.95) | Relapse free survival | GSE7390 | 207783_x_at | −0.4546 |
| Colorectal cancer | 177 | 3.56×10−2 | 0.22 (0.05–0.90) | Overall survival | GSE17536 | 214673_s_at | −1.5180 |
| Colorectal cancer | 55 | 3.20×10−4 | 0.01 (0.00–0.11) | Overall survival | GSE17537 | 214673_s_at | −4.8093 |
| Colorectal cancer | 55 | 2.01×10−4 | 0.00 (0.00–0.06) | Disease free survival | GSE17537 | 214673_s_at | −6.1017 |
| Colorectal cancer | 55 | 2.31×10−2 | 0.02 (0.00–0.58) | Overall survival | GSE17537 | 208599_at | −3.9563 |
| Colorectal cancer | 49 | 7.78×10−3 | 0.01 (0.00–0.32) | Disease specific survival | GSE17537 | 214673_s_at | −4.2781 |
| Esophagus cancer | 34 | 4.49×10−2 | 14.93 (1.06–209.58) | Overall survival | GSE11595 | 66423 | 2.7033 |
| Lung cancer | 104 | 1.09×10−2 | 0.01 (0.00–0.32) | Overall survival | jacob-00182-MSK | 207783_x_at | −4.9303 |
| Lung cancer | 204 | 4.40×10−3 | 18.26 (2.47–134.83) | Overall survival | GSE31210 | 207783_x_at | 2.9050 |
| Lung cancer | 204 | 1.27×10−5 | 28.08 (6.28–25.48) | Relapse free survival | GSE31210 | 207783_x_at | 3.3349 |
| Lung cancer | 204 | 1.44×10−2 | 0.38 (0.17–0.82) | Overall survival | GSE31210 | 236294_at | −0.9750 |
| Lung cancer | 204 | 3.28×10−6 | 0.26 (0.14–0.45) | Relapse free survival | GSE31210 | 236294_at | −1.3652 |
| Lung cancer | 138 | 6.40×10−3 | 0.00 (0.00–0.22) | Relapse free survival | GSE8894 | 236294_at | −5.4148 |
| Lung cancer | 138 | 2.50×10−2 | 0.15 (0.03–0.79) | Relapse free survival | GSE8894 | 208599_at | −1.8913 |
| Ovarian cancer | 133 | 4.32×10−2 | 1.95 (1.02–3.72) | Overall survival | DUKE-OC | 208599_at | 0.6675 |
| Prostate cancer | 281 | 5.88×10−3 | 1.80 (1.19–2.75) | Overall survival | GSE16560 | DAP4_0152 | 0.5900 |
| Skin cancer | 38 | 2.33×10−3 | 11.36 (2.38–54.33) | Overall survival | GSE19234 | 208599_at | 2.4304 |
| Soft tissue cancer | 140 | 6.89×10−4 | 3.20 (1.64–6.27) | Distant Recurrence Free survival | GSE30929 | 208598_s_at | 1.1640 |
HUWE1, E3 ubiquitin-protein ligase; HR, hazard ratio; CI, confidence interval.
Genetic expression levels of HUWE1 and patient survival
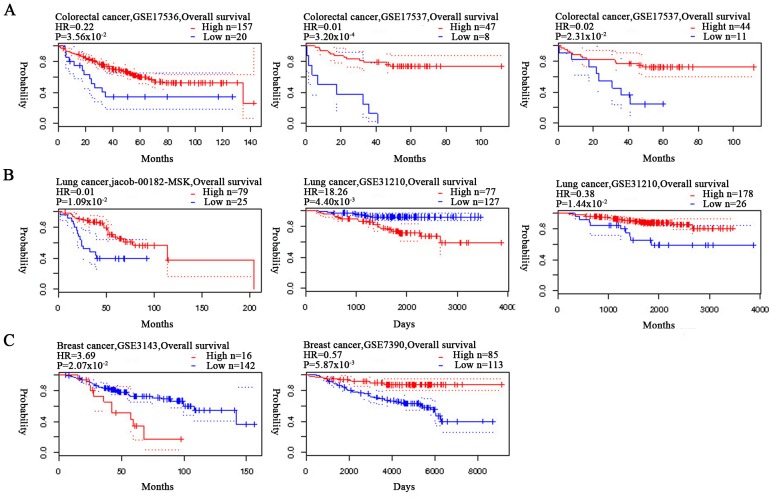
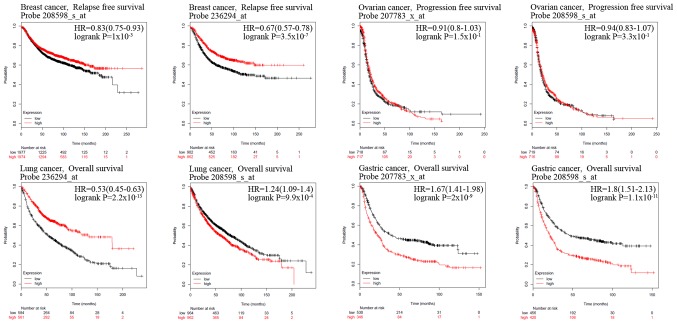
The association between the expression levels of HUWE1 and OS rate of patients with colorectal cancer was analyzed using Kaplan-Meier analysis. Patients with colorectal cancer with a high expression level of HUWE1 exhibited a significantly higher survival rate (Fig. 3). Analysis of gastric cancer revealed an association between a high expression level of HUWE1 and low OS rate. HUWE1 expression and PFS rate of patients with ovarian cancer were not statistically significant (P=1.50×10−1 and P=3.30×10−1; Fig. 4). The significance of the association of HUWE1 expression and survival rates in lung and breast cancer remains inconclusive (Figs. 3B and 4).
Figure 3.
Associations between the overall survival rates of patients with colorectal, lung and breast cancer, and HUWE1 expression levels, according to the PrognoScandatabase. Survival curves were plotted according to HUWE1 expression, comparing patients with high (red) and low (blue) expression levels. Analysis was performed for (A) colorectal, (B) lung and (C) breast cancer. HR, hazard ratio; HUWE1, E3 ubiquitin-protein ligase.
Figure 4.
Kaplan-Meier curves for patients with breast, ovarian, lung and gastric cancer according to HUWE1 expression. Kaplan-Meier curves were generated to compare the survival rates of patients with high (red) and low (black) expression levels of HUWE1. HUWE1, E3 ubiquitin-protein ligase; HR, hazard ratio.
The expression of HUWE1 was evaluated using the cBioPortal database. The data demonstrated lower expression levels of HUWE1 in brain, lymphoma and round cell liposarcoma; however, its roles in lung and breast cancer were uncertain (Table II). Despite this controversy, certain data agreed with previously published reports. Previous studies have demonstrated that HUWE1 is highly expressed in a significant proportion of lung and breast carcinomas (14,15), and lowly expressed in colorectal carcinomas; HUWE1 expression was positively correlated with tumor stage and negatively correlated with p53 protein expression (26).
Table II.
HUWE1 expression in cancer types.
| Cancer type | P-value | Fold-change | Sample size, n | (Refs.) |
|---|---|---|---|---|
| Brain | ||||
| Glioblastoma | 2.70×10−5 | −2.428 | 84 | (30) |
| Glioblastoma | 8.42×10−5 | −3.928 | 54 | (31) |
| Anaplastic oligoastrocytoma | 3.00×10−3 | −2.916 | 54 | (31) |
| Anaplastic oligodendroglioma | 2.80×10−2 | −2.667 | 54 | (31) |
| Glioblastoma | 3.20×10−2 | −3.046 | 38 | (46) |
| Oligoastrocytoma | 4.00×10−2 | −2.045 | 38 | (46) |
| Glioblastoma | 2.92×10−10 | −2.045 | 180 | (32) |
| Breast | ||||
| Mixed lobular and ductal breast carcinoma | 1.70×10−5 | 2.693 | 68 | (60) |
| Invasive breast carcinoma | 3.09×10−19 | −5.100 | 59 | (33) |
| Leukemia | ||||
| Chronic lymphocytic leukemia | 2.00×10−3 | 2.562 | 111 | (47) |
| Lung | ||||
| Squamous cell lung carcinoma | 9.00×10−3 | 3.019 | 203 | (48) |
| Small cell lung carcinoma | 2.80×10−2 | 2.137 | 73 | (59) |
| Lymphoma | ||||
| Diffuse large B-cell lymphoma | 1.00×10−2 | −3.503 | 27 | (50) |
| Marginal zone B-cell lymphoma | 8.00×10−3 | −3.615 | 27 | (50) |
| Other | ||||
| Testicular seminoma | 2.00×10−3 | 11.159 | 30 | (51) |
| Prostate | ||||
| Prostate carcinoma | 5.00×10−3 | 6.016 | 30 | (52) |
| Prostate carcinoma | 3.40×10−2 | −2.207 | 19 | (53) |
| Sarcoma | ||||
| Round cell liposarcoma | 6.00×10−3 | −3.047 | 54 | (54) |
HUWE1, E3 ubiquitin-protein ligase; TCGA, The Cancer Genome Atlas.
Proteins associated with HUWE1
Functional proteins understood to be associated with HUWE1 were selected by STRING analysis tool for the retrieval of interacting genes/proteins. The ten predicted protein partners of HUWE1 were as follows (Fig. 5): non-POU domain-containing octamer-binding protein, transmembrane protein 164, protein Jade-3 (PHF16), TGF-β activated kinase 1 (TAB3), TPP synthase 2 (CTPS2), solute carrier family 9 member A7 (SLC9A7), thyroid hormone receptor interactor 12, zinc finger protein 280C, ubiquitin-like modifier-activating enzyme 1 (UBA1) and RNA binding motif protein 10 (RBM10). The above ten predicted proteins were analyzed by the cBioPortal database, each protein according to the threshold criteria of studies, and datasets ≥100 samples and samples with >20% alteration frequency were selected for further analysis: RBM10, UBA1, PHF16, TAB3, CTPS2 and SLC9A7.
Figure 5.
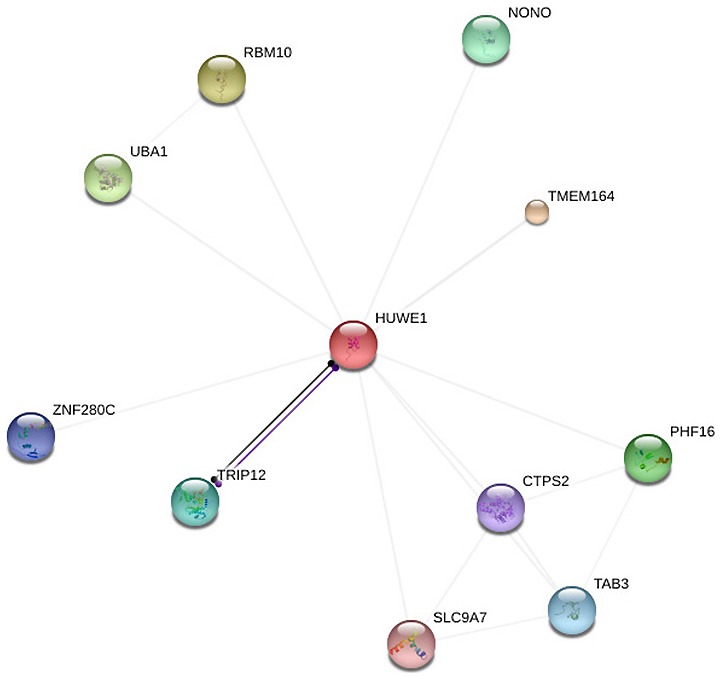
Identification of proteins associated with HUWE1 using STRING. Interacting nodes are presented in colored circles. HUWE1, E3 ubiquitin-protein ligase; RBM10, RNA binding motif protein 10; UBA1, ubiquitin-like modifier-activating enzyme 1; ZNF280C, zinc finger protein 280C; TRIP12, thyroid hormone receptor interactor 12; SLC9A7, solute carrier family 9 member A7; NONO, non-POU domain-containing octamer-binding protein; TMEM164, transmembrane protein 164; PHF16, protein Jade-3; CTPS2, TPP synthase 2; TAB3, TGF-β activated kinase 1.
Alterations of HUWE1 in different cancer types
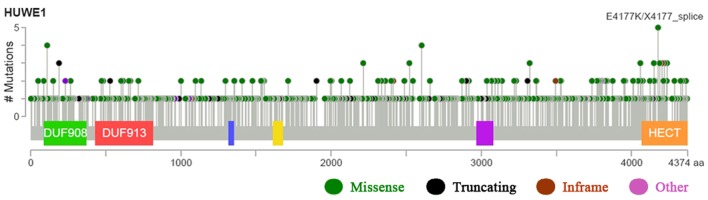
To investigate mutations and CNAs of the HUWE1 gene and the six predicted protein partners in various cancer types, the cBioPortal tool was used to analyze 91 cancer studies. In total, 11 of these studies included ≥100 samples in the dataset and samples with >20% alteration frequency. The alteration frequency ranged between 20.00 and 42.10% (Table III)(39–45). HUWE1 mutations occurred in many domains of the protein, predominantly in the C-terminus. Following database analysis, it was indicated that the most critical site mutation E4177K/X4177 occurs at the C-terminus. The higher the value of the vertical axis, the higher the mutation rate. Therefore, it is suggested that since C-terminus had the highest mutation rate, it may have the greatest impact on the function of HUWE1 (Fig. 6) and the highest mutation frequencies were revealed in neuroendocrine prostate cancer (Fig. 7).
Table III.
Alteration frequency of a six-gene signature (RBM10, UBA1, JADE3, TAB3, CTPS2 and SLC9A7) in different cancer types.
| Cancer type | Data source | n | Frequency, (%) | Amplification, (%) | Deletion, (%) | Mutation, (%) | Multiple alterations, (%) | (Refs.) |
|---|---|---|---|---|---|---|---|---|
| Neuroendocrine prostate cancer | Beltran et al (2016) | 107 | 42.10 | 39.30 | 0.90 | (39) | ||
| Prostate adenocarcinoma | Kumar et al (2016) | 136 | 25.70 | 19.10 | 4.40 | 2.20 | (40) | |
| Skin cutaneous melanoma | Hodis et al (2012) | 121 | 24.00 | 24.00 | (41) | |||
| Uterine corpus endometrial carcinoma | Kandoth et al (2013) | 240 | 23.80 | 2.50 | 0.80 | 20.00 | 0.40 | (42) |
| Lung squamous cell carcinoma | TCGA, Provisional | 177 | 23.70 | 2.80 | 4.50 | 15.80 | 0.60 | |
| Uterine Corpus Endometrial carcinoma | TCGA, Provisional | 242 | 23.60 | 2.50 | 0.80 | 19.80 | 0.40 | |
| Esophageal carcinoma | TCGA, Provisional | 184 | 22.80 | 3.80 | 3.80 | 15.20 | ||
| Lung adenocarcinoma | TCGA, Provisional | 230 | 20.40 | 2.20 | 0.90 | 16.10 | 1.30 | |
| Pan-lung cancer | Campbell et al (2016) | 1144 | 20.40 | 1.70 | 1.70 | 16.30 | 0.70 | (43) |
| Lung adenocarcinoma | Imielinski et al (2012) | 182 | 20.30 | 1.10 | 2.20 | 16.50 | 0.50 | (44) |
| Lung Adenocarcinoma | Collisson et al (2014) | 230 | 20.00 | 1.70 | 0.90 | 15.70 | 1.70 | (45) |
RBM10, RNA binding motif protein 10; UBA1, ubiquitin-like modifier-activating enzyme 1; TAB3, TGF-β activated kinase 1; CTPS2, TPP synthase 2; SLC9A7, solute carrier family 9 member A7; TCGA, The Cancer Genome Atlas.
Figure 6.
Mutation diagram of HUWE1 in different cancer types across protein domains. The diagram presents the HUWE1 mutation sites and HUWE1 mutation frequencies. Mutation diagram circles are colored with respect to the corresponding mutation types. In case of different mutation types at a single position, color of the circle is determined with respect to the most frequent mutation type. The blue, yellow and magenta boxes on the gene represent different protein domains. The higher the value of the vertical axis, the higher the mutation rate. HUWE1, E3 ubiquitin-protein ligase; aa, amino acid.
Figure 7.
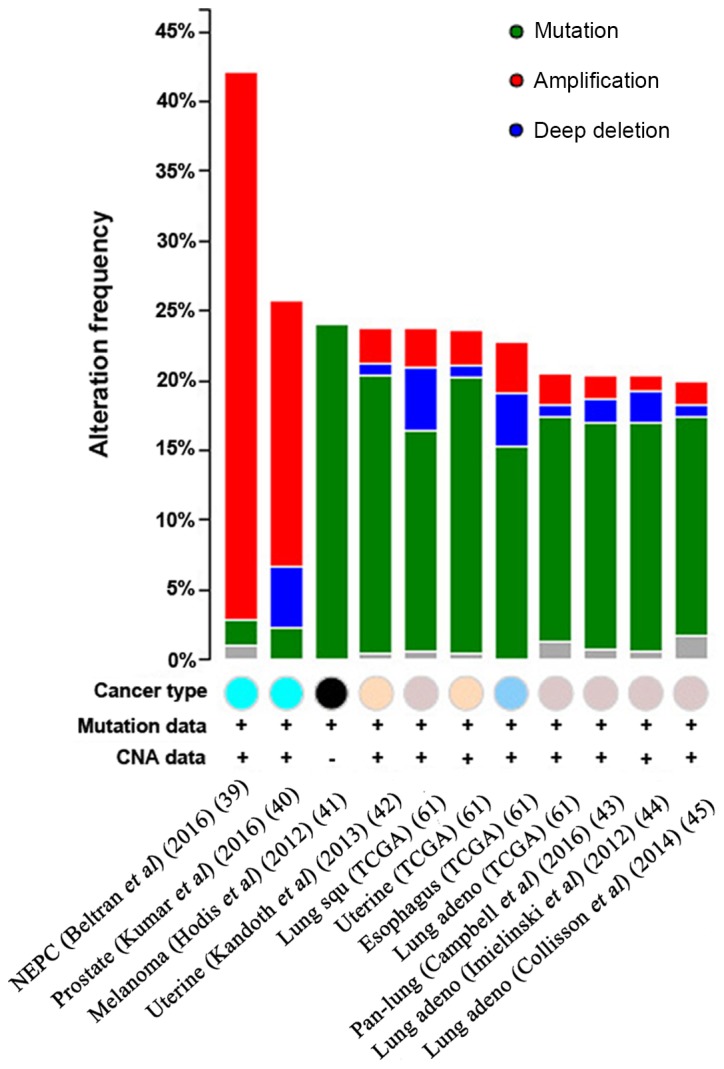
CNAs of the HUWE1 gene in different cancer subtypes. The alteration frequency of HUWE1 was determined using the cBioPortal database. Cancer types containing >100 samples were selected and only alteration frequencies >20% are presented. The different colored circles represent different types of tumors. The alterations included amplifications (red), deletions (blue), multiple alterations (grey) or mutations (green). HUWE1, E3 ubiquitin-protein ligase; NEPC, neuroendocrine prostate cancer; Prostate, prostate cancer; Melanoma, Skin cutaneous melanoma cancer; Uterine, Uterine corpus endometrial carcinoma cancer; Lung squ, Lung squamous cell carcinoma cancer; Esophagus, Esophageal carcinoma cancer; Lung adeno, Lung adenocarcinoma cancer; Pan-lung, Pan-lung cancer; TCGA, The Cancer Genome Atlas; CNA, copy number alteration.
Subsequently, OncoPrint was used to query for alterations in the RBM10, UBA1, JADE3, TAB3, CTPS2 and SLC9A7 genes. The proportion of alterations in these genes among prostate cancer varied between 1.8 and 2.5% for individual genes (RBM10, 2%; UBA1, 2.5%; JADE, 3.2%; TAB3, 1.7% CTPS2, 1.9%; and SLC9A7, 1.8%; Fig. 8). The UBA1 gene demonstrated a high level of amplification in prostate cancer.
Figure 8.
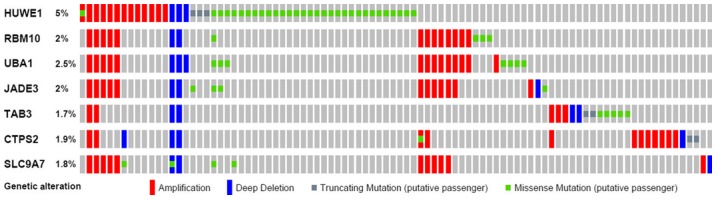
Mutation of HUWE1 and associated genes in prostate cancer. The Oncoprint feature of cBioPortal was used to determine the copy number alteration frequency of each individual gene. The percentages of alterations in HUWE1, RBM10, UBA1, JADE3, TAB3, CTPS2 and SLC9A7 genes are presented for prostate cancer. HUWE1, E3 ubiquitin-protein ligase; RBM10, RNA binding motif protein 10; UBA1, ubiquitin-like modifier-activating enzyme 1; SLC9A7, solute carrier family 9 member A7; CTPS2, TPP synthase 2; TAB3, TGF-β activated kinase 1.
A co-expression gene profile for HUWE1 in breast cancer was generated using Oncomine (Fig. 9). HUWE1 was revealed to be co-expressed with roundabout guidance receptor 1 (ROBO1), ectodysplasin A (EDA), spalt like transcription factor 1 (SALL1), p21 (RAC1) activated kinase 2 (PAK2) and glutamyl aminopeptidase (ENPEP).
Figure 9.
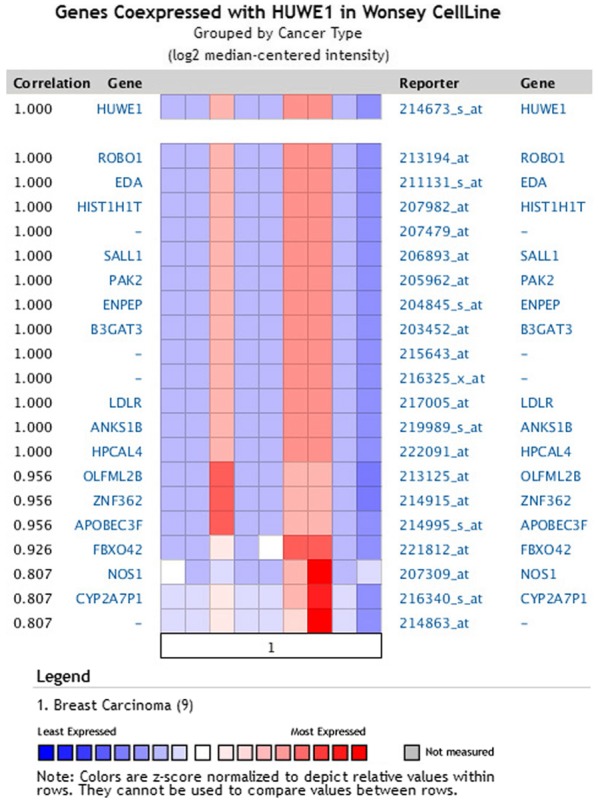
HUWE1-associated genes in breast carcinoma. HUWE1 is co-expressed in breast carcinoma tissues with the indicated genes. HUWE1, E3 ubiquitin-protein ligase; ROBO1, roundabout guidance receptor 1; EDA, ectodysplasin A; SALL1, spalt like transcription factor 1; PAK2, p21 (RAC1) activated kinase 2; ENPEP, glutamyl aminopeptidase; HIST1H1T, histone cluster 1 H1 family member T; B3GAT3, β-1,3-glucuronyltransferase 3; LDLR, low-density lipoprotein receptor; ANKS1B, ankyrin repeat and sterile alpha motif domain-containing protein 1B; HPCAL4, hippocalcin like 4; OLFML2B, olfactomedin like 2B; ZNF362, zinc finger protein 362; APOBEC3F, apolipoprotein B mRNA editing enzyme catalytic subunit 3F; FBXO42, F-box protein 42; NOS1, nitric oxide synthase 1; CYP2A7P1, cytochrome p450 family 2 subfamily member 7 pseudogene 1.
Discussion
HUWE1 is a HECT E3 ligase that serves a critical role in proteasomal degradation of several proteins and participates in cell cycle control, DNA damage response and tumorigenesis (1,11–22). However, to the best of our knowledge, whether any of these existing or unidentified substrates, including p53, MCL-1 and cdc6 ect, mediate the predominant function of ARF-BP1, also named MULE, HUWE1, in cell proliferation remains unclear. HUWE1 was first identified in 2005 (14); however, it remains unclear whether it can serve as a biomarker for cancer diagnosis or prognosis. To investigate this, the present study selected data according to the expression of several genes with clearly defined parameters between cancer and normal tissues. In the Oncomine analysis, HUWE1 was revealed to be overexpressed in leukemia and lung cancer, but was downregulated in brain cancer, lymphoma, sarcoma and testicular seminoma, compared with normal tissue. To further investigate the survival rates of patients with different expression levels of HUWE1, the associations between the expression levels of HUWE1 and the survival rates of patients were analyzed using Kaplan-Meier analysis and PrognoScan. In general, high expression levels of HUWE1 were associated with lower survival rates for patients with ovarian cancer (30–33,46–55); however, the results for lung cancer were not clear.
The main four causes of cancer are somatically-acquired genetic, epigenetic, transcriptomic and proteomic alterations in cells. These alterations occur in specific genomic regions, which can result in tumor suppressive or oncogenic effects (56). The cBioPortal analysis identified cancer types with significant CNAs in the selected HUWE1-gene signature. In total, 11 cancer studies representing 681 samples were analyzed, which contained >20% alteration frequency and ≥100 samples in the dataset. The alteration frequency ranged between 20.00 and 42.10%, with the dominance hierarchy.
The cBioPortal was used for interactive analysis and visualization of the HUWE1-associated network. The network was generated based on pathways and interactions from the Human Protein Reference database, Reactome Pathway database, National Cancer Institute Pathway Interaction database and the MSKCC Cancer Cell Map (56,57). The generated network improves understanding of the molecular mechanisms of HUWE1 in cancer.
The Oncomine database presents a potentially significant list of co-expressed genes that are critical in defining pathways. HUWE1 was identified to be co-expressed with ROBO1 in breast carcinoma, as well as with EDA, SALL1, PAK2 and ENPEP. This analysis may assist future studies regarding the function of HUWE1.
However, the precise role of HUWE1 remains controversial due to conflicting evidence. To investigate the role of HUWE1 in various cancer types, the Oncomine platform, which includes data from ~90,000 microarray experiments, was used to assess gene expression in different cancer types (28,29). In addition, the survival of patients with cancer was assessed using Kaplan-Meier plots and the PrognoScan database (34,38). Co-expressed data show that HUWE1 may regulate proteins and the signaling pathways involved in these proteins, which may assist in the examination of the molecular mechanism by which HUWE1 regulates cellular biological functions. The function and regulatory mechanisms of genes can be identified by STRING (36).
An aim of the present study was to determine whether CNAs of the HUWE1 network were associated with aggressive cancer subtypes, based on both the cBioPortal and Tumorscape (37,38,58). The role of HUWE1 in different cancer types was first analyzed. It was revealed that HUWE1 may serve an oncogenic role in breast, brain, central nervous system and prostate cancer types, and may serve a tumor suppressive role in colorectal cancer and certain types of lung cancer. In addition, the associations between HUWE1 expression levels and patient survival rates were investigated. According to STRING analysis, co-expression analysis demonstrated that HUWE1 was co-expressed with ROBO1 in breast carcinoma, as well as with EDA, HIST1S1T, SALL1, PAK2, ENPEP, B3GAT3 and ROBO1. The relationship between the expression of HUWE1 and prognosis remains unclear. By contrast, a high HUWE1 expression level was associated with a poor prognosis for gastric and ovarian cancer. The cBioPortal and Tumorscape analyses identified a HUWE1 alteration frequency of 20–42%, with the highest frequency observed in prostate cancer. In summary, HUWE1-coexpressed proteins may be used to further elucidate the function of HUWE1 in specific types of cancer.
The present study interpreted multidimensional oncogenic data. The use of databases contributes to improved understanding of cancer molecular etiology and epidemiology, which may assist with the translation of genomic understanding into clinical practice (59). The current study aimed to use extensive oncogenic databases to improve understanding regarding the molecular mechanisms mediated by HUWE1. In conclusion, HUWE1 may serve as a target for treatment strategies and act as a biomarker for certain cancer types.
Acknowledgements
We would like to acknowledge all of the data contributors of the TCGA, Oncomine, Kaplan-Meier plotter, Prognoscan, STRING and cBioPortal databases, which generously providing cyber resources for the scientists for the exploration, visualization and analysis of the multidimensional genomics data of cancer.
Funding
This study was supported by grants from the National Natural Science Foundation of China (grant nos. 31501114 and 81502035), the Foundation of Xiamen City (grant no. 3502Z20174080) and the Foundation of Fujian Province (grant no. 2018-ZQN-87).
Availability of data and materials
The data that support the findings of this study are available from Oncomine, Kaplan-Meier plotter, Prognoscan, STRING and cBioPortal databases, but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Oncomine, Kaplan-Meier plotter, Prognoscan, STRING and cBioPortal databases.
Authors' contributions
JH and CS conceived the study. CS, TW, JZ and JC analyzed the data. JH, CS and TW wrote and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber A, Peter M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim Biophys Acta. 2014;1843:163–181. doi: 10.1016/j.bbamcr.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 3.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 4.Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nat Chem Biol. 2009;5:815–822. doi: 10.1038/nchembio1209-954c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teixeira LK, Reed SI. Ubiquitin ligases and cell cycle control. Annu Rev Biochem. 2013;82:387–414. doi: 10.1146/annurev-biochem-060410-105307. [DOI] [PubMed] [Google Scholar]
- 6.Aleidi SM, Howe V, Sharpe LJ, Yang A, Rao G, Brown AJ, Gelissen IC. The E3 ubiquitin ligases, HUWE1 and NEDD4-1, are involved in the post-translational regulation of the ABCG1 and ABCG4 lipid transporters. J Biol Chem. 2015;290:24604–24613. doi: 10.1074/jbc.M115.675579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clague MJ, Liu H, Urbé S. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev Cell. 2012;23:457–467. doi: 10.1016/j.devcel.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Scheffner M, Kumar S. Mammalian HECT ubiquitin-protein ligases: Biological and pathophysiological aspects. Biochim Biophys Acta. 2014;1843:61–74. doi: 10.1016/j.bbamcr.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 9.Sun Y. Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol Ther. 2003;2:623–629. doi: 10.4161/cbt.2.6.677. [DOI] [PubMed] [Google Scholar]
- 10.Scheffner M, Staub O. HECT E3s and human disease. BMC Biochem. 2007;8(Suppl 1):S6. doi: 10.1186/1471-2091-8-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leboucher GP, Tsai YC, Yang M, Shaw KC, Zhou M, Veenstra TD, Glickman MH, Weissman AM. Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol Cell. 2012;47:547–557. doi: 10.1016/j.molcel.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou X, Li TT, Feng X, Hsiang E, Xiong Y, Guan KL, Lei QY. Targeted polyubiquitylation of RASSF1C by the Mule and SCFβ-TrCP ligases in response to DNA damage. Biochem J. 2012;441:227–236. doi: 10.1042/BJ20111500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Kan S, Huang B, Hao Z, Mak TW, Zhong Q. Mule determines the apoptotic response to HDAC inhibitors by targeted ubiquitination and destruction of HDAC2. Genes Dev. 2011;25:2610–2618. doi: 10.1101/gad.170605.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 15.Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Hall JR, Kow E, Nevis KR, Lu CK, Luce KS, Zhong Q, Cook JG. Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol Biol Cell. 2007;18:3340–3350. doi: 10.1091/mbc.e07-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herold S, Hock A, Herkert B, Berns K, Mullenders J, Beijersbergen R, Bernards R, Eilers M. Miz1 and HectH9 regulate the stability of the checkpoint protein, TopBP1. EMBO J. 2008;27:2851–2861. doi: 10.1038/emboj.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, Do H, Tian X, Zhang C, Liu X, Dada LA, Sznajder JI, Liu J. E3 ubiquitin ligase Mule ubiquitinates Miz1 and is required for TNFalpha-induced JNK activation. Proc Natl Acad Sci USA. 2010;107:13444–13449. doi: 10.1073/pnas.0913690107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsons JL, Tait PS, Finch D, Dianova, Edelmann MJ, Khoronenkova SV, Kessler BM, Sharma RA, McKenna WG, Dianov GL. Ubiquitin ligase ARF-BP1/Mule modulates base excision repair. EMBO J. 2009;28:3207–3215. doi: 10.1038/emboj.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Miao D, Xia Q, Hermo L, Wing SS. Regulated expression of the ubiquitin protein ligase, E3(Histone)/LASU1/Mule/ARF-BP1/HUWE1, during spermatogenesis. Dev Dyn. 2007;236:2889–2898. doi: 10.1002/dvdy.21302. [DOI] [PubMed] [Google Scholar]
- 21.Zhao X, Heng JI, Guardavaccaro D, Jiang R, Pagano M, Guillemot F, Iavarone A, Lasorella A. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi CF, Kim YS, Xiang S, Abdullaev Z, Torrey TA, Janz S, Kovalchuk AL, Sun J, Chen D, Cho WC, et al. Characterization of ARF-BP1/HUWE1 Interactions with CTCF, MYC, ARF and p53 in MYC-Driven B Cell Neoplasms. Int J Mol Sci. 2012;13:6204–6219. doi: 10.3390/ijms13056204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E, Farina A, Martinez E. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol. 2005;25:10220–10234. doi: 10.1128/MCB.25.23.10220-10234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vervoorts J, Lüscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K, Austen M, Lüscher B. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003;4:484–490. doi: 10.1038/sj.embor.embor821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: Multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Myant KB, Cammareri P, Hodder MC, Wills J, Von Kriegsheim A, Győrffy B, Rashid M, Polo S, Maspero E, Vaughan L, et al. HUWE1 is a critical colonic tumour suppressor gene that prevents MYC signalling, DNA damage accumulation and tumour initiation. EMBO Mol Med. 2017;9:181–197. doi: 10.15252/emmm.201606684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peter S, Bultinck J, Myant K, Jaenicke LA, Walz S, Müller J, Gmachl M, Treu M, Boehmelt G, Ade CP, et al. Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO Mol Med. 2014;6:1525–1541. doi: 10.15252/emmm.201403927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, et al. Stem cell-related ‘self-renewal’ signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 31.Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, Sikic BI. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 2005;65:8679–8689. doi: 10.1158/0008-5472.CAN-05-1204. [DOI] [PubMed] [Google Scholar]
- 32.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 34.Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 35.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9. 1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41 (Database Issue) 2013:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, Etzioni R, Bolouri H, Montgomery B, White T, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22:369–378. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas Research Network. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Comprehensive molecular profiling of lung adenocarcinoma, corp-author. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, Lamborn KR, Berger MS, Botstein D, Brown PO, Israel MA. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci USA. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haslinger C, Schweifer N, Stilgenbauer S, Döhner H, Lichter P, Kraut N, Stratowa C, Abseher R. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J Clin Oncol. 2004;22:3937–3949. doi: 10.1200/JCO.2004.12.133. [DOI] [PubMed] [Google Scholar]
- 48.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storz MN, van de Rijn M, Kim YH, Mraz-Gernhard S, Hoppe RT, Kohler S. Gene expression profiles of cutaneous B cell lymphoma. J Invest Dermatol. 2003;120:865–870. doi: 10.1046/j.1523-1747.2003.12142.x. [DOI] [PubMed] [Google Scholar]
- 51.Skotheim RI, Lind GE, Monni O, Nesland JM, Abeler VM, Fosså SD, Duale N, Brunborg G, Kallioniemi O, Andrews PW, Lothe RA. Differentiation of human embryonal carcinomas in vitro and in vivo reveals expression profiles relevant to normal development. Cancer Res. 2005;65:5588–5598. doi: 10.1158/0008-5472.CAN-05-0153. [DOI] [PubMed] [Google Scholar]
- 52.Luo JH, Yu YP, Cieply K, Lin F, Deflavia P, Dhir R, Finkelstein S, Michalopoulos G, Becich M. Gene expression analysis of prostate cancers. Mol Carcinog. 2002;33:25–35. doi: 10.1002/mc.10018. [DOI] [PubMed] [Google Scholar]
- 53.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 54.Detwiller KY, Fernando NT, Segal NH, Ryeom SW, D'Amore PA, Yoon SS. Analysis of hypoxia-related gene expression in sarcomas and effect of hypoxia on RNA interference of vascular endothelial cell growth factor A. Cancer Res. 2005;65:5881–5889. doi: 10.1158/0008-5472.CAN-04-4078. [DOI] [PubMed] [Google Scholar]
- 55.Klonowska K, Czubak K, Wojciechowska M, Handschuh L, Zmienko A, Figlerowicz M, Dams-Kozlowska H, Kozlowski P. Oncogenomic portals for the visualization and analysis of genome-wide cancer data. Oncotarget. 2016;7:176–192. doi: 10.18632/oncotarget.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie S, Shen C, Tan M, Li M, Song X, Wang C. Systematic analysis of gene expression alterations and clinical outcomes of adenylate cyclase-associated protein in cancer. Oncotarget. 2017;8:27216–27239. doi: 10.18632/oncotarget.16111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 60.National Cancer Institute. Genomic Data Commons Data Portal. https://portal.gdc.cancer.gov/ [Aug 11;2017 ];
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from Oncomine, Kaplan-Meier plotter, Prognoscan, STRING and cBioPortal databases, but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Oncomine, Kaplan-Meier plotter, Prognoscan, STRING and cBioPortal databases.