Abstract
Chloride channel accessory (CLCA) is a gene family that encode Ca2+ activated chloride channels, which make a substantial contribution to various diseases. The aim of the present study was to investigate the prognostic value of CLCA expression in colon cancer. In an attempt to elucidate the value of CLCA mRNA expression in the prognosis of patients with colon cancer, the gene expression data of 438 patients with colon cancer were analyzed. The source of the data was The Cancer Genome Atlas, and it was identified that high expression levels of CLCA1 and CLCA2 were associated with a favorable overall survival (OS) time in patients with colon cancer. As revealed by joint effects analysis, the co-occurrence of high expression levels of CLCA1 and CLCA2 was associated with a favorable OS time in patients with colon cancer. CLCA genes were investigated using gene set enrichment analysis. The results of the bioinformatics analysis demonstrated that high expression levels of CLCA1 and CLCA2 were associated with the prognosis of colon cancer. These findings suggest that CLCA1 and CLCA2 are potential prognostic biomarkers for patients with colon cancer. Furthermore, combining CLCA1 and CLCA2 can enhance the sensitivity of the prediction of the OS time of patients with colon cancer.
Keywords: chloride channel accessory, mRNA, prognosis
Introduction
Cancer is one of the most pivotal public health concerns worldwide and has therefore been the focus of an increasing amount of attention. Colorectal cancer is one of the most common causes of cancer-associated mortality, and the USA statistics for 2018 reported that the estimated number of new colorectal cancer cases will reach 140,250 (1). With increased understanding and improved treatment options, the 5- and 10-year survival rates for colorectal cancer have reached 65 and 58%, respectively (2). The primary treatment for colorectal cancer is surgery, accompanied by chemotherapy, immunotherapy and radiation (3). Therefore, the molecular detection of colorectal cancer provides a non-invasive diagnosis, which has been widely accepted by the majority of patients and clinicians (4).
Chloride channel accessory (CLCA) is a gene family of Ca2+ activated chloride channels, which includes four genes in humans and a minimum of six genes in mice (5,6). Each member of this gene family map to a similar location on the chromosome 1p31-p22, share a similar hypothesized structure and share a similar homology; however, these genes substantially differ in their tissue locations (7,8). The human genome encodes three functional CLCA proteins, which are encoded by CLCA1, CLCA2 and CLCA4. CLCA3 is considered a shortened pseudogene, as it does not encode a protein due to the fact that it possesses premature stop codons, and is therefore not likely to be expressed at detectable levels (9). CLCA1, which was the first reported human CLCA family member, is primarily expressed in the large and small intestines, particularly in the crypt cells (6). Previous studies have demonstrated that increased expression of CLCA1 decreases the aggressiveness of colorectal cancer cells in vitro and in vivo (10,11). A high expression of CLCA2 has been observed in the trachea and mammary glands, and has been reported to serve a role in breast cancer by acting as a p53 family target (12,13). Predominant expression of CLCA4 and CLCA1 has been observed in colon tissue (14). Similar to CLCA2, it has been identified that ectopic expression of CLCA4 results in the inhibition of breast cancer cell growth (15).
A previous study reported that CLCA1 inhibited the growth of colon cancer cell lines (11). However, to the best of our knowledge, few studies have investigated the prognostic value of CLCA family expression in patients with colon cancer. Therefore, the present study investigated the prognostic value of CLCA mRNA expression levels using data from 438 patients with colon cancer in The Cancer Genome Atlas (TCGA) database. Bioinformatics analysis demonstrated that elevated expression levels of CLCA1 and CLCA2 were associated with a favorable prognosis in colon cancer.
Materials and methods
Data preparation
TCGA (cancergenome.nih.gov) was employed for obtaining the survival data, including sex, age, tumor-node-metastasis (TNM) stage, events, survival time, death status, and the mRNA expression levels of CLCA1, CLCA2 and CLCA4. All datasets contained clinical and follow-up data. Datasets without prognostic outcome information were excluded from the current study. The patients were divided into low and high expression groups based on the median value of gene expression of each CLCA and the patients' survival data were extracted.
Association and bioinformatics analysis
The present study investigated the functions and associations of CLCA genes by multiple bioinformatics approaches. The relative degrees of expression of CLCA genes in multiple normal tissue samples were calculated using the GTEx Portal (www.gtexportal.org/home) (16). The Metabolic gEne RApid Visualizer (MERAV) tool (merav.wi.mit.edu) (17) was searched for the purpose of constructing a boxplot of the expression levels of CLCA genes in normal tissues and primary tumors of colon cancer. Co-expression analysis was performed using GeneMANIA (www.genemania.org) (18). The Database for Annotation, Visualization, and Integrated Discovery (version 6.7; david.ncifcrf.gov/tools.jsp) (19,20) was employed for analyzing the functional enrichment, which included gene ontology (GO) functional analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. P<0.05 was considered to indicate a statistically significant difference.
Survival analysis
With regard to each CLCA mRNA expression, division of patients into high- and low-expression cohorts was performed using median gene expression level as the cut-off value. Evaluation of the prognosis of colon cancer was based on overall survival (OS) time. Kaplan-Meier curves with a log-rank test were used for the identification of associations between the mRNA expression levels of the three CLCAs and patient survival. Adjustments for age, sex and TNM stage were made using the Cox proportional hazards regression framework.
Joint-effects analysis
The present study also performed a joint-effects analysis for combining the genes identified as significant with the help of the survival analysis. Groups were formulated through the summarization of the selected expression of genes associated with the improved OS time in one group (Group 1), a worse OS time in another group (Group 3) and others in the last group (Group 2). The grouping is presented in Table III.
Table III.
Grouping information for the combination among CLCA genes.
| Group | Patients (n=438) | Composition |
|---|---|---|
| 1 | 170 | High CLCA1 + high CLCA2 |
| 2 | 98 | High CLCA1 + low CLCA2 or low CLCA1 + high CLCA2 |
| 3 | 170 | Low CLCA1 + low CLCA2 |
CLCA, chloride channel accessory.
Nomogram
In accordance with the clinical information, coupled with the findings of the survival analysis subsequent to adjusting it with Cox proportional hazards regression model, a risk model for tumor stage, CLCA1 and CLCA2 expression levels was established. Nonetheless, the points against each factor were scored and 1-, 5- and 10-year survival rates were computed.
Gene set enrichment analysis (GSEA)
GSEA 3.0 (http://software.broadinstitute.org/gsea/msigdb/index.jsp) was employed for the purpose of analyzing disparities in the levels of the gene expression of the biological pathways in the low and high expression groups for each gene with reference gene sets from the Molecular Signatures Database of c2 (KEGG gene sets: c2.all.v6.1.symbols.gmt), c5 (GO gene sets: c5.all.v6.1.symbols.gmt) and c6 (oncogenic signatures gene sets: c6.all.v6.2.symbols.gmt) (21). In addition, the number of permutations was set at 1,000. Enrichment findings that satisfied a typically significance cutoff of P<0.05 with a false discovery rate (FDR)<0.25 were regarded as statistically significant.
Statistical analysis
Kaplan-Meier survival analysis with a log-rank test was used for the calculation of the OS time and P-values for all associations. Cox proportional hazards regression analysis was also employed to calculate the crude or attuned hazard ratio (HR) and 95% confidence interval (CI) in the univariate and multivariate analyses. Statistical analyses of gene expressions in different groups were performed using one-way analysis of variance followed by Student-Newman-Keuls multiple comparison test. P<0.05 was considered to indicate a statistically significant difference. The Kaplan-Meier and scatter curves were generated using GraphPad software (version 7.0; GraphPad Software, Inc., La Jolla, CA, USA). Analysis of the data was performed with SPSS 20.0 software (IBM Corp., Armonk, NY, USA). The correlation plots and nomogram were produced by R platform (version 3.5.1; www.r-project.org).
Results
Clinical features and outcomes
An aggregate of 438 patients with colon cancer, who had accomplished follow-up profiles, were recruited for the purpose of investigation. The clinical characteristics of the patients have been summarized in Table I. TNM stage was significantly associated with the median survival time (MST; P<0.001; Table I).
Table I.
Demographic and clinical data for 438 patients with colon cancer.
| Variable | Patients (n=438) | No. of mortalities, % | MST, days | HR (95% CI) | Log-rank P-value |
|---|---|---|---|---|---|
| Age, years | 0.398 | ||||
| <60 | 122 | 81.1 | 3,039 | Ref. | |
| ≥60 | 316 | 76.3 | 2,535 | 1.223 (0.766–1.952) | |
| Sex | 0.545 | ||||
| Female | 204 | 78.4 | 2,990 | Ref. | |
| Male | 234 | 76.9 | 2,320 | 1.131 (0.759–1.686) | |
| TNM stage | <0.001 | ||||
| I | 73 | 94.5 | 3,234 | Ref. | |
| II | 167 | 83.8 | 2,838 | 2.24 (0.781–6.421) | |
| III | 126 | 75.4 | 2,856 | 4.068 (1.434–11.538) | |
| IV | 61 | 49.2 | 1,114 | 11.291 (3.980–32.026) | |
| Unknown | 11 |
MST, median survival time; HR, hazard ratio; CI, confidence interval; TNM, tumor-node-metastasis.
Bioinformatics analysis
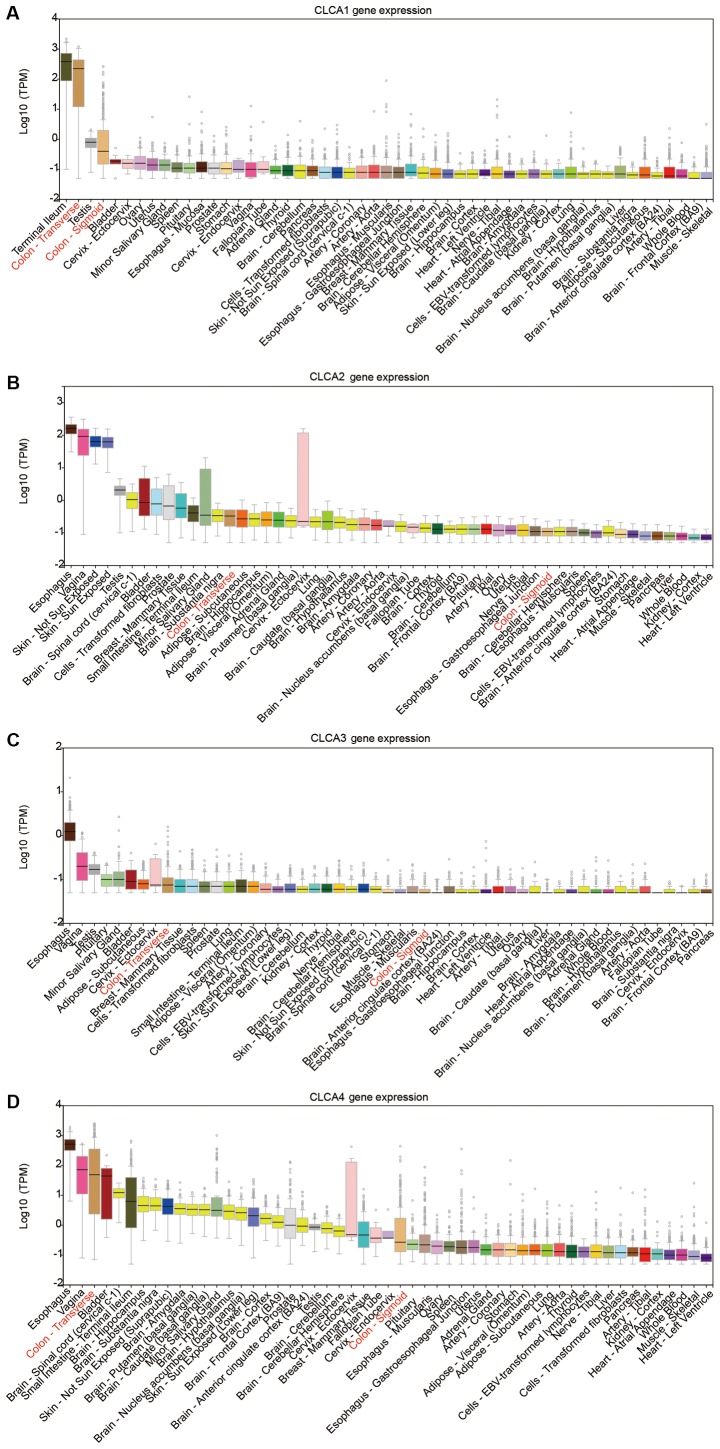
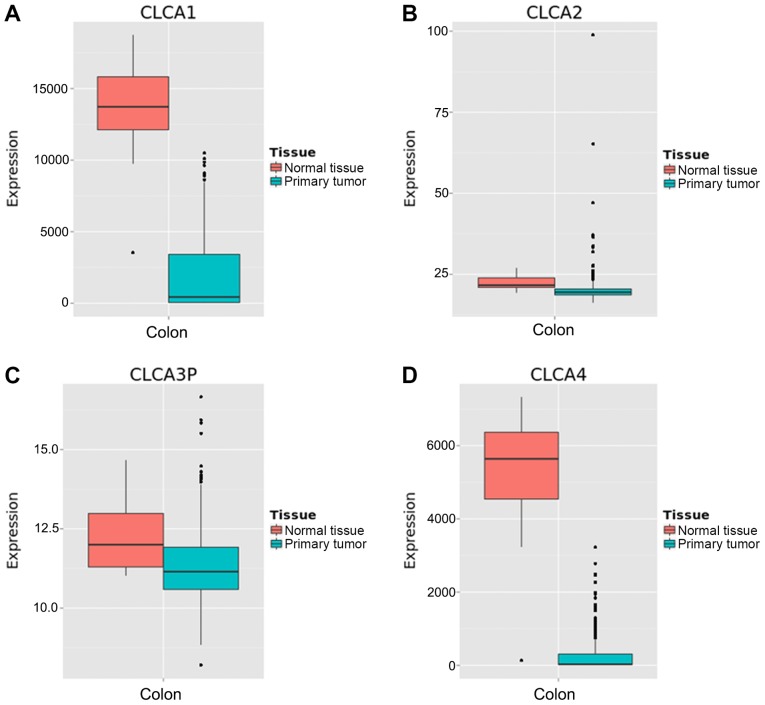
The CLCA family consists of four members (9). The bioinformatics analysis revealed that CLCA1, CLCA2, CLCA3 and CLCA4 were expressed at high levels in the human colon tissue. (Fig. 1A-D). The expression levels of CLCA1, CLCA2, CLCA3 and CLCA4 in colon tissue were higher than the majority of the other organs (Fig. 1). Boxplots illustrating dissimilarities in the expression of the four CLCA genes in normal colon tissue compared with primary colon cancer tissue were generated by MERAV (Fig. 2). The median expression levels of CLCA1, CLCA2, CLCA3 and CLCA4 were higher in the normal colon tissue when compared with that in the primary colon cancer tissue. Scatter plots for the expression of CLCA1, CLCA2 and CLCA4 in accordance with the 50th percentile cutoff are presented in Fig. 3A. The expression level of CLCA3 is not provided because CLCA3 is a truncated pseudogene and does not encode a protein (9). Scatter plots for the expression of the three genes in different TNM stages have been presented in Fig. 3B. A gene co-expression interaction analysis revealed that all the CLCA genes were co-expressed (Fig. 3C), with the exclusion of CLCA3 as it was not recognized by GeneMANIA. The assessment of biological functions of the CLCA genes, excluding CLCA3, was performed in accordance with the biological process, molecular function and cellular component categories for the GO functional analysis, and the results of KEGG pathway analysis are presented in Fig. 3D.
Figure 1.
CLCA genes in multiple normal tissues in the GTEx Portal database. The expression of CLCA genes in colon normal tissue are highlighted in red. (A) CLCA1 gene expression in multiple normal tissues. (B) CLCA2 gene expression in multiple normal tissues. (C) CLCA3 gene expression in multiple normal tissues. (D) CLCA4 gene expression in multiple normal tissues. CLCA, chloride channel accessory.
Figure 2.
Metabolic gEne RApid Visualizer boxplots for CLCA gene expression in colon normal tissue and primary colon cancer tissue. (A) CLCA1 expression. (B) CLCA2 expression. (C) CLCA3 expression. (D) CLCA4 expression. CLCA, chloride channel accessory.
Figure 3.
Intergroup differences of gene expression, gene interaction network, GO and KEGG enrichment results. (A) Scatter plots for CLCA1, CLCA2 and CLCA4 gene expression levels according to data from The Cancer Genome Atlas. (B) Scatter plots for CLCA1, CLCA2 and CLCA4 gene expression levels at different TNM stages. (C) Gene interaction network among CLCA genes produced by the GeneMANIA. (D) Analysis of the enriched GO terms and KEGG pathways for CLCA genes using the Database for Annotation, Visualization, and Integrated Discovery. *P<0.05, **P>0.05. CLCA, chloride channel accessory; TNM, tumor-node-metastasis; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Survival influence of differential CLCA gene expression
The key findings of the univariate survival analysis are presented in Fig. 4A-C. The results revealed that high expression levels of CLCA1, CLCA2 and CLCA4 are significantly associated with an improved OS time for patients with colon cancer (P<0.001, P=0.012 and P=0.039, respectively). As identified by the multivariate Cox proportional hazards regression analysis, there were associations of TNM stage with the prognosis of patients with colon cancer (Table I). The multivariate survival analysis suggested that, individually, elevated expression levels of CLCA1 (HR, 0.577; 95% CI, 0.376–0.885; adjusted P=0.012) and CLCA2 (HR, 0.647; 95% CI, 0.427–0.982; adjusted P=0.041) were associated with a favorable OS time (Table II).
Figure 4.
Prognostic value of CLCA expression for OS time. Kaplan-Meier survival curves for patients with colon cancer according the expression of (A) CLCA1, (B) CLCA2, (C) CLCA4 (n=438). (D) Joint-effects analysis of the impact of the combined CLCA gene expression on OS time with stratification in accordance with CLCA1 and CLCA2 genes. CLCA, chloride channel accessory; OS, overall survival; HR, hazard ratio.
Table II.
Prognostic survival analysis according to high or low expression of CLCA family genes.
| Gene expression | Patients (n=438) | No. of events, % | MST, days | Crude HR (95% CI) | Crude P-value | Adjusted HR (95% CI)a | Adjusted P-valuea |
|---|---|---|---|---|---|---|---|
| CLCA1 | <0.001 | 0.012 | |||||
| Low | 219 | 71.2 | 2,236 | Ref. | Ref. | ||
| High | 219 | 84.0 | 3,125 | 0.500 (0.331–0.757) | 0.577 (0.376–0.885) | ||
| CLCA2 | 0.012 | 0.041 | |||||
| Low | 219 | 73.1 | 2,301 | Ref. | Ref. | ||
| High | 219 | 82.2 | 3,070 | 0.596 (0.397–0.893) | 0.647 (0.427–0.982) | ||
| CLCA4 | 0.039 | 0.161 | |||||
| Low | 219 | 72.6 | 2,274 | Ref. | Ref. | ||
| High | 219 | 82.6 | 3,305 | 0.651 (0.433–0.978) | 0.743 (0.491–1.125) |
Adjustment for sex, age and TNM stage. CLCA, chloride channel accessory; MST, median survival time; HR, hazard ratio; CI, confidence interval; TNM, tumor-node-metastasis.
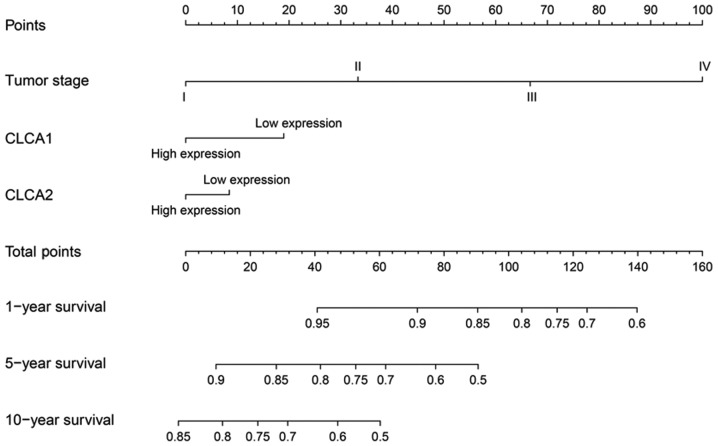
Subsequently, a joint-effects model was constructed for the determination of the cumulative impacts of CLCA genes on the OS time of patients with colon cancer. Furthermore, different groups for this analysis were generated in accordance with the expression of CLCA1 and CLCA2 (Table III). In the analysis, high CLCA1 and CLCA2 expression levels were observed to be significantly associated with a favorable OS time (P=0.003; Fig. 4D). The nomogram prognostic evaluation model was developed on the basis of the multivariate analysis (Fig. 5).
Figure 5.
A nomogram for the prediction of the overall survival was established for patients with colon cancer. The predictor points of each variable were observed on the upper-most point axis. A total points line is presented at the top of the nomogram, and each variable score was summed for the purpose of providing the aggregate points. A vertical line down to the 1-, 5- and 10-year survival lines allowed for the determination of probabilities for each individual. CLCA, chloride channel accessory.
Gene set enrichment analysis (GSEA)
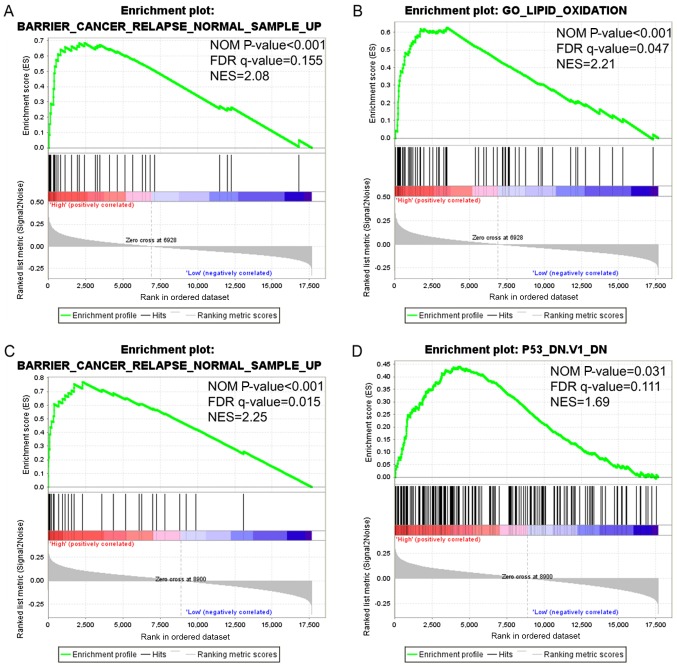
GSEA analysis was performed in order to investigate the potential biological processes that a high expression of CLCA1 and CLCA2 is likely to exert an impact on. The findings obtained from GSEA demonstrated that gene set differences in patients with high vs. low CLCA1 and CLCA2 expression levels indicated that CLCA1 and CLCA2-regulated gene sets were primarily associated with colon cancer relapse (P<0.001, FDR=0.155, Fig. 6A; P<0.001, FDR=0.015, Fig. 6C; respectively). The data also revealed that the elevated expression of CLCA1 was associated with lipid oxidation (P<0.001, FDR=0.047, Fig. 6B), while CLCA2 was associated with the p53 pathway (P=0.031, FDR=0.111, Fig. 6D). In summary, the data suggest that CLCA1 and CLCA2 are likely to be associated with the prognosis of colon cancer.
Figure 6.
Gene set enrichment analysis of CLCA genes expressed in patients with colon cancer. Results have been presented for the high CLCA1 and high CLCA2 expression groups. (A) CLCA1 high expression enriched cancer relapse. (B) CLCA1 high expression enriched lipid oxidation. (C) CLCA2 high expression enriched cancer relapse. (D) CLCA2 high expression enriched p53. CLCA, chloride channel accessory; FDR, false discovery rate; NES, normalized enrichment score; NOM, normalized.
Discussion
Recently, an increasing number of studies have investigated the function of ion channels in the pathogenesis of various cancer types (22,23). The chloride channel serves important roles in tumorigenesis (24). In addition, the CLCA proteins were primarily isolated in the year 1991 and have subsequently been revealed to be an intricate family (9). Each human CLCA family member identified thus far has been located on the short arm of the chromosome 1 (1p22-31), in addition each CLCA protein shares a similar homology and hypothesized structure; however, significant differences exist in their tissue location (8). Numerous functions of the CLCA family have been identified, including involvement in mucus secretion and tumor metastasis, and regulation of apoptosis, cell cycle and blood pressure (10,25,26).
The bioinformatics analysis of the present study demonstrated that the most notable molecular functions of CLCA were intracellular calcium activated chloride channel activity, chloride channel activity and chloride transmembrane transport. An increasing number of studies have suggested that CLCA proteins, acting on calcium-dependent chloride channels and facilitating chloride conductance, are closely associated with tumor progression (12,27). With regard to colon cancer, CLCA family members exert a pivotal function in the regulation of cell development, invasion and metastasis. Note that CLCA4 was misidentified as CLCA2 in a study conducted by Bustin et al (28).
In the present study, elevated expression levels of CLCA1, CLCA2 and CLCA4 in normal tissue were observed. Kaplan-Meier curves from univariate survival analysis revealed that elevated expression levels of CLCA1, CLCA2 and CLCA4 in tumor tissues were associated with a favorable OS time in all patients with colon cancer, which suggests that CLCA1, CLCA2 and CLCA4 act as tumor suppressors in colon cancer. In addition, the multivariate survival analysis validated the results of the univariate survival analysis, except for CLCA4. The univariate survival analysis revealed that the elevated expression of CLCA4 was associated with a favorable prognosis, whereas, in the multivariate survival analysis, neither low nor elevated expression of CLCA4 was observed to be associated with OS time. This is likely due to the adjustment in the Cox proportional hazards regression model, which suggested that CLCA4 is not likely to be an independent risk factor.
CLCA1 is primarily expressed in the large and small intestines, in particular in the crypt cells (6). CLCA1 has also been reported to be involved in the formation of the mucus and mucins of the goblet cells, providing the foremost protection line of the gastrointestinal tract, together with interacting with the immune system (29,30). There have been several studies to date, which have hypothesized that CLCA1 functions a tumor suppressor in colorectal cancer (10,11,28). For example, it has been reported that the elevated expression level of CLCA1 has the potential to suppress the incidence of colorectal cancer not only in vitro but also with in vivo experiments, as it was associated with inhibiting the Wnt signaling pathway, as well as EMT (10). In addition, CLCA1 is likely to control the proliferation to the differentiation transition of enterocytes through the regulation of the Wnt/β-catenin signaling pathway, accordingly acting as a tumor suppressor in colorectal tumorigenesis (11,31). Similar to CLCA1, CLCA2 has also been confirmed to suppress migration and invasiveness in breast and colorectal cancer cell lines (13,32). Despite the fact that CLCA1 and CLCA2 have a similar domain structure, their amino acid conservation is only almost 40%, and CLCA2 is primarily expressed in the trachea (9,33). The tumor suppressor protein p53 substantially contributes to the prevention of human cancer and to a transcription element activating and/or repressing its target genes (34). As reported in the present study, CLCA2 is a p53 target gene that regulates the p53-induced apoptotic pathway (27). CLCA2 has also been hypothesized to function as a p53-inducible senescence mediator in regulating the senescence pathway, together with carcinogenesis (35). Tumor relapse is regarded as a pivotal prognostic factor for patients with cancer (36). Early relapse substantially reduced OS rates when compared with the non-early recurrence cases in colon cancer (37). In the present study, patients in the GSEA dataset with colon cancer revealed that high CLCA1 and CLCA2 expressions were associated with cancer relapse, and that CLCA2 had a significant association with the p53 pathway, which was in line with previous research (21,38). Meanwhile, GSEA will help in linking prior knowledge to the newly generated data. This will ultimately aid in elucidating the collective behavior of genes in states of health and disease (21). In addition, the GSEA dataset demonstrated that high expression levels of CLCA1 and CLCA2 were associated with colon cancer relapse and not survival. This may be associated with other prior studies conducted on CLCA genes and relapse (15,31). With the increase of newly generated data, the association between CLCA and prognosis will appear in GSEA. The joint-effects analysis in the present study revealed that high expression levels of CLCA1 and CLCA2 were associated with a favorable OS time in patients with colon cancer. The nomogram prognostic evaluation framework was capable of assessing the patient survival time that was advantageous to the individualized therapy. In addition, the expression of CLCA1 and CLCA2 was previously observed in colon cancer, which indicated that the upregulated expression of these two genes is likely to indicate a favorable prognosis in colon cancer.
However, there were limitations to the present investigation. Firstly, the medical information in the public databases was not detailed, as not all of the 438 cases included data on the specific tumor size and type. Therefore, confounding factors that may impact the prognosis of patients were not factored into the Cox proportional hazards regression model. Secondly, the patient data used in the present study were entirely obtained from a single source. Therefore, it is imperative to validate the prognostic value of these genes in colon cancer with the help of the independent external validation datasets that contain the complete clinical information. Despite these limitations, the present study, to the best of our knowledge, is the first to report that the upregulation of CLCA1 and CLCA2 in colon cancer is associated with a favorable prognosis, and that CLCA1 and CLCA2 are potential prognostic biomarkers for patients with colon cancer.
In conclusion, the present study demonstrated that high expression levels of CLCA1 and CLCA2 were individually and mutually associated with a favorable prognosis for colon cancer. CLCA1 and CLCA2 exhibit potential as prognostic biomarkers for patients with colon cancer. However, these results require confirmation in further investigations.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CLCA
chloride channel accessory
- OS
overall survival
- EMT
epithelial-mesenchymal transition
Funding
The present study was supported by the Science Universities Foundation (Guangxi, China; grant no. 2013ZD015).
Availability of data and materials
The datasets analyzed in the present study are all available in TCGA (cancergenome.nih.gov).
Authors' contributions
XP and JG conceived and designed the study. XP and QW processed the data and performed the statistical analysis. CX, LY and SP wrote and revised the manuscript and helped to perform the analysis and interpretation of data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 3.Feo L, Polcino M, Nash GM. Resection of the primary tumor in stage IV colorectal cancer: When is it necessary? Surg Clin North Am. 2017;97:657–669. doi: 10.1016/j.suc.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruber AD, Elble RC, Ji HL, Schreur KD, Fuller CM, Pauli BU. Genomic cloning, molecular characterization, and functional analysis of human CLCA1, the first human member of the family of Ca2+-activated Cl-channel proteins. Genomics. 1998;54:200–214. doi: 10.1006/geno.1998.5562. [DOI] [PubMed] [Google Scholar]
- 6.Gandhi R, Elble RC, Gruber AD, Schreur KD, Ji HL, Fuller CM, Pauli BU. Molecular and functional characterization of a calcium-sensitive chloride channel from mouse lung. J Biol Chem. 1998;273:32096–32101. doi: 10.1074/jbc.273.48.32096. [DOI] [PubMed] [Google Scholar]
- 7.Pauli BU, Abdel-Ghany M, Cheng HC, Gruber AD, Archibald HA, Elble RC. Molecular characteristics and functional diversity of CLCA family members. Clin Exp Pharmacol Physiol. 2000;27:901–905. doi: 10.1046/j.1440-1681.2000.03358.x. [DOI] [PubMed] [Google Scholar]
- 8.Loewen ME, Forsyth GW. Structure and function of CLCA proteins. Physiol Rev. 2005;85:1061–1092. doi: 10.1152/physrev.00016.2004. [DOI] [PubMed] [Google Scholar]
- 9.Patel AC, Brett TJ, Holtzman MJ. The role of CLCA proteins in inflammatory airway disease. Annu Rev Physiol. 2009;71:425–449. doi: 10.1146/annurev.physiol.010908.163253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Hu W, Zhou J, Huang Y, Peng J, Yuan Y, Yu J, Zheng S. CLCA1 suppresses colorectal cancer aggressiveness via inhibition of the Wnt/beta-catenin signaling pathway. Cell Commun Signal. 2017;15:38. doi: 10.1186/s12964-017-0192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang B, Cao L, Liu B, McCaig CD, Pu J. The transition from proliferation to differentiation in colorectal cancer is regulated by the calcium activated chloride channel A1. PLoS One. 2013;8:e60861. doi: 10.1371/journal.pone.0060861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruber AD, Pauli BU. Tumorigenicity of human breast cancer is associated with loss of the Ca2+-activated chloride channel CLCA2. Cancer Res. 1999;59:5488–5491. [PubMed] [Google Scholar]
- 13.Sasaki Y, Koyama R, Maruyama R, Hirano T, Tamura M, Sugisaka J, Suzuki H, Idogawa M, Shinomura Y, Tokino T. CLCA2, a target of the p53 family, negatively regulates cancer cell migration and invasion. Cancer Biol Ther. 2012;13:1512–1521. doi: 10.4161/cbt.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agnel M, Vermat T, Culouscou JM. Identification of three novel members of the calcium-dependent chloride channel (CaCC) family predominantly expressed in the digestive tract and trachea. FEBS Lett. 1999;455:295–301. doi: 10.1016/S0014-5793(99)00891-1. [DOI] [PubMed] [Google Scholar]
- 15.Yu Y, Walia V, Elble RC. Loss of CLCA4 promotes epithelial-to-mesenchymal transition in breast cancer cells. PLoS One. 2013;8:e83943. doi: 10.1371/journal.pone.0083943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J, Gelfand ET, et al. A novel approach to high-quality postmortem tissue procurement: The GTEx project. Biopreserv Biobank. 2015;13:311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaul YD, Yuan B, Thiru P, Nutter-Upham A, McCallum S, Lanzkron C, Bell GW, Sabatini DM. MERAV: A tool for comparing gene expression across human tissues and cell types. Nucleic Acids Res. 2016;44(D1):D560–D566. doi: 10.1093/nar/gkv1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38 (Web Server Issue) 2010:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pardo LA, Stühmer W. The roles of K(+) channels in cancer. Nat Rev Cancer. 2014;14:39–48. doi: 10.1038/nrc3635. [DOI] [PubMed] [Google Scholar]
- 23.Cuddapah VA, Sontheimer H. Ion channels and transporters [corrected] in cancer. 2. Ion channels and the control of cancer cell migration. Am J Physiol Cell Physiol. 2011;301:C541–C549. doi: 10.1152/ajpcell.00102.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peretti M, Angelini M, Savalli N, Florio T, Yuspa SH, Mazzanti M. Chloride channels in cancer: Focus on chloride intracellular channel 1 and 4 (CLIC1 AND CLIC4) proteins in tumor development and as novel therapeutic targets. Biochim Biophys Acta. 2015;1848:2523–2531. doi: 10.1016/j.bbamem.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Hoshino M, Morita S, Iwashita H, Sagiya Y, Nagi T, Nakanishi A, Ashida Y, Nishimura O, Fujisawa Y, Fujino M. Increased expression of the human Ca2+-activated Cl- channel 1 (CaCC1) gene in the asthmatic airway. Am J Respir Crit Care Med. 2002;165:1132–1136. doi: 10.1164/ajrccm.165.8.2107068. [DOI] [PubMed] [Google Scholar]
- 26.Kim JA, Kang YS, Lee YS. Role of Ca2+-activated Cl-channels in the mechanism of apoptosis induced by cyclosporin A in a human hepatoma cell line. Biochem Biophys Res Commun. 2003;309:291–297. doi: 10.1016/j.bbrc.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Walia V, Ding M, Kumar S, Nie D, Premkumar LS, Elble RC. hCLCA2 Is a p53-inducible inhibitor of breast cancer cell proliferation. Cancer Res. 2009;69:6624–6632. doi: 10.1158/0008-5472.CAN-08-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bustin SA, Li SR, Dorudi S. Expression of the Ca2+-activated chloride channel genes CLCA1 and CLCA2 is downregulated in human colorectal cancer. DNA Cell Biol. 2001;20:331–338. doi: 10.1089/10445490152122442. [DOI] [PubMed] [Google Scholar]
- 29.Pelaseyed T, Bergström JH, Gustafsson JK, Ermund A, Birchenough GM, Schütte A, van der Post S, Svensson F, Rodríguez-Piñeiro AM, Nyström EE, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260:8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nyström EEL, Birchenough GMH, van der Post S, Arike L, Gruber AD, Hansson GC, Johansson MEV. Calcium-activated chloride channel regulator 1 (CLCA1) controls mucus expansion in colon by proteolytic activity. EBioMedicine. 2018;33:134–143. doi: 10.1016/j.ebiom.2018.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang B, Cao L, Liu J, Xu Y, Milne G, Chan W, Heys SD, McCaig CD, Pu J. Low expression of chloride channel accessory 1 predicts a poor prognosis in colorectal cancer. Cancer. 2015;121:1570–1580. doi: 10.1002/cncr.29235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramena G, Yin Y, Yu Y, Walia V, Elble RC. CLCA2 interactor EVA1 is required for mammary epithelial cell differentiation. PLoS One. 2016;11:e0147489. doi: 10.1371/journal.pone.0147489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma A, Ramena G, Yin Y, Premkumar L, Elble RC. CLCA2 is a positive regulator of store-operated calcium entry and TMEM16A. PLoS One. 2018;13:e0196512. doi: 10.1371/journal.pone.0196512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanikawa C, Nakagawa H, Furukawa Y, Nakamura Y, Matsuda K. CLCA2 as a p53-inducible senescence mediator. Neoplasia. 2012;14:141–149. doi: 10.1593/neo.111700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herr HW, Donat SM. Prostatic tumor relapse in patients with superficial bladder tumors: 15-year outcome. J Urol. 1999;161:1854–1857. doi: 10.1016/S0022-5347(05)68826-X. [DOI] [PubMed] [Google Scholar]
- 37.Lu CY, Uen YH, Tsai HL, Chuang SC, Hou MF, Wu DC, Juo SH, Lin SR, Wang JY. Molecular detection of persistent postoperative circulating tumour cells in stages II and III colon cancer patients via multiple blood sampling: Prognostic significance of detection for early relapse. Br J Cancer. 2011;104:1178–1184. doi: 10.1038/bjc.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrier A, Lemoine A, Boelle PY, Tse C, Brault D, Chiappini F, Breittschneider J, Lacaine F, Houry S, Huguier M, et al. Colon cancer prognosis prediction by gene expression profiling. Oncogene. 2005;24:6155–6164. doi: 10.1038/sj.onc.1208984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed in the present study are all available in TCGA (cancergenome.nih.gov).