Abstract
Cervical cancer pathogenesis is regulated by numerous factors, including microRNAs. MicroRNA 1246 (miR-1246) has been shown to serve a role in cervical cancer tumorigenesis. However, the mechanisms through which miR-1246 exerts its oncogenic effects are largely unknown. The aim of the current study was to evaluate the effects of lentivirus-mediated miR-1246 knockdown on the biological characteristics and behavior of cervical cancer cells, and to identify the downstream signaling pathways affected by miR-1246 knockdown. Short hairpins inhibiting miR-1246 were synthesized and cloned into a recombinant lentiviral vector (LV-miR-1246-Inh), which was then used to infect SiHa cervical cancer cells. The effects of LV-miR-1246-Inh infection on cell invasion, proliferation and apoptosis were evaluated by Transwell assay, Cell Counting Kit-8 assay and flow cytometry, respectively. Thrombospondin-2 (THBS2), matrix metalloproteinase 2 (MMP2), MMP9 and extracellular matrix (ECM) component expression levels were evaluated, and the growth of xenograft tumors formed following injection of SiHa cells with knockdown of miR-1246 was assessed. miR-1246 downregulation in SiHa cells decreased proliferation, induced apoptosis and upregulated THBS2 expression. Furthermore, MMP2 and MMP9 levels were downregulated, whereas components of the ECM were upregulated subsequent to miR-1246 knockdown, indicating that this miRNA regulates cervical cancer cell pathogenesis via the THBS2/MMP/ECM pathway. Notably, SiHa cells with miR-1246 downregulation had a markedly decreased ability to form tumors in vivo. These results suggest that miR-1246 functions during cervical cancer pathogenesis and tumor formation via the THBS2/MMP/ECM signaling pathway. These findings support the future use of miR-1246 suppression in the treatment of cervical cancer.
Keywords: thrombospondin-2, microRNA, cervical cancer, SiHa cells, apoptosis
Introduction
Cervical squamous cell carcinoma (CSCC) is a leading cause of cancer-associated mortality in women worldwide and has significant societal and economic consequences (1). At present, surgery and radiotherapy are standard treatments for cervical cancer (2). However, the clinical outcomes of these treatments vary dramatically and are difficult to predict. Therefore, it is necessary to identify new, effective treatment approaches, particularly for patients with resistance to radiotherapy and those susceptible to relapse.
MicroRNAs (miRNAs) are small noncoding RNA molecules that target protein-coding genes and downregulate their expression (3). They are of 19–25 nucleotides in length and are cleaved by 70–100 nt long hairpin precursor miRNAs (pre-miRNAs) (4). Numerous essential processes are regulated by miRNAs, including viral infection, cell growth, apoptosis and cancer development (5). However, few studies have focused on the role of miRNAs in cervical cancer (6–8), and the association between miRNAs and cervical cancer transformation remains poorly understood. Among the miRNAs investigated in cervical cancer, miR-1246 has oncogenic effects (9), and is elevated in the serum and tumor tissues isolated from patients with CSCC with lymph node metastasis (10).
Thrombospondin-2 (TSP2 or THBS2) is a member of the thrombospondin family. In a previous study in SiHa cervical cancer cells, THBS2 was shown to be a target of miR-1246 (11). THBS2 regulates cell adhesion and migration via hydrolysis of the extracellular matrix (ECM) (12). THBS2 also may serve a role in inhibiting angiogenesis by regulating matrix metalloproteinases (MMPs) and ECM proteins (13). While it is possible that the miR-1246/THBS2/ECM signaling cascade is involved in cervical cancer metastasis, direct evidence for a role of this pathway has not been reported.
In the present study, the effects of miR-1246 downregulation via lentiviral transfection, on the THBS2/MMPs/ECM pathway in the human cervical cancer cell line SiHa were evaluated.
Materials and methods
Synthesis of recombinant lentiviral particles (LV-miR-1246-Inh)
Lentivirus particles expressing oligonucleotides against hsa-miR-1246 (miRBase accession number, MIMAT0005898) were constructed by Shanghai GeneChem Co., Ltd. Briefly, double-stranded oligonucleotides encoding a miR-1246-inhibitor (miR-1246-Inh) was annealed and inserted into the linearized eukaryotic hU6-MCS-Ubiquitin-EGFR-puro vector (Shanghai GeneChem Co., Ltd.). The identity of vector was confirmed by sequencing. An empty vector control was used to examine if the transfection reagents or the transfection process itself had any cytotoxic effects on the target cells. The recombinant vectors or empty expression vector and packaging vectors (pHelper 1.0 and pHelper 2.0; Shanghai GeneChem Co., Ltd.) were then co-transfected into 293T cells (Invitrogen; Thermo Fisher Scientific, Inc.) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The culture supernatants were collected at 48 h following transfection. All lentiviral vectors expressed the enhanced green fluorescent protein, which enabled the measurement and titration of expression efficiency in infected cells. The cells were screened with puromycin (2 µg/ml) for 3 days.
Lentiviral infection of SiHa cells
SiHa cells were purchased from the American Type Culture Collection and cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 2 mM glutamine and 0.1 mM nonessential amino acids at 37°C in a humidified chamber with 5% CO2. The cells were divided into three groups: No viral infection (NC), infection with control virus (LV-NC; empty vector) and infection with lentiviral particles expressing miR-1246-Inh (LV-miR-1246-Inh; multiplicity of infection, 50).
Cell proliferation assay
Cell proliferation was assayed using a Cell Counting Kit-8 (CCK-8; Promega Corporation), according to the manufacturer's protocol. SiHa cells were seeded in 96-well plates at a density of 1,000–2,000 cells/well in 100 µl complete RPMI-1640 with 10% FBS. Cell proliferation was assayed every 24 h. Briefly, the CCK-8 solution (10 µl) was added to each well and incubated for 2 h at 37°C. Next, the absorbance of each well was measured spectrophotometrically at 450 nm using a TriStar microplate reader (LB941; Berthold Technologies).
Detection of apoptosis by flow cytometry
Cells (~2.5×106) were trypsinized, collected, washed with phosphate buffered saline (PBS) and stained with Annexin V-phycoerythrin (PE) and 7-aminoactinomycin D staining using an PE Annexin V kit (cat. no. 556421; BD Biosciences) for 10 min at 4°C, according to the manufacturer's protocol. Apoptotic cells were determined using a flow cytometer (FACScan; Becton Dickinson; BD Biosciences). Flow cytometry data were analyzed using ModFit LT software (version 2.0; Verity Software House, Inc.).
Cell cycle assay
Cells were fixed in 70% ethanol at −20°C for 24 h. After washing with PBS, cells were treated with RNase A (50 µg/ml) and stained with propidium iodide (25 µg/ml) in PBS for 30 min at 37°C. Samples were analyzed using a flow cytometer, and cell cycle distribution was determined using ModFit LT software (version 2.0; Verity Software House, Inc.). The proliferative index was calculated as the percentage of cells in S/G2/M phase.
Matrigel invasion assay
The invasion activity of SiHa cells was measured using Transwell chambers (Costar; Corning Inc.) and Matrigel (BD Biosciences). In total, 1.5–2×104 cells/well in RPMI-1640 with 10% FBS were seeded in the upper chamber that had previously been coated with 1 mg/ml Matrigel. The lower chamber was filled with RPMI-1640 containing 20% serum. Following a 24-h incubation, the non-migrating cells in the upper chamber were removed by gentle scraping, and the adherent cells on the lower surface of the insert were stained with crystal violet for 15 min at room temperature and imaged using a fluorescence microscope (Olympus TH4-200). Automated quantification and cell counting were performed using ImageJ Fiji software (National Institutes of Health).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cultured cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse transcription was performed using a Takara PrimeScript RT reagent kit (Takara Bio, Inc.). Each RT reaction included 150 ng template RNA and a pool of RT primers. qPCR was performed using TaKaRa SYBR® Premix Ex Taq™ II kit (Takara Bio) and the following primer sequences (Shanghai GenePharma Co., Ltd.): miR-1246 forward, 5′-TACGAAATGGATTTTTGGAGCAGG-3′ and U6 forward, 5′-ATTGGAACGATACAGAGAAGATT-3′, along with the universal reverse primer, 5′-GTCCTTGGTGCCCGAGTG-3′. The reaction conditions were 40 amplification cycles of 95°C for 3 min, 95°C for 15 sec and 62°C for 30 sec using Applied Biosystems ABI 7500 (Applied Biosystems; Thermo Fisher Scientific, Inc.) quantitative Real-Time PCR System. U6 was used as a reference for miRNAs. Each sample was analyzed in triplicate. Comparative threshold cycle method-fold change (2−ΔΔCq) was used to analyze relative changes (14).
Western blotting
Xenograft tumor tissues were lysed with radioimmunoprecipitation assay lysis and extraction buffer (Thermo Fisher Scientific, Inc.). Proteins (20 µg/lane) were separated by 8% SDS-PAGE and transferred to polyvinylidene difluoride membranes (EMD Millipore) by electroblotting. Membranes were blocked with 5% BSA in TBST (10 mM Tris-HCl, pH 8.0, 150 mM NaCl and 0.05% Tween-20) for 1–1.5 h, and then incubated with the following primary antibodies (Abcam) overnight at 4°C: THBS2 (1:500 dilution; cat. no. ab84469), MMP2 (1:5,000 dilution; cat. no. ab37150), MMP9 (1:5,000 dilution; cat. no. ab38898), ECM components (1:1,000 dilution; cat. no. ab130585) and β-actin (1:5,000 dilution; cat. no. ab228001). Subsequent to washing, the blots were incubated with goat anti-rabbit immunoglobulin G (H+L)-horseradish peroxidase secondary antibodies (1:5,000 dilution; cat. no. BS13278; Bioworld Technology, Inc.) and visualized using super enhanced chemiluminescence detection reagent (Amersham; GE Healthcare). Band intensity was scanned using Bio-Rad Universal Hood II (Bio-Rad Laboratories, Inc.) and analyzed using the Image Lab™ software (version 5.1; Bio-Rad Laboratories, Inc.) and normalized to the expression of β-actin.
Tumor xenograft model
Athymic BALB/c nude female mice (4–6 weeks old; body weight, 15–18 g) were obtained and raised in the Animal Experimental Center of Guangxi Medical University (Guangxi, China) in solid-bottomed cages. Cages were sanitized once weekly. Environmental conditions were maintained at 21±2°C with 50±20% relative humidity and 15 air changes hourly. Animals were kept on a 12-h light/dark cycle and provided ad libitum access to water and food. Mice were randomly assigned to one of three groups (16 mice/group). SiHa cells (NC, LV-NC or LV-miR-1246-Inh) were trypsinized and resuspended to a final concentration of 5×106 cells/0.1 ml PBS. The cells were then injected subcutaneously into the left flanks of the mice. Four weeks later, mice were euthanized by exposure to carbon dioxide with a flow rate of 9 l/min (volume of the chamber, 30 l). Death was confirmed by physical examination. Lack of breath, heartbeat and eyelid reflex was observed for 10 min. Tumor size was monitored every 2 days by measuring length and width with calipers, and volume was calculated according to the formula: Volume (mm3)=width2 (mm2) × length (mm)/2 (15). Tumor growth rate is expressed as the change in tumor volume over the number of days from the initial cell injection. Animal experiments were conducted according to the institutional guidelines of the National Research Council's Guide for the Care and Use of Laboratory Animals (China) and were approved by the Institutional Experimental Animals Review Board of Guangxi Medical University (Guangxi, China).
Immunohistochemistry
The Ki-67 protein is expressed in all phases of the cell cycle except G0 and serves as a good marker for proliferation (16). The mice were sacrificed and the tumors were fixed in 10% formalin at 4°C overnight. Immunohistochemistry was performed using 3-µm-thick paraffin sections. Paraffin sections were dewaxed and rehydrated through xylene and a graded alcohol series. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 15 min at 20–25°C. Following washing in water, nonspecific binding sites were blocked with 5% bovine serum albumin (cat. no. A2153; Sigma-Aldrich; Merck KGaA) in PBS for 10 min at 20–25°C. The slides were then incubated with a primary polyclonal antibody against Ki67 (cat. no. ab15580; 1:1,000; Abcam) at 4°C overnight. The slides were then gently rinsed with PBS and developed using the Envision system/HRP (cat. no. K400911; Dako; Agilent Technologies, Inc.) for 30 min and substrate-chromogen (3,3′-diaminobenzidine) at room temperature. The nuclei were counterstained with Mayer's hematoxylin for 3 min at room temperature. The percentage of positive stains under a fluorescent microscope (Olympus X71) was recorded. The Ki-67 proliferation index was automatically evaluated using the ImageJ plugin ‘IHC profiler’ (17).
To quantify angiogenesis, microvessel density (MVD) was assessed by immunostaining with an anti-CD31 antibody (no dilution; cat. no. MAB-0031; Fuzhou Maixin Biotech Co., Ltd.), as previously described (18). The sections were observed at a low magnification (×100), and the densest area of microvessels was selected and counted at a high magnification. Microvessel number was recorded by counting any positively stained endothelial cell or endothelial cell cluster as a single, countable microvessel in a high-power field (HPF; ×200). The 10 most neovascularized regions were selected for each sample. The mean of the top three counts was used as the microvessel count for each case.
Statistical analysis
Data are expressed as the mean ± standard deviation of three independent experiments, each performed in triplicate. Data were expressed as means with standard error. SPSS software was used for statistical analysis (version 23.0; IBM Corp.). Kolmogorov-Smirnov was used to test for normality. Differences between groups were assessed by two-way repeated measures ANOVA and multiple comparisons by least significant difference (LSD) post hoc test when the data had a normal distribution (tumor xenograft model). The remaining data did not display normal distribution, and was therefore analyzed with non-parametric methods; Kruskal-Wallis H test comparing three groups and Mann-Whitney U test as a post hoc test between any two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulation of miR-1246 inhibits proliferation, and induces apoptosis of SiHa cells
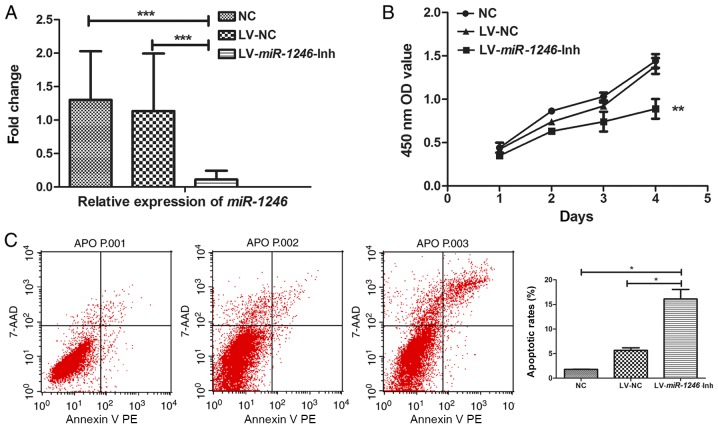
SiHa cells were infected with LV-miR-1246-Inh and a stable anti-miR-1246 SiHa cell line was established. Control lentiviruses were used to infect SiHa cells as control. Compared with the expression in the control group, LV-miR-1246-Inh-infected SiHa cells showed significant downregulation of miR-1246 (P=0.001; Fig. 1A), indicating stable knockdown of miR-1246.
Figure 1.
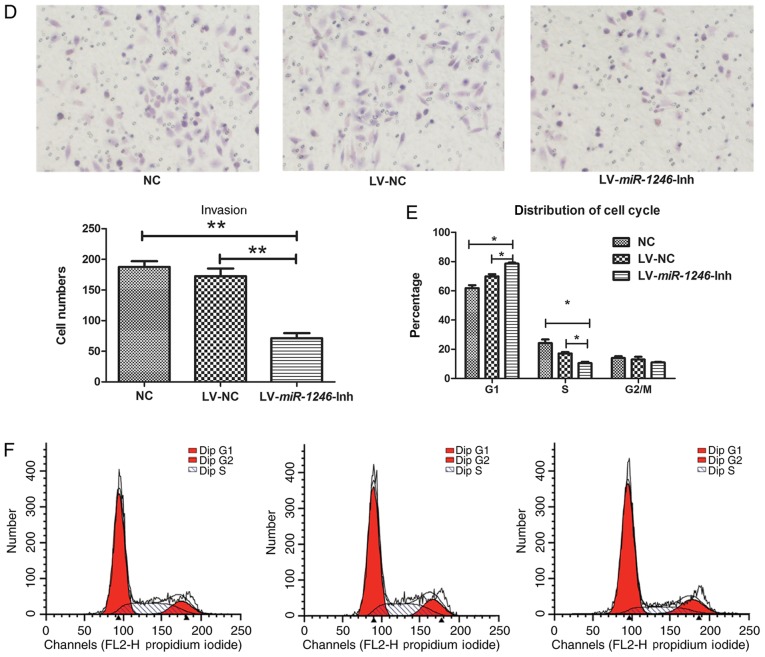
SiHa cell growth, cell cycle and invasion following miR-1246 downregulation. (A) miR-1246 expression in SiHa cells following infection with LV-miR-1246-Inh or LV-NC compared with untreated NC. (B) Proliferation of SiHa cells following miR-1246 downregulation. (C) Cell apoptosis detected by Annexin V-PE and 7-AAD labelling and flow cytometry. (D) Matrigel cell invasion assay showing the effects of LV-miR-1246-Inh on cell invasion. Magnification, ×200. (E and F) Cell cycle distribution determined by propidium iodide staining and flow cytometry highlighting the effects of LV-miR-1246-Inh on cell populations in the G1 and S phases. *P<0.05, **P<0.01, ***P<0.001. miR-1246, microRNA-1246; LV, lentivirus; Inh, inhibitor; NC, negative control; PE, phycoerythrin; 7-AAD, 7-aminoactinomycin D; OD, optical density.
Next, the role of miR-1246 in various cell processes, including proliferation, apoptosis and invasion was evaluated. In miR-1246-Inh-infected SiHa cells, proliferation was significantly suppressed compared with that in the control groups (NC and LV-NC; Fig. 1B). Flow cytometry revealed average apoptotic rates of 1.78, 6.00 and 16.10% in the NC, LV-NC and LV-miR-1246-Inh groups, respectively. The apoptotic rate of the LV-miR-1246-Inh group was increased compared with that in the NC and LV-NC groups (both P=0.040; Fig. 1C), indicating that miR-1246 downregulation significantly increased apoptosis in this cervical cancer cell line. Furthermore, in the LV-miR-1246-Inh group, the number of cells that invaded the Transwell Matrigel-coated membrane was 71.3±4.27, while that in the NC and LV-NC groups were 187.5±4.79 and 162.5±4.78, respectively (Fig. 1D), demonstrating altered cell invasion upon miR-1246 knockdown (P=0.013; Fig. 1D).
Downregulation of miR-1246 induces cell cycle arrest in the G1/S phase
Given that miR-1246 downregulation inhibited SiHa cell proliferation, the effect of decreased miR-1246 on cell cycle progression was determined. Analysis of the cell cycle distribution of SiHa cells indicated that the average number of cells in G1 phase increased in cells infected with LV-miR-1246-Inh (78.59%) compared with LV-NC (71.52%, P=0.042) and NC (61.79%, P=0.042; Fig. 1E). Additionally, the cell population in the S phase was reduced sharply in the LV-miR-1246-Inh group (10.48%) compared with the LV-NC (24.21%, P=0.027) and NC groups (15.49%, P=0.027; Fig. 1F).
Downregulation of miR-1246 inhibits tumor growth
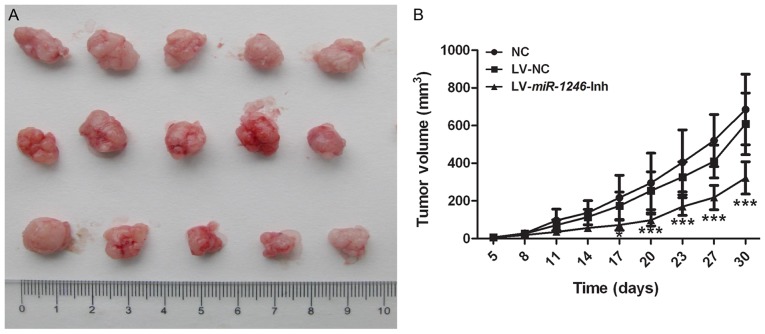
To elucidate the role of miR-1246 in tumor growth, SiHa cells of each treatment group (NC, LV-NC and LV-miR-1246-Inh) were injected into nude mice, and the growth of the xenograft tumors was monitored. Each subgroup contained 5–6 mice, and the experiment was repeated three times (n=16) for each group. Mice were euthanized when the average tumor volume was >500 mm3. During the four weeks of observation, three mice in the NC group and one mouse in the LV-NC group <5 weeks old died of anorexia and injury in the third week, although the skin was disinfected and the mice were shifted to another cage to ensure sufficient access to food. The tumor volume in the NC group was lower on day 17 following inoculation compared with that in the LV-miR-1246-Inh group (Fig. 2B; P<0.05). The xenograft tumors derived from cells with miR-1246 downregulation grew slowly compared with those in the two control groups, indicating that downregulation of miR-1246 markedly inhibited tumor development (Fig. 2A and B).
Figure 2.
Knockdown of miR-1246 inhibits xenograft tumor growth. (A) Images of xenograft tumors following surgical removal. Top row: Tumors formed following injection of untreated cells (NC). Middle row: Tumors formed after injection of LV-NC-infected cells. Bottom row: Tumors formed subsequent to injection of LV-miR-1246-Inh-infected cells. (B) Growth curve of SiHa xenografts in athymic BALB/c nude mice. *P<0.05, ***P<0.001 vs. NC and LV-NC groups. miR-1246, microRNA-1246; LV, lentivirus; Inh, inhibitor; NC, negative control.
Antiproliferative and antiangiogenic effect of miR-1246 downregulation in xenograft tumors
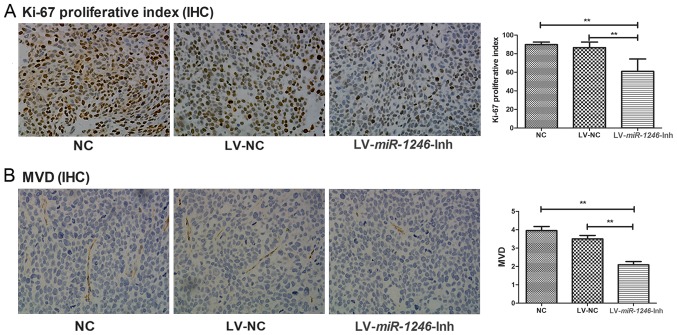
To investigate the growth of the xenograft tumors from the different groups, the Ki-67 proliferative index was used as a measure of the proliferative activity. Fig. 3A shows representative immunostaining images for the three groups. In the LV-miR-1246-Inh group, Ki67 level (60.93±13.36%) was significantly lower compared with that in the control groups (NC, 89.8±2.76%; LV-NC, 86.54±6.03%; P<0.001).
Figure 3.
Antiproliferative and antiangiogenic effects of miR-1246 downregulation in tumor xenografts. (A) Expression of Ki67 proliferation marker in the following groups: NC, LV-NC and LV-miR-1246-Inh. Magnification, ×200. (B) CD-31 expression in the following groups: NC, LV-NC and LV-miR-1246-Inh. Magnification, ×200. **P<0.01. miR-1246, microRNA-1246; LV, lentivirus; IHC, immunohistochemistry; Inh, inhibitor; MVD, microvessel density; NC, negative control.
To investigate the effect of miR-1246 downregulation on angiogenesis, MVD was assessed following CD31 staining. The MVD of the NC and LV-NC group were similar (3.95±1.48 and 3.58±1.19 microvessels/HPF, respectively; Fig. 3B). However, the MVD was significantly lower in the LV-miR-1246-Inh group (2.08±1.20 microvessels/HPF; P<0.001) than in the NC and LV-NC groups.
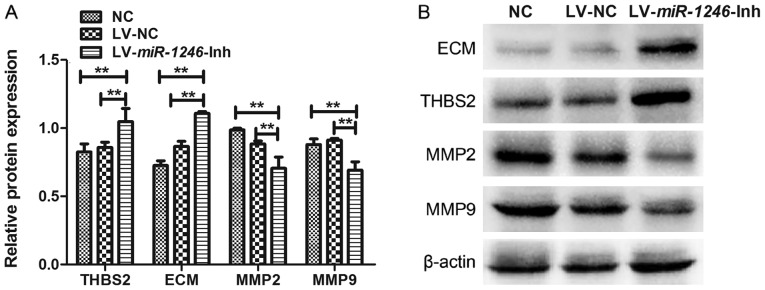
Downregulation of miR-1246 increases THBS2 protein expression and induces changes in the THBS2/MMP signaling pathway
THBS2 is a predicted miR-1246 target gene (10). To evaluate whether THBS2 was affected by miR-1246 downregulation, the protein level of THBS2 was examined by western blotting. THBS2 protein expression in cells infected with LV-miR-1246-Inh was significantly higher than that observed in the control groups (Fig. 4A). These data suggest that downregulation of miR-1246 leads to increased expression of THBS2 in SiHa cells.
Figure 4.
Effect of miR-1246 downregulation on THBS2, MMP2, MMP9 and ECM protein expression in SiHa cells. (A) Western blot analysis of protein expression following miR-1246 downregulation. Protein expression was compared with that of the control groups. (B) Representative western blot image. **P<0.01. miR-1246, microRNA-1246; LV, lentivirus; Inh, inhibitor; NC, negative control; THBS2, thrombospondin 2; MMP, matrix metalloproteinase; ECM, extracellular matrix.
The expression levels of ECM, MMP2 and MMP9 proteins were also examined to further investigate the role of miR-1246 in regulating THBS2 signaling (Fig. 4). The expression of ECM components was increased following miR-1246 knockdown compared with that in the LV-NC and NC groups. In contrast, MMP2 and MMP9 levels were decreased in the miR-1246-silenced cells compared with the LV-NC and NC groups. There were no significant differences in the expression levels of these proteins between the NC and LV-NC groups. Thus, downregulation of miR-1246 in SiHa cervical cancer cells significantly elevated the expression of THBS2 and ECM, and decreased expression of MMP2 and MMP9, suggesting that miR-1246 inhibits the THBS2 signaling pathway.
Discussion
A number of tumor-suppressor genes and oncogenes are aberrantly expressed in CSCC, including the pro-apoptotic protein Bax, which is frequently inactivated in carcinoma, and tumor protein p53, which serves a key role in cervical cancer (19). Previous studies (20–22) have provided significant new information concerning the function of noncoding RNAs, including miRNAs, in cervical cancer pathogenesis. In the present study, the role of miR-1246 in SiHa cervical cancer cells and tumor xenografts was investigated. Lentiviruses expressing shRNA against miR-1246 were used to downregulate miR-1246 expression, and the effects of miR-1246 downregulation on cellular processes and tumor growth were evaluated, with particular emphasis on the THBS2/ECM/MMP signaling pathway.
miR-1246 was identified in 2008 during microRNA profiling of human stem cells (23). In 2015, Krissansen et al (24) revealed that miR-1246 was significantly upregulated in the sera of patients with inflammatory bowel disease, Crohn's disease and ulcerative colitis compared with healthy individuals. Furthermore, in a study evaluating 168 patients with cancer and 65 healthy controls, Todeschini et al (25) identified miR-1246 as a potential diagnostic biomarker for high-grade serous ovarian carcinoma, with a sensitivity of 87%, a specificity of 77%, and an accuracy of 84%. The link between miR-1246 and cancer pathogenesis has been investigated in other studies. Zhang et al (26) showed that miR-1246 confers tumorigenicity and is required for lung cancer metastasis in non-small cell lung cancer (NSCLC) via the tumor-initiating cell model. Inhibition of miR-1246 appeared to decrease stemness and epithelial-mesenchymal transition in NSCLC, in addition to suppressing proliferation, sphere-formation, colony formation and invasion (27). Additionally, Hasegawa et al (28) demonstrated that miR-1246 increased the tumor-initiating potential of cells and induced drug resistance in vivo, and it was showed that high miR-1246 expression correlated with poor prognosis in patients.
Notably, the present study is not the first to investigate the molecular mechanism of miR-1246 in cervical cancer. In our own laboratory, the full miRNA profile of patients with CSCC was screened and it was demonstrated that miR-1246 is expressed at similar levels in the serum and cervical tumor tissues of patients with lymph node metastasis (10). These data are consistent with the study by Takeshita et al (29), which reported that miR-1246 expression can be used to differentiate patients with esophageal squamous cell carcinoma from healthy individuals, indicating that this miRNA may also have applications as a marker for lymph node metastasis in CSCC. Despite significant efforts to understand miR-1246 function in CSCC and other type of cancer, its mechanism remains unclear.
In the present study, miRNA knockdown was performed to evaluate the role of miR-1246 in CSCC tumorigenesis and progression. The role of miR-1246 in these processes has been previously investigated in a similar study using the same cell line. The expression of miR-1246 has been transiently upregulated or downregulated by transfection of analogs and antagonists with Lipofectamine® 2000 (11). THBS2 has been characterized as a functional target of miR-1246 by luciferase reporter gene assays and western blot analysis (11). However, in the present study, the effects of LV-miR-1246-Inh infection in SiHa cells were evaluated in more depth, focusing on cell proliferation, apoptosis and invasion following stable miR-1246 knockdown. Additionally, the present study investigated the role of the THBS2/ECM signaling pathway in cervical cancer using a tumor xenograft model. The results of the current study indicated that, compared with control cells, the growth rate of cells infected with LV-miR-1246-Inh was reduced, as was the capacity for invasion. Apoptosis of the cervical cells was increased when miR-1246 was knocked down. Moreover, downregulation of miR-1246 arrested the cell cycle in the G1 phase. Further studies of the molecular mechanisms underlying this phenomenon are required to identify the changes in regulatory proteins that control the G1/S transition. These results indicate that miR-1246 may function as an oncogene in CSCC development and progression, and that miR-1246 downregulation inhibits tumor growth and impairs cervical cancer cell invasion.
Notably, the identification of miR-1246-specific target genes has provided new insights into the mechanisms underlying its role in cancer development. THBS2 (also called TSP2) is an miR-1246 target gene (11). THBS gene and/or protein expression is significantly correlated with decreased microvessel count in oral cancer (30), gastric cancer (31), colorectal cancer (32), NSCLC (33), glioma (34) and pancreatic cancer (35). In cervical cancer, the microvessel count in patients lacking THBS2 expression is higher than that observed in patients expressing it, although this difference is not statistically significant (36). In the present study, inhibition of miR-1246 was associated with THBS2 upregulation. The tumor growth index was decreased in the xenograft tumors of the LV-miR-1246-Inh group, consistent with the in vitro results showing increased apoptosis and cell cycle arrest in SiHa cells with miR-1246 knocked down. In addition, in the LV-miR-1246-Inh group, MVD was decreased and THBS2 expression was increased. It has been suggested that THBS2 serves a unique role in the control of angiogenesis, as well as tumor growth and metastasis, and downregulation of THBS2 has been associated with tumor progression and poor prognosis in cervical cancer (36). In the present study, miR-1246 downregulation resulted in increased expression of THBS2, but decreased expression of MMP2 and MMP9. These data are consistent with a previous study showing that THBS2 overexpression downregulated MMP2 and MMP9 expression in a human colon carcinoma cell line (37). However, a study by Chen et al (38) found conflicting results, as THBS2 was observed to promote prostate cancer bone metastasis via MMP2 upregulation. These data suggest that the role of THBS2 may differ among different cancer types.
Degradation of the basement membrane and remodeling of the ECM by matrix metalloproteinases, including MMP2 and MMP9 is essential in tumor invasion and metastasis (39). Hirose et al (40) previously demonstrated that decreased THBS2 affects the levels of MMP2 and MMP9, and the THBS2/MMP interaction directly modulates the metabolism of the ECM. This observation was supported by the results of the present study, as ECM proteins levels, potentially upregulated due to THBS2/MMP interaction, were altered with miR-1246 downregulation. Taken together, these data show that miR-1246 may serve a distinct role in cervical cancer cell growth and tumor pathogenesis by regulating THBS2 expression, which subsequently alters the expression and function of MMPs. These changes have a profound impact on the ECM and ECM-related processes, including invasion, proliferation and apoptosis.
In summary, the present study provides evidence of the essential biological role of miR-1246 in cervical cancer pathogenesis and identifies the THBS2/MMP/ECM pathway as a potential target in cervical cancer cells. These findings provide a hypothesis for the future use of miR-1246 suppression in the treatment of cervical cancer. Although large-scale and long-term follow-up studies are needed to confirm the role of miR-1246 and THBS2 in cervical cancer, the present study firmly supports the essential role of miR-1246 in cervical cancer cell invasion and metastasis.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CSCC
cervical squamous cell carcinoma
- THBS2
TSP2, thrombospondin-2
- MMPs
matrix metalloproteinases
- FBS
fetal bovine serum
- NC
negative control
- NSCLC
non-small cell lung cancer
Funding
The study was supported by the National Natural Science Foundation of China (grant no. 81460398).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
DSY, JYC and PD contributed to the study design and conception. PD, YHL and ND contributed to the collection, analysis and interpretation of data, and PD contributed to the writing of the manuscript.
Ethics approval and consent to participate
Animal experiments were conducted according to the institutional guidelines of the National Research Council's Guide for the Care and Use of Laboratory Animals (China) and were approved by the Institutional Experimental Animals Review Board of Guangxi Medical University (Guangxi, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Cibula D, Abu-Rustum NR, Fischerova D, Pather S, Lavigne K, Slama J, Alektiar K, Ming-Yin L, Kocian R, Germanova A, et al. Surgical treatment of ‘intermediate risk’ lymph node negative cervical cancer patients without adjuvant radiotherapy-A retrospective cohort study and review of the literature. Gynecol Oncol. 2018;151:438–443. doi: 10.1016/j.ygyno.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarz DS, Hutvagner G, Du T, Xu ZS, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/S0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 4.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 5.Pedroza-Torres A, Lopez-Urrutia E, Garcia-Castillo V, Jacobo-Herrera N, Herrera LA, Peralta-Zaragoza O, López-Camarillo C, De Leon DC, Fernández-Retana J, Cerna-Cortés JF, et al. MicroRNAs in cervical cancer: Evidences for a miRNA profile deregulated by HPV and its impact on radio-resistance. Molecules. 2014;19:6263–6281. doi: 10.3390/molecules19056263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu X, Schwarz JK, Lewis JS, Jr, Huettner PC, Rader JS, Deasy JO, Grigsby PW, Wang X. A microRNA expression signature for cervical cancer prognosis. Cancer Res. 2010;70:1441–1448. doi: 10.1158/0008-5472.CAN-09-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribeiro J, Sousa H. MicroRNAs as biomarkers of cervical cancer development: A literature review on miR-125b and miR-34a. Mol Biol Rep. 2014;41:1525–1531. doi: 10.1007/s11033-013-2998-0. [DOI] [PubMed] [Google Scholar]
- 8.Fiannaca A, La Rosa M, La Paglia L, Rizzo R, Urso A. Analysis of miRNA expression profiles in breast cancer using biclustering. BMC Bioinformatics. 2015;16(Suppl 4):S7. doi: 10.1186/1471-2105-16-S4-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao JM, Zhou X, Zhang Y, Lu H. MiR-1246: A new link of the p53 family with cancer and Down syndrome. Cell Cycle. 2012;11:2624–2630. doi: 10.4161/cc.20809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Yao D, Li Y, Chen H, He C, Ding N, Lu Y, Ou T, Zhao S, Li L, Long F. Serum microRNA expression levels can predict lymph node metastasis in patients with early-stage cervical squamous cell carcinoma. Int J Mol Med. 2013;32:557–567. doi: 10.3892/ijmm.2013.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Yao D, Zhao S, He C, Ding N, Li L, Long F. MiR-1246 promotes SiHa cervical cancer cell proliferation, invasion, and migration through suppression of its target gene thrombospondin 2. Arch Gynecol Obstet. 2014;290:725–732. doi: 10.1007/s00404-014-3260-2. [DOI] [PubMed] [Google Scholar]
- 12.Bao Y, Wang L, Shi L, Yun F, Liu X, Chen Y, Chen C, Ren Y, Jia Y. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell Mol Biol Lett. 2019;24:38. doi: 10.1186/s11658-019-0162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Kyriakides TR, Bornstein P. Matricellular proteins as modulators of cell-matrix interactions: Adhesive defect in thrombospondin 2-null fibroblasts is a consequence of increased levels of matrix metalloproteinase-2. Mol Biol Cell. 2000;11:3353–3364. doi: 10.1091/mbc.11.10.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Naito S, von Eschenbach AC, Giavazzi R, Fidler IJ. Growth and metastasis of tumor cells isolated from a human renal cell carcinoma implanted into different organs of nude mice. Cancer Res. 1986;46:4109–4115. [PubMed] [Google Scholar]
- 16.Menon SS, Guruvayoorappan C, Sakthivel KM, Rasmi RR. Ki-67 protein as a tumour proliferation marker. Clin Chim Acta. 2019;491:39–45. doi: 10.1016/j.cca.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Varghese F, Bukhari AB, Malhotra R, De A. IHC Profiler: An open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS One. 2014;9:e96801. doi: 10.1371/journal.pone.0096801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis-correlation in invasive breast carcinoma. N Engl J Med. 1991;324:1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 19.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network-another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Liu Y, Li G, Li L, Geng P, Song H. microRNA-214 suppresses the growth of cervical cancer cells by targeting EZH2. Oncol Lett. 2018;16:5679–5686. doi: 10.3892/ol.2018.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedroza-Torres A, Campos-Parra AD, Millan-Catalan O, Loissell-Baltazar YA, Zamudio-Meza H, Cantú de León D, Montalvo-Esquivel G, Isla-Ortiz D, Herrera LA, Ángeles-Zaragoza Ó, et al. MicroRNA-125 modulates radioresistance through targeting p21 in cervical cancer. Oncol Rep. 2018;39:1532–1540. doi: 10.3892/or.2018.6219. [DOI] [PubMed] [Google Scholar]
- 22.González-Quintana V, Palma-Berré L, Campos-Parra AD, López-Urrutia E, Peralta-Zaragoza O, Vazquez-Romo R, Pérez-Plasencia C. MicroRNAs are involved in cervical cancer development, progression, clinical outcome and improvement treatment response (Review) Oncol Rep. 2016;35:3–12. doi: 10.3892/or.2015.4369. [DOI] [PubMed] [Google Scholar]
- 23.Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krissansen GW, Yang Y, McQueen FM, Leung E, Peek D, Chan YC, Print C, Dalbeth N, Williams M, Fraser AG. Overexpression of miR-595 and miR-1246 in the sera of patients with active forms of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:520–530. doi: 10.1097/MIB.0000000000000285. [DOI] [PubMed] [Google Scholar]
- 25.Todeschini P, Salviato E, Paracchini L, Ferracin M, Petrillo M, Zanotti L, Tognon G, Gambino A, Calura E, Caratti G, et al. Circulating miRNA landscape identifies miR-1246 as promising diagnostic biomarker in high-grade serous ovarian carcinoma: A validation across two independent cohorts. Cancer Lett. 2017;388:320–327. doi: 10.1016/j.canlet.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 26.Zhang WC, Chin TM, Yang H, Nga ME, Lunny DP, Lim EK, Sun LL, Pang YH, Leow YN, Malusay SR, et al. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun. 2016;7:11702. doi: 10.1038/ncomms11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim G, An HJ, Lee MJ, Song JY, Jeong JY, Lee JH, Jeong HC. Hsa-miR-1246 and hsa-miR-1290 are associated with stemness and invasiveness of non-small cell lung cancer. Lung Cancer. 2016;91:15–22. doi: 10.1016/j.lungcan.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 28.Hasegawa S, Eguchi H, Nagano H, Konno M, Tomimaru Y, Wada H, Hama N, Kawamoto K, Kobayashi S, Nishida N, et al. MicroRNA-1246 expression associated with CCNG2-mediated chemoresistance and stemness in pancreatic cancer. Br J Cancer. 2014;111:1572–1580. doi: 10.1038/bjc.2014.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:644–652. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishi M, Nakamura M, Nishimine M, Ishida E, Shimada K, Kirita T, Konishi N. Loss of heterozygosity on chromosome 6q correlates with decreased thrombospondin-2 expression in human salivary gland carcinomas. Cancer Sci. 2003;94:530–535. doi: 10.1111/j.1349-7006.2003.tb01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ao R, Guan L, Wang Y, Wang JN. Silencing of COL1A2, COL6A3, and THBS2 inhibits gastric cancer cell proliferation, migration, and invasion while promoting apoptosis through the PI3k-Akt signaling pathway. J Cell Biochem. 2018;119:4420–4434. doi: 10.1002/jcb.26524. [DOI] [PubMed] [Google Scholar]
- 32.Tokunaga T, Nakamura M, Oshika Y, Abe Y, Ozeki Y, Fukushima Y, Hatanaka H, Sadahiro S, Kijima H, Tsuchida T, et al. Thrombospondin 2 expression is correlated with inhibition of angiogenesis and metastasis of colon cancer. Br J Cancer. 1999;79:354–359. doi: 10.1038/sj.bjc.6690056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weng TY, Wang CY, Hung YH, Chen WC, Chen YL, Lai MD. Differential expression pattern of THBS1 and THBS2 in lung cancer: Clinical outcome and a systematic-analysis of microarray databases. PLoS One. 2016;11:e161007. doi: 10.1371/journal.pone.0161007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fears CY, Grammer JR, Stewart JE, Jr, Annis DS, Mosher DF, Bornstein P, Gladson CL. Low-density lipoprotein receptor-related protein contributes to the antiangiogenic activity of thrombospondin-2 in a murine glioma model. Cancer Res. 2005;65:9338–9346. doi: 10.1158/0008-5472.CAN-05-1560. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura M, Oida Y, Abe Y, Yamazaki H, Mukai M, Matsuyama M, Chijiwa T, Matsumoto H, Ueyama Y. Thrombospondin-2 inhibits tumor cell invasion through the modulation of MMP-9 and uPA in pancreatic cancer cells. Mol Med Rep. 2008;1:423–427. [PubMed] [Google Scholar]
- 36.Kodama J, Hashimoto I, Seki N, Hongo A, Yoshinouchi M, Okuda H, Kudo T. Thrombospondin-1 and −2 messenger RNA expression in invasive cervical cancer: Correlation with angiogenesis and prognosis. Clin Cancer Res. 2001;7:2826–2831. [PubMed] [Google Scholar]
- 37.Kamochi J, Tokunaga T, Tomii Y, Abe Y, Hatanaka H, Kijima H, Yamazaki H, Watanabe N, Matsuzaki S, Ueyama Y, Nakamura M. Overexpression of the thrombospondin 2 (TSP2) gene modulated by the matrix metalloproteinase family expression and production in human colon carcinoma cell line. Oncol Rep. 2003;10:881–884. [PubMed] [Google Scholar]
- 38.Chen PC, Tang CH, Lin LW, Tsai CH, Chu CY, Lin TH, Huang YL. Thrombospondin-2 promotes prostate cancer bone metastasis by the up-regulation of matrix metalloproteinase-2 through down-regulating miR-376c expression. J Hematol Oncol. 2017;10:33. doi: 10.1186/s13045-017-0390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies KJ. The complex interaction of matrix metalloproteinases in the migration of cancer cells through breast tissue stroma. Int J Breast Cancer. 2014;2014:839094. doi: 10.1155/2014/839094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirose Y, Chiba K, Karasugi T, Nakajima M, Kawaguchi Y, Mikami Y, Furuichi T, Mio F, Miyake A, Miyamoto T, et al. A functional polymorphism in THBS2 that affects alternative splicing and MMP binding is associated with lumbar-disc herniation. Am J Hum Genet. 2008;82:1122–1129. doi: 10.1016/j.ajhg.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.