Abstract
Colorectal cancer is a common gastrointestinal cancer ranking in third place of all cancers. Downregulation of miR-148a has been observed in many tumors, and miR-148a was found to be an oncogene in colorectal cancer. The aim of our study was to investigate the molecular mechanisms by which miR-148a and ErbB3 proliferate and migrate in colorectal cancer. The expression of miR-148a and ErbB3 were measured by western blot analysis and RT-qPCR. MTT and transwell assays were performed to analyze the proliferative and migratory abilities. The dual luciferase reporter assay was employed to confirm miR-148a regulated the expression of ErbB3 in colorectal cancer. It was discovered that miR-148a was overexpressed while ErbB3 expression was low in colorectal cancer, and the mRNA level of miR-148a had a negative correlation with the expression of ErbB3. Upregulation of miR-148a suppressed the proliferation and migration in colorectal cancer cells. Furthermore, ErbB3 was identified as a direct target of miR-148a, which suppressed the proliferation and migration through directly binding to the 3′UTR of ErbB3 mRNA. This study established that miR-148a inhibited the proliferative and migratory abilities through mediating the expression of ErbB3. The newly identified miR-148a/ErbB3 axis provides novel insight into the pathogenesis of colorectal cancer, and represents a potential target for treatment of colorectal cancer.
Keywords: miR-148a, colorectal cancer, proliferation, migration, ErbB3
Introduction
Colorectal cancer (CRC) is a common gastrointestinal cancer that affects more than 900,000 patients each year, and its incidence is ranked third of all cancers (1,2). Colorectal cancer is mainly divided into adenocarcinoma, mucinous adenocarcinoma and undifferentiated carcinoma (3). It is necessary to further investigate the pathogenesis and biological features of colorectal cancer.
MicroRNAs (miRNAs) are a class of non-coding RNAs that mediate gene expression at the post-transcriptional level (4,5). Growing evidence demonstrates that miRNAs could act as oncogenes or tumor suppressors in diverse tumors including colorectal cancer (2,6,7). Furthermore, increasing miRNAs has been reported to play an important role in colorectal cancer, such as miR-144, miR-495, miR-590, miR-6803 (8–11). miR-148a has been reported to be downregulated in many kinds of tumors including breast cancer, renal cell carcinoma and endometrial cancer (12–14). According to Li et al (12), miR-148a promotes apoptosis and inhibits growth of breast cancer. Feng et al (15) reported similar findings that miR-148a suppressed the proliferation and migration in pancreatic cancer cells. In addition, miR-148a could directly bind to 3′UTR of target mRNAs to effect cell progression, and the target genes including BCL-2, AKT2, IQGAP1 and ErbB3 (12–15). Our study explored the pivotal roles of miR-148a and ErbB3 on the proliferation and migration of colorectal cancer.
ErbB3, known as HER3, is a transmembrane tyrosine kinase receptor that is the only member of the ErbB receptor family that lacks tyrosine kinase activity, and containing other three members: ErbB1 (EGFR, HER1), ErbB2 (Neu, HER2), and ErbB4 (HER4) (16–19). Inactivation of ErbB3 promotes cell apoptosis and inhibits the growth and invasiveness of lung adenocarcinoma cell (20). Similarly, Appert-Collin et al (21) found that ErbB3 was overexpressed and promoted migration and invasion in gliomas and in non-small cell lung carcinoma (21). Silence of ErbB3 reduced the proliferation and tumor growth in osteosarcoma cells. Considering all the findings, we verified the hypothesis that miR-148a regulated the proliferation and migration through suppressing the expression of ErbB3 in CRC cells. miR-148a has been found to be downregulated in CRC tissues and cell lines, and it has an inverse correlation between the expression of miR-148a and ErbB3 in CRC tissues. We subsequently found that miR-148a targeted the 3′UTR of ErbB3 mRNA and regulated the expression of ErbB3 in CRC cells. Moreover, we discovered that downregulation of miR-148a caused repression of ErbB3, thereby suppressing the proliferation and migration of colorectal cancer cells.
Patients and methods
Patients and clinical samples
Fifty-one pairs of human colorectal cancer tissue samples and corresponding paracancerous tissues were obtained from colorectal cancer patients at the First Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China) from 2015 to 2017. The tissues were immediately snap-frozen in liquid nitrogen and stored in a −80°C freezer. None of these patients had local or systemic treatment before operation.
The present study was approved by the Ethics Committee of The First Affiliated Hospital of Xi'an Jiaotong University (Human ethic no. 2015-06). Patients who participated in this research had complete clinical data. The signed informed consents were obtained from the patients or the guardians.
Cell lines and culture condition
Human colorectal cancer cell lines LoVo (cat. no. CCL-229) and SW480 (cat. no. CCL-228) and the normal colon cell line CCD-18Co (cat. no. CRL-1459) were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). All the cancer and normal cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C and 5% CO2.
RNA isolation and RT-qPCR
Total RNAs (from tissues and cell lines) were extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Waltham, MA, USA) containing total miRNAs, following the manufacturers instructions of MagMAX™ and mirVana™ total RNA isolation kits (both from Thermo Fisher Scientific, Inc.). The temperature conditions for reverse transcription were as follows: 37°C for 15 min and 85°C for 5 sec. Thermoscript RT-qPCR system (Takara Biotechnology Co., Ltd., Dalian, China) were utilized to perform reverse-transcription. SYBR Prime Script miRNA RT-qPCR kit or SYBR premix kit (Takara Biotechnology Co.) were applied to analyze qPCR on an Applied Biosystems 7300 sequence detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). Relative expression levels of mRNA and miRNA were calculated using 2−ΔΔCq method (22). The relative quantification of ErbB3 mRNA was normalized by GAPDH. The premier sequences were: For miR-148a, F: 5′-ACACTCCAGCTGGGTCAGTGCACTACA-GAA-3′, and R: 5′-TGGTGTCGTGGAGTCG-3′; for U6, F: 5′-TCCGATCGTGAAGCGTTC-3′, and R: 5′-GTGCAGGGTCCGAGGT-3′; for ErbB3, F: 5′-TTCCGAGATGGGCAACTCTC-3′, and R: 5′-CTTGCAGACTTCGTGACAGG-3′; for GADPH, F: 5′-GAAGGTGAAGGTCGGAGTC-3′ and R: 5′-ATCCAGTGCAGGGTCCGAGG-3′. The reactions were incubated in a 96-well plate at 95°C for 5 min, followed by 40 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec.
Protein extraction and western blot analysis
Total proteins were extracted from tissue specimens and cell lines on ice using RIPA lysis buffer containing 1% proteinase inhibitor (Beyotime Institute of Biotechnology, Haimen, China). Same amount of proteins (50 µg) were separated using 8% SDS polyacrylamide gels, which were quantitated by BCA protein assay kit (Solarbio, Beijing, China); and then the protein blots were transferred onto polyvinylidene difluoride membrane (PVDF; Bio-Rad Laboratories, Inc., Hercules, CA, USA). After blocking in 5% at room temperature for 1.5 h non-fat milk, the membrane was incubated with rabbit anti-ErbB3 monoclonal antibody (cat. no. 32121; dil, 1:1,000; Abcam, Cambridge, UK) and anti-GAPDH mouse monoclonal antibody (cat. no. 8795; 1:4,000; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), which was used as internal reference. We visualized the proteins using ECL detection kit (GE Healthcare, Chicago, IL, USA) on Bio-Rad Gel Doc XR instrument (Bio-Rad Laboratories, Inc.).
Cell proliferation assay
The cellular proliferative capacity was performed using 3-(4,5-dimethylthiazol-2-yl)-2,5-dipheny-ltetrazolium bromide (MTT; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and dimethyl sulfoxide (DMSO; Beijing Solarbio Science & Technology Co., Ltd.) solutions. The applicable cells were seeded into 96-well plates and cultured in 37°C incubator for 24, 48, 72 and 96 h. The cells were added with 10 µl MTT solution for 4 h and then 150 µl DMSO in order to solubilize the formazan crystals. The optical density was measured by a microplate reader (BioTek China, Beijing, China) at wavelength of 490 nm.
Transwell assay
Transwell assay utilized 8 µm pore inserts covered with or without Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) to detect the invasion and migratory abilities in a 24-well plate, in upper and lower chambers. Subsequently, cell suspension prepared in serum-free medium was added into the upper chamber, whereas 500 µl RPMI-1640 medium was added into the lower chamber. After migrating for 24 h, the non-migrating cells were removed using cotton swab. The migratory or invasive cells were fixed in 4% paraformaldehyde and then stained with 0.1% crystal violet. The cell number in five random fields were counted under a microscope (Olympus, Tokyo, Japan) and the average number calculated.
Transfection
Cells were transfected with miR-148a mimic, and miR-148a inhibitor to up or down regulate the miR-148a expression, and pcDNA3.1-ErbB3 vectors were employed to overexpress ErbB3 (Shanghai GenePharma Co., Ltd., Shanghai, China).
LoVo cells (4×106 cells/well) were seeded into a 6-well plate at 80% density, using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After transfected for 48 h, the cells were harvested for western blot analysis or RT-qPCR.
Plasmid construction and luciferase reporter assay
TargetScan (http://www.targetscan.org/vert_71/) was utilized to predict the target genes of miR-148a and to confirm that ErbB3 was a target gene of miR-148a. ErbB3 mRNA containing the putative miR-148a binding sites on 3′UTR segment was cloned into firefly luciferase vector pmirGlo (pmirGlo-ErbB3-WT, WT). QuickChange Site-Directed Mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA, USA) was applied to mutate the binding sites from UGCACUG to ACGUGAC (pmirGlo-ErbB3-MUT, MUT).
LoVo cells were seeded into 6-well plates and cultured overnight, and then co-transfected with miR-148a or control and pmirGlo-ErbB3-WT or pmirGlo-ErbB3-MUT vectors with Renilla luciferase report vector as the internal control. After 48 h, dual-luciferase assay kit (Promega Corporation, Madison, WI, USA) was employed to perform the luciferase activity.
Statistical analysis
Statistical analysis was executed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). Statistical analysis was performed using Student's t-test between two groups and Kruskall-Wallis test with Bonferonni's post hoc test for multiple groups. Differences were considered statistically significant at P<0.05.
Results
The relationship of miR-148a and ErbB3 in colorectal cancer
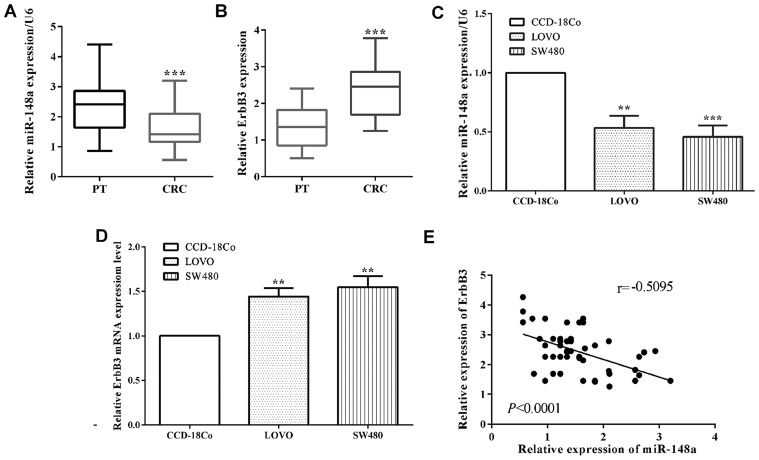
The mRNA level of miR-148a in colorectal cancer tissues was determined to be lower than that in corresponding paracancerous tissue samples (P<0.0001) (Fig. 1A). Due to the limitation of the experiment condition, we did not evaluate the expression of ErbB3 in paraffin block. However, RT-qPCR was performed to calculate the expression of ErbB3 in colorectal cancer tissue. The expression of ErbB3 was higher in colorectal cancer tissues than that in the corresponding paracancerous tissues (P<0.0001) (Fig. 1B). Moreover, in colorectal cancer LoVo cell lines (P=0.0014) and SW480 (P=0.0006), the mRNA level of miR-148a was also reduced versus the normal colon cells CCD-18Co performed by RT-qPCR (Fig. 1C). On the contrary, the mRNA level of ErbB3 of CRC cell lines LoVo (P=0.0012) and SW480 (P=0.0016) were upregulated compared to CCD-18Co cells (Fig. 1D). In addition, the mRNA level of miR-148a had a negative relationship with ErbB3 (P<0.0001; r=0.5095) in colorectal cancer tissues (Fig. 1E). Our results showed that miR-148a is downregulated while ErbB3 was upregulated in NPC cells.
Figure 1.
Expression of miR-148a and ErbB3 in CRC tissues and cell lines. (A) The expression of miR-148a in 51 paired colorectal cancer and paracancerous tissue samples were evaluated by RT-qPCR. (B) The expression of ErbB3 in 51 paired colorectal cancer and paracancerous tissue samples evaluated by RT-qPCR. (C) The expression level of miR-148a was also measured in CRC cells and the normal colon CCD-18Co cells. (D) The expression level of ErbB3 in CRC cell lines and the normal colon CCD-18Co cells was detected. (E) Correlation between the expression of miR-148a and ErbB3 in CRC tissues. **P<0.01; ***P<0.001; PT, paracancerous tissues; CRC, colorectal cancer.
miR-148a inhibits proliferation of colorectal cancer LoVo cells
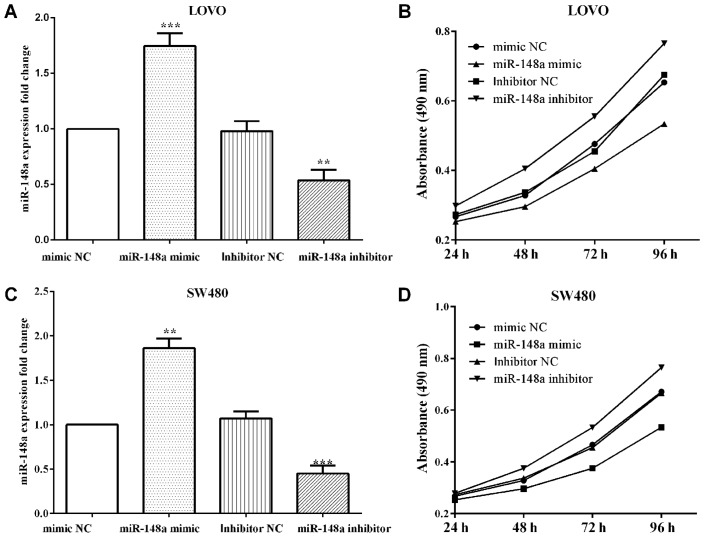
To overexpress or knockdown miR-148a, miR-148a mimics (P=0.0004 and P=0.0026) or inhibitor (P=0.0043 and P=0.0008) were transfected, as well as their negative control in LoVo and SW480 cells and then MTT and Transwell assays were performed (Fig. 2A and C). The proliferative absorbancy was reduced (P<0.0001) when transfected with miR-148a mimic, while increased (P<0.0001) when transfected with miR-148a inhibitor in LoVo cells (Fig. 2B). Similar findings to the results in LoVo cells were found where miR-148a mimic decreased the proliferation (P<0.0001), and miR-148a inhibitor increased in SW480 (P<0.0001) (Fig. 2D).
Figure 2.
miR-148a inhibits the proliferation and migration in CRC cells (A and C) After transfection with the miR-148a mimic or inhibitor to up/down regulate miR-148a in LoVo and SW480 cells. (B and D) The absorbancy of proliferation was attenuated when transfected with miR-148a mimic, whereas it was promoted by transfecting with miR-148a inhibitor in LoVo and SW480 cells. **P<0.01; ***P<0.001.
miR-148a inhibits migration and invasion of colorectal cancer LoVo and SW480 cells
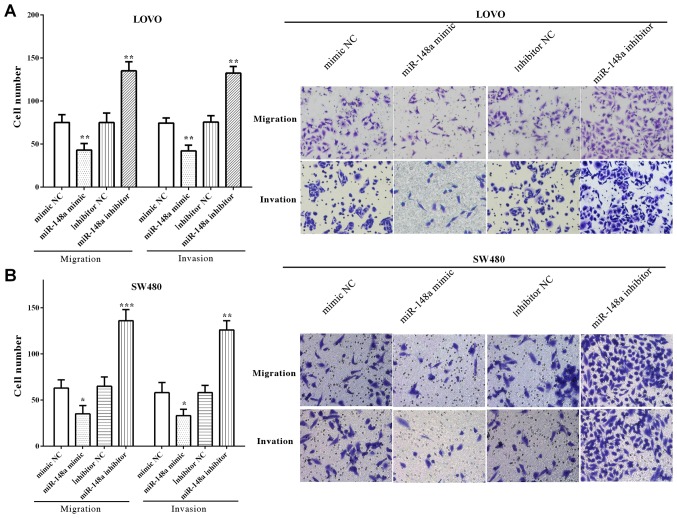
The migratory and invasive number of LoVo cells was decreased (P=0.0095 and P=0.0052) after upregulated miR-148a, whereas enhanced (P=0.0024 and P=0.0035) when downregulated with miR-148a (Fig. 3A). Similarly, cell migration and invasion were reduced by miR-148a mimic (P=0.0342 and P=0.0284), whereas increased by miR-148a inhibitor (P=0.0002 and P=0.0075) (Fig. 3B). All the results demonstrated that miR-148a inhibited the migration and invasion of LoVo and SW480 cells.
Figure 3.
miR-148a inhibits migration and invasion of colorectal cancer LoVo and SW480 cells. (A) Migration and invasion were reduced after upregulating miR-148a, while it was increased when downregulated miR-148a in LoVo cells (×200). (B) The migratory and invasive abilities were decreased by miR-148a mimic, whereas increased by miR-148a inhibitor in SW480 cells (×200). *P<0.05; **P<0.01; ***P<0.001.
miR-148a targets ErbB3 and inhibits its expression
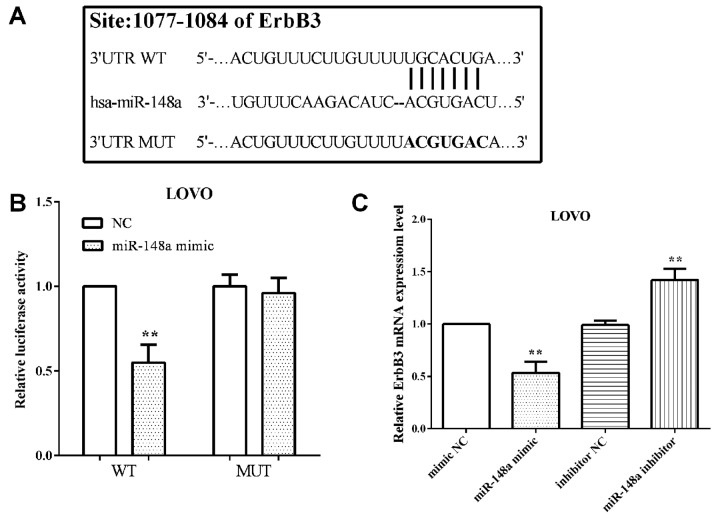
TargetScan software predicted that ErbB3 was a direct target gene of miR-148a and the binding sequence was UGCACUGA, which was located at 1077 to 1084 on 3′UTR of mRNA. The binding sequences were then mutated from 5′-…ACUGUUUCUUGUUUUUGCACUGA…-3′ to 5′-…ACUGUUUCUUGUUUUACGUGACA…-3′, as shown in Fig. 4A.
Figure 4.
ErbB3 is a target gene of miR-148a and mediated by miR-148a in CRC cells. (A) The complementary sequences of miR-148a and ErbB3, as well as the mutant sequences of ErbB3 binding site. (B) ErbB3 WT binds to miR-148a while the MUT does not. (C) ErbB3 expression changes along with miR-148a exogenous changes. **P<0.01; NC, negative control; WT, wild-type of ErbB3 3′UTR; MUT, mutant of ErbB3 3′UTR.
The luciferase reporter assay was performed to confirm that ErbB3 was a direct target of miR-148a. We co-transfected pmirGlo-miR-148a mimic or negative control and pmirGlo-ErbB3-WT or pmirGlo-ErbB3-MUT into LoVo cells and measured the luciferase reporter activity. The results showed that miR-148a suppressed (P=0.0018) pmirGlo-ErbB3-WT reporter activity, but not pmirGlo-ErbB3-MUT (P=0.05762) (Fig. 4B), which illustrated ErbB3 wild-type binding to miR-148a, whereas not that of the mutant.
The miR-148a mimic or inhibitor was transfeted to alter the expression of miR-148a and to determined the mRNA level of ErbB3. As expected, transfection of miR-148a mimic decreased (P=0.0016) the mRNA level of ErbB3 in LoVo cells. Simultaneously, the mRNA level of ErbB3 was increased (P=0.0027) with miR-148a in LoVo cells (Fig. 4C).
ErbB3 reverses partial function of miR-148a
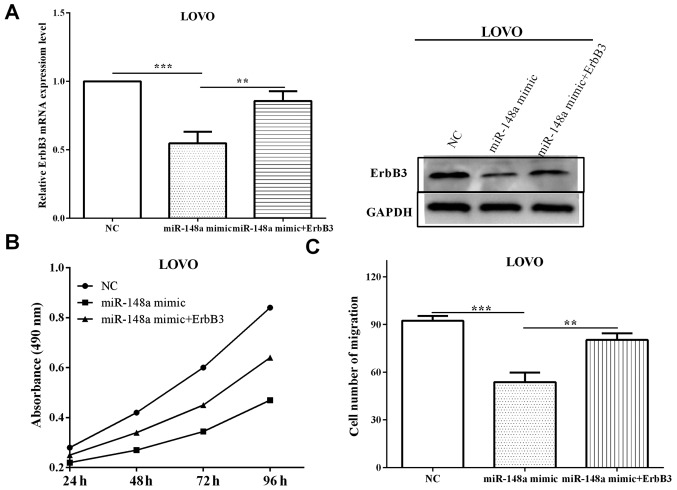
To explore whether ErbB3 re-expression could reverse the inhibitory effects of miR-148a on ErbB3 and cell proliferation and migration miR-148a mimic was transfected into LoVo cells, along with ErbB3 overexpression plasmid (pcNA31-ErbB3) or negative control. The cells transfected with miR-148a mimic and pcNA31-ErbB3 exhibited higher (P=0.0084) ErbB3 level compared with cells transfected with miR-148a in mRNA and protein levels (Fig. 5A), suggesting that ErbB3 rescued the suppression by miR-148a. Subsequently, the changes of proliferation and migration affected by miR-148a mimic and ErbB3 were measured. The proliferative absorbancy was increased (P=0.0032) when re-transfected with ErbB3, compared with only transfected with miR-148a mimic, which was decreased (P=0.00006) versus negative control (Fig. 5B). Migration was reduced (P=0.00009) when transfected with miR-148a mimic, and the roles were partially reversed (P=0.0079) after re-transfected with ErbB3 (Fig. 5C). Taken together, these results suggest that miR-148a affects cell proliferative and migratory abilities through directly mediating ErbB3.
Figure 5.
ErbB3 partly abrogates the impact of miR-148 on CRC cell proliferation and migration. (A) The mRNA and protein levels of ErbB3 were increased when re-expressed in LoVo cells. (B) The proliferative absorbancy was determined when co-transfected with ErbB3 and miR-148a mimic or only transfected with miR-148a mimic. (C) The number of migration was determined when co-transfected with ErbB3 and miR-148a mimic or only transfected with miR-148a mimic. **P<0.01; ***P<0.001.
Discussion
Colorectal cancer has a high incidence, and is divided into adenocarcinoma, mucinous adenocarcinoma and undifferentiated carcinoma (1,3). Colorectal cancer has a high mortality rate and due to drug resistance and relapse, the outcome is poor. Therefore, it is urgent to identify new biomarkers for effective therapeutics of CRC. MicroRNAs are a class of non-coding RNAs, which mediate gene expression at post-transcriptional level (4,5). Increasing evidence demonstrates that miRNAs play important roles in cancer inhibition and promotion of colorectal cancer (2,6,7). miR-148a is downregulated and are associated with several cellular processes including development, differentiation, growth, migration and apoptosis (12–14). In breast cancer, Li et al discovered that miR-148a promoted apoptosis and inhibited growth (12). Similarly, Feng et al demonstrated that miR-148a suppressed the proliferation and migration in pancreatic cancer cells (15). Our results were consistent with these findings, miR-148a was found to be downregulated in colorectal cancer tissue samples and cells, while ErbB3 was upregulated. In addition, miR-148a suppressed the proliferation and migration through directly targeting ErbB3 in LoVo cells. However, there are likely to be many factors governing proliferation of CRC cells, thus, cell proliferation was partially suppressed by miR-148a.
ErbB3 is a transmembrane tyrosine kinase receptor, which is a member of ErbB receptor family (16–19). In lung adenocarcinoma, ErbB3 promotes cell apoptosis and inhibits cell growth and invasiveness (20). Similar findings were found by Appert-Collin et al (21), ErbB3 was usually overexpressed and promoted cell migration and invasion in gliomas and non-small cell lung carcinoma. However, in colorectal cancer miR-148a downregulation contributed to carcinogenesis and cell invasion (2). Our findings were consistent with these findings. ErbB3 was overexpressed in colorectal cancer tissues and cell lines LoVo and SE480. In CRC tissues, the mRNA level of ErbB3 had a negative correlation with the expression of miR-148a. Furthermore, Yu et al (23) elucidated that miR-148a inhibited tumor angiogenesis through targeting ErbB3 and its downstream signaling molecules. Similar findings were found by Feng et al as miR-148 suppressed the proliferation and migration through downregulating ErbB3 in pancreatic cancer cells (15). Our results were consistent with the above findings, ErbB3 was a direct target of miR-148a and it reversed partial function of miR-148a on cell proliferation and migration. We propose that miR-148a impact the proliferation and migration through directly targeting ErbB3 in colorectal cancer cells.
In conclusion, we established that miR-148a was downregulated while ErbB3 was upregulated in colorectal cancer tissues and cell lines. miR-148a was found to suppress the proliferative and migratory abilities by directly regulating the expression of ErbB3 in colorectal cancer cells. The newly identified miR-148a/ErbB3 axis provides novel insight into pathogenesis of colorectal cancer, and represents a potential target for colorectal cancer treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XS as the corresponding author contributed to the conception of the study. WZ and JZ, contributed significantly to analysis and manuscript preparation. GW, performed the data analyses and wrote the manuscript. KY and GW, helped perform the analysis with constructive discussions. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of The First Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China) (Human ethic no. 2015-06). Patients who participated in this research had complete clinical data. The signed informed consents were obtained from the patients or the guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Peng YC, Lin CL, Hsu WY, Chang CS, Yeh HZ, Liao SC, Kao CH. The risk of colorectal cancer is related to frequent hospitalization of IBD in an Asian population: Results from a nationwide study. QJM. 2015;108:457–463. doi: 10.1093/qjmed/hcu225. [DOI] [PubMed] [Google Scholar]
- 2.Hibino Y, Sakamoto N, Naito Y, Goto K, Oo HZ, Sentani K, Hinoi T, Ohdan H, Oue N, Yasui W. Significance of miR-148a in colorectal neoplasia: Downregulation of miR-148a contributes to the carcinogenesis and cell invasion of colorectal cancer. Pathobiology. 2015;82:233–241. doi: 10.1159/000438826. [DOI] [PubMed] [Google Scholar]
- 3.Jass JR, Love SB, Northover JM. A new prognostic classification of rectal cancer. Lancet. 1987;1:1303–1306. doi: 10.1016/S0140-6736(87)90552-6. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol Med. 2010;16:257–267. doi: 10.1016/j.molmed.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doleshal M, Magotra AA, Choudhury B, Cannon BD, Labourier E, Szafranska AE. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2008;10:203–211. doi: 10.2353/jmoldx.2008.070153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao R, Li C, Chai B. miRNA-144 suppresses proliferation and migration of colorectal cancer cells through GSPT1. Biomed Pharmacother. 2015;74:138–144. doi: 10.1016/j.biopha.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Yan L, Yao J, Qiu J. miRNA-495 suppresses proliferation and migration of colorectal cancer cells by targeting FAM83D. Biomed Pharmacother. 2017;96:974–981. doi: 10.1016/j.biopha.2017.11.138. [DOI] [PubMed] [Google Scholar]
- 10.Kim CW, Oh ET, Kim JM, Park JS, Lee DH, Lee JS, Kim KK, Park HJ. Hypoxia-induced microRNA-590-5p promotes colorectal cancer progression by modulating matrix metalloproteinase activity. Cancer Lett. 2018;416:31–41. doi: 10.1016/j.canlet.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 11.Yan S, Jiang Y, Liang C, Jin C, Duan Q, Xu D, Yang L, Zhang X, Ren B, Jin P. Exosomal miR-6803-5p as potential diagnostic and prognostic marker in colorectal cancer. J Cell Biochem. 2018;119:4113–4119. doi: 10.1002/jcb.26609. [DOI] [PubMed] [Google Scholar]
- 12.Li Q, Ren P, Shi P, Chen Y, Xiang F, Zhang L, Wang J, Lv Q, Xie M. MicroRNA-148a promotes apoptosis and suppresses growth of breast cancer cells by targeting B-cell lymphoma 2. Anticancer Drugs. 2017;28:588–595. doi: 10.1097/CAD.0000000000000498. [DOI] [PubMed] [Google Scholar]
- 13.Cao H, Liu Z, Wang R, Zhang X, Yi W, Nie G, Yu Y, Wang G, Zhu M. miR-148a suppresses human renal cell carcinoma malignancy by targeting AKT2. Oncol Rep. 2017;37:147–154. doi: 10.3892/or.2016.5257. [DOI] [PubMed] [Google Scholar]
- 14.Dong P, Ihira K, Xiong Y, Watari H, Hanley SJ, Yamada T, Hosaka M, Kudo M, Yue J, Sakuragi N. Reactivation of epigenetically silenced miR-124 reverses the epithelial-to-mesenchymal transition and inhibits invasion in endometrial cancer cells via the direct repression of IQGAP1 expression. Oncotarget. 2016;7:20260–20270. doi: 10.18632/oncotarget.7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng H, Wang Y, Su J, Liang H, Zhang CY, Chen X, Yao W. MicroRNA-148a suppresses the proliferation and migration of pancreatic cancer cells by down-regulating ErbB3. Pancreas. 2016;45:1263–1271. doi: 10.1097/MPA.0000000000000677. [DOI] [PubMed] [Google Scholar]
- 16.Campbell MR, Amin D, Moasser MM. HER3 comes of age: new insights into its functions and role in signaling, tumor biology, and cancer therapy. Clin Cancer Res. 2010;16:1373–1383. doi: 10.1158/1078-0432.CCR-09-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guy PM, Platko JV, Cantley LC, Cerione RA, Carraway KL., III Insect cell-expressed p180erbB3 possesses an impaired tyrosine kinase activity. Proc Natl Acad Sci USA. 1994;91:8132–8136. doi: 10.1073/pnas.91.17.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sierke SL, Cheng K, Kim HH, Koland JG. Biochemical characterization of the protein tyrosine kinase homology domain of the ErbB3 (HER3) receptor protein. Biochem J. 1997;322:757–763. doi: 10.1042/bj3220757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc Natl Acad Sci USA. 2009;106:21608–21613. doi: 10.1073/pnas.0912101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sithanandam G, Fornwald LW, Fields J, Anderson LM. Inactivation of ErbB3 by siRNA promotes apoptosis and attenuates growth and invasiveness of human lung adenocarcinoma A549. Oncogene. 2005;24:1847–1859. doi: 10.1038/sj.onc.1208381. [DOI] [PubMed] [Google Scholar]
- 21.Appert-Collin A, Hubert P, Crémel G, Bennasroune A. Role of ErbB receptors in cancer cell migration and invasion. Front Pharmacol. 2015;6:283. doi: 10.3389/fphar.2015.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Yu J, Li Q, Xu Q, Liu L, Jiang B. MiR-148a inhibits angiogenesis by targeting ERBB3. J Biomed Res. 2011;25:170–177. doi: 10.1016/S1674-8301(11)60022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.