Abstract
Whereas the structures of small to medium-sized well folded RNA molecules often can be determined by either X-ray crystallography or NMR spectroscopy, obtaining structural information for large RNAs using experimental, computational, or combined approaches remains a major interest and challenge. RNA is very sensitive to small-angle X-ray scattering (SAXS) due to high electron density along phosphate-sugar backbones, whose scattering contribution dominates SAXS intensity. For this reason, SAXS is particularly useful in obtaining global RNA structural information that outlines backbone topologies and, therefore, molecular envelopes. Such information is extremely valuable in bridging the gap between the secondary structures and three-dimensional topological structures of RNA molecules, particularly those that have proven difficult to study using other structure-determination methods. Here we review published results of RNA topological structures derived from SAXS data or in combination with other experimental data, as well as details on RNA sample preparation for SAXS experiments.
Introduction
It was recognized in the 1980s that small-angle X-ray scattering (SAXS) data contain information about the overall shape of biomacromolecules in solution [1,2]. Applications of SAXS for biomolecule studies exploded after the third generation of synchrotron became operational and a number of critical computer programs and software tools became available. Over the years, those software tools have been developed to mine and interpret this information such as to reconstruct the low-resolution molecular envelopes of several macromolecules or to extract structural parameters mostly for proteins or protein complexes (Figure 1) [3•,4,5,6••,7,8•,9,10]. In almost the same way, SAXS has also been applied to determine dimensional and physical parameters of RNAs (Figure 1), such as radius of gyration (Rg) [11], maximum particle diameter (Dmax), molecular weight, Porod volume (Vp) [12••], and molecular compactness [13•, 14]. There are a number of excellent reviews on the fundamental physics of SAXS [2,15], computational tools and their applications [5,6••,9,16•]. There are also several reviews covering various aspects of applications of SAXS to study RNA [17,18•,19,20]. This current review specifically focuses on the application of SAXS for determining RNA 3D topological structures.
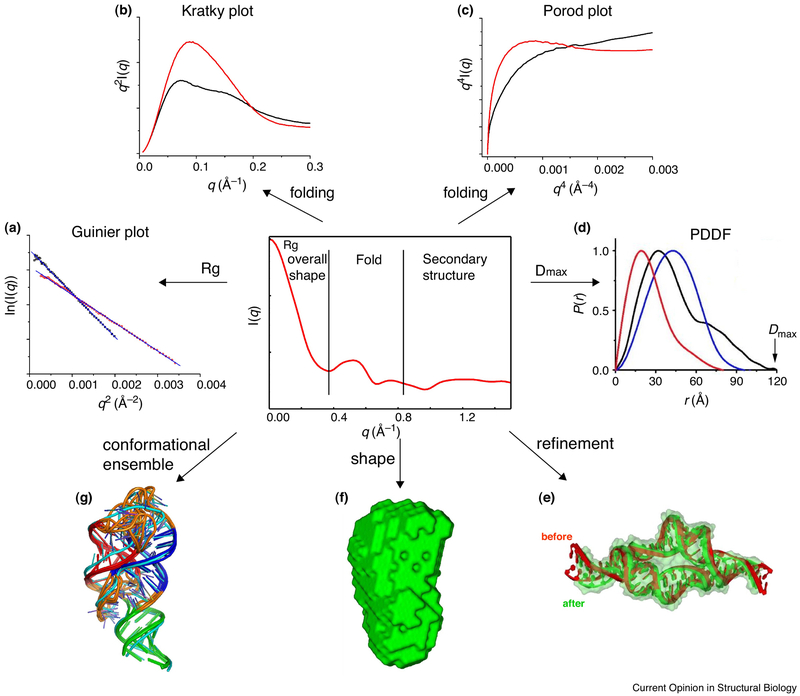
Figure 1.
A schematic illustration of useful information that can be extracted from SAXS data. (a). Guinier plot from which one can obtain radius of gyration and approximate molecular weight; (b) & (c) Kratky and Porod plots to characterize dynamics and folding [12••]; (d) Pair distance distribution function (PDDF) to derive maximum dimension, Dmax and weighted distance distribution tally; (e) Refining RNA structure by restraining global dimension [56•]; (f) generating global molecular shapes [6••]; (g) calculating conformational ensembles.
The sugar-phosphate backbone of RNA makes it more electron-rich and, therefore, a stronger scatterer of X-rays than protein (Figure 2). In the analysis of experimental SAXS data, RNA has the following advantageous features compared to its protein counterpart: (a) Secondary structures of RNAs based on predictions [21,22] and/or experimental determination [23] are usually known prior to structure determination; (b) RNA structure tends to be relatively modular with a limited variety of structural elements and motifs [24••]; and (c) RNA is usually non-spherical in overall shape [25••]. In essence, RNA tertiary structures can be generally understood as being “packed” predominantly with A-form duplexes, which are linked by “single-stranded” linkers in junctions [26••,27••,28••], interrupted by asymmetrical internal loops/bulges or “capped” with terminal structured or non-structured loops. The junctions themselves are prevalent structural elements, where intricate interactions among the junction residues dictate overall folding of RNAs [26••,27••,28••]. Major structural features such as long duplexes, which have a cross-sectional diameter of ~22 Å, can be readily recognized in a SAXS-derived molecular envelope, such as in the Varkud satellite (VS) ribozyme [29••]. Consequently, SAXS has been utilized as a valuable low-resolution technique to determine the 3D topological structures of large RNAs.
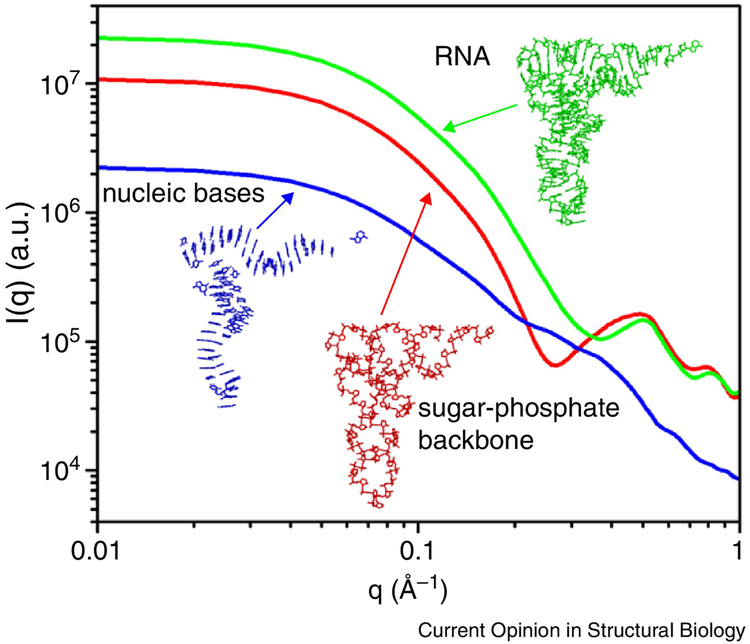
Figure 2.
A schematic illustration of contributions to the SAXS profiles from nucleobases (blue) and sugar-phosphate backbone (red) of a tRNAphe. The electron-rich sugar-phosphate backbone dominates the shape of the SAXS profile [62].
Constructing global structures of RNA
It is recently recognized that RNA 3D structure and folding pathways are encoded in RNA secondary structure [30••,31] and that RNA coarse structures may simplistically be viewed as a collection of duplexes that are branched out at junctions for multi-domain RNA [28••]. The RNA conformation space, which consists of all possible global arrangements of duplexes, is restricted to a subspace due to covalent linkages imposed by linkers at junctions [28••,30••,31]. This subspace is further reduced to a few possible conformers because of preferential folding patterns of various types of junctions and tertiary interactions [26••,27••,28••,30••]. Thus, in principle, one might be able to identify a correct fold based on secondary structural information, given an approximate global shape and a few tertiary contacts.
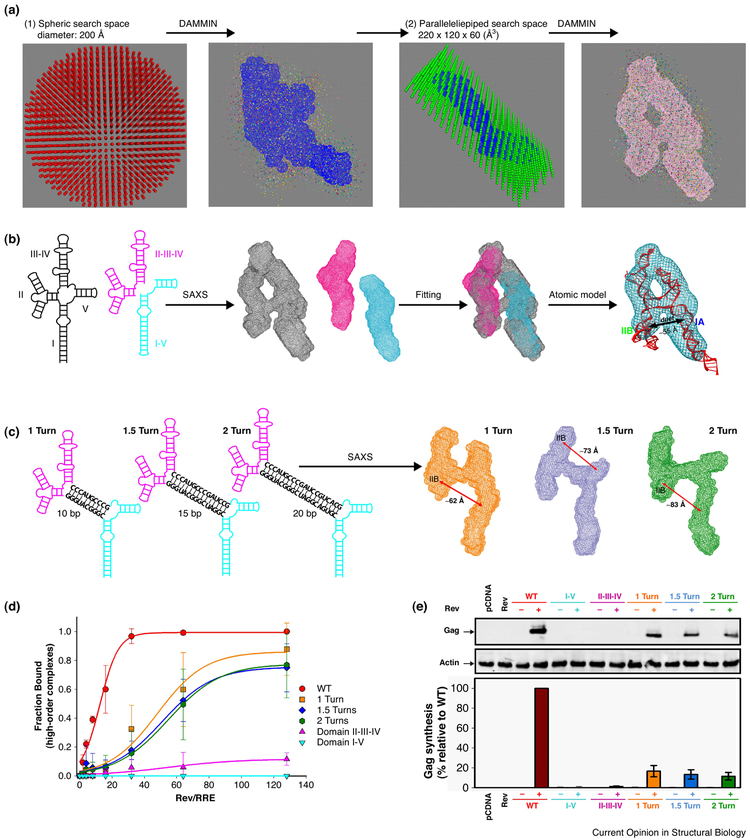
The first RNA whose molecular shape was outlined using SAXS was a tRNA in 1964 by J. Witz [32]. In this study the tRNA was depicted as a boomerang-shaped molecule composed of two double-helices connected by a sharp kink. This model approximately agreed with the later-determined L-shaped high-resolution structure of the tRNAphe [33]. Around the year 2000, several software programs were developed for the reconstruction of biomacromolecular shapes from SAXS data. Among these is the most popular ab-initio 3D-shape reconstruction software, DAMMIN [6••]. The program’s algorithm is to search through a defined volume of densely packed beads (dummy atoms) and find a model(s) that best fits the experimental data using simulated annealing [6••]. The initial search space is usually a bead-filled sphere whose diameter is defined by Dmax. The search calculation is usually performed several tens of times with random number seeding, and accepted sets of values are used to compute an average shape.
The first RNA 3D-shape generated by ab-initio reconstruction was the free Thermus flavus 5S ribosomal RNA (rRNA) at 13 Å resolution, and a putative 5S rRNA atomic model was also presented [34]. The calculated molecular envelope of the 5S rRNA based on the SAXS data was remarkably similar to that in crystal structures, even though the proposed putative atomic model was not completely correct [34]. More widespread use of SAXS for RNA shape reconstruction did not start until the late 2000s [25••,29••,35,36,37•,38,39.40••,41•]. For many RNAs, the 3D molecular shapes determined using ab-initio reconstruction have been highly valuable since they are often the only source of 3D structural information available and are sufficient to answer some key biological questions.
SAXS-aided divide-and-conquer strategies have also been employed to solve rather complex global topological structures of RNA. The first example of this approach is the VS ribozyme, a 160-nucleotide (nt) RNA consisting of multiple double-helices, junctions, and hairpins. In this pioneering and very clever work, Lipfert el al. used SAXS and a divide-and-conquer approach to define the locations of the various domain segments within the molecular envelope (Figure 3) [29••]. The topological structure of VS ribozyme is consistent with previous results of chemical accessibility probing [42] and later activity studies [43]. Such a strategy appears to be generally applicable in many other cases. Another example is the HIV-1 5' untranslated region (5' UTR), in which nucleotides 1-334 were divided into three segments and studied using SAXS and computational modeling [36]. This structure revealed that the primer binding site (PBS) folds into an L-shape, similar to tRNA, thus providing a structural basis for interaction with tRNA-binding cellular factors.
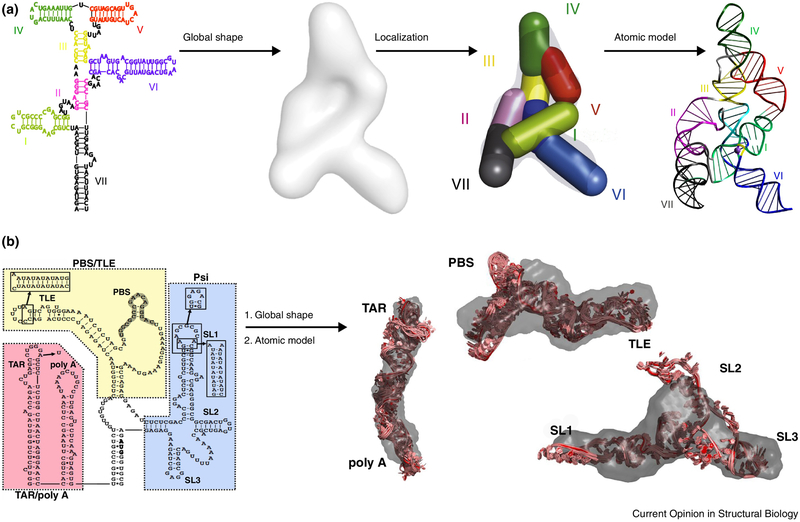
Figure 3.
Two examples of applying the “divide-and-conquer” strategy and SAXS to derive three-dimensional topological structures of large RNAs. (a) Varkud satellite (VS) ribozyme; [29••] (b) HIV-1 5' untranslated region (5' UTR) RNA [36]. In the VS ribozyme case, it was the first demonstration of the strategy to delineate the three-dimensional fold of an RNA using SAXS and the secondary structure information.
Global shape analysis using SAXS has become a useful tool to study conformation “switching” of riboswitch RNAs [25••,44,45]. Riboswitches are structural elements of mRNA involved in regulation of gene expression. Binding of their cognate ligands is believed to induce global conformational changes besides local structural change in binding pockets. However, a recent systematic study of aptamer domains of eight different riboswitches reveals considerable diversity amongst the range of conformation changes taking place in the ligand binding domains [25••,44]. Those changes, revealed by SAXS envelope shapes, range from distinct global conformation switching, such as in the thiamine pyrophosphate (TPP) and cyclic diguanylate (c-di-GMP) riboswitches, to a complete lack of switching such as in the flavin mononucleotide (FMN) riboswitch [44]. Combined with thermodynamic data, these studies used SAXS to unveil a wide spectrum of ligand-interaction modes in response to the environment. In these cases, SAXS becomes indispensable, and is in fact particularly useful when crystal structures of riboswitches in their apo form often are not available.
RNA structure-function relationship
Structure-function relationship studies of large RNAs often only rely on secondary structure information or three-dimensional structures of smaller RNA fragments. Previous research on the HIV-1 Rev response element (RRE) RNA, which plays a key role in viral recognition of its own mRNA for nuclear export, is a good example. The secondary structure of RRE [46-49] and the detailed local structural contacts between the viral Rev protein and the high-affinity binding site of RRE [50] have been known for more than a decade. In addition, the second Rev binding site was also discovered [51]. However, because of lack of binding specificity of either sites, neither two pieces of the local structural information alone could resolve the long-standing mystery—the structural basis for specific viral RNA recognition. This long-standing mystery can only be solved with the knowledge of the overall 3D topological structure of the whole RRE RNA where the relative positions of the two binding sites are known. A recent study using SAXS corroborated with biological assays resolved the mystery [52••]. The shape-reconstruction of the RRE RNA resulted in an uncanny “A”-shaped molecular envelope, where the two known Rev binding sites are about 55 Å apart, a distance coinciding with that of the two arginine rich motifs (ARMs) in the Rev dimer [53,54] (Figure 4). The structural model and the specific mode of binding were validated by experiments using SAXS as well as both in vitro and in vivo assays. It was shown that the specificity of viral RNA recognition is achieved through a dual “lock-and-key” mechanism between the two binding sites on RRE and the two ARMs of a Rev dimer.
Figure 4.
An illustration of a combined use of SAXS and in vitro and in vivo assays to study the structure-function relationship of the HIV-1 Rev response element (RRE) RNA [52••]. (a) an illustration of the two-step molecular shape derivation; (b) an illustration of divide-and-identify of domain locations in the RRE RNA; (c) extending the central helix to confirm the global topology of RRE; (c) & (e) corroborating the in vitro and in vivo assay results with the topological structural information.
Besides its biological significance, the SAXS analysis of RRE as a case study illustrates several useful hints. First, large RNA structures often consist of modular structural segments [55••]. But whether a specific RNA is made of modular building blocks can only be validated by experimental means, and SAXS might be useful approach. Second, the secondary structures of large RNAs and some local structural details of small RNA fragments are often known. However, information derived from the global topological structures of large RNAs may be necessary to answer fundamental biological questions. While high-resolution structures of large, full-length RNAs in their functional forms are still sought-after prizes, relatively accessible 3D topological structures determined by SAXS may be a useful alternative. Such topological structures can be critical to understanding known biochemical and local structural information about a system in a global structural context, as illustrated in the recent study [52••].
Combined use of SAXS and NMR measurements for global structure determination
By nature of physics, SAXS provides global structural information whereas NMR provides local structural details. One might consider the combination of these two methods to be a “perfect marriage” for RNA structure determination [56•,57•]. The first demonstration of such an approach was determining the structure of a 30 kDa homodimeric tetraloop-receptor RNA complex [56•] (Figure 1). In this study, the SAXS data and residual dipolar couplings (RDCs) were used to define the global shape and relative orientation between the two domains, respectively. The SAXS-refined structures had a slightly improved backbone root-mean-square deviation (RMSD) from the average of the ensemble, ~1.3 Å, as compared to ~1.8 Å for the non-SAXS-refined structures. More importantly, the SAXS data complemented the NMR restraints in defining the global shape of the dimer, and the impact on the global structure is noticeable with an RMSD between the two structures of about 3.2 Å [56•]. The pair distance distribution functions (PDDF) of the SAXS-refined structures are more narrowly dispersed and better matched to the experimental values in real space, and the Rg value of the structure changes from 25.1 Å (before) to 23.0 Å after the SAXS refinement, agreeing well with the experimental value of 23.2 Å.
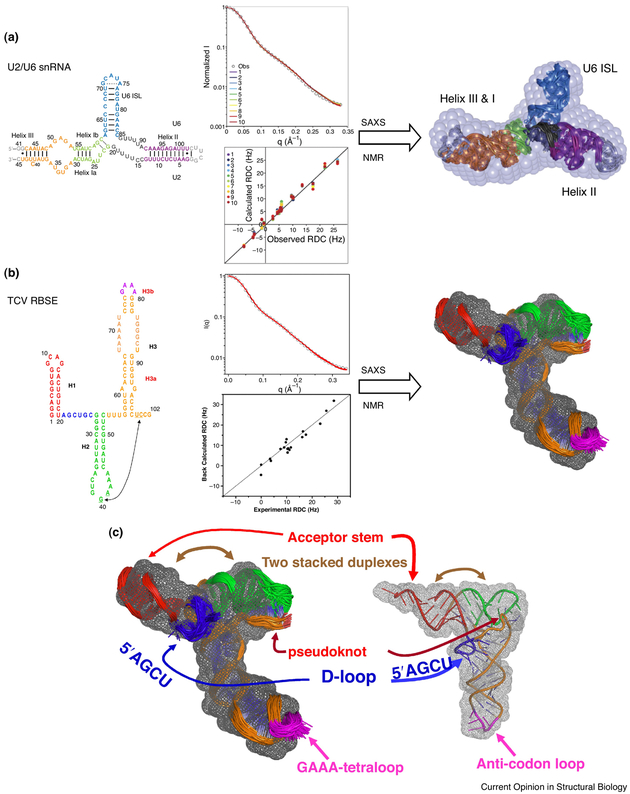
Since the chemical-shift assignments and RDCs of RNA imino groups as well as SAXS data are relatively easy to obtain, combined use of these data has become a useful approach for rapid ab initio global structure determination of RNA [37•,41•,58•,59-62]. An example of this approach is the structure determination of the U2/U6 snRNA complex [37•]: a conserved and essential component of the active spliceosome that interacts with the pre-mRNA substrate and protein splicing factors to promote splicing catalysis. Using a combined approach that integrates SAXS, NMR, and molecular modeling, the three-dimensional solution structure of the 111-nt U2/U6 complex was determined that best fits both SAXS and NMR data (Figure 5). The conformation of the coaxial stacking of Helix Ib on the U6 ISL is similar to that in the Domain V structure in group II introns [37•]. This U2/U6 structure is the largest RNA solution structure determined using the combined approach. Previously, the same method was applied to determine the 3D topological structure of the 102-nt ribosome-binding structural element (RBSE) of the 3' UTR of turnip crinkle virus (TCV) genomic RNA using SAXS and NMR RDC data and software package G2G [41•]. The RBSE RNA shares similarities with almost all of the structural elements and features of a tRNA (Figure 5). The RBSE structure represents a prototype structure of a new class of RNA and provides a structural basis for understanding its involvement in viral translation/replication processes. In general, since RNA is mostly made of duplexes, more than 70% of total residues in an RNA structure, as well as backbones of duplexes, differ from each other on average within only 1 Å RMSD [60]. The resolution of the structures determined using only RDCs of imino groups and SAXS as a global constraint, as discussed in this review, could be better than 3.5 Å [60].
Figure 5.
Illustration of combined use of SAXS and NMR measurements to determine three-dimensional structures of U2/U6 snRNA [37•] (a) and TCV RNA [41•] (b). On the left are the secondary structures that are color-coded the same as in the three-dimensional models. The central panels are the fits to the experimental SAXS and NMR RDC data. The grey beads in (a) and mesh in (b) are molecular envelopes that are superimposed with the ensembles of NMR structures. (c) the 102-nt TCV RNA shares the identical folding topology with tRNA, even though the two RNAs have different sequences and sizes.
Conformation space of RNA
RNAs are inherently dynamic biomolecules and undergo conformational changes in response to a diverse set of cellular signals and environmental cues, such as recognition of proteins, nucleic acids, and metabolites, changes in physiochemical conditions, chemical reactions, and thermal and mechanical triggers [63••,64]. RNA conformational changes occur through complex and often multilayered motions at different temporal and spatial scales. These changes can be rapid, local, small-scale atomic fluctuations around an average structure, or involve slow, global, large-scale rearrangements of molecular machinery [64]. Such motions are not random; they are linked to the topological constraints encoded at the secondary structural level, dominated, if not fully determined, by intra- and intermolecular interactions, which in turn depend on the three-dimensional structure [30••]. RNA dynamics form the basis for the functional complexity of a vast universe of non-coding RNAs performing regulation and guide roles in multiple cellular processes [63••], as witnessed in the past two decades [65]. Detailed characterization of the structural-dynamical properties of RNAs is crucial for a deeper understanding of their wide variety of functions and in rational RNA-targeted drug design [66].
The number of potential conformers in which a structured RNA molecule such as RRE may exist can be very large. This is made possible in part by structural features such as bulges, internal loops and junctions that serve as pivot points of flexibility [63••,67••]. Not only may multiple conformations coexist, but they may also change with time [68,69]. Therefore, a description of large RNAs with a single conformation may not be sufficient for understanding their behavior. Furthermore, for cases in which a large number of RNA conformations exists, consideration of each of them individually would be a task of scope beyond all possibility of realization, given such potentially large conformation space. This brings us at once to the necessity of adopting a statistical point of view.
When characterizing a heterogeneous and flexible system using experimental observables such as SAXS, the simplest approach is to use the SAXS-restrained ensemble of structures, where the scattering intensity is calculated as
Where wi is the weight of ensemble member i, taken as 1/Ne here, and Ii(q) is the contribution to the scattering intensity from ensemble member i. If the surface-bound solvent contribution is omitted, the Ii(q) of an individual member i can be calculated using the Debye formula [70]:
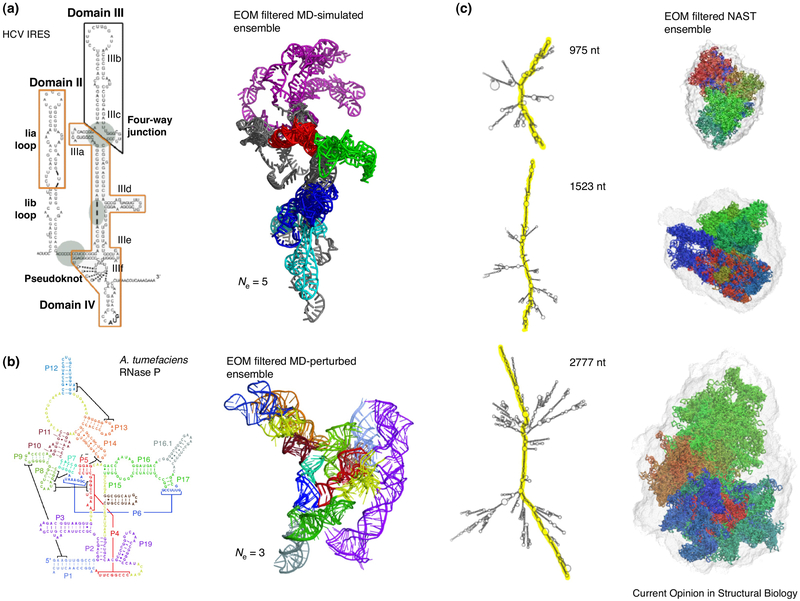
where n is the number of atoms in the molecule, fj(q) and fk(q) are the form factors for atoms j and k, respectively, q is the momentum transfer and rjk is the distance between atoms j and k. There are three computation programs that calculate conformation space using SAXS data [71•,72••,73••]. In the Xplor-NIH, simulated annealing under restraints of various pseudo-potential functions, including a nucleic-acid database function, is used to calculate ensembles that best fit experimental SAXS data. The population weights can be set equal or different for conformation subsets [71•]. The program was first applied to sample the conformation space of a Dickenson DNA dodecamer [71•] and later applied to characterize the conformation space of the HIV-1 RRE RNA [52••]. The Ensemble Optimization Method (EOM) [72••] uses a metaheuristic algorithm to search for ensembles that best fit experimental data [74••,75,76•,77]. The Minimal Ensemble Search (MES) uses a high-temperature molecular dynamics (MD) simulation to search conformation space by a genetic algorithm [73••,77]. In all of these methods, duplexes are considered rigid, whereas linkers, junctions, and loops serve as pivot points of flexibility [31,63••]. Using the ensemble approach, Pérard et al. found that collective motions dominate the flexibility of the full-length internal ribosome entry site (IRES) of the hepatitis C virus genome, suggesting that a large-scale conformation change could take place when forming the initiation complex [76•] (Figure 6). In the case of RNase P, the crystal structure of the free RNA in the RNase P is significantly different from that observed in the ternary complex, and the RNase P RNA in solution can only be described by an ensemble of four conformations that could best fit SAXS data (Figure 6) [75].
Figure 6.
Characterizing RNA conformation space using SAXS. (a) SAXS-filtered MD-simulated ensemble of the HCV IRES RNA [76•]. An ensemble of five can best fit the experimental SAXS data; (b) In the case of RNase P it requires an ensemble of three to best fit experimental SAXS data [75]; (c) SAXS-filtered ensembles of models generated using a nucleic acid simulation tool (NAST) [95].
With the exception of rRNAs in ribosomes, overall structures or molecular shapes of kilobase-sized RNAs are relatively unknown, and it is of a great interest to demonstrate if those kilo-base sized RNA transcripts are largely linear—like what we often see conceptually in biologists’ presentations—or form complex 3D folds. A recent study by Gupal et al. [74••] investigated the global folds of three large RNA transcripts (975-, 1523- and 2777-nt long) under two different buffer conditions: Tris-EDTA or an assembly buffer that contained Mg2+. Using SAXS, together with cryo-EM and the ensemble MD calculations, they showed: (1) under neither condition are the RNAs linear; and (2) Mg2+ promotes folding of the RNAs into a prolate geometry [74••] (Figure 6). This study provides the first peek into the global three-dimensional shapes of kilobase-long RNA transcripts.
Practical considerations for RNA sample preparation
RNA molecules exhibit some unique behavior in solution. Thus it is informative to provide a more detailed account regarding RNA sample preparation for SAXS experiments. The first consideration is sample homogeneity. Typically, RNA samples used for SAXS experiments are usually produced using in vitro transcription by the T7 polymerase, purified by polyacrylamide gel electrophoresis (PAGE) under denaturing conditions, and then refolded. While this purification method works for small RNAs with simple structural elements (e.g., a single hairpin), larger RNAs with more complex folds often cannot be refolded into their native states once they have been denatured. This is true for the RRE RNA, where the refolded RNA under various folding conditions exhibits multiple bands on a native gel [52••]. Such a sample containing a mixture of conformers may complicate SAXS data analysis and interpretation. Whenever possible, purification using size exclusion chromatography under a native condition is highly recommended.
Another important consideration is buffer selection and background subtraction. While phosphate buffer systems are convenient for NMR spectroscopy because they generate no proton NMR signals, these buffers should be avoided in SAXS experiments due to high background scattering and the promotion of free-radical formation. Organic buffer systems (e.g., Tris, HEPES), on the other hand, are highly recommended because they contribute very little to background scattering and also serve as radical scavengers to reduce radiation damage.
One critical aspect of processing SAXS data is the subtraction of the background scattering produced by the solvent. As water itself is a scatter of X-rays, SAXS of very dilute biomacromolecules is detected against a very strong background of water. The scattering profile for the biomacromolecule can then be calculated using the equation below:
For this reason, it is critical that the buffer used to measure the background scattering exactly matches that of the sample. In our experiments, first the wide angle X-ray scattering (WAXS) profile was obtained using the above equation and tuning the value of α to eliminate the water peak around 2.0 AÅ−1. Then, the resulting WAXS profile was used as a guide for the SAXS background subtraction by tuning the value of α in the SAXS subtraction and overlaying the resulting SAXS profile with the WAXS profile at the overlapping q range (i.e., 0.1-0.28 Å−1). The final scattering data were obtained by piecing the resulting SAXS and WAXS profiles together to achieve scattering q values of 0.006 < q < 2.3Å−1 [41,60,62].
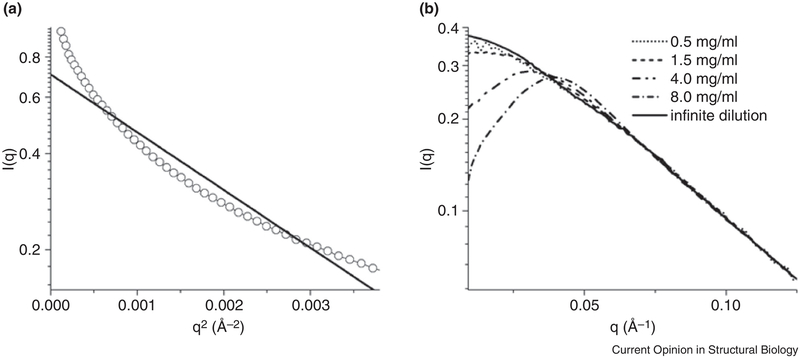
Lastly, RNAs are heavily charged polyanions and strongly interact with cations, water molecules, and sometimes anions (including themselves), repulsively or attractively, depending on the ionic conditions and types of counter ions in solution [62,78••]. The interactions among macromolecules are referred to as the structure factor, s(q), and may have an effect on the quality of the SAXS data and skew interpretation. More specifically, s(q) is the ratio of the scattering under a given condition relative to that under an “ideal” condition, where the macromolecules do not interact with each other (i.e., monodispersed) (Figure 7) [79,80]. A non-uniform structure factor leads to non-linearity in the Guinier region. A linear plot in the Guinier region is critical for extracting accurate dimension- and mass-related information, such as Rg and molecular weight. In the absence of interactions, s(q)=1; when attractive interactions exist that cause aggregation (Figure 7), s(q) > 1 at q close to 0; and when repulsive interactions exist, s(q) < 1 at q close to 0 (Figure 7 ).
Figure 7.
(a) A curve-up scattering profile of RNA, indicative of high order aggregation; (b) A curve-down scattering profile, indicative of repulsive interaction among RNA molecules in low-salt conditions [62].
Commonly used software tools
The wide application of SAXS for characterizing biological molecules is not possible without a suite of indispensable software tools. The tools were originally developed for proteins and protein complexes but are largely applicable for RNA and RNA containing complexes as well. In this section, we provide a brief review of computation software tools and packages that are commonly used.
Software Primus [81], developed by Svergun’s group in early 2000, is widely used for data averaging and for extracting Rg and I0 at the initial stage of SAXS data processing and analysis. Often, one can also accomplish the same task by programming using a commercial program package Igor Pro (Wavemetrics, Lake Oswego, OR, USA). For extracting some basic structural parameters from SAXS data, such as pair-distance-distribution-function (PDDF), maximum dimension (Dmax) as well as Rg, the program GNOM is widely used [7]. Two programs on web-based servers, SAXS MoW (http://www.if.sc.usp.br/~saxs/) and Scatter (http://www.bioisis.net/tutorial), have been used to calculate molecular weights. Programs and packages DAMMINDI [6••], DAMMIF [8•] and MOSA [6••] are widely used for ab initio shape reconstruction. Programs BUNCH, SASREF and CORAL in the ATSAS package [82] or Xplor-NIH [83] are widely used for rigid body modeling using SAXS. Programs of the EOM [72], MES [73] methods and Xplor-NIH are widely used for ensemble calculations to characterize RNA conformation space using SAXS data. The latter can also take combined data of both NMR measurements and SAXS to calculate ensembles [52••,71•] or to refine structures [37•,41•,56•,60]. Often, coordinates of a structure are known, and one needs to compare the SAXS profile of the structure with the experimentally measured SAXS profile of the same molecule. Programs CRYSOL [84], FoXS [85] and Xplor-NIH are widely used for that purpose. The latter two can also accurately calculate the profile of wide-angle X-ray scattering (WAXS).
Conclusions and outlook
Our understanding of the roles of RNA in cellular processes has undergone a tremendous expansion over the past two decades. In contrast, our knowledge about RNA 3D structures and structure-function relationships is still very limited. Significant progress has been made, such as mapping the secondary structure of the entire HIV-1 genomic RNA, which is similar to rRNA in size. However, the “gap” between RNA secondary and tertiary structures is enormous. SAXS has become an ideal tool to bridge this gap, as has been illustrated in this review. As synchrotron beams become more accessible to users, and with sensitive in-house scattering instruments now commercially available, we expect a much broader application of SAXS in RNA structural biology. Besides what has been discussed in this review, the future directions of applying SAXS to RNA research may include: 1) time-resolved SAXS to study dynamics of RNA folding and conformation space [86•,87-89], 2) studying RNA-protein complexes [90-92] and 3) determining 3D topological structures of large RNAs using RNA secondary structures and SAXS data as input. The latter is feasible since RNA secondary structure encodes 3D structure and folding pathway [30••,31]. With global shape restraints and a few long-range contacts, topological structures of RNA may be computed.
The rapid development of X-ray free electron laser (XFEL) technology and its applications may revolutionize biological research, including RNA structural biology [93••]. An XFEL beam is approximately one billion times brighter than the latest third-generation synchrotron sources, and pulses on a femtosecond time-scale. The time-resolved “snap shot” SAXS using XFEL has been demonstrated in a protein case to observe molecular motions on a much shorter time scale [94••]. In particular, on the femtosecond time scale, any events that occur at the diffusion limit, ~10−9 sec., is steady still. Even photosynthesis processes that occur at a sub-picosecond time scale can be captured. Thus, the brilliance of XFEL and its femtosecond time frame potentially make it possible to image a single molecule in action (true molecular movies) in the very near future.
Highlights.
SAXS is used to determine global topological structures of RNA
SAXS is used to study structure-function relationship of RNA
SAXS is used to study conformation space of RNA
Acknowledgements
We sincerely thank David Lilley, Samuel Butcher, Marc Jamin, Norman Pace, Alexei Kazanstev, Ajaykumar Gopal and William Gelbart for providing materials for figures. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute (NCI) Center for Cancer Research. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357 and by NCI under the PUP-77 Agreement between NCI and Argonne National Laboratory.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, are highlighted as:
• of special interest
•• of outstanding interest
- 1.Feigin LA, Svergun DI: Structure Analysis by Small Angle X-Ray and Neutron Scattering. New York: Plenum Press; 1987. [Google Scholar]
- 2.Glatter O, Kratky O: Small Angle X-ray Scattering. New York: Academic Press; 1982. [Google Scholar]
- 3. •.DI Svergun, Volkov VV, Kozin MB, Stuhrmann HB: New Developments in Direct Shape Determination from Small-Angle Scattering. 2. Uniqueness. In Acta Crystallographica Section A. Edited by; 1996:419–426. vol 52. [Google Scholar]; This article presents further progress in the shape determination method based on multipole expansion, including fast algorithms for evaluating shape-scattering correlations and the resolution in real and reciprocal space. Various types of shapes were tested and the study demonstrated that low-resolution shapes can be generated from SAXS data.
- 4.DI Svergun, Volkov VV, Kozin MB, Stuhrmann HB, Barberato C, Koch MHJ: Shape determination from solution scattering of biopolymers. J Appl Cryst 1997, 30:798–802. [Google Scholar]
- 5.DI Svergun, Petoukhov MV, Koch MH: Determination of domain structure of proteins from X-ray solution scattering. Biophys J 2001, 80:2946–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. ••.Svergun DI: Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing). Biophys J 1999, 77:2896–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article presents an algorithm to restore ab initio low resolution shape using simulated annealing. Densely packed dummy atoms in a spherical initial space is characterized by a configuration vector assigning the atom to a specific phase or to the solvent. Simulated annealing is employed to find a configuration that fits the data while minimizing the interfacial area. Application of the method is illustrated by the restoration of a ribosome-like model structure and more realistically by the determination of the shape of several proteins from experimental x-ray scattering data.
- 7.Svergun DI: Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst 1992, 25:495–503. [Google Scholar]
- 8. •.Franke D, Svergun DI: DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. In Journal of Applied Crystallography. Edited by; 2009:342–346. vol 42. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article presents improved implementation of the previous ab-initio shape-determination program DAMMIN. The improvements are: 1. Eliminating the limit on search volume; 2. Option to impose symmetry and anisometry constraints; 3. 25-40 times faster than DAMMIN on a single CPU and capability to run a multiple CPU platform.
- 9.Chacon P, Moran F, Diaz JF, Pantos E, Andreu JM: Low-resolution structures of proteins in solution retrieved from X-ray scattering with a genetic algorithm. Biophys J 1998, 74:2760–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walther D, Cohen FE, Doniach S: Reconstruction of low-resolution three-dimensional density maps from one-dimensional small-angle X-ray solution scattering data for biomolecules. J Appl Cryst 2000, 33:350–363. [Google Scholar]
- 11.Lake JA: Yeast transfer RNA: a small-angle x-ray study. Science 1967, 156:1371–1373. [DOI] [PubMed] [Google Scholar]
- 12. ••.Rambo RP, Tainer JA: Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 2011, 95:559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article presents the first discussion of using SAXS and the Porod-Debye law to characterize flexible protein and RNA systems. The application of the law allows distinguishing discrete conformation change from the localized flexibility.
- 13. •.Russell R, Millett IS, Tate MW, Kwok LW, Nakatani B, Gruner SM, Mochrie SG, Pande V, Doniach S, Herschlag D, et al. : Rapid compaction during RNA folding. Proc Natl Acad Sci U S A 2002, 99:4266–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article presents one of the first studies of RNA comformation space with time trajectory using SAXS aided by computer simulations. The study provides a time-resolved picture of the global folding process of the Tetrahymena group I RNA over a time window of more than five orders of magnitude. The folding of the RNA takes place on the low millisecond timescale, and the overall global shape changes are complete within one second.
- 14.Russell R, Millett IS, Doniach S, Herschlag D: Small angle X-ray scattering reveals a compact intermediate in RNA folding. Nat Struct Biol 2000, 7:367–370. [DOI] [PubMed] [Google Scholar]
- 15.Feigin LA, Svergun DI: Structure Analysis by Small-Angle X-ray Scattering and Neutron Scattering. Edited by Fiegin LA, Svergun DI New York: Plenum Press; 1987. [Google Scholar]
- 16. •.Hura GL, Budworth H, Dyer KN, Rambo RP, Hammel M, McMurray CT, Tainer JA: Comprehensive macromolecular conformations mapped by quantitative SAXS analyses. Nat Methods 2013, 10:453–454. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first report of the concept, namely SAXS structural comparison map (SCM) and volatility of ratio (VR) difference metric. SCM and VR allow use of SAXS to quantify conformational similarities and distinguish different assembly states.
- 17.Lipfert J, Doniach S: Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annu Rev Biophys Biomol Struct 2007, 36:307–327. [DOI] [PubMed] [Google Scholar]
- 18. •.Lipfert J, Herschlag D, Doniach S: Riboswitch conformations revealed by small-angle X-ray scattering. Methods Mol Biol 2009, 540:141–159. [DOI] [PMC free article] [PubMed] [Google Scholar]; SAXS is a convenient solution method to outline the global shape of an RNA molecule and was applied to study the conformation change of a riboswitch, a glycine-binding tandem aptamer from Vibrio cholerae, in response to change in concentrations of ions and ligands.
- 19.Burke JE, Butcher SE: Nucleic acid structure characterization by small angle X-ray scattering (SAXS). Curr Protoc Nucleic Acid Chem 2012, Chapter 7:Unit7 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Lau MW, Ferre-D'Amare AR: Ribozymes and riboswitches: modulation of RNA function by small molecules. Biochemistry 2010, 49:9123–9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathews DH, Turner DH, Zuker M: RNA secondary structure prediction. Curr Protoc Nucleic Acid Chem 2007, Chapter 11:Unit 11 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster P: Fast folding and comparison of RNA secondary structures. Monatsh. Chem. Monatshefte fur Chemie 1994, 125:167–188. [Google Scholar]
- 23.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM: RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 2005, 127:4223–4231. [DOI] [PubMed] [Google Scholar]
- 24. ••.Holbrook SR: Structural principles from large RNAs. Annual review of biophysics 2008, 37:445–464. [DOI] [PubMed] [Google Scholar]; This paper reports a survey of large RNA structures (> 100 nt) and provides insightful observation about the characteristics of folded RNA structures. These structures “are dominated by long continuous interhelical base stacking, tend to segregate into domains, and are planar in overall shape as opposed to their globular protein counterparts”. We believe that these obeservations serve as several basic parametrics for RNA shape reconstruction using SAXS data.
- 25. ••.Zhang J, Jones CP, Ferre-D'Amare AR: Global analysis of riboswitches by small-angle X-ray scattering and calorimetry. Biochim Biophys Acta 2014, 1839:1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]; Monitoring global shape change in solution without knowledge of structure coordinates is one of the useful applications of SAXS. In riboswitch cases, often the structures of riboswitch in the free state are unknown. Using the SAXS shape reconstruction and analysis of thermodynamics data, the authors discovered a wide range of shape changes in repsonse to ligand binding, from distinct shape transition to almost none. This is the first comprehensive survey of mode of switching using SAXS.
- 26. ••.Laing C, Wen D, Wang JT, Schlick T: Predicting coaxial helical stacking in RNA junctions. Nucleic acids research 2012. 40 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]; RNA consists of multiple helices that are connected by junctions. The configuration at junctions often determines global folding of RNA. This paper introduces a computation method to predict coaxial stacking arrangements from secondary structural information. This work provides a basic guidance to topological structure construction based on SAXS-derived molecular envelopes of junction-containing RNA.
- 27. ••.de la Pena M, Dufour D, Gallego J: Three-way RNA junctions with remote tertiary contacts: a recurrent and highly versatile fold. RNA 2009, 15:1949–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]; The three-way junction is one of the widely-seen high-order structural elements in RNAs. This paper provides a survey of structures with three-way junctions together with a key observation: “a recurrent Y shape when two of the helices form a coaxial stack and the third helix establishes one or more tertiary contacts several base pairs away from the junction”.
- 28. ••.Lescoute A, Westhof E: Topology of three-way junctions in folded RNAs. Rna 2006, 12:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports the first survey of RNA three-way junctions. The authors observed a) two helices approximately coaxially stacked; b) three-way junctions can be divided into three main families.
- 29. ••.Lipfert J, Ouellet J, Norman DG, Doniach S, Lilley DM: The complete VS ribozyme in solution studied by small-angle X-ray scattering. Structure 2008, 16:1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper presents the first molecular coordinates of large VS ribozyme RNA based on SAXS-derived molecular envelopes and secondary structural information using a divide-and-conquer approach. The structural model was later further corroborated with activity studies.
- 30. ••.Bailor MH, Mustoe AM, Brooks CL, Al-Hashimi HM 3rd: Topological constraints: using RNA secondary structure to model 3D conformation, folding pathways, and dynamic adaptation. Current opinion In structural biology 2011, 21:296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]; Whether RNA secondary structure or tertiary structure determines RNA folding was an unsettled question until this article. In this paper, the authors convincingly demostrated that it is the RNA secondary structure that predefines RNA conformation space and folding pathway, and tertiary interactions helps to narrow the structure down to a few specific conformations. This paper is of fundamental and conceptual importance. It provides the basis for a possibility of determining 3D topological structures of RNA given global and a few local restraints.
- 31.Bailor MH, Sun X, Al-Hashimi HM: Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science 2011, 327:202–206. [DOI] [PubMed] [Google Scholar]
- 32.Witz J: 1964: The first model for the shape of a transfer RNA molecule. An account of an unpublished small-angle X-ray scattering study. Biochimie 2003, 85:1265–1268. [DOI] [PubMed] [Google Scholar]
- 33.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A: Structure of yeast phenylalanine tRNA at 3 A resolution. Nature 1974, 250:546–551. [DOI] [PubMed] [Google Scholar]
- 34.Funari SS, Rapp G, Perbandt M, Dierks K, Vallazza M, Betzel C, Erdmann VA, Svergun DI: Structure of free Thermus flavus 5 S rRNA at 1.3 nm resolution from synchrotron X-ray solution scattering. J Biol Chem 2000, 275:31283–31288. [DOI] [PubMed] [Google Scholar]
- 35.Lipfert J, Chu VB, Bai Y, Herschlag D, Doniach S: Low-resolution models for nucleic acids from small-angle X-ray scattering with applications to electrostatic modeling. J Appl Cryst 2007, 40:S229–S234. [Google Scholar]
- 36.Jones CP, Cantara WA, Olson ED, Musier-Forsyth K: Small-angle X-ray scattering-derived structure of the HIV-15' UTR reveals 3D tRNA mimicry. Proc Natl Acad Sci U S A 2014, 111:3395–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. •.Burke JE, Sashital DG, Zuo X, Wang YX, Butcher SE: Structure of the yeast U2/U6 snRNA complex. RNA 2012, 18:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]; The U2/U6 snRNA complex was one of the largest RNA structures determined using a combination of NMR and SAXS. It consists of a three-way junction that forms an extended "Y" shape.
- 38.Bai Y, Tambe A, Zhou K, Doudna JA: RNA-guided assembly of Rev-RRE nuclear export complexes. Elife 2014, 3:e03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hammond JA, Rambo RP, Filbin ME, Kieft JS: Comparison and functional implications of the 3D architectures of viral tRNA-like structures. RNA 2009, 15:294–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. ••.Rambo RP, Tainer JA: Improving small-angle X-ray scattering data for structural analyses of the RNA world. Rna 2010, 16:638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article outlines an experimental protocol, which includes sample preparation, characterization of RNA samples such as detecting heterogeneity, SAXS analysis for shape reconstruction, and investigating flexibility.
- 41. •.Zuo XB, Wang JB, Yu P, Eyler D, Xu H, Starich MR, Tiede DM, Simon AE, Kasprzak W, Schwieters CD, et al. : Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3 ' UTR of turnip crinkle virus. Proc Natl Acad Sci U S A 2010, 107:1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article presents a tRNA-like structure of a structural element in turnip crinkle virus 3’ UTR RNA. The structure was determined using a combination of NMR RDC measurements and SAXS data.
- 42.Hiley SL, Collins RA: Rapid formation of a solvent-inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J 2001, 20:5461–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson TJ, Lilley DM: Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid-base catalysis? A critical assessment of available experimental data. RNA 2011, 17:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baird NJ, Kulshina N, Ferre-D'Amare AR: Riboswitch function: flipping the switch or tuning the dimmer? RNA Biol 2010, 7:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haller A, Souliere MF, Micura R: The dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc Chem Res 2011, 44:1339–1348. [DOI] [PubMed] [Google Scholar]
- 46.Mann DA, Mikaelian I, Zemmel RW, Green SM, Lowe AD, Kimura T, Singh M, Butler PJ, Gait MJ, Karn J: A molecular rheostat. Co-operative rev binding to stem I of the rev-response element modulates human immunodeficiency virus type-1 late gene expression. J Mol Biol 1994, 241:193–207. [DOI] [PubMed] [Google Scholar]
- 47.Heaphy S, Finch JT, Gait MJ, Karn J, Singh M: Human immunodeficiency virus type 1 regulator of virion expression, rev, forms nucleoprotein filaments after binding to a purine-rich "bubble" located within the rev-responsive region of viral mRNAs. Proc Natl Acad Sci U S A 1991, 88:7366–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dayton ET, Powell DM, Dayton AI: Functional analysis of CAR, the target sequence for the Rev protein of HIV-1. Science 1989, 246:1625–1629. [DOI] [PubMed] [Google Scholar]
- 49.Kjems J, Brown M, Chang DD, Sharp PA: Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc Natl Acad Sci U S A 1991, 88:683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Battiste JL, Mao H, Rao NS, Tan R, Muhandiram DR, Kay LE, Frankel AD, Williamson JR: Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science 1996, 273:1547–1551. [DOI] [PubMed] [Google Scholar]
- 51.Daugherty MD, D'Orso I, Frankel AD: A solution to limited genomic capacity: using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol Cell 2008, 31:824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. ••.Fang X, Wang J, O'Carroll IP, Mitchell M, Zuo X, Wang Y, Yu P, Liu Y, Rausch JW, Dyba MA, et al. : An unusual topological structure of the HIV-1 Rev response element. Cell 2013, 155:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]; It has been a long-standing mystery about how HIV-1 virus recognizes its own genomic RNA from more abundant host RNAs in the nucleus for export. There have been a large number of biochemical, biological and structural studies on the subject and it is well known that the interaction between the Rev protein and RRE is key to RNA recognition. However, the structural basis for such a recognition has been elusive due to lack of the global structure of RRE, until now. This paper presents an example of how SAXS analysis, combined with functional studies, can be used to answer an important biological question.
- 53.DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM: Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc Natl Acad Sci U S A 2010, 107:5810–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daugherty MD, Liu B, Frankel AD: Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat Struct Mol Biol 2010, 17:1337–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. ••.Cruz JA, Westhof E: The dynamic landscapes of RNA architecture. Cell 2009, 136:604–609. [DOI] [PubMed] [Google Scholar]; This is an excellent review on some basic structural principles of RNA architectures: a). RNA architechture is modular and hirearchical; b). RNA architecture is dominated by long helices and often continued with coaxial stacking; c). helices are often packed parallel and orthogonal to each other; d). Junctions play the central role in RNA architecture.
- 56. •.Zuo XB, Wang JB, Foster TR, Schwieters CD, Tiede DM, Butcher SE, Wang YX: Global molecular structure and interfaces: Refining an RNA : RNA complex structure using solution X-ray scattering data. J Am Chem Soc 2008, 130:3292–3294. [DOI] [PubMed] [Google Scholar]; Determining the global architecture of multicomponent systems is a key problem in understanding the biological interactions on a molecular level. This paper demonstrates that given structures of components, the global architecture of multicomponent systems can be accurately determined using global orientation and shape restraints derived, respectively, from NMR and SAXS data.
- 57. •.Grishaev A, Ying J, Canny MD, Pardi A, Bax A: Solution structure of tRNAVal from refinement of homology model against residual dipolar coupling and SAXS data. J Biomol NMR 2008, 42:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper presents a procedure to obtain an acurate structure based on a homology model using orientation and shape restraints derived, respectively, from NMR and SAXS measurements.
- 58. •.Wang J, Zuo X, Yu P, Xu H, Starich MR, Tiede DM, Shapiro BA, Schwieters CD, Wang YX: A Method for Helical RNA Global Structure Determination in Solution Using Small-Angle X-Ray Scattering and NMR Measurements. J Mol Biol 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]; RNA structures predominantly consist of A-form helices with a backbone RMSD varying around 1 A in all high resolution structures in the PDB database. This paper presents a method and a software tool for determining RNA 3D structures.
- 59.Chen B, Zuo X, Wang YX, Dayie TK: Multiple conformations of SAM-II riboswitch detected with SAXS and NMR spectroscopy. Nucleic Acids Res 2012, 40:3117–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X, Wang J, Zuo X: A Top-Down Approach to Determining Global Structures of RNAs in Solution Using NMR and Small-Angle X-ray Scattering Measurements In RNA 3D Structure Analysis and Prediction: 27 (Nucleic Acids and Molecular Biology). Edited by Leontis N, Westhof E: Springer; 2012. vol 27. [Google Scholar]
- 61.Wang YX, Zuo XB, Wang JB, Yu P, Butcher SE: Rapid global structure determination of large RNA and RNA complexes using NMR and small-angle X-ray scattering. Methods 2010, 52:180–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zuo X, Wang J, Wang X: NMR and SAXS: A Perfect Marriage and New Approach for RNA Structure Determination. In Biomolecular NMR Spectroscopy. Edited by Dingley AJ, Pascal SM: IOS Press; 2011. [Haris PI (Series Editor): Advances in Biomedical Spectroscopy, vol 3.] [Google Scholar]
- 63. ••.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM: Functional complexity and regulation through RNA dynamics. Nature 2012, 482:322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]; Conformation changes of RNA molecules is fundamentally important to RNA functions. This paper illustrates that RNA information contents are encoded beyond its primary sequence and single static structure. Multiple conformations and changes among them on biological time scales form spatial and temporal bases of RNA functional complexity and divergence.
- 64.Al-Hashimi HM, Walter NG: RNA dynamics: it is about time. Curr Opin Struct Biol 2008, 18:321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharp PA: The centrality of RNA. Cell 2009, 136:577–580. [DOI] [PubMed] [Google Scholar]
- 66.Stelzer AC, Kratz JD, Zhang Q, Al-Hashimi HM: RNA dynamics by design: biasing ensembles towards the ligand-bound state. Angew Chem Int Ed Engl 2012, 49:5731–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. ••.Bailor MH, Sun X, Al-Hashimi HM: Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science 2010, 327:202–206. [DOI] [PubMed] [Google Scholar]; This is another conceptual break-through article on the subject that RNA folding is encoded in secondary structure. This time, the authors demonstrated, using simple two-way junction structural elements, that topological contraints in the junctions define global folding. It is known that junctions are at the center of RNA architectures.
- 68.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM: Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature 2007, 450:1263–1267. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Q, Sun X, Watt ED, Al-Hashimi HM: Resolving the motional modes that code for RNA adaptation. Science 2006, 311:653–656. [DOI] [PubMed] [Google Scholar]
- 70.Debye P: Zerstreuung von Roentgenstrahlen. Annalen der Physik 1915, 46:809–823. [Google Scholar]
- 71. •.Schwieters CD, Clore GM: A physical picture of atomic motions within the Dickerson DNA dodecamer in solution derived from joint ensemble refinement against NMR and large-angle X-ray scattering data. Biochemistry 2007, 46:1152–1166. [DOI] [PubMed] [Google Scholar]; This is the first study of DNA conformation space using both NMR and SAXS/WAXS data.
- 72. ••.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI: Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc 2007, 129:5656–5664. [DOI] [PubMed] [Google Scholar]; This article presents a new approach, the ensemble optimization method (EOM), to characterize flexible proteins in solution using SAXS.
- 73. ••.pelikan M, Hura GL, Hammel M: Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys 2009, 28:174–189. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article presents a genetic algorithm, (minimal ensemble search, MES), to identify the minimal ensemble required to best fit the experimental SAXS data.
- 74. ••.Gopal A, Zhou ZH, Knobler CM, Gelbart WM: Visualizing large RNA molecules in solution. RNA 2012, 18:284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article presents an exciting comparative study of genomic-size RNAs using both SAXS and cryo-EM. The results clearly show 1). Genomic-sized RNAs are non-linear and folded; 2). Mg2+ induced marked global structure changes; 3). Dynamic nature of large RNA structures from SAXS as compared with the static pictures determined by cryo-EM.
- 75.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR: Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci U S A 2005, 102:13392–13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. •.Perard J, Leyrat C, Baudin F, Drouet E, Jamin M: Structure of the full-length HCV IRES in solution. Nat Commun 4:1612. [DOI] [PubMed] [Google Scholar]; This article reports a characterization of possible conformation space of a large RNA using SAXS and ensemble calculation. The study indicates that the HCV IRES is very dynamic and no single model can fit the experimental data.
- 77.Ribeiro Ede A, Beich-Frandsen M Jr., Konarev PV, Shang W, Vecerek B, Kontaxis G, Hammerle H, Peterlik H, Svergun DI, Blasi U, et al. : Structural flexibility of RNA as molecular basis for Hfq chaperone function. Nucleic Acids Res 40:8072–8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. ••.Pollack L: SAXS studies of ion-nucleic acid interactions. Annu Rev Biophys 2011, 40:225–242. [DOI] [PubMed] [Google Scholar]; This paper illustrates unique solution behaviors of nucleic acids characterized using SAXS. The balance between negative charges on nucleic acid backbones and counter ions in solution dictates the structure factor and has a practical implication when preparing scattering samples.
- 79.Ducruix A, Guilloteau JP, Riès-Kautt M, Tardieu A: Protein interactions as seen by solution X-ray scattering prior to crystallogenesis. J Crystal Growth 1996, 168:28–39. [Google Scholar]
- 80.Putnam CD, Hammel M, Hura GL, Tainer JA: SAXS combined with crystallography and computation: defining accurate macromolecular structures conformations and assemblies in solution. Q Rev Biophys 2007, 40:191–285. [DOI] [PubMed] [Google Scholar]
- 81.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI: PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Cryst 2003, 36:1277–1282. [Google Scholar]
- 82.Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI: New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Cryst 2012, 45:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schwieters CD, Kuszewski JJ, Clore GM: Using Xplor-NIH for NMR molecular structure determination. Prog Nucl Magn Reson Spectrosc 2006, 48:47–62. [Google Scholar]
- 84.Svergun D, Barberato C, Koch MHJ: CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst 1995, 28:768–773. [Google Scholar]
- 85.Schneidman-Duhovny D, Hammel M, Sali A: FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res 2010, 38:W540–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. •.Pollack L, Doniach S: Time-resolved X-ray scattering and RNA folding. Methods Enzymol 2009, 469:253–268. [DOI] [PubMed] [Google Scholar]; This article presents a pioneering work on characterizing RNA folding processes using time-resolved SAXS, which provides low resolution spatial and temporal information.
- 87.Pollack L: Time resolved SAXS and RNA folding. Biopolymers 2011, 95:543–549. [DOI] [PubMed] [Google Scholar]
- 88.Kwok LW, Shcherbakova I, Lamb JS, Park HY, Andresen K, Smith H, Brenowitz M, Pollack L: Concordant exploration of the kinetics of RNA folding from global and local perspectives. J Mol Biol 2006, 355:282–293. [DOI] [PubMed] [Google Scholar]
- 89.Graceffa R, Nobrega RP, Barrea RA, Kathuria SV, Chakravarthy S, Bilsel O, Irving TC: Sub-millisecond time-resolved SAXS using a continuous-flow mixer and X-ray microbeam. J Synchrotron Radiat 2013, 20:820–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Madl T, Gabel F, Sattler M: NMR and small-angle scattering-based structural analysis of protein complexes in solution. J Struct Biol 2011, 173:472–482. [DOI] [PubMed] [Google Scholar]
- 91.Hennig J, Wang I, Sonntag M, Gabel F, Sattler M: Combining NMR and small angle X-ray and neutron scattering in the structural analysis of a ternary protein-RNA complex. J Biomol NMR 2013, 56:17–30. [DOI] [PubMed] [Google Scholar]
- 92.Gupta K, Contreras LM, Smith D, Qu G, Huang T, Spruce LA, Seeholzer SH, Belfort M, Van Duyne GD: Quaternary arrangement of an active, native group II intron ribonucleoprotein complex revealed by small-angle X-ray scattering. Nucleic Acids Res 2014, 42:5347–5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. ••.Chapman HN: X-ray imaging beyond the limits. Nat Mater 2009, 8:299–301. [DOI] [PubMed] [Google Scholar]; The briliant, brief pulses of XFEL will revolutionized the macromolecules without crystallization.
- 94. ••.Arnlund D, Johansson LC, Wickstrand C, Barty A, Williams GJ, Malmerberg E, Davidsson J, Milathianaki D, DePonte DP, Shoeman RL, et al. : Visualizing a protein quake with time-resolved X-ray scattering at a free-electron laser. Nat Methods 2014, 11:923–926. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is one of the first studies using XFEL SAXS/WAXS to study ultrafast protein conformation changes upon perturbation by multiphoton excitation. “We describe a method to measure ultrafast protein structural changes using time-resolved wide-angle X-ray scattering at an X-ray free-electron laser. “ The study provides direct structural evidence for a 'protein quake' hypothesis.
- 95.Jonikas MA, Radmer RJ, Laederach A, Das R, Pearlman S, Herschlag D, Altman RB: Coarsegrained modeling of large RNA molecules with knowledge-based potentials and structural filters. RNA 2009, 15:189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]