Abstract
Nongenetic resistance has recently been described as a major impediment to effective cancer therapy. Nongenetic resistance is challenging to study since it occurs nonuniformly, even in cell lines, and can involve the interplay of multiple survival pathways. Until recently, no technology allowed measurement of large-scale alterations in survival pathways with single-cell resolution. Mass cytometry, a flow-based technique in which the activation of up to 50 proteins can be measured simultaneously in single-cell, now provides the ability to examine nongenetic resistance on the functional level on a cell-by-cell basis. The application of mass cytometry, in combination with new bioinformatic techniques, will allow fundamental questions on nongenetic resistance to be addressed: Is resistance caused by selection of cells with a pre-existing survival phenotype or induction of a survival program? Which survival pathways are necessary for nongenetic resistance and how do they interact? Currently, mass cytometry is being used to investigate the mechanism of nongenetic resistance to TRAIL-induced apoptosis. The approaches being developed to understand resistance to TRAIL will likely be applied to elucidate the mechanisms of nongenetic resistance broadly and in the clinic.
1. Introduction
The advent of targeted therapeutic agents was hailed as a major break through in the treatment of cancer. However, in most cases, initial promise is thwarted by the rapid development drug resistance. For targeted therapies to have sustained patient benefit, it will be necessary to understand and develop methods to combat resistance mechanisms. Canonically, resistance has been understood as a genetic process driven solely by mutations in therapeutic targets and associated regulators (Hanahan and Weinberg 2000). Nongenetic resistance has recently been implicated as an additional and significant hindrance to therapeutic efficacy (Marusyk et al. 2012). A host of mechanisms including upregulation of survival proteins, epigenetic modifications, regulated “noise” in gene expression, and selective activation of drug pumps have been proposed to account for nongenetic resistance (Brock et al. 2009; Sharma et al. 2010; Pisco et al. 2013). These mechanisms may operate in a coordinated manner or be activated selectively in different cancers or in response to distinct biological or chemical challenges. An ongoing question is whether nongenetic resistance is driven by the induction or by selection of survival mechanisms.
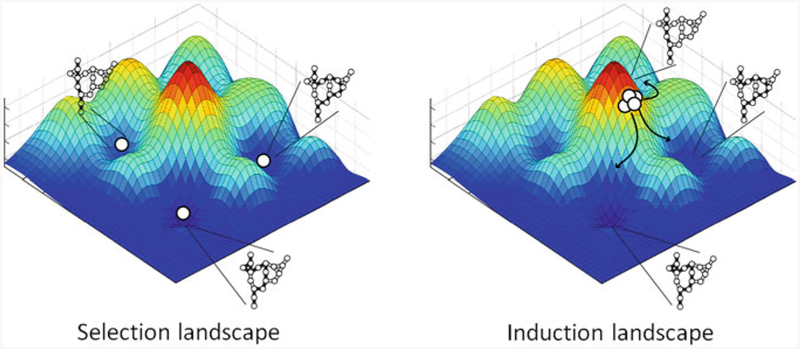
Theoretic and computational studies on the nature of the epigenetic (nongenetic) landscape suggest that either inductive or selective mechanisms could be at play in maintaining resistance (Waddington 1957; Pujadas and Feinberg 2012). In a conceptualization of the network state of the cell, known as a Waddington Landscape, the cellular signaling network state is depicted as a gravity well in which there is an interplay between unstable network states (visualized as being on the side of a hill) and stable network states (visualized as being on the floor of a valley). In the case of induction of a survival program, cells with unstable network states (hill states) are induced by the perturbation to assume more differentiated, stable network states (valley states) that encode a survival phenotype. In the selective case, cells in the unperturbed state are already distributed into survival network states (valley states) by virtue of nongenetic heterogeneity, and the perturbation does not significantly alter the diversity of survival phenotypes. Rather, the addition of a perturbation acts like a strainer to select cells with the network states that are resilient to the perturbation (Fig. 1).
Fig. 1.
Depiction of Waddington Landscape schematics for selection and induction of survival mechanisms. Peaks correspond to unstable network states; valleys correspond to stable network states encoding survival phenotype. Chain link diagrams represent signaling networks transducing survival signal via different pathways in each example. (left) If nongenetic resistance is achieved by selection of cells with resistant network states, then cells in the basal state are already distributed into survival network states (valley states) and the perturbation acts as sieve to select the resistant phenotypes. (right) Alternatively, if resistance is achieved by induction of survival network states, cells with unstable network states (hill states) are induced by the perturbation to assume more differentiated, resistant network states (valley states)
2. Difficulties in the Study of Nongenetic Resistance
The study of the interplay between induction and selection of resistance states has been challenging. In order to examine the induction of survival pathways, it is necessary to measure a large number of proteins simultaneously, and in order to measure the selection of cells with survival phenotypes, it is necessary to use a single-cell approach. Before the advent of mass cytometry, it was not possible to simultaneously detect a large number of proteins with single-cell resolution. Early studies seeking to understand the contribution of induction to nongenetic resistance utilized bulk methods to measure the large number of proteins necessary to assess network state. These methods include reverse-phase protein microarray (Lee et al. 2012), receptor tyrosine kinase arrays (Rodrik-Outmezguine et al. 2011), quantitative western blotting (Lee et al. 2012), and multiplexed kinase inhibitor beads/mass spectroscopy (MIB/MS). In MIB/MS beads are used to enrich receptor tyrosine kinases from a population of cells and then the activity state of individual kinases is assayed by MALDI TOF/TOF (Oppermann et al. 2009; Duncan et al. 2012).
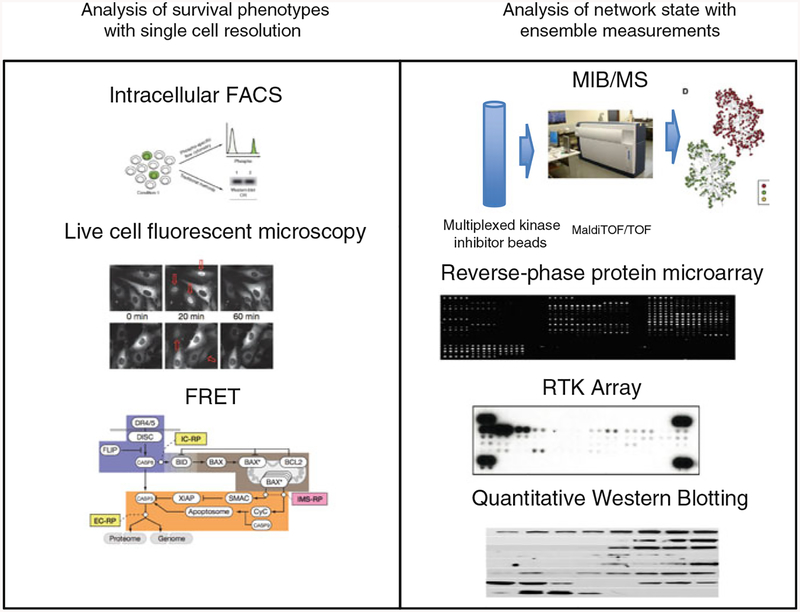
Studies on the contribution of selection to nongenetic resistance have employed the tools of single-cell analysis, chiefly fluorescent microscopy and fluorescently activated cell sorting (FACS), which are practically limited to measuring no more than a half dozen intracellular proteins simultaneously except for a few unique cases (Spencer and Sorger 2011; Sachs 2005) (Fig. 2).
Fig. 2.
Pre-dating mass cytometry there was no method to examine a large number of proteins (20+) with single-cell resolution. A variety of single-cell methods have been used to examine selection of cells with survival phenotypes. These include intracellular FACS and phosphoflow (Krutzik and Nolan 2003; Spencer et al. 2009), live cell fluorescent microscopy (Cohen et al. 2008), and Förster resonance energy transfer (FRET) (Albeck et al. 2008). A number of distinct methods that take ensemble (i.e., bulk) measurements have been used to assay a large number of proteins to examine network state. These methods include MIB/MS (Oppermann et al. 2009; Duncan et al. 2012), reverse-phase protein microarrays (Lee et al. 2012), receptor tyrosine kinase arrays (Rodrik-Outmezguine et al. 2011), and quantitative western blotting
In order to understand the relative contributions of induction and selection of survival phenotypes to nongenetic resistance, it is necessary to use a technology, such as mass cytometry, that is able to measure the activation states of a large number of proteins in response to drug exposure, simultaneously, and on a cell-by-cell basis. Mass cytometry is a flow-based technique in which the activation of up to 50 proteins can be measured simultaneously in a single cell. Data from 1,000 cells can be collected per second, making the analysis of millions of cells possible in a routine workflow (Bendall et al. 2011).
3. Features of Induction and Selection of Nongenetic Resistance
Induction of nongenetic resistance is most commonly referred to in the literature as network rewiring or compensatory signaling (Ryoo et al. 2004; Lee et al. 2012). In a basic example of induction, blockage of one survival pathway leads to the enhancement of an alternate survival pathway, which correlates with the presence of a resistant population (Rodrik-Outmezguine et al. 2011). Induction can act systematically and involve changes in the activation states of dozens of proteins throughout the cell leading to the increased prevalence of new network configurations that circumvents therapeutic intervention (Duncan et al. 2012).
The mechanism underlying induction of new phenotypic states has yet to be described and studies that have claimed to have induced a new network configuration may potentially be selecting for a subpopulation with a network state that is distinct from the modal network state before perturbation. Since these studies have not been performed with single-cell resolution, it is difficult to ascertain if a selective mechanism is involved. One study, however, has demonstrated that by sequential, but not simultaneous, application of targeted inhibitors it is possible to suppress the survivor phenotype, suggesting that it is possible to induce a change in the network state that affects phenotypic outcome (Lee et al. 2012).
Overall, the literature describing selection of nongenetic resistance demonstrates that particular network states that exist before perturbation can confer nongenetic resistance. Resistant cells can be detected based on their basal mitogenic protein expression before addition of therapy, implicating selection as a driver of resistance through nongenetic heterogeneity (Slack et al. 2008). It has been established that a diverse distribution of protein levels and protein activation potential can lead to increased survival (Slack et al. 2008; Singh et al. 2010). In normal tissues, variability in network state can be regulated and contribute to a population in which any single-cell may be susceptible to therapeutic intervention, but the population as a whole is unlikely to be completely ablated (Yuan et al. 2011). Diversity generating mechanisms may be directly selected for, as is seen in a number of other areas, such as ecology and microbiology, in which diversity has been shown to lead to system-wide robustness (Raser 2004; Flynn et al. 2011).
Diversity in general may help promote survival by allowing for the selection of cells occupying widespread survival niches. In addition, cells with a cancer stem cell phenotype can enable survival by occupying a refractory, quiescent state. The addition of chemotherapy induces apoptosis in faster growing cells and, therefore, selects for the quiescent cells (Reya et al. 2001). Surviving cells are positive for the cancer stem cell markers CD133 and CD24 and can be ablated by the use of histone deacetylase inhibitors, suggesting that DNA modifications enable the existence of this population (Sharma et al. 2010).
Induction of survival phenotypes can play a role in maintaining resistance after selection, suggesting that these two paradigms are not mutually exclusive in promoting nongenetic resistance. Several transcriptional regulators are selectively activated in surviving cells after treatment with the chemotherapeutic camptothecin, which activates downstream survival proteins (Cohen et al. 2008). Following death receptor activation with TRAIL and the initial selection of cells with survivor phenotypes, there is evidence that upstream survival signaling is induced in a subset of the survivors to maintain resistance (Spencer et al. 2009).
4. Use of Mass Cytometry to Decipher the Role of Inductive and Selective Mechanisms in Nongenetic Drug Resistance
Mass cytometry overcomes the limitations of previous technologies by providing parametric breadth with single-cell resolution. It is uniquely suited to analyzing complex, network processes that occur heterogeneously. Mass cytometry is currently being used by the Nolan group to determine whether selection or induction of survival network states underlies nongenetic drug tolerance to TRAIL-induced apoptosis. TRAIL-induced apoptosis is an ideal model process for understanding the mechanisms underlying nongenetic resistance. It occurs as the result of a directed and physiologically relevant stimulus, therefore, its induction does not involve significant off-target and complicating effects, as chemotherapy and even targeted therapy can. Most of the effectors of TRAIL-induced apoptosis are well studied, and sensitive markers of TRAIL induction are available (Johnstone et al. 2008). Finally, cell cycle state and positional effects have been ruled out as drivers of nongenetic resistance in this system (Spencer et al. 2009; Flusberg et al. 2013; Flusberg and Sorger 2013). A panel of antibodies against over 3 dozen proteins implicated in survival response from TRAIL-induced apoptosis, including multiple markers of activation in the MAPK, JNK, p38, and NFjB pathways have been validated. These pathways in combination with TRAIL are necessary for survival (Sah et al. 2003; Ohtsuka et al. 2003; Frese et al. 2003; Weldon et al. 2004).
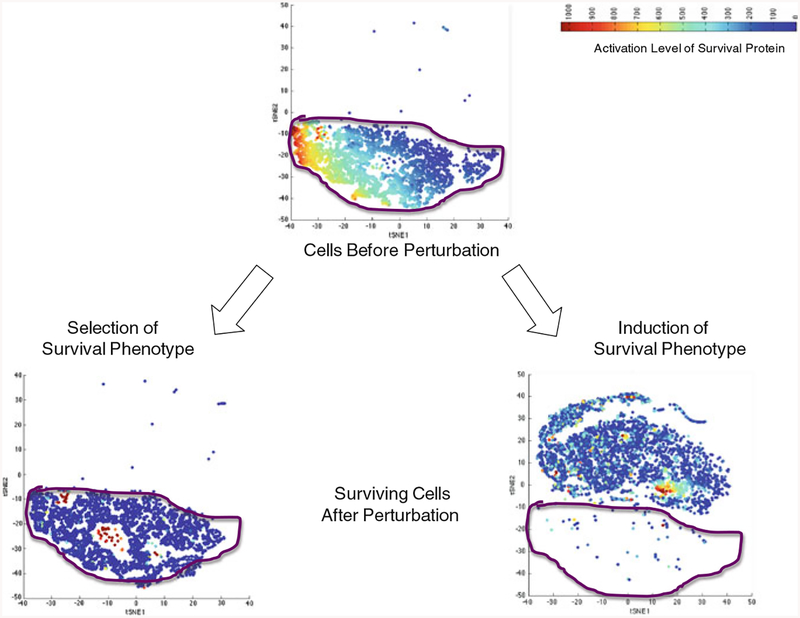
Mass cytometry is a destructive technology, so survivors cannot be tracked to determine whether a particular network state is induced by a perturbation. However, the network state of cells can be traced on the subpopulation level using bioinformatics clustering approaches. New algorithms, such as viSNE, allow high-dimensional data to be compressed into a 2-D map of phenotypic space (Amir et al. 2013). Therefore, one can determine whether the signaling state of survivors fits within the survival niche of cells in the basal state, suggesting a selection paradigm, or whether the survivors create a novel niche that did not exist in the basal state, suggesting an induction paradigm (Fig. 3).
Fig. 3.
Simulated data showing network state of survivors in viSNE plot. Cells are clustered based on functional alterations in 20 survival proteins. Each circle represents a single-cell. Cells in close proximity possess similar network states. Cells are colored based on activation level of one of the survival proteins. Area within purple line represents signaling state of cells before perturbation. If nongenetic resistance results from a selection mechanism, the surviving cells will fit within the signaling space of cells in the basal state. If nongenetic resistance results from an inductive mechanism, the surviving cells will occupy a signaling space that is distinct from the signaling space of cells in the basal state. If nongenetic resistance results from a hybrid mechanism (not pictured), cells will occupy both the pre-existing signaling space and a new signaling space
The network state of survivors can also be probed to determine whether induction or selection of the survivor phenotype underlies survival. The simultaneous measurement of the functional states of dozens of proteins in single-cell allows construction of network maps using the correlations of each protein in the network. Using straightforward statistical methods, how network features vary with survival can be determined. If the perturbation induces selective pressure that result in a survival phenotype, those perturbations that increase apoptosis (and decrease survival) should show a high level of net change in the network. In this case, the perturbation effectively acts as a sieve, and a small subset of cells with network states that differ substantially from the modal cell network state survive. Conversely, if the survival phenotype is induced, then conditions with the greatest level of net change in the network should correlate with perturbations that do not increase apoptosis. In this situation, the perturbation acts as a road block, forcing cells to drive further out of their way (i.e., alter their network state more profoundly) in order to reach the same destination (a survival network state).
By combining bioinformatics clustering to map the phenotypic state of the survivors and network deconvolution methods to dissect the level of network alteration in the survivors, it will be possible to determine whether induction, selection, or some combination of the two mechanisms, supports nongenetic resistance to TRAIL. Furthermore, these methods represent a generalizable approach that can be expanded to other systems to determine how these mechanisms of nongenetic resistance interact more broadly.
5. Conclusion
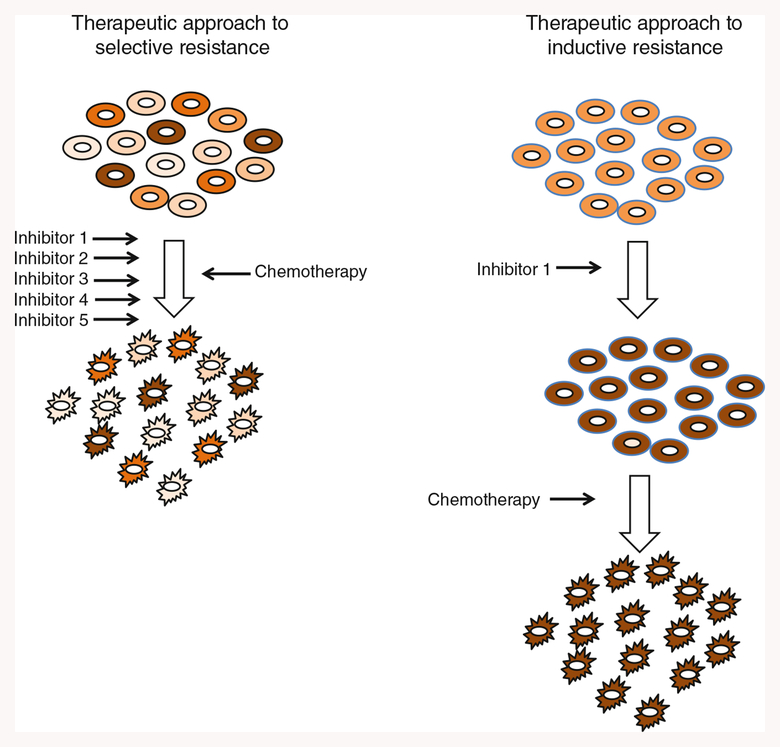
Despite great promise, no TRAIL-based therapy is used clinically, even though at least nine therapeutics targeting TRAIL having entered clinical trials (Newsom-Davis et al. 2009; Dimberg et al. 2012). Nongenetic drug tolerance may be a significant contributor to the lackluster clinical trial results. In cancers such as chronic myelogenous leukemia, nongenetic drug tolerance appears to act in concert with genetic mutations to render therapy ineffective (Okabe et al. 2008; Brock et al. 2009). Disentangling whether induction or selection underlies nongenetic resistance in a particular cancer may allow design of smarter and more effective cancer therapies (Fig. 4). If a survival phenotype is induced, then it may be possible to push the survivors into a less refractory state by the application of targeted inhibitors prior to the use of chemotherapy (Lee et al. 2012). If a selective process is at work, the simultaneous application of poly-specific inhibitors could be used to narrow the possible range of survival niches and lead to an ablation of resistant populations.
Fig. 4.
Schematics representing methods to exploit selective or inductive mechanisms of nongenetic resistance with targeted therapy. In cases in which selective resistance occurs, low-dose multidrug combinations could be used to decrease the number of network states that are resistant to therapy. In cases of inductive resistance, pretreatment with targeted inhibitors could induce cells to assume a network configuration that is more susceptible to follow-on therapy
References
- Albeck JG, Burke JM, Aldridge BB et al. (2008) Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell 30:11–25. doi: 10.1016/j.molcel.2008.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir E-AD, Davis KL, Tadmor MD et al. (2013) viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 31:545–552. doi: 10.1038/nbt.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P et al. (2011) Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332:687–696. doi: 10.1126/science.1198704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock A, Chang H, Huang S (2009) Non-genetic heterogeneity—a mutation-independent driving force for the somatic evolution of tumours. Nature 10:1–7 [DOI] [PubMed] [Google Scholar]
- Cohen AA, Geva-Zatorsky N, Eden E et al. (2008) Dynamic proteomics of individual cancer cells in response to a drug. Science 322:1–6 [DOI] [PubMed] [Google Scholar]
- Dimberg LY, Anderson CK, Camidge R et al. (2012) On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene 32:1341–1350. doi: 10.1038/onc.2012.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JS, Whittle MC, Nakamura K et al. (2012) Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149:307–321. doi: 10.1016/j.cell.2012.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg DA, Roux J, Spencer SL, Sorger PK (2013) Cells surviving fractional killing by TRAIL exhibit transient but sustainable resistance and inflammatory phenotypes. Mol Biol Cell. doi: 10.1091/mbc.E12-10-0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg DA, Sorger PK (2013) Modulating cell-to-cell variability and sensitivity to death ligands by co-drugging. Phys Biol 10:035002. doi: 10.1088/1478-3975/10/3/035002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn DFB, Mirotchnick N, Jain M et al. (2011) Functional and phylogenetic diversity as predictors of biodiversity–ecosystem-function relationships. Ecology 92:1573–1581 [DOI] [PubMed] [Google Scholar]
- Frese S, Pirnia F, Miescher D et al. (2003) PG490-mediated sensitization of lung cancer cells to Apo2L/TRAIL-induced apoptosis requires activation of ERK2. Oncogene 22:5427–5435. doi: 10.1038/sj.onc.1206842 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70 [DOI] [PubMed] [Google Scholar]
- Johnstone RW, Frew AJ, Smyth MJ (2008) The TRAIL apoptotic pathway incancer onset, progression and therapy. Nat Rev Cancer 8:782–798. doi: 10.1038/nrc2465 [DOI] [PubMed] [Google Scholar]
- Krutzik PO, Nolan GP (2003) Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry 55A:61–70. doi: 10.1002/cyto.a.10072 [DOI] [PubMed] [Google Scholar]
- Lee MJ, Ye AS, Gardino AK et al. (2012) Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell 149:780–794. doi: 10.1016/j.cell.2012.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, Almendro V, Polyak K (2012) Intra-tumour heterogeneity: a looking glass for cancer? Nat Publ Group 12:323–334. doi: 10.1038/nrc3261 [DOI] [PubMed] [Google Scholar]
- Newsom-Davis T, Prieske S, Walczak H (2009) Is TRAIL the holy grail of cancer therapy? Apoptosis 14:607–623. doi: 10.1007/s10495-009-0321-2 [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Buchsbaum D, Oliver P et al. (2003) Synergistic induction of tumor cell apoptosis by death receptor antibody and chemotherapy agent through JNK/p38 and mitochondrial death pathway. Oncogene 22:2034–2044. doi: 10.1038/sj.onc.1206290 [DOI] [PubMed] [Google Scholar]
- Okabe S, Tauchi T, Ohyashiki K (2008) Characteristics of dasatinib- and imatinib-resistant chronic myelogenous leukemia cells. Clin Cancer Res 14:6181–6186. doi: 10.1158/1078-0432.CCR-08-0461 [DOI] [PubMed] [Google Scholar]
- Oppermann FS, Gnad F, Olsen JV et al. (2009) Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics 8:1751–1764. doi: 10.1074/mcp.M800588-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisco AO, Brock A, Zhou J et al. (2013) Non-Darwinian dynamics in therapy-induced cancer drug resistance. Nat Commun 4:2467. doi: 10.1038/ncomms3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujadas E, Feinberg AP (2012) Regulated noise in the epigenetic landscape of development and disease. Cell 148:1123–1131. doi: 10.1016/j.cell.2012.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM (2004) Control of stochasticity in eukaryotic gene expression. Science 304:1811–1814. doi: 10.1126/science.1098641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL (2001) Stem cells, cancer, and cancer stem cells : abstract : nature. Nature 414:105–111. doi: 10.1038/35102167 [DOI] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC et al. (2011) mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov 1(3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Gorenc T, Steller H (2004) Apoptotic cells can induce compensatory cell proliferation through the JNK and the wingless signaling pathways. Dev Cell 7:491–501. doi: 10.1016/j.devcel.2004.08.019 [DOI] [PubMed] [Google Scholar]
- Sachs K (2005) Causal protein-signaling networks derived from multiparameter single-cell data.Science 308:523–529. doi: 10.1126/science.1105809 [DOI] [PubMed] [Google Scholar]
- Sah NK, Munshi A, Kurland JF et al. (2003) Translation inhibitors sensitize prostate cancer cells to apoptosis induced by tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) by activating c-Jun N-terminal kinase. J Biol Chem 278:20593–20602. doi: 10.1074/jbc.M211010200 [DOI] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B et al. (2010) A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141:69–80. doi: 10.1016/j.cell.2010.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Ku C-J, Wichaidit C et al. (2010) Patterns of basal signaling heterogeneity can distinguish cellular populations with different drug sensitivities. Mol Syst Biol 6:1–10. doi: 10.1038/msb.2010.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack MD, Martinez ED, Wu LF, Altschuler SJ (2008) Characterizing heterogeneous cellular responses to perturbations. Proc Natl Acad Sci 105(49):1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Sorger PK (2011) Measuring and modeling apoptosis in single cells. Cell 144:926–939. doi: 10.1016/j.cell.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Gaudet S, Albeck JG et al. (2009) Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459:428–432. doi: 10.1038/nature08012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH (1957) The strategy of the genes. A discussion of some aspects of theoretical biology With an appendix by Kacser H. Allen & Unwinpp, London, pp. ix 262 [Google Scholar]
- Weldon CB, Parker AP, Patten D et al. (2004) Sensitization of apoptotically-resistant breast carcinoma cells to TNF and TRAIL by inhibition of p38 mitogen-activated protein kinase signaling. Int J Oncol 24:1473–1480 [PubMed] [Google Scholar]
- Yuan TL, Wulf G, Burga L, Cantley LC (2011) Cell-to-cell variability in PI3 K protein level regulates PI3 K-AKT pathway activity in cell populations. Curr Biol 21:173–183. doi: 10.1016/j.cub.2010.12.047 [DOI] [PMC free article] [PubMed] [Google Scholar]