Abstract
The cerebral cortex may be organized into anatomical genetic modules, communities of brain regions with shared genetic influences via pleiotropy. Such modules could represent novel phenotypes amenable to large-scale gene discovery. This modular structure was investigated with network analysis of in vivo MRI of extended pedigrees, revealing a “multiscale” structure where smaller and larger modules exist simultaneously and in partially overlapping fashion across spatial scales, in contrast to prior work suggesting a specific number of cortical thickness modules. Inter-regional genetic correlations, gene co-expression patterns and computational models indicate that two simple organizational principles account for a large proportion of the apparent complexity in the network of genetic correlations. First, regions are strongly genetically correlated with their homologs in the opposite cerebral hemisphere. Second, regions are strongly genetically correlated with nearby regions in the same hemisphere, with an initial steep decrease in genetic correlation with anatomical distance, followed by a more gradual decline. Understanding underlying organizational principles of genetic influence is a critical step towards a mechanistic model of how specific genes influence brain anatomy and mediate neuropsychiatric risk.
Keywords: gene expression, genetic correlations, modularity, network analysis, structural MRI
Introduction
A primary aim of imaging genomics is to identify specific genes that influence neuroanatomical variation (Glahn et al. 2007). As genetic influences on human brain anatomy are profound (Pennington et al. 2000; Posthuma et al. 2000; Baaré et al. 2001; Schmitt et al. 2007a), it is likely that specific genes influence individual differences observed in morphometric analyses. Furthermore, these genes are strong candidate genes for illnesses associated with neuropathology (Glahn et al. 2014). Heritability, the proportion of population variability due to genetic factors, has been quantified for univariate brain traits such as volume, thickness or surface area at whole-brain (Bartley et al. 1997), lobar (Geschwind et al. 2002), gyral (Peper et al. 2007) and subgyral resolutions (Stein et al. 2010). Typically, these analyses involve parcellating an MRI image into anatomically distinct regions (e.g., Fischl et al. 2004b). Large-scale genome wide association studies (GWAS) have been conducted on these parcellated traits, providing a number of replicable loci (Bis et al. 2012; Stein et al. 2012; Hibar et al. 2015; 2017). Additionally, single-gene mutations cause specific cortical dysmorphologies (Gaitanis and Walsh 2004; Guerrini and Marini 2006). However, specific genes that influence normal anatomical variation have yet to be identified.
This limited progress could reflect the complexity of genetic influences. Individual brain traits are thought to be influenced by a complex relationship of environmental factors, common variants of small effects and rare variants of large effects (Thompson et al. 2013). While it is theoretically possible that the genetic influences on different parcellated brain regions are independent of one another, the evidence for such genetic autonomy is limited. Rather, multiple morphometrically distinct brain regions have common genetic roots (Baaré et al. 2001; Schmitt et al. 2007b, 2010; Glahn et al. 2015; McKay et al. 2015), suggesting pleiotropic effects whereby the same genes influence distributed neuroanatomical networks. Ontogenetically, it is possible that shared genetic variance results when neurons that comprise 2 regions derive from similar germinal zones at similar developmental periods. Genetically unique brain systems could provide novel phenotypes amenable to large-scale genome-wide search, which would constitute a fundamental advance in the genetics of human neuroanatomy. The application of formal network analysis (Bullmore and Sporns 2009; Sporns 2010) based upon genetic correlation provides a method for delineating neuroanatomic networks defined by common genetic factors. Therefore, we used graph theoretical techniques to investigate any potential genetic network architecture of the adult human brain.
In a series of recent studies, Chen and colleagues (Chen et al. 2011a, 2012, 2013), applied various network methods in ~200 twin pairs to investigate patterns of inter-regional genetic correlations, reporting the genetic modularity of neuroanatomic networks. More specifically, Chen and colleagues identified 12 specific modules (or clusters). Although cortical surface area and cortical thickness were not genetically correlated (Panizzon et al. 2009; Winkler et al. 2010), these 13 fundamental modules were largely overlapping between these morphometric indices. Identified modules were largely bilaterally symmetric (left and right hemispheres were mirror images) and spatially contiguous (within hemispheres modules were composed of anatomically adjacent regions). Given that brain networks across multiple modalities tend to have a scale-free organizational structure (Meunier et al. 2009; Bassett et al. 2010), evidence for a specific modular resolution (a single “natural” number of modules) is particularly striking and has profound implications for studies of the regional specificity of brain function, pathophysiology and gene discovery. Unfortunately, the modular networks identified by Chen and colleagues have yet to be replicated in an independent sample.
The present study extends this prior work on the genetics of brain cortical architecture in several ways. First, this study is one of the largest studies to date of genetic correlations in cortical thickness between brain regions, using a sample of ~1400 individuals in large, extended pedigrees. Second, we use network theory and machine learning techniques to explore whether the brain is organized into a “natural” number of genetic correlation modules. Third, we investigate the relationship between genetic correlations and the anatomical distance between brain regions, as spatial constraints are known to influence network properties (Vértes et al. 2012). Finally, we compare the imaging-genetic findings with correlations in gene expression from postmortem microarray studies (Hawrylycz et al. 2012). Our findings suggest a multiscale, multiresolution modular structure consistent with a hierarchical organization, where smaller components are iteratively connected into larger components and systems. Relatively simple organizational principles, including contralateral homology and greater genetic overlap with greater anatomical proximity, underlie much of the apparent complexity observed in the network of genetic correlations.
Methods
Imaging Acquisition and Processing
Since 2006, the Genetics of Brain Structure and Function study (GOBS), has recruited randomly ascertained extended pedigrees of Mexican American descent living in San Antonio (McKay et al. 2014). High-resolution structural MRI scans were acquired on a Siemens 3T TIM Trio at UTHSCSA. The sequence was composed of multiple T1-weighted 3D turbo-flash sequences with an adiabatic inversion contrast pulse (0.8 mm isotropic voxels, TE 3 ms, TR 2100 ms, TI 785 ms, flip angle 13°, FOV 200 mm). The sample included 1443 individuals (836 female) with mean age of 40.7 years (SD = 15.5, range = 18–85).
Image processing used FreeSurfer version 5.3 (Dale et al. 1999; Fischl et al. 1999; 2001; Fischl et al. 2004a, 2004b; Desikan et al. 2006). Briefly, after normalization and skull stripping, white-matter voxels were identified based on intensity and covered in a topologically corrected, tessellated mesh. This gray-white surface was then expanded to fit the pial surface. Surfaces were examined manually and edited if necessary. Cortical curvature was used to register surfaces to a common atlas (Desikan atlas) and identify 68 regions interest (Desikan et al. 2006). Cortical thickness was calculated as the distance between the gray-white surface and the pial surface at each vertex.
A replication dataset was also analyzed, using publically available data from the human connectome project (HCP; http://www.humanconnectome.org/), which comprised MRI data from 1113 individuals (606 female) from 457 unique families (including 170 dizygotic twins, 286 monozygotic twins, 576 non-twin siblings, and 25 non-sibling familial relations) with mean age 28.8 years (SD = 3.7, range = 22–37). As previously described (Glasser et al. 2013; Van Essen et al. 2013), T1-weighted and T2-weighted structural images were acquired on a 3T Siemens Skyra employing a 32-channel head coil. T1 scans used a magnetization prepared rapid gradient echo sequence (0.7 mm isotropic voxels, TE 2.14 ms, TR 2240 ms, TI 1000 ms, flip angle 8°, ES 7.6 ms, GRAPPA = 2.). T2 scans used a variable flip angle turbo spin-echo sequence (0.7 mm isotropic voxels, TE 565 ms, TR 3200 ms, BW = 744 Hz per pixel, no fat suppression pulse, GRAPPA = 2). Preprocessing was performed using version 3 of the HCP pipeline (Glasser et al. 2013), including co-registration of T1 and T2 scans, B1 (bias field) correction, and segmentation and surface reconstruction using FreeSurfer version 5.3-HCP to estimate cortical thickness in 68 regions interest (Desikan et al. 2006).
Genetic Correlation Network
Variance-components genetic analysis was performed to differentiate genetic and environmental contributions to the phenotypic covariance between cortical thicknesses of different brain regions (Williams et al. 1999). For an individual trait such as the cortical thickness of one region, heritability is estimated using the polygenic model (Almasy and Blangero 1998; Lynch and Walsh 1998)
| (1) |
where is the trait vector, X is a matrix of covariates, is a vector of coefficients, g is a vector reflecting the random additive effect of genetics, and e is a vector of residuals (which includes environmental effects not modeled). The vector e follows a multivariate normal distribution with zero mean and variance–covariance matrix , where is a scalar reflecting the additive genetic variance, and is twice the kinship matrix (quantifying the genetic relationships between subjects). The vector e follows a multivariate normal distribution with zero mean and variance–covariance matrix , where is the residual variance, and is the identity matrix. The vector y is distributed as where , , and are free parameters. The heritability of the trait is given by . Maximum likelihood estimation was performed with SOLAR software (www.solar-eclipse-genetics.org) (Almasy and Blangero 1998).
The univariate polygenic model described above can be extended to estimate the genetic and environmental variances of 2 traits, A and B, as well as the genetic and environmental correlations between them, using the bivariate polygenic model
| (2) |
where is a concatenated vector containing both traits. The variance–covariance matrix of is , where is the Kronecker product, and is a 2-by-2 matrix whose diagonal elements are the genetic variances of A and B ( and , respectively) and whose off-diagonal elements are the covariance between the genetic effects . The variance–covariance matrix of is , where contains the residual variances and covariance (, , and ). After maximum likelihood estimation with SOLAR, the genetic correlation is calculated as . Age, age2, sex, and their interactions, as well as intracranial volume, were included as covariates in all analyses.
Genetic correlations were calculated between all 2 278 pairs of macro-anatomic brain regions. The absolute value of these ρg formed the elements of a 68 × 68 association matrix, and weighted network models were constructed by thresholding these matrices at different edge densities (the proportion of elements of the association matrix included in the network as edges) including minimum spanning trees to ensure all networks were connected (Alexander-Bloch et al. 2010). Networks were constructed at each density between 0 and 0.2 at 0.001 intervals. Note that at this range of connection densities, the magnitude of positive correlations always exceeded that of negative correlations, so the results were robust to taking or not taking the absolute value of ρg.
The modularity, Q, of these weighted networks was estimated as , where m is the total weight of the edges in the network; if there is an edge between i and j (otherwise ); the delta function if i and j are in the same module M (otherwise ); and is the expected weight of an edge between i and j, where is the total weight of i’s edges. At each density between 0 and 0.2, the optimal modular partition was estimated using Newman’s algorithm (Clauset et al. 2004).
Representative consensus partitions across densities were then calculated by successively matching partitions by solving the linear assignment problem (Dimitriadou et al. 2002) in order of their average normalized mutual information with other partitions (Strehl and Ghosh 2002). Network analysis was performed in R using iGraph (Csardi and Nepusz 2006), Clue (Hornik 2005) and Cluster (Maechler et al. 2012) software packages. All networks were compared with randomly generated networks with same number of nodes, connection densities and degree distributions (Viger and Latapy 2005).
Two additional analyses were performed to specifically query the modular structure across scales or resolutions. Newman’s method may be biased against smaller modules with fewer nodes (Fortunato and Barthélemy 2007), which was addressed by the addition of a resolution or scale parameter, , to the modularity quality function which was optimized alternatively with simulated annealing (Reichardt and Bornholdt 2004) and with the generalized Louvain method (Blondel et al. 2008; De Meo et al. 2011) (see Supplementary Materials). A plateau in the optimal number of modules across a range of values for this parameter, or a peak in the similarity of the modular structure across multiple instantiations of the algorithm, may indicate a more natural number of modules or specific spatial scale (Arenas et al. 2008; Betzel et al. 2017) as opposed to there being no preference for a specific resolution or scale. In addition, partitioning around medoids (PAM) clustering was performed on the genetic correlation matrix. When queried across a range of possible clusters (the number of clusters, k, is an input parameter for this clustering method), a sharp peak in a distinct quality function such as the silhouette width can indicate a more or less “natural” split into a number of homogenous clusters (Kaufman and Rousseeuw 1987). Alternatively, a lack of such a peak can be interpreted as evidence of a lack of a specific and distinct number of clusters.
The relationship between anatomical distance and ρg was estimated using general linear models in R statistics (R Core Team 2016). For polynomial models
| (3) |
where d is a vector of the Euclidean between all pairs of regions (distance between the centroids of the regions on the pial surface) and e is a vector of residuals. This model was fit separately for intra- and inter-hemispheric pairs of regions, where for interhemispheric regions, distance was estimated as the distance from a region’s contralateral homolog. Linear, quadratic, cubic and higher order polynomial models were compared using likelihood ratio tests and Akaike Information Criterion (Venables and Ripley 2002). Exponential decay models
| (4) |
were compared with polynomial models using the Akaike Information Criterion. The significance of the relationship with distance was also tested with Mantel permutation tests.
Gene-Expression Networks
Gene-expression networks were derived from publicly available microarray data from the Allen Human Brain Atlas (http://human.brain-map.org/) (Hawrylycz et al. 2012). Postmortem brain tissue was collected from 6 individuals, with ~500 samples collected from each hemisphere; left hemisphere data was collected in all 6 subjects, while right hemisphere data was collected in 2 subjects. Expression levels for ~60 000 RNA probes were collected for each sample. The data were averaged across probes corresponding to the same gene, resulting in 20 739 genes from 3702 samples. MNI coordinates corresponding to each sample were matched to the FreeSurfer average cortical surface, allowing for comparisons with the imaging-genetics data. Only the 1745 cortical samples were included in subsequent analysis, with the subcortical samples excluded for consistency with the analysis of cortical thickness in the brain imaging data. Data within each gene were standardized, and gene expression correlations (rGE) were calculated using Pearson’s r between every pair of cortical samples, providing a measurement of the similarity in the samples’ gene-expression profiles.
The within-module and between-module rGE was calculated by assigning each cortical sample to its nearest ρg module. The within-module and between-module rGE was calculated for each subject individually and then averaged across subjects. The significance of within module rGE was assessed by comparing these data to permutations where the module membership of each region was randomized. The anatomical distance between brain samples was estimated using the Euclidean distance between the MNI coordinates of each sample. Contralateral homology was inferred from the anatomical labels for the two subjects with bilateral data. Differential expression between cortical modules for each gene was calculated using lme4 in R (Bates et al. 2014), using subject and region as random effects and module as a fixed effect. P-values were obtained by likelihood ratio tests of the full model including module membership compared with the reduced model without module membership. The results of this test were entered into a gene ontology (GO) analysis using TopGO in R (Alexa et al. 2006) with Entrez gene annotations and Kolmogorov–Smirnov test of enrichment. FDR correction for multiple comparisons was performed on the results of the enrichment test (Benjamini et al. 2006).
Computational Model
We used a simple computational model to investigate whether the modularity of the network was related to or driven by the constraints of anatomical distance and contralateral homology, by generating null networks where these properties were preserved but wiring was otherwise generated at random. For the genetic correlation network, the relationship between anatomical distance and ρg was estimated with polynomial models as described above. As described in the Results, the best-fit model was a quadratic polynomial model for intrahemispheric pairs of regions, and a cubic polynomial model for interhemispheric pairs of regions. For each pair of regions, the predicted from these models, , was an estimate of the genetic correlation expected based on anatomical distance constraints and contralateral homology alone. The final model of genetic correlation, , also included a stochastic element
| (5) |
where was a vector of noise, derived by adding or subtracting 0.05 (equivalent to 5% of the highest magnitude genetic correlation in the real data) from pairs of regions randomly chosen with replacement. The number of pairs of regions chosen was varied such that the stochastic element accounted for 0–50% of the weight of the total genetic correlation matrix (in 1% intervals) in the modeled networks. As this number increases, modeled networks are predicted to increasingly reflect topological characteristics of random networks.
The modular characteristics of these modeled networks were compared with those of the actual ρg network to determine whether a given “value” of (the proportion of the modeled network that was based on the stochastic process) generated model networks with similar modularity as the actual network. For this purpose, the mean modularity was calculated across the range of density thresholds (from 0.0 to 0.2 in 0.001 intervals). Finally, for model networks with the same modularity as the actual network, the within module gene expression correlations, rGE, were calculated to determine whether the network model could account for the relationship between genetic correlation modules and inter-regional correlations in gene expression profiles.
Results
Genetic Correlations Reveal Modular Structure
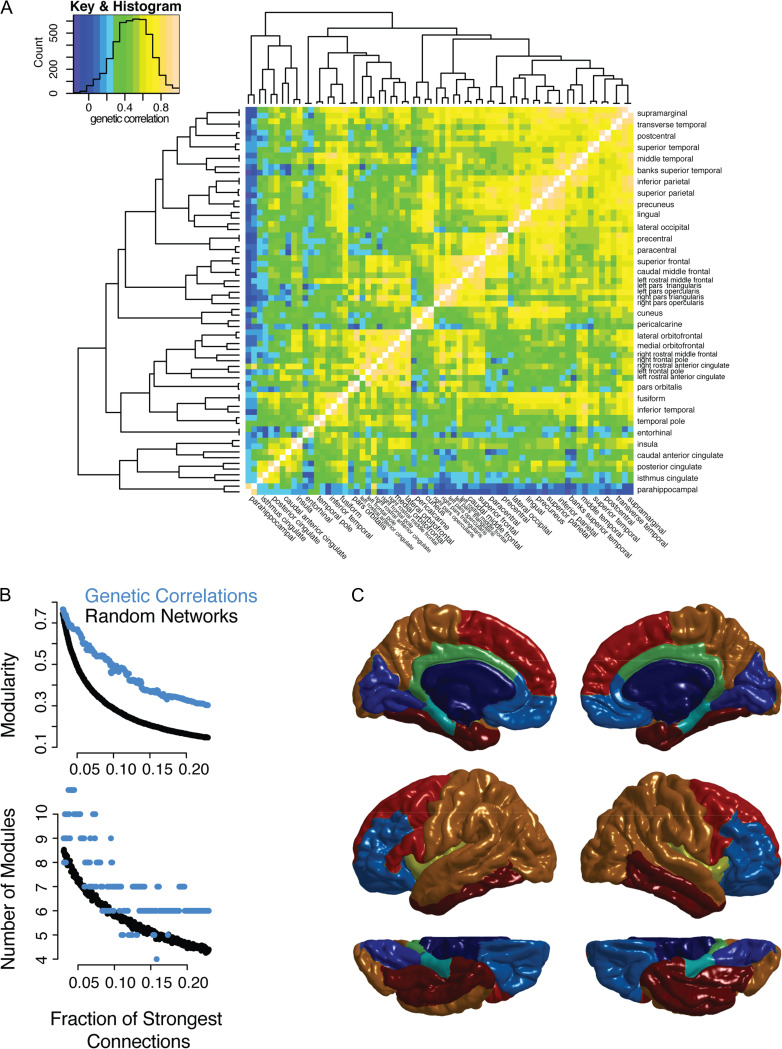
The pattern of genetic correlations (ρg) formed a complex, heterogeneous network (Fig. 1A). A greater proportion of the shared variance in cortical thickness between regions was due to shared genetic factors (mean ρg = 0.48, SD = 0.19, range = −0.17–1.0) than to environmental factors (mean ρe = 0.22, SD = 0.13, range = −0.18–0.74, see Supplementary Fig. 1). Although there were some negative genetic correlations, the magnitude of the high positive correlations exceeded those of the negative correlations, so the negative correlations were not included in the networks derived from the inter-regional genetic correlations. This network had significant modular structure (Fig. 1B) compared with randomized, connected networks with the same degree distribution, at every connection density tested (P < 0.01, 1000 random networks per connection density).
Figure 1.
Network model of cortical thickness genetic correlations showing multiresolution modular structure. (A) All correlations between brain regions are shown with color scale indicating strength of genetic correlation. The regions are organized by complete linkage hierarchical clustering, with the dendrogram shown to the left and to the top of the heat map. Names of regions are shown to the right and to the bottom of the heat map; hemisphere (left or right) is specified only for regions that were not clustered such that they are adjacent to their contralateral homolog. Color key is shown to the top left, as well as a histogram showing the number of pairs of regions (count) with genetic correlations at each strength. (B) Network models (graphs) were constructed at each connection density from 0.0 to 0.2 in 0.001 intervals, where the connections density refers to the fraction of the strongest connections that are included in the graph. The genetic correlation networks are shown in addition to random control networks (see Methods). The top plot shows modularity, a measure of the strength of the community structure of the network, which is significantly higher in the genetic correlation network compared with the random network. The bottom plot shows the optimal number of modules at each density. (C) An illustration of the consensus partition (modular community structure) with 8 cortical modules (see Supplementary Fig. 3 for illustrations of the partition at different resolutions).
In contrast, there was not a significant difference in the optimum number of modules for the ρg network compared with random networks (Fig. 1B). Across graph densities, optimal partitions are found across a wide range in terms of the number of modules, suggesting that there is no single number of modules that satisfactorily represents the structure of the network (Fig. 1B). Supplementary analysis, including silhouette plots of k-medoid (PAM) clusters and the addition of a resolution parameter, , to identify modules across spatial scales, further confirmed this multiscale modular structure (see Supplementary Fig. 2).
The modular partitions appeared to largely respect a priori functional boundaries. In particular, medial occipital, dorsal–lateral prefrontal, ventral-medial prefrontal and the limbic system were relatively well delineated (Fig. 1C). At a coarser resolution (2 and 5 module solutions in Supplementary Fig. 3) the brain was subdivided principally along a dorsal-ventral axis. It is notable that, although the algorithms were in no way constrained to cluster spatially adjacent regions, nor to cluster contralateral homologous regions, modules were largely composed of laterally symmetric, spatially contiguous regions of the cortical surface. Contralateral symmetry has previously been observed in modules derived from functional connectivity (Meunier et al. 2009) and structural covariance (Chen et al. 2011b) (which combines genetic and environmental correlation). Notable exceptions to contralateral symmetry were regions within the inferior frontal gyrus, which in the right hemisphere were located in the dorsal–lateral (red) and ventral medial (light blue) module; while in the left hemisphere these regions were all located within the dorsal–lateral module. There was only limited overlap between pairs of regions with high genetic correlations and those with relatively high environmental correlations (see Supplementary Fig. 1).
Quantitatively, genetic modules were compared with the recent multimodal parcellation generated from HCP data (Glasser et al. 2016). Although the limitations of this kind of comparison should be noted (Alexander-Bloch et al. 2017; see Supplementary Materials), there appeared to be a high correspondence, such that in each hemisphere the 180 HCP parcels tended to lie within genetic modules. Using the 8-module partition for example, nearly half of the HCP-parcels were >90% within a single genetic module, respectively (mean = 84% within a single genetic module, median 89%, range = 36–100%).
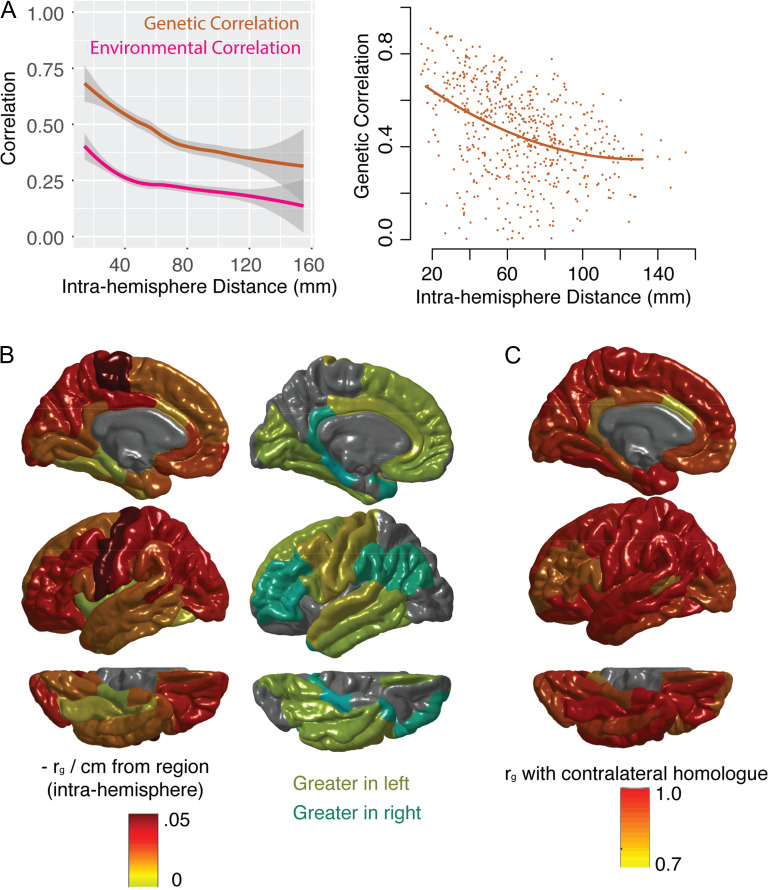
The Influence Anatomical Distance and Contralateral Homology
As suggested by the modular structure, the genetic correlations between contralateral homologs were greater on average than the genetic correlations between other regions (mean genetic correlation with contralateral homolog, ρg = 0.94, SD = 0.06, range = 0.74–1.0). Of all 68 regions, only 5 regions had higher genetic correlations with any other region than with their homolog in the opposite hemisphere; exceptions included canonical language areas with known functional asymmetry (left bank of the superior temporal sulcus, left pars opercularis, left pars triangularis) as well as left rostral middle frontal gyrus and right caudal anterior cingulate. This reflects a general pattern where the genetic correlations with contralateral homologs were relatively lower in the cingulate and in inferior prefrontal regions compared with the rest of the cortex (Fig. 2C). The magnitude of environmental correlations between contralateral homologs was lower than that of genetic correlations, both in absolute terms and relative to environmental correlations with non-homologous regions (see Supplementary Fig. 1).
Figure 2.
The relationship between genetic correlation (ρg), anatomical distance and contralateral homology. (A) The left-hand plot shows the relationship between anatomical distance and phenotypic (total) correlation, as well as genetic correlation (ρg, the genetic component of the phenotypic correlation), and environmental correlation (the non-genetic component of the phenotypic correlation). Lines represent smoothed local averages, with standard error silhouettes (ggplot2 package in R). The right-hand plot focuses on the genetic correlation (ρg), showing the fit of a quadratic model of the relationship with distance, which is a better fit than either a linear model or an exponential decay model (see Results). These plots show correlations within the left hemisphere only. (B) For each region, the linear relationship between ρg between that region and every other region was estimated. At the regional level, the model fits were not improved by adding quadratic components. Color scheme represents the estimated decrease in ρg per cm from the region of origin. The hemispheres were averaged for the left-hand plot. The right-hand plot whether the slope of the linear relationship with distance was greater in the left hemisphere (yellows) or right hemisphere (aqua), for each region. (C) The ρg between each region and its contralateral homolog in the opposite hemisphere.
Genetic correlations were strongly influenced by the anatomical distance between brain regions (Fig. 2A, B). For intrahemispheric genetic correlations, this relationship was approximated by a quadratic polynomial model (residual difference in sum of squares compared with a linear model = 0.67, Chi square test (1 df) P < 1e–5). There was no evidence for a significant cubic relationship with intrahemispheric anatomical distance (residual difference in sum of squares compared with quadratic model = 0.07, Chi square test (1 df) P = 0.12). The quadratic model also outperformed an exponential decay model (adjusted r2-quadratic = 0.15; adjusted r2-quadratic exponential = 0.04; Akaike information criterion quadratic = −737; Akaike information criterion exponential = 2097).
The relationship between genetic correlations and anatomical distance was regionally heterogeneous. This relationship also diverged from that between anatomical distance and the environmental correlations between regions. Although there was a negative relationship between anatomical distance and environmental correlations, this relationship largely plateaued around 4 cm, while the relationship with genetic correlations extended to longer distances (Fig. 2A). When the relationship of ρg with distance was estimated for each region separately, lateral and dorsal aspects of the cortex appeared to have stronger negative relationships with distance; relatively weaker relationships with distance existed in ventral and temporal regions (Fig. 2B). Although largely convergent between hemispheres, certain regions diverged from their contralateral homologs in terms of the strength of there with distance, including canonical language areas (Fig. 2B).
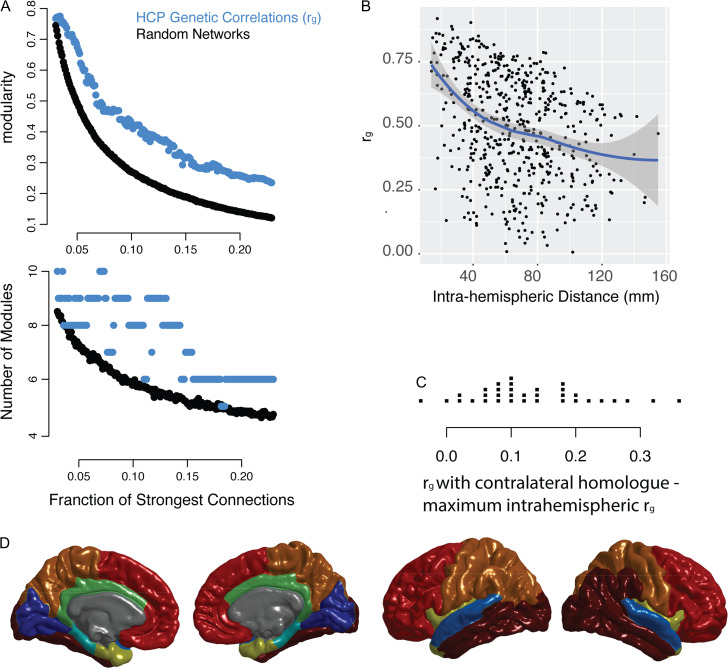
Replication Data
A similar network structure was observed in the ρg network derived from the HCP data. The network demonstrated significant modularity at every connection density, but did not demonstrate a single optimal number of modules (Fig. 5A). The exact modular partitions derived for the GOBS and HCP data were similar but not identical. For example, prefrontal cortical regions that belonged to different modules in the GOBS data (Fig. 1C) belong to the same module in the HCP data (Fig. 5D). Similar relationships with anatomical distance and contralateral homology were also observed in the HCP data. The HCP genetic correlations were strongly influence by the anatomical distance between brain regions (Fig. 5B). In addition, the HCP genetic correlations between contralateral homologs were greater on average than the genetic correlations between other regions (Fig. 5C), recapitulating the results from the GOBS data.
Figure 5.
Replication of key results using data from Human Connectome Project. (A) Network models (graphs) were constructed at intervals of 0.001 connections density, where the connections density refers to the fraction of the strongest connections that are included in the graph. The genetic correlation networks are shown in addition to random control networks (see Methods). The top plot shows modularity, a measure of the strength of the community structure of the network, which is significantly higher in the genetic correlation network compared to the random network. The bottom plot shows the optimal number of modules at each density. For comparison with GOBS data, see Figure 1B. (B) An illustration of the consensus partition (modular community structure) with 8 cortical modules (For comparison with GOBS data, see Fig. 1C). (C) The relationship between anatomical distance and the genetic correlation between brain regions, shown for the left hemisphere. The line represents smoothed local averages, with standard error silhouettes (ggplot2 package in R). The right-hand plot focuses on the genetic correlation (ρg), showing the fit of a quadratic model of the relationship with distance, which is a better fit than either a linear model or an exponential decay model (see results). For comparison with GOBS data, see Figure 2A. (D) For each of the FreeSurfer regions, the strip chart shows the difference between ρg with the contralateral homolog and the maximum within-hemisphere ρg.
Gene Expression, Genetic Correlation
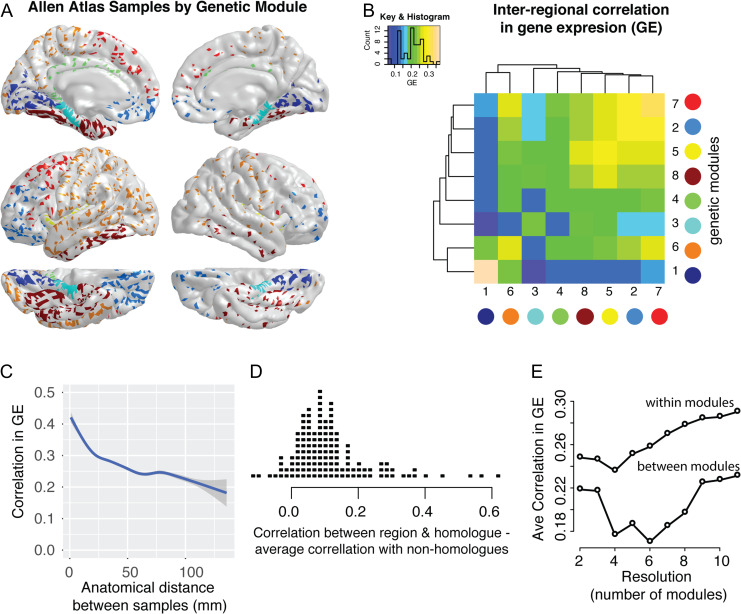
Correlations in gene expression, rGE, were higher within ρg modules as opposed to between ρg modules, despite the fact that these modules were derived independently from the gene expression data (Fig. 3A, B). Compared with a null model where samples were randomly assigned to modules, the within-module genetic correlation was significantly higher when regions were assigned to their actual module (P < 0.001, 1000 permutations). Particularly strong correlations in gene expression profiles occurred within the occipital module (dark blue) and within the dorsal prefrontal module (red), although the prefrontal module had higher between-module rGE with other modules than did the occipital module (Fig. 3B).
Figure 3.
Correlations in cortical gene expression (rGE) influenced by genetic correlation (ρg) modules, anatomical distance and contralateral homology. (A) Location of cortical samples available for 6 subjects in Allen Atlas, color-coded based on ρg module (as per Fig. 1C). (B) The intersample correlations in expression pattern across genes were calculated for each pair of samples (within subjects only). The heat map illustrates the average within-module rGE (diagonal elements) and the average between-module rGE (off-diagonal elements). (C) The relationship between anatomical distance and rGE (smoothed local average with standard error silhouettes). (D) For each of the FreeSurfer regions (for subjects with samples from both left and right hemispheres), the strip chart shows the difference between rGE with the contralateral homolog and the average rGE with non-homologous regions. (E) The correlation in gene expression patterns across genes across a range of modular resolutions from 2 to 11 genetic correlation modules, showing the average within-module and the average between module rGE across all modules.
Similar to the ρg networks, rGE networks also revealed the influence of anatomical distance and contralateral homology. For the 2/6 subjects with gene expression data from both cortical hemispheres, the rGE between a brain region and its contralateral homolog was greater than the average rGE between that region and other brain regions in >90% of cases (Fig. 3D). Comparable to the ρg data, a biphasic relationship also existed between rGE and anatomical distance, with a steep initial decrease in correlation strength followed by a more gradual decrease at longer anatomical distances (Fig. 3C).
GO analysis showed that the differential expression of genes between cortical genetic modules were linked to plausible biological processes. In total, 66 GO terms survived correction for multiple comparison (FDR-corrected P < 0.05), including nervous system development, brain development, telencephalon development, regulation of stem cell proliferation, chromatin regulation and protein targeting. Supplementary Figure 4 shows the top 5 GO terms and their hierarchical relationships with other biological processes, and Supplementary Table 1 shows all 66 significant GO terms.
The elevated rGE, within—as opposed to between—ρg modules was consistent across a range of modular resolutions (Fig. 3E). For the consensus modular structures from n = 2 (33 regions per module on average) to n = 11 (6 regions per module on average), the average within-module rGE (mean across resolutions = 0.264, SD = 0.019) always exceeded the average between-module (mean across resolutions = 0.204, SD = 0.023).
Modeling Genetic-correlation Networks
The constraints of anatomical distance and contralateral homology on genetic correlations appeared to be directly related to the modularity of this network. Models were constructed where the genetic correlation between regions was predicted by these relationships alone, and these models adequately predicted the modularity of the actual ρg network (Fig. 4A,B). As noted above, quadratic models were appropriate for the intrahemispheric relationship with distance, while cubic models were necessary to accurately model the interhemispheric relationship with distance because of the disproportionately high correlations between contralateral homologs.
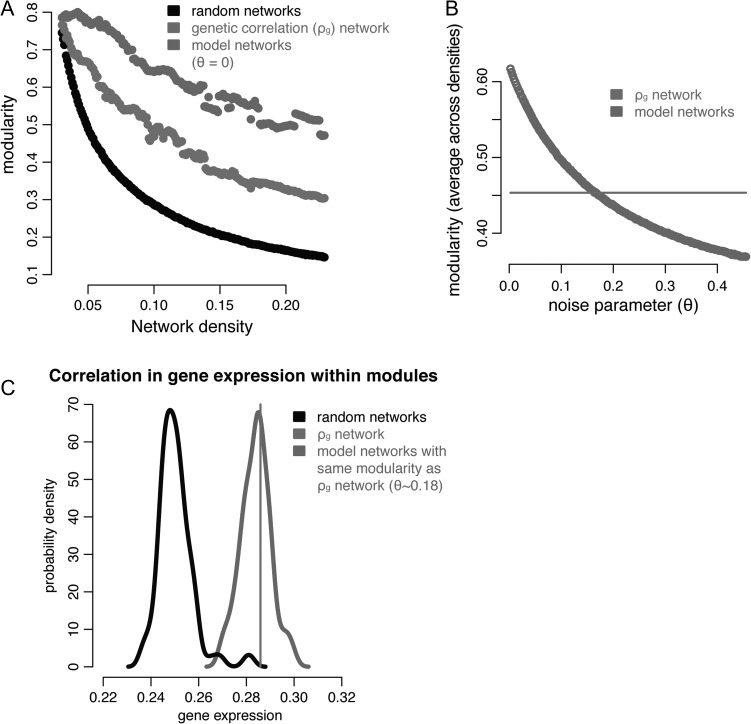
Figure 4.
Modeling the effect of anatomical distance and contralateral homology. (A) Modularity for genetic correlation networks and random networks as per Figure 1B. For model networks, inter-regional correlation is based on what would be predicted based on the statistical relationship with anatomical distance within and between hemispheres (Fig. 2) and a noise parameter ( in this subplot). (B) The modularity (averaged across densities) of the model networks at different values of , which superimposes noise on top of the predicted relationships. For , the modularity of the model networks intersects with that of the actual ρg network. (C) Probability density plots of the average within-module correlation in gene expression (rGE, see Fig. 3): for the actual ρg network modules (black line); for modules derived from 1000 randomly generated networks (light gray distribution); and for modules derived from 1000 modeled networks with the same modularity as the actual ρg network (dark gray distribution).
The modules predicted by the computational model also had significantly elevated within-module rGE, suggesting that the influence of distance and contralateral homology may drive the observed convergence between rGE and ρg network organization (Fig. 4C). Importantly, the model’s success in predicting within-module rGE was not due entirely to samples within the same FreeSurfer anatomical region. For networks that respected this cortical parcellation but whose between-region connections were randomized, the within-module rGE was significantly lower than for the rGE within the actual ρg modules (P = 0.001, 1000 random networks). In contrast, for models with network modularity the same as the actual ρg network, the within-module rGE was similar to the within-module rGE for the actual ρg modules (P = 0.4, 1000 network models).
Discussion
Network analysis of genetic correlations in human cortical thickness reveals a multiscale modular architecture. Different modular organizations, with greater or smaller numbers of regions per module, exist at different scales (or resolutions). In contrast to previous reports (Chen et al. 2012, 2013), we do not find evidence for a more “natural” number of clusters or modules at a certain spatial resolution. While perhaps not as intuitive as a single, unique modular structure, multiscale modularity has been reported by several brain functional and anatomical studies (Betzel and Bassett 2016). This structure may point to a hierarchical organization where larger modules are at least partially composed of smaller modules across spatial scales (Simon 1962).
Two fundamental principles appear to underlie this multiscale modular structure. First, regions are strongly genetically correlated with their counterparts in the opposite cerebral hemisphere (contralateral homology; Levin 2005). Notably, heterogeneity in this contralateral homology, with lower homology in inferior prefrontal language areas, suggests a connection between functional and genetic asymmetry. Second, regions are highly genetically correlated with nearby regions, with an initial steep decrease with anatomical distance followed by a more gradual decline. These findings are replicated in an independent dataset (Glasser et al. 2013) and largely convergent with postmortem correlations in gene expression (Hawrylycz et al. 2012). Computational models demonstrate that, together, the constraints of contralateral homology and anatomical distance predict the degree of modularity of the actual genetic correlation network.
Physically embedded networks like the brain tend to have spatial constraints, with increased connectivity at shorter connection distance (Bullmore and Sporns 2012). Both brain functional (Alexander-Bloch et al. 2013) and diffusion imaging (Gong et al. 2012) networks are spatially constrained, which acts to decrease network wiring cost, although long distance connections do exist within functional modules (such as anterior and posterior regions of the default mode network). A possible molecular model of spatial constraints in genetic correlations is suggested by animal studies, with signaling molecules emitted by patterning centers in the developing cortex in a gradated fashion, and subsequently refined into disjunctive cortical maps based on sensory, thalamocortical connections (O’Leary et al. 2007). It should be noted that confounds such as motion artifact (Blumenthal et al. 2002; Power et al. 2012) and spatial smoothing by image processing pipelines could augment this signal, although this kind of artifact should not have differential impact on genetic as opposed to environmental correlations, and the regions we report with the strongest spatial constraints do not converge with those predicted to be differentially impacted by motion artifact in terms of cortical thickness estimates (Reuter et al. 2015; Alexander-Bloch et al. 2016).
Modules discovered by community detection techniques, which split heterogeneous data into relatively homogenous subgroups, are not necessarily significant in a statistical or biological sense (Lancichinetti et al. 2008; Porter et al. 2009). Robustness across a range of methods, and correspondence with known functional-anatomical boundaries, are some indicators of high-quality modular partitions. Evidence for a particular resolution of the community structure may be compelling (a specific, preferential number of modules), but multiscale systems by definition lack a specific resolution or scale. An advantage of network analysis is that the degree of modularity can be quantified relative to a null model. In the present case, for example, modularity is significantly higher than in random networks but not significantly higher than networks with the same anatomical distance constraints as the actual network, suggesting that the relationship with anatomical distance could account for the network’s modularity.
Arguably this discussion begs the question of what counts as a “modular” network. Early work on modularity suggested standardized cut-offs for network modularity, e.g., Q > 0.3; however, because networks generated at random can have elevated modularity, current work stresses the importance of comparing the modularity of a network to that of a null model (Guimerà et al. 2004). While we interpret the present results as suggestive that anatomical constraints may account for the modularity of the genetic correlation network, a reasonable alternative conclusion is that relative to the computational null model (which preserves the spatial constraints and the constraints of contralateral symmetry of the real data), the genetic correlation networks were not found to be modular.
The comparisons between neuroimaging data and gene expression analysis provides a potential link to biological mechanisms. For example, recent studies have shown an association between imaging-derived longitudinal changes in myelination and expression of genes related to synaptic activity in the same regions (Whitaker et al. 2016), as well as increased correlations in gene expression within resting-state fMRI networks for a subset of genes preferentially expressed in human upper cortical layers (Krienen et al. 2016). The fact that correlations in gene expression show analogous spatial constraints and relationships with contralateral homologs, compared to patterns of genetic correlation, also suggests that these relationships are not merely an artifact of imaging or postprocessing techniques. Similarly, the fact that modules derived from the imaging-genetics data show differential expression of genes related to nervous-system and telencephalon development is suggestive of an underlying biological mechanism. However, gene ontological analysis may be strongly influenced by publication bias and should be interpreted as hypothesis-generating rather than confirmatory analyses. Correspondence between genetic cortical thickness modules and other ways of dividing the cortex into functionally or anatomically relevant subsections, such as the recent multimodal parcellation of the HCP data (Glasser et al. 2016), suggests the possibility of similar genetic principles of organization across modalities. Although beyond the scope of this paper, this hypothesis should be tested in future work.
Several additional and related methodological issues should be highlighted. (1) We do not directly assess the question of brain parcellation, i.e., how best to divide the brain into distinct regions. Although similar methods can be used to address this question, it is a mistake to conflate community detection with parcellation, as modules can be composed of spatially discontiguous regions. In addition, the degree of spatial smoothing necessary to make intersubject comparisons in morphology at the millimeter scale can bias inter-regional comparisons. (2) While the majority of published work on genetic correlations has used twin designs, analyses of extended pedigrees are superior in many ways including increased power and ability to distinguish genetic and environmental effects (Blangero 2004), as shared environment is necessarily reduced in extended pedigrees relative to twin pairs. The fact that the HCP data (unlike GOBS) is largely made up of twin pairs and siblings raises particular questions about the importance of distinguishing shared environment from genetic effects in these data. Because the GOBS data comes from extended pedigrees (Olvera et al. 2011), they do not share the same level of environmental overlap as sibling or twin samples, which makes replication in both datasets particularly compelling. Empirically, the fact that the genetic correlations are not notably higher in the HCP data than in the GOBS data (Figs. 1–2 and Fig. 5) also argues against the possibility of inflated estimates of genetic correlation due to these additional shared environmental factors. This is consistent with prior imaging-genetics work reporting similar genetic effects in “AE” structural equation models (which model additive genetic and individual environmental effects) compared to “ACE” models (which also model shared environmental effects), as estimates of common environmental variance tended to be at or near zero (Chen et al. 2012, 2013).
Another limitation (3) relates to the precision of the genetic correlations. The precision of an estimate of genetic correlation depends on the heritabilities of both traits, such that it is estimated poorly when the traits have low heritabilities. Although mean regional cortical thicknesses of all regions were significantly heritable, there was some variability in their heritability values, and consequently, some variability in the precisions of their estimated genetic correlations. (This concerns also applies to prior published work on genetic correlations.) Finally, (4) there is no single correct way to account for both negative and positive connections in brain networks. Depending on the context, it may be appropriate to use network measures that can be estimated with both positive and negative edges (Traag and Bruggeman 2009), to include only positive edges, or to take the absolute value of edges (for example to focus on the magnitude of the shared genetic effect rather than the direction of the effect). In the present case, the results were robust to this methodological choice as the negative correlations were all lower in magnitude than the positive correlations which were included in the networks used to derive genetic modules.
In conclusion, it is worth stressing that the analysis of the genetic architecture of cortical thickness has potentially large clinical implications. Determined by the thickness of specific horizontal layers, which differ between regions in terms neuronal populations as well as neuropil (Rakic 1988), cortical thickness decreases in normal adolescence, in neurodegenerative disease (where loss of thickness is thought to represent neuronal death) and in schizophrenia (where loss of thickness may correspond to loss of neuropil in accordance with the overpruning hypothesis). Decreased thickness in schizophrenia, in particular, has been suggested to target specific “developmental modules,” which develop in synchrony during normative adolescence (Alexander-Bloch et al. 2014) and are likely composed of regions influenced by overlapping sets of genes. In addition, schizophrenia is associated with alterations in the normative spatial constraints on anatomical (Bassett et al. 2008) and function connectivity (Alexander-Bloch et al. 2013), as well as disruption in normative constraints on the contralateral symmetry of morphology between hemispheres (Crow et al. 1989; Oertel-Knöchel and Linden 2011). Neuropsychiatric risk is thus likely to be expressed in over- or under-realization of normative organizational principles.
Supplementary Material
Notes
Conflict of interest: None declared.
References
- Alexa A, Rahnenfuhrer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 22:1600–1607. [DOI] [PubMed] [Google Scholar]
- Alexander-Bloch A, Clasen L, Stockman M, Ronan L, Lalonde F, Giedd J, Raznahan A. 2016. Subtle in-scanner motion biases automated measurement of brain anatomy from in vivo MRI. Hum Brain Mapp. 37:2385–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A, Vandekar SN, Shinohara RT, Liu S, Satterthwaite TD, Glahn DC, Raznahan A. 2017. The correspondence problem: which brain maps are significantly similar? bioRxiv. 203083.
- Alexander-Bloch AF, Gogtay N, Meunier D, Birn R, Clasen L, Lalonde F, Lenroot R, Giedd J, Bullmore ET. 2010. Disrupted modularity and local connectivity of brain functional networks in childhood-onset schizophrenia. Front Syst Neurosci. 4:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch AF, Reiss PT, Rapoport J, McAdams H, Giedd JN, Bullmore ET, Gogtay N. 2014. Abnormal cortical growth in schizophrenia targets normative modules of synchronized development. Biol Psychiatry. 76:438–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch AF, Vértes PE, Stidd R, Lalonde F, Clasen L, Rapoport J, Giedd J, Bullmore ET, Gogtay N. 2013. The anatomical distance of functional connections predicts brain network topology in health and schizophrenia. Cereb Cortex. 23:127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J. 1998. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 62:1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas A, Fernandez A, Gomez S. 2008. Analysis of the structure of complex networks at different resolution levels. New J Phys. 10:053039. [Google Scholar]
- Baaré WF, Hulshoff Pol HE, Boomsma DI, Posthuma D, de Geus EJ, Schnack HG, van Haren NE, van Oel CJ, Kahn RS. 2001. Quantitative genetic modeling of variation in human brain morphology. Cereb Cortex. 11:816–824. [DOI] [PubMed] [Google Scholar]
- Bartley AJ, Jones DW, Weinberger DR. 1997. Genetic variability of human brain size and cortical gyral patterns. Brain.. 120(Pt 2):257–269. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Bullmore E, Verchinski BA, Mattay VS, Weinberger DR, Meyer-Lindenberg A. 2008. Hierarchical organization of human cortical networks in health and schizophrenia. J Neurosci. 28:9239–9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Greenfield DL, Meyer-Lindenberg A, Weinberger DR, Moore SW, Bullmore ET. 2010. Efficient physical embedding of topologically complex information processing networks in brains and computer circuits. PLoS Comput Biol. 6:e1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. 2014. Fitting Linear Mixed-Effects Models using lme4.
- Benjamini Y, Krieger AM, Yekutieli D. 2006. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 93:491–507. [Google Scholar]
- Betzel RF, Bassett DS. 2016. Multi-scale brain networks. NeuroImage. [DOI] [PMC free article] [PubMed]
- Betzel RF, Medaglia JD, Papadopoulos L, Baum GL, Gur R, Gur R, Roalf D, Satterthwaite TD, Bassett DS. 2017. The modular organization of human anatomical brain networks: accounting for the cost of wiring. Netw Neurosci. 1:42–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bis JC, DeCarli C, Smith AV, van der Lijn F, Crivello F, Fornage M, Debette S, Shulman JM, Schmidt H, Srikanth V, et al. , Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium . 2012. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet. 44:545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangero J. 2004. Localization and identification of human quantitative trait loci: king harvest has surely come. Curr Opin Genet Dev. 14:233–240. [DOI] [PubMed] [Google Scholar]
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. 2008. Fast unfolding of communities in large networks. J Stat Mech. 2008:P10008. [Google Scholar]
- Blumenthal JD, Zijdenbos A, Molloy E, Giedd JN. 2002. Motion artifact in magnetic resonance imaging: implications for automated analysis. NeuroImage. 16:89–92. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 10:186–198. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. 2012. The economy of brain network organization. Nat Rev Neurosci. 13:336–349. [DOI] [PubMed] [Google Scholar]
- Chen C-H, Fiecas M, Gutierrez ED, Panizzon MS, Eyler LT, Vuoksimaa E, Thompson WK, Fennema-Notestine C, Hagler DJ, Jernigan TL, et al. . 2013. Genetic topography of brain morphology. Proc Natl Acad Sci U S A. 110:17089–17094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-H, Gutierrez ED, Thompson W, Panizzon MS, Jernigan TL, Eyler LT, Fennema-Notestine C, Jak AJ, Neale MC, Franz CE, et al. . 2012. Hierarchical genetic organization of human cortical surface area. Science. 335:1634–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-H, Panizzon MS, Eyler LT, Jernigan TL, Thompson W, Fennema-Notestine C, Jak AJ, Neale MC, Franz CE, Hamza S, et al. . 2011. a. Genetic influences on cortical regionalization in the human brain. Neuron. 72:537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, He Y, Rosa-Neto P, Gong G, Evans AC. 2011. b. Age-related alterations in the modular organization of structural cortical network by using cortical thickness from MRI. NeuroImage. 56:235–245. [DOI] [PubMed] [Google Scholar]
- Clauset A, Newman M, Moore C. 2004. Finding community structure in very large networks. Phys Rev E Stat Nonlin Soft Matter Phys. 70:066111. [DOI] [PubMed] [Google Scholar]
- Crow TJ, Ball J, Bloom SR, Brown R, Bruton CJ, Colter N, Frith CD, Johnstone EC, Owens DG, Roberts GW. 1989. Schizophrenia as an anomaly of development of cerebral asymmetry. A postmortem study and a proposal concerning the genetic basis of the disease. Arch Gen Psychiatry. 46:1145–1150. [DOI] [PubMed] [Google Scholar]
- Csardi G, Nepusz T. 2006. The igraph software package for complex network research. InterJournal Complex Systems:1695. http://igraph.org. [Google Scholar]
- Dale AM, Fischl B, Sereno MI. 1999. Cortical surface-based analysis. I. Segmentation and surface reconstruction. NeuroImage. 9:179–194. [DOI] [PubMed] [Google Scholar]
- De Meo P, Ferrara E, Fiumara G, Provetti A. 2011. Generalized Louvain method for community detection in large networks. In: Presented at the 2011 11th International Conference on Intelligent Systems Design and Applications (ISDA). IEEE. p. 88–93.
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, et al. . 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 31:968–980. [DOI] [PubMed] [Google Scholar]
- Dimitriadou E, Weingessel A, Hornik K. 2002. A combination scheme for fuzzy clustering. Int J Patt Recogn Artif Intell. 16:901–912. [Google Scholar]
- Fischl B, Liu A, Dale AM. 2001. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans Med Imaging. 20:70–80. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, van der Kouwe AJW, Makris N, Ségonne F, Quinn BT, Dale AM. 2004. a. Sequence-independent segmentation of magnetic resonance images. NeuroImage. 23(Suppl 1):S69–S84. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. 1999. Cortical surface-based analysis. II: inflation, flattening, and a surface-based coordinate system. NeuroImage. 9:195–207. [DOI] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, et al. . 2004. b. Automatically parcellating the human cerebral cortex. Cereb Cortex. 14:11–22. [DOI] [PubMed] [Google Scholar]
- Fortunato S, Barthélemy M. 2007. Resolution limit in community detection. Proc Natl Acad Sci U S A. 104:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanis JN, Walsh CA. 2004. Genetics of disorders of cortical development. Neuroimaging Clin N Am. 14(2):219–229. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Miller BL, DeCarli C, Carmelli D. 2002. Heritability of lobar brain volumes in twins supports genetic models of cerebral laterality and handedness. Proc Natl Acad Sci U S A. 99:3176–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Knowles EEM, McKay DR, Sprooten E, Raventós H, Blangero J, Gottesman II, Almasy L. 2014. Arguments for the sake of endophenotypes: examining common misconceptions about the use of endophenotypes in psychiatric genetics. Am J Med Genet B Neuropsychiatr Genet. 165B:122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Paus T, Thompson PM. 2007. Imaging genomics: mapping the influence of genetics on brain structure and function. Hum Brain Mapp. 28:461–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Sprooten E, Knowles E, McKay DR, Mathias SR, Almasy L, Blangero J. 2015. Imaging genetics. In: Toga AW, editor. Brain mapping. Boston, MA: Elsevier. p. 929–932. [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, et al. . 2016. A multi-modal parcellation of human cerebral cortex. Nat Neurosci. 536:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, et al. , WU-Minn HCP Consortium . 2013. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage. 80:105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, He Y, Chen ZJ, Evans AC. 2012. Convergence and divergence of thickness correlations with diffusion connections across the human cerebral cortex. NeuroImage. 59:1239–1248. [DOI] [PubMed] [Google Scholar]
- Guerrini R, Marini C. 2006. Genetic malformations of cortical development. Exp Brain Res. 173:322–333. [DOI] [PubMed] [Google Scholar]
- Guimerà R, Sales-Pardo M, Amaral L. 2004. Modularity from fluctuations in random graphs and complex networks. Phys Rev E Stat Nonlin Soft Matter Phys. 70:025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. . 2012. An anatomically comprehensive atlas of the adult human brain transcriptome. Nat Neurosci. 489:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Adams HHH, Jahanshad N, Chauhan G, Stein JL, Hofer E, Renteria ME, Bis JC, Arias-Vasquez A, Ikram MK, et al. . 2017. Novel genetic loci associated with hippocampal volume. Nat Commun. 8:13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivières S, Jahanshad N, Toro R, Wittfeld K, Abramovic L, Andersson M, et al. . 2015. Common genetic variants influence human subcortical brain structures. Nat Neurosci. 520:224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornik K. 2005. A CLUE for CLUster ensembles. J Stat Softw. 14:65–72. [Google Scholar]
- Kaufman L, Rousseeuw P. 1987. Clustering by means of medoids. In: Dodge Y, editor. Statistical data analysis based on the L1–norm and related methods. Amsterdam: North Holland. p. 405–416. [Google Scholar]
- Krienen FM, Yeo BTT, Ge T, Buckner RL, Sherwood CC. 2016. Transcriptional profiles of supragranular-enriched genes associate with corticocortical network architecture in the human brain. Proc Natl Acad Sci U S A. 113:E469–E478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancichinetti A, Fortunato S, Radicchi F. 2008. Benchmark graphs for testing community detection algorithms. Phys Rev E Stat Nonlin Soft Matter Phys. 78:046110. [DOI] [PubMed] [Google Scholar]
- Levin M. 2005. Left-right asymmetry in embryonic development: a comprehensive review. Mech Dev. 122:3–25. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B. 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates Incorporated.
- Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. 2012. cluster: Cluster Analysis Basics and Extensions [WWW Document]. CRANR-projectorg. URL http://CRAN.R-project.org/package=cluster
- McKay DR, Knowles EEM, Winkler AAM, Sprooten E, Kochunov P, Olvera RL, Curran JE, Kent JW, Carless MA, Göring HHH, et al. . 2014. Influence of age, sex and genetic factors on the human brain. Brain Imaging Behav. 8:143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DR, Winkler AM, Kochunov P, Knowles EEM, Sprooten E, Fox PT, Blangero J, Glahn DC. 2015. Genetic influence on the human brain. In: Genome mapping and genomics in human and non-human primates. Berlin, Heidelberg: Springer Berlin Heidelberg. p. 247–258. [Google Scholar]
- Meunier D, Lambiotte R, Fornito A, Ersche KD, Bullmore ET. 2009. Hierarchical modularity in human brain functional networks. Front Neuroinform. 3:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary DDM, Chou S-J, Sahara S. 2007. Area patterning of the mammalian cortex. Neuron. 56:252–269. [DOI] [PubMed] [Google Scholar]
- Oertel-Knöchel V, Linden DEJ. 2011. Cerebral asymmetry in schizophrenia. Neuroscientist. 17:456–467. [DOI] [PubMed] [Google Scholar]
- Olvera RL, Bearden CE, Velligan DI, Almasy L, Carless MA, Curran JE, Williamson DE, Duggirala R, Blangero J, Glahn DC. 2011. Common genetic influences on depression, alcohol, and substance use disorders in Mexican-American families. Am J Med Genet B Neuropsychiatr Genet. 156B:561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizzon MS, Fennema-Notestine C, Eyler LT, Jernigan TL, Prom-Wormley E, Neale M, Jacobson K, Lyons MJ, Grant MD, Franz CE, et al. . 2009. Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex. 19:2728–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington BF, Filipek PA, Lefly D, Chhabildas N, Kennedy DN, Simon JH, Filley CM, Galaburda A, DeFries JC. 2000. A twin MRI study of size variations in human brain. J Cogn Neurosci. 12:223–232. [DOI] [PubMed] [Google Scholar]
- Peper JS, Brouwer RM, Boomsma DI, Kahn RS, Hulshoff Pol HE. 2007. Genetic influences on human brain structure: a review of brain imaging studies in twins. Hum Brain Mapp. 28:464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter MA, Onnela J-P, Mucha PJ. 2009. Communities in networks. Not Am Math Soc. 56:1082–1097. [Google Scholar]
- Posthuma D, de Geus EJ, Neale MC, Hulshoff Pol HE, Baaré WFC, Kahn RS, Boomsma DI. 2000. Multivariate genetic analysis of brain structure in an extended twin design. Behav Genet. 30:311–319. [DOI] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage. 59:2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core Team R. 2016. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rakic P. 1988. Specification of cerebral cortical areas. Science. 241:170–176. [DOI] [PubMed] [Google Scholar]
- Reichardt J, Bornholdt S. 2004. Detecting fuzzy community structures in complex networks with a Potts model. Phys Rev Lett. 93:218701. [DOI] [PubMed] [Google Scholar]
- Reuter M, Tisdall MD, Qureshi A, Buckner RL, van der Kouwe AJW, Fischl B. 2015. Head motion during MRI acquisition reduces gray matter volume and thickness estimates. NeuroImage. 107:107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JE, Eyler LT, Giedd JN, Kremen WS, Kendler KS, Neale MC. 2007. a. Review of twin and family studies on neuroanatomic phenotypes and typical neurodevelopment. Twin Res Hum Genet. 10:683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JE, Wallace GL, Lenroot RK, Ordaz SE, Greenstein D, Clasen L, Kendler KS, Neale MC, Giedd JN. 2010. A twin study of intracerebral volumetric relationships. Behav Genet. 40:114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JE, Wallace GL, Rosenthal MA, Molloy EA, Ordaz S, Lenroot R, Clasen LS, Blumenthal JD, Kendler KS, Neale MC, et al. . 2007. b. A multivariate analysis of neuroanatomic relationships in a genetically informative pediatric sample. NeuroImage. 35:70–82. [DOI] [PubMed] [Google Scholar]
- Simon HA. 1962. The architecture of complexity. Proc Am Philos Soc. 106:467–487. [Google Scholar]
- Sporns O. 2010. Networks of the brain. Cambridge, MA: The MIT Press. [Google Scholar]
- Stein JL, Hua X, Lee S, Ho AJ, Leow AD, Toga AW, Saykin AJ, Shen L, Foroud T, Pankratz N, et al. , Alzheimer’s Disease Neuroimaging Initiative . 2010. Voxelwise genome-wide association study (vGWAS). NeuroImage. 53:1160–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Toro R, Appel K, Bartecek R, Bergmann Ø, et al. , Enhancing Neuro Imaging Genetics through Meta-Analysis Consortium . 2012. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 44:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehl A, Ghosh J. 2002. Cluster ensembles — a knowledge reuse framework for combining multiple partitions. J Mach Learn Res. 3:583–617. [Google Scholar]
- Thompson PM, Ge T, Glahn DC, Jahanshad N, Nichols TE. 2013. Genetics of the connectome. NeuroImage. 80:475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag VA, Bruggeman J. 2009. Community detection in networks with positive and negative links. Phys Rev E Stat Nonlin Soft Matter Phys. 80:036115. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K, WU-Minn HCP Consortium . 2013. The WU-Minn Human Connectome Project: an overview. NeuroImage. 80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. 2002. Modern applied statistics with S. 4th ed.. New York: Springer. [Google Scholar]
- Vértes PE, Alexander-Bloch AF, Gogtay N, Giedd JN, Rapoport JL, Bullmore ET. 2012. Simple models of human brain functional networks. Proc Natl Acad Sci U S A. 109:5868–5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viger F, Latapy M. 2005. Random generation of large connected simple graphs with prescribed degree distribution. In: Wang L, editor. Computing and combinatorics. Lecture notes in computer science. Berlin, Germany: Springer. p. 440–449. [Google Scholar]
- Whitaker KJ, Vértes PE, Romero-Garcia R, Váša F, Moutoussis M, Prabhu G, Weiskopf N, Callaghan MF, Wagstyl K, Rittman T, et al. , NSPN Consortium, Bullmore ET . 2016. Adolescence is associated with genomically patterned consolidation of the hubs of the human brain connectome. Proc Natl Acad Sci U S A. 113:9105–9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JT, Van Eerdewegh P, Almasy L, Blangero J. 1999. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. I. Likelihood formulation and simulation results. Am J Hum Genet. 65:1134–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Kochunov P, Blangero J, Almasy L, Zilles K, Fox PT, Duggirala R, Glahn DC. 2010. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. NeuroImage. 53:1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.