Watch a video presentation of this article
Watch the interview with the author
Abbreviations
- AIH
autoimmune hepatitis
- DI‐AIH
drug‐induced AIH
- FDR
first‐degree relative
- GWAS
genome‐wide association study
- HLA
human leukocyte antigen
- PBC
primary biliary cholangitis
- PSC
primary sclerosing cholangitis
- SDR
second‐degree relative
Autoimmune hepatitis (AIH) is a prototypical autoimmune disease, characterized by robust genetic associations with human leukocyte antigen (HLA) alleles, female predominance, and presence of serum autoantibodies; yet all AIH cases are not the same. This heterogeneous disease affects all population demographics and can lead to cirrhosis, liver failure, and liver transplantation depending on the diagnosis, treatment, and response to therapy. Similar to other autoimmune diseases, AIH is a result of immune intolerance and failure of immunological homeostasis. The etiology remains largely unresolved but is likely the result of various environmental exposures in the background of a permissive genetic architecture (Fig. 1).
Figure 1.
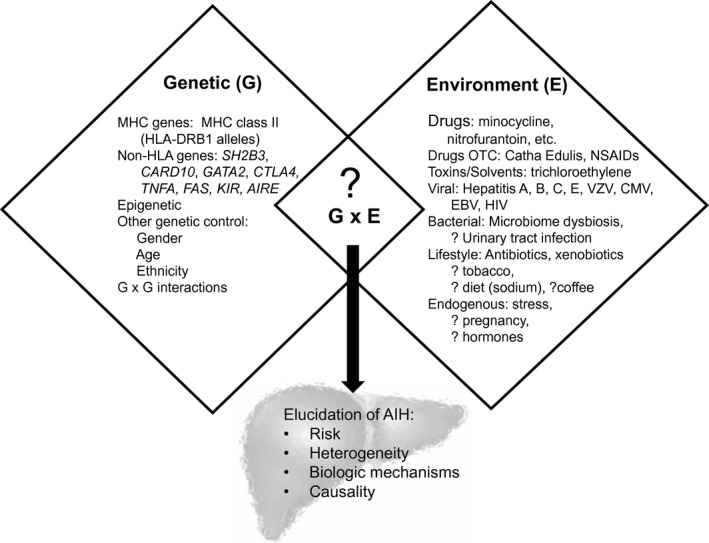
Genes, environment and their interactions alter the risk of developing AIH.
Genetic Susceptibility of AIH
Reports have documented AIH clustering in families, but no formal heritability estimate exists. A large family study of patients with AIH, including first‐degree relatives (FDRs), second‐degree relatives (SDRs), and twin pairs supports genetic contributions to disease.1 Family risk remains low, as 10‐year cumulative risk for AIH was 0.1% in FDRs and 0% in SDRs. Furthermore, monozygotic twin concordance was observed in only 1 out of 22 twin pairs, suggesting AIH is a polygenic disease with possibly low penetrance.
HLA associations with AIH were first observed with serological typing in 1972.2 The only AIH genome‐wide association study (GWAS) to date has revealed a strong association rooted in the major histocompatibility complex in the HLA region (rs2187668) with HLA‐DRB1*0301 as the primary susceptibility genotype and HLA‐DRB1*0401 as a secondary susceptibility genotype.3 These alleles encode the antigen‐binding cleft of HLA class II molecules and may alter its configuration, resulting in a more diverse repertoire of self or pathogen‐derived peptide antigens selected by different HLA allotypes. However, not all HLA‐associated AIH risk can be explained by DRB1 alone because other alleles have been associated with both disease risk and protection in different ethnic populations (Fig. 2). In addition, the top risk alleles from the AIH GWAS encode identical sequences at positions DRβ67‐72, with lysine at position (DRβ71), and may also be associated with different clinical outcomes.4 There were no genetic associations outside the HLA locus meeting strict genome‐wide significance or replicated signals observed in prior candidate gene studies in the AIH GWAS study. However, allele rs3184504 in SH2B3 was consistent in both the discovery and replication cohort. Expression of this risk allele with genes linked to the adaptive immune response and observation in other immune‐mediated diseases support a pleotropic role in autoimmunity.
Figure 2.
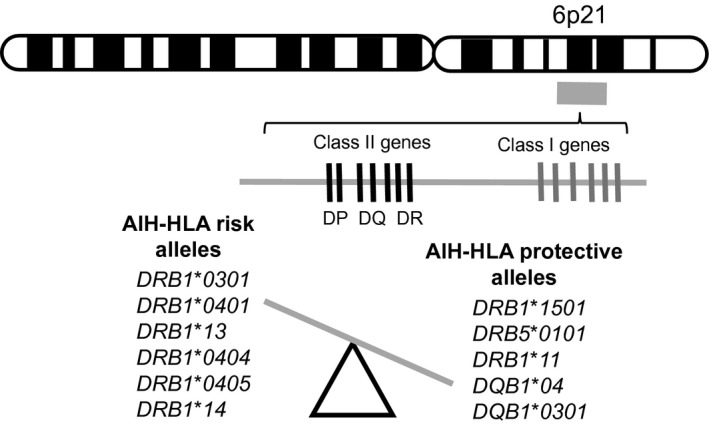
HLA allelic associations with AIH can be associated with disease risk and protection.
Environment Exposures in AIH
In the other autoimmune liver diseases (primary biliary cholangitis [PBC] and primary sclerosing cholangitis [PSC]), approximately 50% of disease susceptibility may be related to environmental contributions. Viral infections and pharmaceutical exposures provide the strongest support of environmental contribution to the development of AIH (Fig. 1). Case series and small retrospective studies have linked a variety of viruses including Epstein‐Barr, varicella zoster, and hepatitis A, B, C, and E to AIH. The finding of autoantibodies in these hepatic insults is well reported, and mechanistic triggering of AIH may include molecular mimicry, neoantigen formation, unmasking of self‐antigens, and bystander T cell activation. Drug exposures, commonly but not limited to nitrofurantoin and minocycline, have also been described as causative triggers for AIH (drug‐induced AIH [DI‐AIH]) and commonly display clinical autoimmune features.5 However, patients with DI‐AIH are not enriched with the common risk alleles observed in idiopathic AIH (HLA‐DRB1*0301 or HLA‐DRB1*0401), thus implicating other underlying risks.5 The onset of immune self‐reactivity triggered by exogenous compounds could be similar to the change in HLA‐peptide repertoire as seen in abacavir and carbamazepime.6
Beyond viral and compound‐specific AIH triggers, investigation of other lifestyle risks remain incomplete because there are no widespread environmental factors consistently linked to AIH. This contrasts with PBC and PSC, where disease risk has been linked to factors such as tobacco smoke and urinary tract infections.7 Antibiotic exposure in the 12 months prior to AIH diagnosis is an independent risk factor for AIH, whereas exposure to wood heating and alcohol consumption are protective.8 Environmental exposures (diet, lifestyle, medications) also help shape the intestinal microbiome, which has been implicated in the pathogenesis of a variety of inflammatory liver diseases. Dysbiosis has been observed in AIH and is linked to alteration in intestinal barrier permeability.9
Gene and Environment Interactions Comprise Substantial Disease Risk
In studies of complex diseases, genetic variation alone generally accounts for less than one‐third of disease susceptibility. Interactions between gene (G) variants and environmental (E) exposures likely comprise a sizable component of the overall remaining disease risk. These interactions occur when the presence or absence of an environmental factor alters the phenotypic expression of a disease genotype (Fig. 3). The increased risk for celiac disease according to the presence of both risk alleles in HLA‐DQB1 and dietary gluten is a classic example of enhanced disease risk resulting from a G × E interaction.10 Such interactions have not been studied in AIH; yet pursuit of this integrative approach in AIH will require high‐quality exposure data with longitudinal measures, detailed phenotyping, and inclusion of diverse ethnic and geographic populations. This approach will help clarify AIH heterogeneity, demarcate possible biological mechanisms, and provide evidence of causality.
Figure 3.
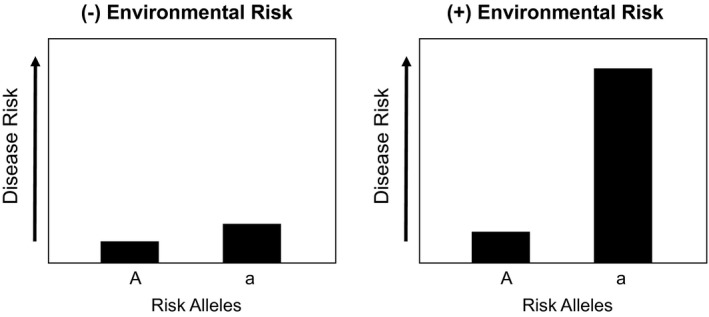
Risk alleles may only influence disease risk when environmental interactions are present.
Conclusion
The mosaic of AIH risk is complex and poorly understood. Clarification of genetic and environmental risks, as well as the disease contributions as a result of their interactions, will eventually transform our understanding of AIH pathogenesis, treatment, and outcomes.
Potential conflict of interest: Nothing to report.
References
- 1. Gronbaek L, Vilstrup H, Pedersen L, et al. Family occurrence of autoimmune hepatitis: a Danish nationwide registry‐based cohort study. J Hepatol 2018;69:873‐877. [DOI] [PubMed] [Google Scholar]
- 2. Mackay IR, Morris PJ. Association of autoimmune active chronic hepatitis with HL‐A1,8. Lancet 1972;2:793‐795. [DOI] [PubMed] [Google Scholar]
- 3. de Boer YS, van Gerven NM, Zwiers A, et al. Genome‐wide association study identifies variants associated with autoimmune hepatitis type 1. Gastroenterology 2014;147:443‐452.e445. [DOI] [PubMed] [Google Scholar]
- 4. van Gerven NM, de Boer YS, Zwiers A, et al. HLA‐DRB1*03:01 and HLA‐DRB1*04:01 modify the presentation and outcome in autoimmune hepatitis type‐1. Genes Immun 2015;16:247‐252. [DOI] [PubMed] [Google Scholar]
- 5. de Boer YS, Kosinski AS, Urban TJ, et al. Features of autoimmune hepatitis in patients with drug‐induced liver injury. Clin Gastroenterol Hepatol 2017;15:103‐112.e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Illing PT, Vivian JP, Dudek NL, et al. Immune self‐reactivity triggered by drug‐modified HLA‐peptide repertoire. Nature 2012;486:554‐558. [DOI] [PubMed] [Google Scholar]
- 7. Juran BD, Lazaridis KN. Environmental factors in primary biliary cirrhosis. Semin Liver Dis 2014;34:265‐272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jing H, Ngu RBG, Frampton CM, et al. Autoimmune hepatitis: the role of environmental risk factors: a population‐based study. Hepatol Int 2013;7:869‐875. [DOI] [PubMed] [Google Scholar]
- 9. Lin R, Zhou L, Zhang J, Wang B. Abnormal intestinal permeability and microbiota in patients with autoimmune hepatitis. Int J Clin Exp Pathol 2015;8:5153‐5160. [PMC free article] [PubMed] [Google Scholar]
- 10. Lundin KE, Scott H, Fausa O, et al. T cells from the small intestinal mucosa of a DR4, DQ7/DR4, DQ8 celiac disease patient preferentially recognize gliadin when presented by DQ8. Hum Immunol 1994;41:285‐291. [DOI] [PubMed] [Google Scholar]


