 The complex DZ1 displayed enhanced three-photon absorption activity and could avoid light-quenching and light-bleaching effectively due to its aggregation-induced emission feature.
The complex DZ1 displayed enhanced three-photon absorption activity and could avoid light-quenching and light-bleaching effectively due to its aggregation-induced emission feature.
Abstract
Multiphoton bioimaging benefits from good penetration of tissue, low phototoxicity and high resolution. Hence, development of efficient multiphoton imaging agents is highly desirable but remains challenging. Herein, a novel terpyridine-based Zn(ii) complex bearing a thiophene bridge was designed rationally and fabricated. Thanks to its aggregation-induced emission (AIE), DZ1 emitted bright yellow-green fluorescence (λem = 575 nm) under physiological conditions. The three-photon spectral changes of DZ1 when binding with RNA unambiguously reflected its RNA-specific targeting behaviour, resulting in twofold enhancement in three-photon action cross-sections located at the second near-infrared window (1700 nm).
Introduction
Multiphoton imaging is an effective strategy because its excitation wavelength can be selected in near-infrared (NIR) optical windows and because it has high spatial resolution. With the availability of ultra-fast pulsed lasers in recent years, significant progress in three-photon absorption (3PA)-based applications has been witnessed, including three-photon pumped lasing and 3PA-based optical limiting and stabilization.1,2 Three-photon fluorescent (3PF) materials have attracted extensive attention due to their potential applications in three-dimensional optical-data storage, photodynamic therapy and biological imaging.3,4 Great efforts have been made by researchers to achieve high-3PA materials, including a series of chromophores with D–π–A–π–D, A–π–D–π–A, D–π–D and D–π–A structures.5–8 Among the strategies implemented to obtain improved 3PA responses, organic chromophores with large and extended conjugated systems for enhanced electron transfer have received increasing attention.9–11 Nevertheless, in contrast to the complicated synthesis stated above, use of metal complexes for 3PA activity, which promote electron transfer to enhance nonlinear optical (NLO) activity after coordination with metal ions, remains very rare.
Considering the applications in bioimaging, searching for NLO-active materials which utilise NIR photons as the excitation source to achieve deeper tissue penetration and weaker specimen photodamage is a highly desired target.12 However, most of them suffer from aggregation-caused quenching (ACQ) due to self-aggregation in physiological conditions. To meet these challenges, in contrast to ACQ, Tang and coworkers first proposed the concept of “aggregation-induced emission” (AIE).13 Molecules with AIE properties are more suitable for exploring 3P features because AIE-active chromophores can enhance fluorescence and 3PA action cross-sections at high concentrations.14,15 However, most of the molecules reported have been organic molecules with large conjugated systems and complicated synthetic processes. For practical applications, the search for multiphoton-active metal complexes possessing considerable fluorescence for bioimaging, which involve simple preparation, is highly desired.
A novel D–A type thiophene terpyridine zinc complex (DZ1) was designed and synthesised. Zinc is an essential trace element in organisms. Zinc can be coordinated with terpyridine groups to obtain excellent luminescent properties and high biological activity.16 Thiophene is a five-membered heterocyclic compound with aromaticity, rich electron density and electronic-transmission ability. It is one of the most promising elements for designing new 3PA functional materials.17–19 Herein, we focused mainly on the three photon-absorption properties and AIE activities of DZ1 and its distinct response to RNA in vitro. A large 3PA cross-section (5.28 × 10–82 cm6 s2 per photon2) has been found under 1700 nm femtosecond (fs) laser excitation via three-photon excitation fluorescence (3PEF).20,21 DZ1 was also found to have good chemical stability, excellent biocompatibility and low toxicity, which was utilized as a multi-photon fluorescence probe for targeting mitochondria in HepG2 cells. In the future, DZ1, with its bright green-yellow AIE fluorescence and 3PA, could aid deep-tissue functional in vivo imaging. This is the first report on a metal–organic complex which behaves as a turn-on 3PF RNA-specific probe (Scheme 1).
Scheme 1. Three-photon property of complex DZ1 in the aggregated state or upon interaction with RNA (schematic).
Results and discussion
Characterization and structural features
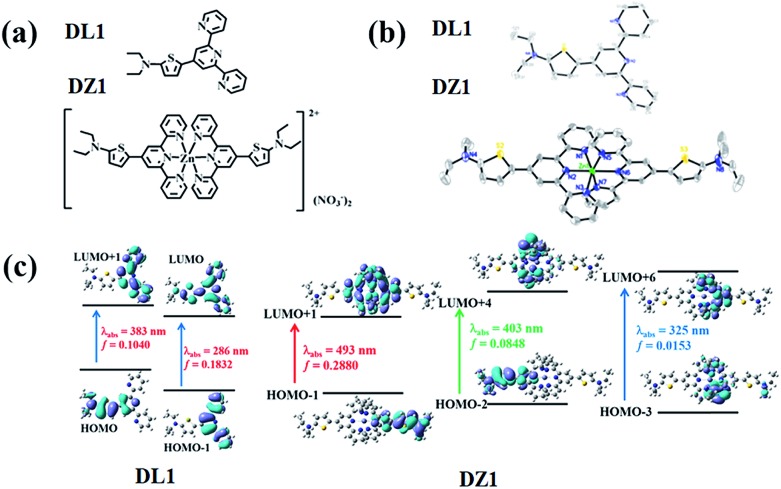
The ligand DL1 and its metal complex DZ1 were prepared according to the synthetic routes shown in Fig. S1.† The corresponding structural features were shown in detail by single-crystal X-ray diffraction (Fig. 1a and b). DL1 (CCDC: ; 1908472) and DZ1 (CCDC: ; 1908473) crystallised in the triclinic crystal system with a P1[combining macron] space group. The terpyridine Zn complex had a classic octahedral configuration (Table S1†). The dihedral angles between the central pyridine group and thiophene ring in DL1 and DZ1 were 122.1° and 120.5° (DZ1 < DL1), respectively. The C–C bonds between the thiophene ring and electron acceptor (pyridine ring) were 1.453 Å and 1.419 Å (DZ1 < DL1), respectively (Table S2†). The coordination behaviour of the Zn(ii) centre, and the enhanced electron-withdrawing effect improve electron separation and fluidity, which favours optimised NLO activity.22,23 In accordance with theoretical calculations (Fig. 1c and S6†), a new absorption band appeared in DZ1 compared with the ligand, which belonged to the intramolecular charge transfer (ICT) process, indicating that DZ1 was more planar than DL1.
Fig. 1. (a) Structures of DL1 and DZ1. (b) Crystal structures of DL1 and DZ1 (H atoms and nitrate ions have been omitted for clarity). (c) Molecular orbital energy diagrams of DL1 and DZ1.
AIE and 3PA properties
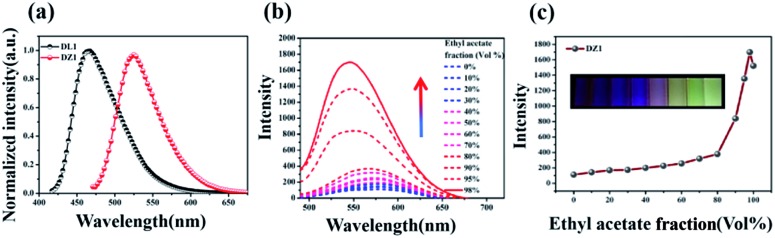
Encouraged by the structural features stated above, we investigated linear optical properties. The enhanced electron-withdrawing ability of complex DZ1 gave rise to a red shift on linear fluorescence compared with the organic ligand DL1 (Fig. 2a).24 In addition, in contrast to the weak fluorescence in good solvent conditions (acetonitrile), the complex DZ1 exhibited strong fluorescence in ethyl acetate (poor solvent), which attracted our attention. We envisioned that DZ1 had AIE in ethyl acetate/acetonitrile mixtures. To test this hypothesis, the fluorescence behaviour of DZ1 in ethyl acetate/acetonitrile mixtures was investigated (Fig. 2b). Quantitative data revealed that, as the fraction of ethyl acetate (fe) reached 98%, the fluorescence intensity of DZ1 was enhanced by 30-fold (Fig. 2c). The fluorescent images of the solvent mixtures at fe = 0% to 98% in Fig. 2c (inset) presented the clear AIE effect of DZ1.25,26 As shown in Fig. S4,† massive C–H···π interactions and hydrogen bonds within DZ1 were found, which could hinder the rotation of single bonds and activate restriction of intramolecular rotation (RIR), thereby resulting in AIE.27
Fig. 2. (a) Normalized fluorescence spectra for DL1, DZ1 in acetonitrile (c = 10 μM); (b) fluorescence spectra of DZ1 (10 μM) in ethyl acetate/acetonitrile mixtures with different ethyl acetate fractions (λex = 480 nm; λem = 568 nm); (c) plot of emission intensity vs. ethyl acetate fraction, inset: photographs of the luminescence of DZ1 in ethyl acetate fractions (0–98%) under UV illumination at 365 nm.
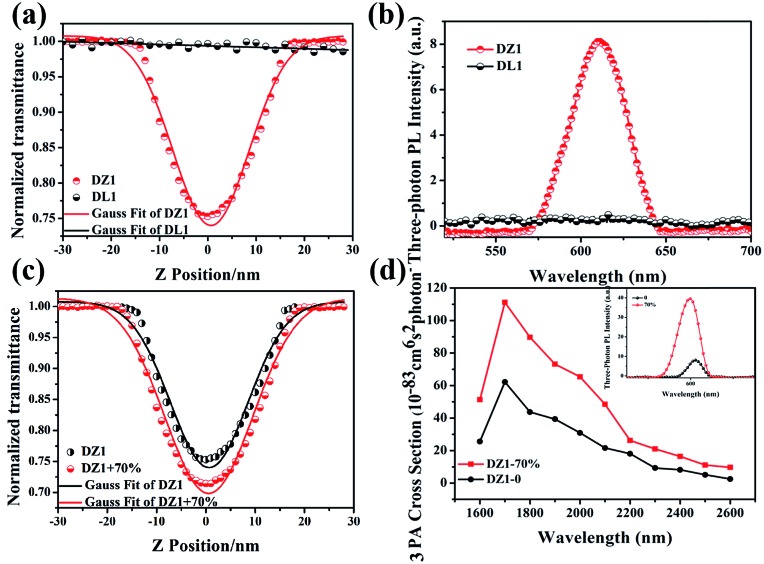
Besides, introduction of a thiophene group afforded the D–A-configurated DZ1 with enhanced ICT behaviour, which motivated us to further evaluate its nonlinear optical property. As expected, totally different from the organic ligand DL1 without significant NLO characteristics, DZ1 exhibited excellent NLO behaviour including two- and 3PA activities (Fig. S6c,† 3a and b). Notably, the two- and 3PA action cross-sections of DZ1 in the aggregated state were enhanced significantly (Fig. S6d,† 3c and d). The 3PA cross-section was calculated to be 1.11 × 10–81 cm6 s2 per photon2 by the 3PEF method (ethyl acetate/acetonitrile (70%)), which was enhanced ∼1.78-times than the molecular state (6.32 × 10–82 cm6 s2 per photon2) (Fig. 3d). The enhanced 3PA properties of DZ1 in aggregated states were confirmed further via open-aperture Z-scan measurements (Fig. 3c, Table S6†).28 The results obtained were consistent with those of 3PEF experiments: DZ1 unveiled an enhanced 3PA cross-section in the aggregated state. Meanwhile, a much larger 3PA cross-section could be obtained via the Z-scan method because it can effectively shield the energy consumed by the thermal effect and molecular rotation during photo-absorption.29 All the results stated above showed that DZ1 could improve NLO activity (2PA and 3PA) based on its AIE, which was an incentive to explore further novel materials with a NLO-response.
Fig. 3. (a) Three-photon absorption spectra of DL1 and DZ1 (c = 1 mM) in DMSO obtained under an open aperture Z-scan. (b) Three-photon fluorescence spectra of DL1 and DZ1 (c = 1 mM) in DMSO; (c) three-photon absorption spectra of DZ1 (c = 0.1 mM) in acetonitrile and ethyl acetate/acetonitrile (70%), obtained under an open-aperture Z-scan. (d) Changes in three-photon absorption action cross-sections of DZ1 (c = 0.1 mM) in acetonitrile and ethyl acetate/acetonitrile (70%), inset: three-photon excited fluorescence spectra.
Cellular localization
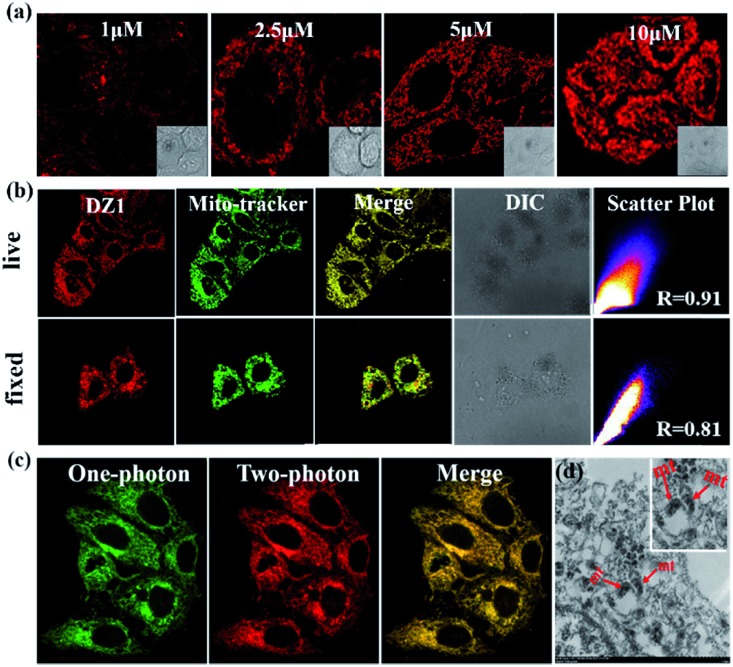
Thanks to the excellent AIE and 3PA activities of DZ1, which can suppress ACQ behaviour in physiological environments and photodamage, the biological application of DZ1 was investigated. As depicted in Fig. S10,† the low toxicity to cells enables DZ1 to be a prominent fluorescent probe. To evaluate the potential biological application of probe DZ1, imaging experiments were carried out in HepG2 cells. First, HepG2 cells hatched for 20 min in different concentrations of DZ1 solution.30 We found that DZ1 passed through the cell membrane and stained mitochondria with an optimum concentration of 10 μM (Fig. 4a). A co-localization experiment was done using the MitoTracker™ (a green fluorescent protein that targets mitochondria). The stained cells emitted red fluorescence from DZ1 and green fluorescence from MitoTracker (Fig. 4b). The merged image showed that the distribution of DZ1 in cells was consistent with that of MitoTracker, indicating the high selectivity of DZ1 towards mitochondria.31 Also, it showed that DZ1 could locate firmly in mitochondria in living and fixed cells (Fig. 4b), indicating that DZ1 is a mitochondrial membrane potential-independent mitochondrial probe. As reported,32,33 the hydrophobicity of cationic probes plays a vital part in mitochondria staining. Therefore, the hydrophobicity (as modelled by log P values) of DZ1 was measured (log P of DZ1 = 0.489), which corroborated the moderate hydrophobicity of DZ1. In addition, the log P of DZ1 was in the reported range (0–5), which further demonstrated its specific targeting effect on mitochondria.34 Hence, the hydrophobic cation of DZ1 could be used to target mitochondria.
Fig. 4. (a) Fluorescence microscopy images of HeLa cells stained with different concentrations of DZ1 (1.0–10 μM) and in aqueous phosphate buffered saline (PBS) buffer. (b) The fluorescence development of DZ1 in fixed cells and living cells, respectively. (c) Confocal one-photon and two-photon fluorescence microscopy image of HepG2 cells incubated with DZ1. (d) TEM micrograph of untreated mitochondria stained solely with DZ1.
Because of its remarkable advantages of increased penetration depth, low tissue autofluorescence, and reduced photodamage, two-photon fluorescence microscopy has drawn considerable attention in non-destructive imaging.35 Bio-imaging studies of DZ1 were carried out using confocal microscopy with a two-photon (2P) excitation wavelength at 760 nm and one-photon (1P) at 405 nm (emission = 450–600 nm), respectively. As shown in Fig. 4c, compound DZ1 could penetrate cell membranes and distribute throughout the cytosolic region from 1P- and 2P channels. All experiments showed that DZ1 could be employed as a two-photon fluorescent (2PF) probe to mark mitochondria in cells. Besides the 2P microscopic imaging results, transmission electron microscopy was used to determine the DZ1 distribution in cells.36 According to Fig. 4d, DZ1-labelled cells showed bright ridges and internal moments on mitochondria, indicating that most zinc substances were present in the mitochondria. To clarify indirectly the advantages of 3PF imaging, we undertook 1PF and 2PF tissue imaging. As shown in Fig. S11,† the 1PF (λex = 405 nm) and 2PF (λex = 780 nm) imaging performance of DZ1 in fixed mouse cardiac tissue was evaluated. Obviously, the 2PF signals with high contrast could be detected at a depth ≤54 μm for DZ1 (the detection depth of 1PF was 30 μm). These results revealed that a deeper penetration depth could be obtained by 2PF imaging. These results implied that DZ1 could be employed for 3PF imaging under excitation of NIR-II light with very deeper penetration depth.37
Studies on RNA binding
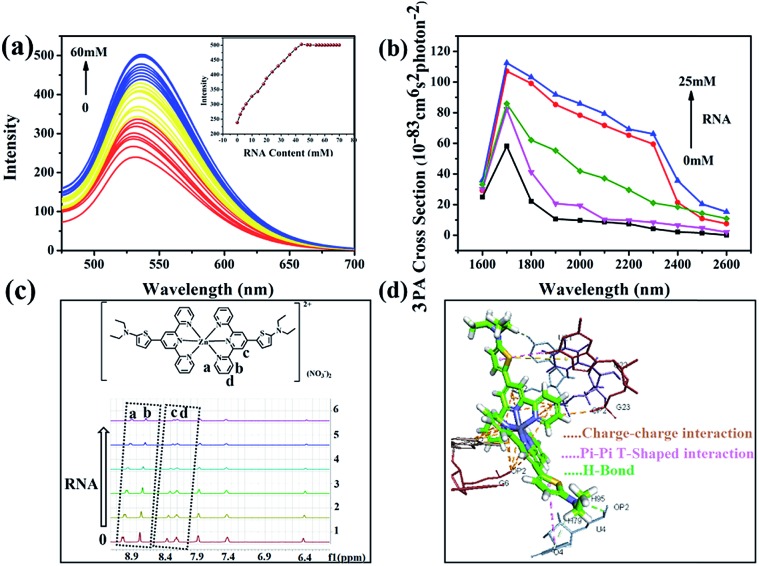
To obtain more insight into how DZ1 targets mitochondria, various amino acids and proteins were screened in vitro. Obviously, DZ1 showed a distinct response to RNA (Fig. S12a†). The interaction of DZ1 with RNA (mitochondria) was tested preliminarily by 1PF (Fig. 5a), 3PF (Fig. S12c†) and 3PA spectroscopy (Fig. S12d†).38 As shown in Fig. 5a, DZ1 (10 μM) was titrated by adding different concentrations of RNA in Tris–HCl buffer (pH = 7.4). The fluorescence intensity reached a maximum and maintained an equilibrium when the amount of RNA reached 44 mM, and the fluorescence intensity increased by 2.1-times compared with that in pure dimethyl sulfoxide (DMSO).39,40 Given that there was weak fluorescence emission for DZ1 in DMSO, the NLO properties of DZ1 with RNA were also measured by open-aperture Z-scanning and 3PF. When RNA was added, the 3PA cross-section (σ) increased obviously (Fig. 5b, Table S6†). The 3PA cross-section increased 1.92-fold (σ = 1.17 × 10–81 cm6 s2 per photon2 at 1700 nm) after 25 mM RNA was added into the system; this may have been due to the interaction between DZ1 and RNA.41 To confirm further that DZ1 was internalised with RNA, 1H-NMR titration experiments were carried out in D2O. Fig. 5c shows that the proton signals Ha, Hb, Hc and Hd on the pyridine ring moved towards high magnetic fields after RNA addition, with the signal intensity decreasing gradually. These data indicated that the location of hydrogen atoms may be the binding sites of RNA.42 Molecular-docking simulation showed that the interaction between the DZ1 molecule and RNA was a charge–charge interaction because the RNA surface was anionic and DZ1 possessed a positive charge (Fig. 5d). As revealed by Horobin and coworkers,34 only probes with moderate hydrophobicity will penetrate mitochondrial membranes passively. Therefore, moderate hydrophobicity enabled DZ1 to penetrate passively through mitochondrial membranes and then recognise mitochondrial RNA specifically by charge action. Considering the results mentioned above, we suggest tentatively that DZ1 can target mitochondria due to its hydrophobic cation, and then bind mitochondrial RNA through electrostatic actions.
Fig. 5. (a) Fluorescence intensity of DZ1 (10 μM) under various amounts of RNA (0–60 mM) in Tris–HCl buffer (pH = 7.4), inset: limit point diagram of RNA titration; (b) three-photon absorption action cross-section changes of 1 mM DZ1 with 25 mM RNA. (c) Partial 1H-NMR titration spectra for DZ1 + RNA in D2O. (d) Models obtained after molecular modeling for the interaction of DZ1 with RNA fragment.
Conclusions
We synthesised a novel AIE-active metal complex DZ1 that could be used to label mitochondria in live and dead cells with distinct multi-photon fluorescence under fs laser excitation. In addition, DZ1 showed an excellent response to RNA, which had a large 3PA action cross-section of 1.17 × 10–81 cm6 s2 per photon2. In addition, compared with commercially multiphoton probes, AIE-active probe DZ1 suppressed ACQ behaviour due to self-aggregation under physiological conditions, but was also synthesised simply and exhibited excellent photo-stability. The unique 3PA of the AIE-active complex may find potential high-tech applications in biomedical theranostics and photonic devices.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by a grant for the National Natural Science Foundation of China (21701160, 51672002, 51432001 and 21871003), Anhui Provincial Natural Science Foundation of China (1708085MC68 and 1908085MB30), Anhui University Doctor Startup Fund (S020118002/026), Returnees Innovation and Entrepreneurship Key Support Program, Focus on Returned Overseas Scholar of Ministry of Education of China, and the Hefei National Laboratory for Physical Sciences at the Microscale (KF2019002).
Footnotes
†Electronic supplementary information (ESI) available. CCDC 1908472 and 1908473. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9sc01705d
References
- Zheng Q. D., He G. S., Lu C., Prasad P. N. J. Mater. Chem. 2005;15:3488–3493. [Google Scholar]
- Manzi A., Tong Y., Feucht J., Yao E. P., Polavarapu L., Urban A. S., Feldmann J. Nat. Commun. 2018;9:1518. doi: 10.1038/s41467-018-03965-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Cao S. L., Huang L. L., Chen L. H., Ouyang X. H. Dyes Pigm. 2017;145:110–115. [Google Scholar]
- Li R., Dong N. N., Ren F., Amekura H., Wang J., Chen F. Sci. Rep. 2018;8:1977. doi: 10.1038/s41598-018-20446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S., Nakano M., Kubo T., Kamada K. J., Ohta K. J., Kishi R., Nakagawa N. J. Phys. Chem. A. 2007;111:3633–3641. doi: 10.1021/jp0713662. [DOI] [PubMed] [Google Scholar]
- Zhu L. Y., Yi Y. P., Shuai Z. G. J. Chem. Phys. 2006;125:044101. doi: 10.1063/1.2355676. [DOI] [PubMed] [Google Scholar]
- Liu W., Wang Y. L., Han X., Lu P., Zhu L., Sun C. W., Qian J., He S. L. Nanoscale. 2018;10:10025–10032. doi: 10.1039/c8nr00066b. [DOI] [PubMed] [Google Scholar]
- Wu P. L., Feng X. J., Tam H. L., Wong M. S., Cheah K. W. J. Am. Chem. Soc. 2009;131:886–887. doi: 10.1021/ja806703v. [DOI] [PubMed] [Google Scholar]
- Vinayakumara D. R., Kumar M., Sreekanth P., Philip R., Kumar S. RSC Adv. 2015;5:26596–26603. [Google Scholar]
- McNitt C. D., Cheng H., Ullrich S., Popik V. V., Bjerknes M. J. Am. Chem. Soc. 2017;139:14029–14032. doi: 10.1021/jacs.7b08472. [DOI] [PubMed] [Google Scholar]
- Qian J., Zhu Z. F., Qin A. J., Qin W., Chu L. L., Cai F. H., Zhang H. Q., Wu Q., Hu R. R., Tang B. Z., He S. L. Adv. Mater. 2015;27:2332–2339. doi: 10.1002/adma.201500141. [DOI] [PubMed] [Google Scholar]
- Zhu Z. F., Leung C. W. T., Zhao X. Y., Wang Y. L., Qian J., Tang B. Z., He S. L. Sci. Rep. 2015;5:15189. doi: 10.1038/srep15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J. D., Xie Z. L., Lam J. W. Y., Cheng L., Chen H. Y., Qiu C. F., Kwork H. S., Zhan X. W., Liu Y. Q., Zhu D. B., Tang B. Z. Chem. Commun. 2001:1740–1741. doi: 10.1039/b105159h. [DOI] [PubMed] [Google Scholar]
- Wang Y. L., Han X., Xi W., Li J. Y., Roe A. W., Lu P., Qian J. Adv. Healthcare Mater. 2017;6:1700685. doi: 10.1002/adhm.201700685. [DOI] [PubMed] [Google Scholar]
- Wang H. B., Liu G. Y. J. Mater. Chem. B. 2018;6:4029–4042. doi: 10.1039/c8tb00674a. [DOI] [PubMed] [Google Scholar]
- Liu D. D., Zhang M. Z., Du W., Hu L., Li F., Tian X. H., Wang A. D., Zhang Q., Zhang Z. P., Wu J. Y., Tian Y. P. Inorg. Chem. 2018;57:7676–7683. doi: 10.1021/acs.inorgchem.8b00620. [DOI] [PubMed] [Google Scholar]
- Agrawal A. K., Jenekhe S. A. Macromolecules. 1991;24:6806–6808. [Google Scholar]
- Feng X. J., Wu P. L., Tam H. L., Li K. F., Wong M. S., Cheah K. W. Chem.–Eur. J. 2009;15:11681–11691. doi: 10.1002/chem.200901460. [DOI] [PubMed] [Google Scholar]
- Raymond J. E., Casado J., Navarrete J. T. L., Takimiya K., Goodson T. J. Phys. Chem. Lett. 2011;2:2179–2183. [Google Scholar]
- He G. S., Tan L. S., Zheng Q. D., Prasad P. N. Chem. Rev. 2008;108:1245–1330. doi: 10.1021/cr050054x. [DOI] [PubMed] [Google Scholar]
- Zhu Z. F., Qian J., Zhao X. Y., Qin W., Hu R. R., Zhang H. Q., Li D. Y., Xu Z. P., Tang B. Z., He S. L. ACS Nano. 2016;10:588–597. doi: 10.1021/acsnano.5b05606. [DOI] [PubMed] [Google Scholar]
- Fang B., Zhu Y. Z., Hu L., Shen Y., Jiang G. Q., Zhang Q., Tian X. H., Li S. L., Zhou H. P., Wu J. Y., Tian Y. P. Inorg. Chem. 2018;57:14134–14143. doi: 10.1021/acs.inorgchem.8b01967. [DOI] [PubMed] [Google Scholar]
- Hu L., Wang H., Xia T. T., Fang B., Shen Y., Zhang Q., Tian X. H., Zhou H. P., Wu J. Y., Tian Y. P. Inorg. Chem. 2018;57:6340–6348. doi: 10.1021/acs.inorgchem.8b00413. [DOI] [PubMed] [Google Scholar]
- Kumari S., Maddipoti K., Das B., Ray S. Inorg. Chem. 2019;58:1527–1540. doi: 10.1021/acs.inorgchem.8b03031. [DOI] [PubMed] [Google Scholar]
- Qin W., Zhang P. F., Li H., Lam J. W. Y., Cai Y. J., Kwok R. T. K., Qian J., Zheng W., Tang B. Z. Chem. Sci. 2018;9:2705–2710. doi: 10.1039/c7sc04820c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Meng F. F., He L. W., Yu X. Q., Lin W. Y. Chem. Commun. 2016;52:8838–8841. doi: 10.1039/c6cc03746a. [DOI] [PubMed] [Google Scholar]
- Sheet S. K., Sen B., Patra S. K., Rabha M., Aguan K., Khatua S. ACS Appl. Mater. Interfaces. 2018;10:14356–14366. doi: 10.1021/acsami.7b19290. [DOI] [PubMed] [Google Scholar]
- Krishnakanth K. N., Seth S., Samanta A., Rao S. V. Nanoscale. 2019;11:945–954. doi: 10.1039/c8nr06969g. [DOI] [PubMed] [Google Scholar]
- Hernandez F. E., Belfield K. D., Cohanoschi I. Chem. Phys. Lett. 2004;391:22–26. [Google Scholar]
- Zhang L. P., Li Y. Y., Che W. L., Zhu D. X., Li G. F., Xie Z. G., Song N., Liu S., Tang B. Z., Liu X. M., Su Z. M., Bryce M. R. Adv. Sci. 2019;6:1802050. doi: 10.1002/advs.201802050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Tian Y. Chem. Commun. 2018;54:12198–12201. doi: 10.1039/c8cc07125j. [DOI] [PubMed] [Google Scholar]
- Horobin R. W., Stockert J. C., Doubell F. R. Histochem. Cell Biol. 2006;126:165–175. doi: 10.1007/s00418-006-0156-7. [DOI] [PubMed] [Google Scholar]
- Zhang R. Y., Niu G. L., Li X. C., Guo L. F., Zhang H. M., Yang R., Chen Y., Yu X. Q., Tang B. Z. Chem. Sci. 2019;10:1994–2000. doi: 10.1039/c8sc05119d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid F., Horobin R. W. Histochemistry. 1990;94:303–308. doi: 10.1007/BF00266632. [DOI] [PubMed] [Google Scholar]
- Kumari P., Verma S. K., Mobin S. M. Chem. Commun. 2019;55:294–297. doi: 10.1039/c8cc07429a. [DOI] [PubMed] [Google Scholar]
- Shewring J. R., Cankut A. J., McKenzie L. K., Crowston B. J., Botchway S. W., Weinstein J. A., Edwards E., Ward M. D. Inorg. Chem. 2017;56:15259–15270. doi: 10.1021/acs.inorgchem.7b02633. [DOI] [PubMed] [Google Scholar]
- Wang S. W., Xi W., Cai F. H., Zhao X. Y., Xu Z. P., Qian J., He S. L. Theranostics. 2015;5:251–266. doi: 10.7150/thno.10396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Shao T., Fang B., Du W., Zhang M. Z., Liu J. J., Liu T. Y., Tian X. H., Zhang Q., Wang A. D., Yang J. X., Wu J. Y., Tian Y. P. Chem. Commun. 2018;54:11288–11291. doi: 10.1039/c8cc06276e. [DOI] [PubMed] [Google Scholar]
- Li H., Li Y. C., Zhang H. H., Xu G. Y., Zhang Y. L., Liu X. H., Zhou H. P., Yang X. Y., Zhang X. J., Tian Y. P. Chem. Commun. 2017;53:13245–13248. doi: 10.1039/c7cc07288k. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Lu X., Wang H., Tian X. H., Wang A. D., Zhou H. P., Wu J. Y., Tian Y. P. Chem. Commun. 2018;54:3771–3774. doi: 10.1039/c8cc00908b. [DOI] [PubMed] [Google Scholar]
- Wang H., Hu L., Du W., Tian X. H., Hu Z. J., Zhang Q., Zhou H. P., Wu J. Y., Uvdal K., Tian Y. P. Sens. Actuators, B. 2018;255:408–415. [Google Scholar]
- Sheet S. K., Sen B., Aguan K., Khatua S. Dalton Trans. 2018;47:11477–11490. doi: 10.1039/c8dt02099j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.