Abstract
Guanine deaminase is a metabolic enzyme, found in all forms of life, which catalyzes the conversion of guanine to xanthine. Despite the availability of several crystal structures, the molecular determinants of substrate orientation and mechanism remains to be elucidated for the amidohydrolase family of guanine deaminase enzymes. Here, we report the crystal structures of E. coli and S. cerevisiae guanine deaminase enzymes (EcGuaD and Gud1, respectively), both members of the amidohydrolase superfamily. EcGuaD and Gud1 retain the overall TIM barrel tertiary structure conserved among amidohydrolase enzymes. Both proteins also possess a single zinc cation with trigonal bipyrimidal coordination geometry within their active sites. We also determined a liganded structure of Gud1 bound to the product, xanthine. Analysis of this structure, along with kinetic data of native and site-directed mutants of EcGuaD, identify several key residues responsible for substrate recognition and catalysis. In addition, after screening a small library of compounds, two guanine derivatives, 8-azaguanine and 1-methylguanine, were identified as EcGuaD substrates. Interestingly, both EcGuaD and Gud1 also exhibit secondary ammeline deaminase activity. Overall, this work details key structural features of substrate recognition and catalysis of the amidohydrolase family of guanine deaminase enzymes in support of our long term goal to engineer these enzymes with altered activity and substrate specificity.
Graphical Abstract

INTRODUCTION
The amidohydrolase superfamily (AHS) is one of the most functionally important and diverse protein superfamilies with over 100,000 members identified to date.1 Members of the AHS superfamily share a highly-conserved (β/α)8 TIM barrel fold and typically utilize a divalent metal center to mediate hydrolysis of amide or ester bond-containing substrates.2, 3 One class of enzymes within the amidohydrolase superfamily is the nucleobase deaminases, which include adenine, cytosine, and guanine deaminase.4
Guanine deaminase plays an important role in purine metabolism, catalyzing the conversion of guanine to xanthine. This enzyme is found ubiquitously in all branches of life, including in humans. Guanine deaminase enzymes belong to one of two protein superfamilies, the amidohydrolase superfamily or the structurally unrelated cytidine deaminase-like superfamily.5 Guanine deaminases from both families utilize a divalent metal cation to activate a water molecule for nucleophilic attack. AHS guanine deaminases typically have three histidines and an aspartate residue as metal coordinating ligands, whereas cytidine deaminase-like guanine deaminases utilize cysteine and histidine metal ligands.5, 6 The guanine deaminase enzymes from E. coli and S. cerevisiae that are the focus of this work are both members of the amidohydrolase superfamily.
To date, three guanine deaminase enzymes from the AHS have been structurally characterized, all via structural genomics efforts. As such, no detailed analysis of the structure or complementary studies into their substrate specificity or catalytic mechanism have been reported. The three structurally characterized guanine deaminase enzymes include guanine deaminase from Bradyrhizobium japonicum (PDB: 2OOD), Homo sapiens (PDB: 2UZ9 and 4AQL), and Clostridium acetobutylicum (PDB: 2I9U). While all of these structures report ligands in their active site, only the human structures containing xanthine (PDB: 2UZ9) and Valacyclovir (PDB: 4AQL) have unambiguous electron density for the ligand. Two of the structures report having the substrate, guanine, bound (PDB: 2I9U and 2OOD). The two structures, however, show the ligand modeled in different orientations. Furthermore, it is unclear whether the enzyme undergoes significant conformational changes upon substrate binding due to a lack of any free enzyme structures. Thus, to clarify the molecular details of substrate recognition and catalysis for this important class of enzymes, we set out to characterize the structures of two representative guanine deaminases, from E. coli (EcGuaD) and S. cerevisiae (Gud1). The amino acid sequences of EcGuaD and Gud1 share 37% and 41% sequence identity, respectively, with human guanine deaminase.
The amidohydrolase superfamily has been classified into clusters of orthologous groups (COGs) by NCBI based on sequence similarity.7 Guanine deaminase is found in COG0402 along with the enzymes cytosine deaminase, isoxanthopterin deaminase, S-adenosylhomocysteine deaminase, 8-oxoguanine deaminase, and N-formimino-L-glutamate deiminase.7, 8 Similar to guanine deaminase, cytosine deaminase is a nucleobase deaminase enzyme which can be found in the amidohydrolase superfamily or the cytidine deaminase-like superfamily.9 E. coli cytosine deaminase (EcCDA) is a member of the AHS that has been well studied and structurally characterized.4, 10 E. coli cytosine deaminase and guanine deaminase share 22% sequence identity, but are expected to display a high degree of structural similarity. Both enzymes share a conserved metal binding HxH motif, an HxxE motif, and conserved His and Asp residues. This suggests that E. coli guanine deaminase likely employs a similar catalytic mechanism to E. coli cytosine deaminase. However, the structural determinants for substrate selectivity and catalysis of AHS guanine deaminase enzymes still remain unresolved. Furthermore, nucleobase deaminases display a remarkable specificity for their cognate nucleobase and show very little promiscuity in their ability to deaminate other nucleobases.4, 11 A careful analysis of the guanine deaminase active site would further illuminate our understanding of its catalytic mechanism and would provide insights into how nucleobase deaminases have evolved their high fidelity for specific nucleobases.
Here, we report high-resolution X-ray crystal structures of E. coli guanine deaminase (to 2.3 Å resolution) and S. cerevisiae guanine deaminase (to 1.8 Å resolution). The structures reveal EcGuaD and Gud1 share the canonical (β/α)8 TIM barrel fold common to all AHS enzymes and contain a single zinc cation with a trigonal bipyramidal coordination geometry. In addition, we solved the structure of Gud1 with xanthine bound. This data, along with complementary kinetic analysis of Gud1, EcGuaD, and EcGuaD mutants clearly defines the organization of the active site and the residues that are involved in substrate binding and catalysis. Additionally, a small library of compounds was tested to investigate substrate specificity. Together, these studies provide a detailed view of the molecular determinants for substrate binding and catalysis of the AHS guanine deaminase enzymes.
MATERIALS AND METHODS
Cloning and site-directed mutagenesis.
Wild-type guanine deaminase from E. coli (GuaD) was expressed from a modified pET-28 vector (pTHT, with a Tobacco Etch Virus protease recognition site in place of the thrombin site) with an N-terminal hexahistidine tag. Wild-type guanine deaminase from S. cerevisiae (Gud1) was expressed as a maltose binding protein (MBP) fusion from the vector pDB.His.MBP (DNASU). Standard methods were used for DNA restriction endonuclease digestion, ligation, and transformation. GuaD was amplified from E. coli by PCR. Gud1 was amplified from S. cerevisiae cDNA provided by Ed Luk of Stony Brook University. Site-directed mutagenesis was performed using OneTaq DNA Polymerase (New England Biolabs, Ipswich, MA) and DpnI (New England Biolabs, Ipswich, MA) was used to digest the methylated parental DNA prior to transformation. All constructs were sequence-verified (Genscript, Piscataway Township, NJ).
Expression and purification of EcGuaD.
E. coli BL21 (DE3) cells were transformed with pTHT_GuaD. Overnight cultures were grown in Luria Broth (LB) medium supplemented with 50 μg/mL kanamycin at 37 °C. The overnight cultures were used to inoculate 1 L of M9 minimal medium supplemented with 50 μg/mL kanamycin. The cells were grown with shaking at 37 °C. When the cells reached an OD600 of 0.3–0.5 the temperature was lowered to 18 °C and 10 μM 2,2’-Bipyridyl was added to sequester metals. To induce protein production, 500 μM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to the media along with 50 μM zinc sulfate. After overnight growth, cells were pelleted by centrifugation at 4 °C for 20 min at 6,000 g and then stored at −20 °C until purification. Cells were resuspended in lysis buffer (20 mM HEPES pH 7.5, 300 mM NaCl) and lysed by sonication. The cell lysate was then cleared by centrifugation at 26,000 g for 30 min at 4 °C. EcGuaD was purified from the cell lysate by immobilized metal affinity chromatography. A Ni-NTA column (Qiagen) was equilibrated in lysis buffer to which the lysate was added. The column was washed with 100 mL of the lysis buffer supplemented with 10 mM imidazole, and EcGuaD was eluted from the column by the addition of nickel binding buffer supplemented with 500 mM imidazole. The sample was subsequently buffer exchanged into 20 mM Tris, pH 7.5 and 100 mM NaCl and concentrated to 33 mg/mL before being snap frozen in liquid nitrogen and stored at −80 °C. Based on SDS-PAGE analysis, the resulting GuaD protein was >90% pure. Mutants of EcGuaD were expressed and prepared in the same manner as described above.
Expression and purification of Gud1.
E. coli BL21 (DE3) cells were transformed with pDB.His.MBP_Gud1. Overnight cultures were grown in Luria Broth (LB) medium supplemented with 50 μg/mL kanamycin at 37 °C. The overnight cultures were used to inoculate 1 L of LB medium supplemented with 50 μg/mL kanamycin. The cells were grown with shaking at 37 °C. When the cells reached an OD600 of 0.3–0.5 the temperature was lowered to 18 °C and protein production was induced by the addition of 500 μM isopropyl β-D-1-thiogalactopyranoside (IPTG). After overnight growth, cells were pelleted by centrifugation at 4 °C for 20 min at 6,000 g and then stored at −20 °C until purification. Cells were resuspended in lysis buffer (20 mM HEPES pH 7.5, 300 mM NaCl) and lysed by sonication. The cell lysate was then cleared by centrifugation at 26,000 g for 30 min at 4 °C. MBP_Gud1 was purified from the cell lysate by immobilized metal affinity chromatography. A Ni-NTA column (Roche cOmplete) was equilibrated in lysis buffer to which the lysate was added. The column was washed with 100 mL of the lysis buffer supplemented with 10 mM imidazole, and MBP_Gud1 was eluted from the column by the addition of nickel binding buffer supplemented with 500 mM imidazole. The MBP tag was cleaved by overnight incubation with hexahistidine-tagged TEV protease at 4 °C. The cleaved protein was flowed over Ni-NTA agarose to remove the protease and His-MBP, followed by size-exclusion chromatography on a GE HiLoad 16/600 Superdex 200PG column. The purified protein was buffer exchanged into 20 mM HEPES pH 7.5 and 50 mM NaCl and concentrated to 7 mg/mL before being snap frozen in liquid nitrogen and stored at −80 °C. Based on SDS-PAGE analysis, the resulting Gud1 protein was >90% pure.
Protein crystallization and data collection.
Initial crystallization conditions were identified with the hanging drop vapor diffusion method using sparse matrix screens at 18 °C. Hanging drops consisted of 1 μL of protein mixed with 1 μL of mother liquor. Needle-shaped crystals of EcGuaD were obtained in a condition containing 0.1 M HEPES pH 7.5, and 10% polyethylene glycol 8000. Further screening of EcGuaD was conducted by microseeding with the initial screening hit. A second crystallization condition that yielded diffraction quality crystals consisted of 0.03 M Citric acid, 0.07 M Bis-Tris Propane pH 7.6, and 20% PEG3350. Crystals of Gud1 were obtained in multiple conditions originating from the Index screen (Hampton Research). Of these, the crystals that gave the best diffraction were found in the condition containing 0.1 M HEPES pH 7.5, 0.2 M Ammonium Sulfate, and 25% polyethylene glycol 3350. Optimization of the crystallization conditions yielded the final condition containing 0.1 M HEPES pH 7.5, 0.2 M Ammonium Sulfate, and 17% polyethylene glycol 3350. Co-crystallization of Gud1 with xanthine was conducted by the addition of 5 mM of guanine (dissolved in 0.1 M NaOH) to the crystallization drop during setup. Crystals were transferred to a cryoprotectant solution composed of the mother liquor supplemented with 33% glycerol or 33% PEG3350 and flash cooled in liquid nitrogen prior to data collection. Data collection was conducted at 100 K at 24-ID-E (NE-CAT) at Advanced Photon Source (APS) and 17-ID-1 (AMX) at National Synchrotron Light Source II (NSLSII). The data collection statistics are summarized in Table 1.
Table 1:
Data collection and refinement statistics for EcGuaD and Gud1
| EcGuaD | EcGuaD-P21 | Gud1 | Gud1-Xan | |
|---|---|---|---|---|
| Data Collection: | ||||
| PDB ID | 6OHC | 6OHB | 6OH9 | 6OHA |
| Beamline | 17-ID-1 AMX | NE-CAT 24-ID-E | NE-CAT 24-ID-E | 17-ID-1 AMX |
| Detector | Eiger 9M | Eiger 16M | Eiger 16M | Eiger 9M |
| Wavelength (Å) | 0.9198 | 0.97918 | 0.97918 | 0.9201 |
| Resolution range (Å) | 48.91–2.30 (2.35–2.30) | 55.99–2.30 (2.35–2.30) | 53.63 – 1.75 (1.81 – 1.75) | 29.07–2.21 (2.29–2.21) |
| Space group | P1 | P21 | P4322 | P4322 |
| Unit cell dimensions | ||||
| a,b,c (Å) | 66.6, 80.6, 101.4 | 66.4, 137.4, 98.5 | 107.3, 107.3, 114.8 | 106.8, 106.8, 114.2 |
| α,β,γ (°) | 104.8, 105.7, 105.8 | 90.0, 102.1, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| No. of measured reflections | 453456 | 173529 | 595058 | 2833330 |
| No. of unique reflections | 79424 (7813) | 73846 (4626) | 67645 (6573) | 33428 (3075) |
| Mean I/sigma(I) | 15.0 (1.9) | 8.3 (1.8) | 14.2 (1.9) | 10.0 (2.0) |
| Completeness (%) | 98.0 (96.0) | 96.2 (97.6) | 99.8 (98.6) | 99.1 (93.6) |
| Redundancy | 5.7 (5.9) | 2.3 (2.4) | 8.8 (8.1) | 8.8 (7.4) |
| Rmerge (%) | 0.062 (0.869) | 0.061 (0.550) | 0.113 (0.970) | 0.146 (0.973) |
| CC1/2 | 0.998 (0.763) | 0.989 (0.731) | 0.998 (0.715) | 0.996 (0.696) |
| Data Refinement | ||||
| Total no. of reflections | 79285 (7746) | 109295 (10947) | 67640 (6572) | 33428 (3075) |
| Test set | 3951 (387) | 5424 (542) | 2003 (194) | 1748 (166) |
| Rwork/Rfree (%) | 22.2/25.5 | 19.9/23.6 | 17.4/19.9 | 18.1/21.0 |
| No. of protein atoms | 14043 | 14040 | 3559 | 3571 |
| No. of ligand atoms | 4 | 4 | 1 | 12 |
| No. of water atoms | 118 | 488 | 514 | 294 |
| R.M.S.D. | ||||
| bonds (Å) | 0.014 | 0.014 | 0.014 | 0.002 |
| angles (deg) | 1.83 | 1.87 | 1.44 | 0.48 |
| Mean B factor (Å2) | 21.88 | 11.82 | 23.49 | 36.64 |
| B factor for water (Å2) | 49.45 | 40.23 | 34.16 | 42.27 |
| Ramachandran plot (%) | ||||
| Favored | 95.73 | 96.77 | 96.41 | 96.42 |
| Outliers | 0.64 | 0.58 | 1.12 | 0.89 |
| MolProbity score (%) | 1.93 (92) | 1.48 (99) | 1.07 (100) | 1.23 (100) |
Structure solution and refinement.
In all cases, the data was indexed, integrated, and scaled using XDS12 and Aimless13. The EcGuaD structure was solved by molecular replacement using Phaser14 with the Bradyrhizobium japonicum protein structure (PDB: 2OOD, 41% sequence identity) as the search model. The EcGuaD structure (33% sequence identity) was used as the search model for molecular replacement using Phaser to solve the Gud1 structure. Initial refinement was carried out with the PDB-REDO server.15 The models were further refined using iterative rounds of manual model building with Coot16, and restrained refinement with REFMAC517 and phenix.refine18. After the refinement had converged, water molecules were added using Coot. Restraints for xanthine were generated using AceDRG and the ligand was added to the model after refinement had converged. The data refinement statistics are summarized in Table 1.
HPLC assay to confirm ligand identity.
The assay was run on an Agilent 1100 series HPLC using a 4.6 × 100 mm Eclipse Plus C18 column with a 3.5 μm pore size. The samples were run in a phosphate buffer (300 mM KH2PO4, pH 4.0, Sigma) with a linear gradient of acetonitrile, from 2% to 10%, over 10 minutes. Authentic standards of guanine and xanthine (>98% pure, Sigma) were run to determine retention times. Control reactions included the following: 1) a drop containing crystallization solution and guanine, but lacking Gud1 protein; 2) a co-crystallization experiment with guanine added to the crystallization solution – a 1 μL drop of the solution was removed (without disturbing the crystals) after crystals were observed in the drop (approximately 48 hours); and 3) a co-crystallization experiment with xanthine added to the crystallization solution – a 1 μL drop of the solution was removed (without disturbing the crystals) after crystals were observed in the drop (approximately 48 hours). The controls were injected on the HPLC for analysis as described above. The experimental samples were collected as follows. Co-crystallization experiments were set up using guanine or xanthine added to a mixture of crystallization solution and Gud1. After crystals were observed (approximately 48 hours), several crystals were removed from the drop, washed twice by placing the crystals in a fresh drop of crystallization solution lacking a ligand, and then dissolved in phosphate buffer. The solution containing the dissolved crystals were boiled for 15 minutes to denature the protein and release any bound ligand. After boiling, the samples were centrifuged to remove precipitate and an aliquot was injected on the HPLC for analysis.
Enzyme activity assays.
Deaminase activity was measured for EcGuaD and Gud1 using a previously described assay that couples ammonia release to nicotinamide adenine dinucleotide (NADH) oxidation.19, 20 All kinetic assays were performed at 37°C. Substrate screening was carried out at two concentrations, 200 μM and 1 mM. The coupled assay was typically run with 20 mM HEPES, pH 7.5, 100 mM NaCl, 0.2 mM NADH, 1 mM α-ketoglutarate, and 4 units of glutamate dehydrogenase (Sigma). The reaction was initiated by the addition of enzyme and the resulting decrease in absorbance at 340 nm was monitored using a Synergy Neo2 microplate reader (Biotek). The enzyme concentration ranged between 0.4 μM and 3.7 μM. The kinetic parameters kcat, kcat/Km and Km were determined by plotting the initial rates (less than 10% reaction completion) as a function of substrate concentration and fitting the data using the Michaelis−Menten equation. All kinetic data were analyzed using Graphpad Prism 4 or KaleidaGraph (Synergy Software). Enzyme concentrations were determined using the Bradford assay.
RESULTS AND DISCUSSION
Overall structure of EcGuaD and Gud1.
EcGuaD is a 439-amino acid protein with a molecular mass of 50.2 kDa. The crystal structure of EcGuaD was solved to 2.3 Å using molecular replacement (MR) with the Bradyrhizobium japonicum guanine deaminase (PDB: 2OOD, 41% identity) as the search model. EcGuaD initially crystallized in the triclinic space group P1. Subsequent screening using microseeding yielded a second crystal form in the higher symmetry monoclinic space group P21. The overall structure of EcGuaD was unchanged in the P21 crystal form. However, electron density corresponding to a flexible loop region (residues 92–102) that was disordered in the P1 crystal form was observed in the P21 crystal form. Four molecules of EcGuaD were found in the asymmetric unit of both crystal forms. The P21 crystal form had a Matthews coefficient of 2.1 Å3/Da corresponding to a solvent content of 39%. The final model contains residues 6–439 with a final crystallographic R-work of 19.9% (Rfree = 23.6%). There is no significant difference observed between the four chains in the asymmetric unit. The overall data collection and refinement statistics are presented in Table 1.
Gud1 is a 489-amino acid protein with a molecular mass of 55.2 kDa. The structure of Gud1 was solved to 1.75 Å using molecular replacement (MR) with the structure of EcGuaD (33% sequence identity) as the search model. Gud1 crystallized in the tetragonal space group P4322 with one monomer in the asymmetric unit and a Matthews coefficient of 3.0 Å3/Da corresponding to 59% solvent content. The final model contains residues 13–489, with a final crystallographic R-work of 17.4% (Rfree = 19.9%). Residues 81–87 and 436–453 were not modeled due to gaps in electron density. The data collection and refinement statistics for Gud1 are presented in Table 1.
A sequence alignment of EcGuaD and Gud1 with other structurally characterized guanine deaminase enzymes is presented in Figure S1. A gap in the alignment of Gud1 is caused by an insertion of about 20 residues near the C-terminus (residues 433–456). However, this region was not observed in the electron density map, which suggests it is disordered in the protein. Consistent with sequence homology, EcGuaD and Gud1 adopt an overall tertiary structure similar to that of other AHS guanine deaminase enzymes (Fig. 1). The active site is located centrally within a distorted TIM-barrel composed of eight β-strands interspersed with α-helices connected by flexible loop regions.
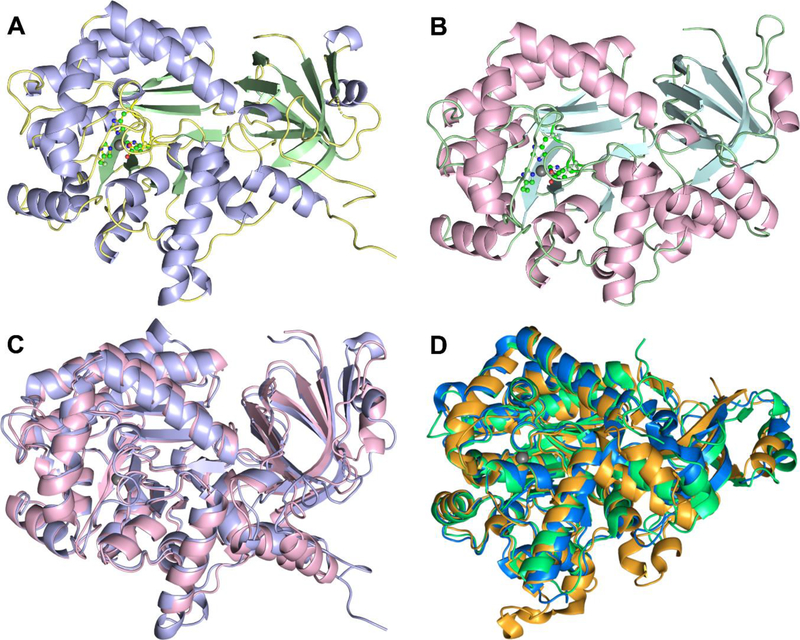
Figure 1. EcGuaD and Gud1 display the conserved TIM barrel fold characteristic of AHS members.
The structure of the EcGuaD protomer (A), and the Gud1 protomer (B) are shown. The metal ion (grey sphere) and the metal coordinating residues (shown in ball and stick) at the active site are given for reference. Superposition (C) of EcGuaD (purple) and Gud1 (pink) illustrates the structural similarity between the two (overall R.M.S.D. of 1.5 Å). Similarly, the superposition of the known structurally characterized guanine deaminases (D) demonstrates the high degree of structural conservation within this class of enzyme. Shown are the guanine deaminase from H. sapiens (PDB: 2UZ9, green); B. japonicum (PDB: 2OOD, orange); and C. acetobutylicum (PDB: 2I9U, blue). The metal ion (gray sphere) in the active site is shown for reference
Despite sharing only 33% sequence identity, structural superposition between EcGuaD and Gud1 by PDBeFOLD21 produced an overall root-mean-square-deviation (RMSD) of 1.5 Å. The most significant differences between the two structures lie outside the highly conserved TIM barrel region. This suggests that the two enzymes are likely to have evolved from a common ancestor. As expected, a structural similarity search of EcGuaD using the SSM server21 identified three structurally characterized AHS guanine deaminases as the closest homologues (Table S1): B. japonicum with an overall RMSD of 1.1 Å (42% sequence identity, PDB: 2OOD), C. acetobutylicum with an overall RMSD of 1.5 Å (33% sequence identity, PDB: 2I9U), and H. sapiens with an overall RMSD of 1.5 Å (35% sequence identity, PDB: 2UZ9). The three closest non-guanine deaminase homologues are P. aeruginosa adenosine deaminase with an overall RMSD of 1.9 Å (25% sequence identity, PDB: 4GBD), an amidohydrolase from Sargasso Sea environmental sample with an overall RMSD of 1.8 Å (29% sequence identity, PDB: 3HPA), and A. aurescens atrazine chlorohydrolase with an overall RMSD of 2.0 Å (23% sequence identity, PDB: 3LSB). All three of the non-guanine deaminase structural homologues are also clustered within COG0402 by NCBI. Additionally, a structural superposition between EcGuaD and E. coli cytosine deaminase (22% sequence identity, PDB: 3O7U) produced an overall RMSD of 2.6 Å, which suggests cytosine and guanine deaminase may be related evolutionarily, but have since diverged significantly. The SSM server returned similar results for Gud1 (Table S1).
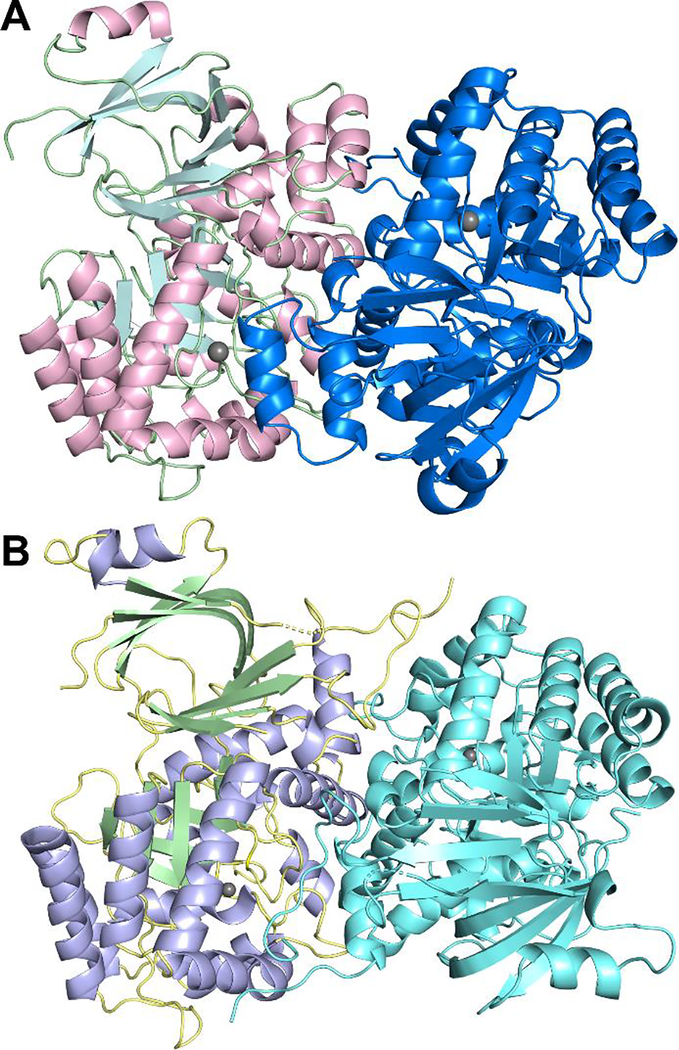
The three structurally characterized AHS guanine deaminases are predicted to be dimeric. Size-exclusion chromatography suggested that EcGuaD and Gud1 also exist as dimers in solution (data not shown). Analysis by PISA identified a dimeric interface, with two guanine deaminase protomers arranged in an antiparallel orientation (Fig. 2).22 For both enzymes, the dimer interface is formed by interactions involving residues in the helix between β-strand 1 and 2 (see Figure S2 for strand numbering), residues in the helix following β-strand 8, and residues at the C-terminus which lie outside the central barrel. The predicted buried dimer interface area by PISA is 2040 Å2 per monomer for EcGuaD (12% of total surface area) and is 2233 Å2 per monomer for Gud1 (12% of total surface area).
Figure 2. Structures of EcGuaD and Gud1 Oligomers.
Both EcGuaD (A) and Gud1 (B) appear as dimers in the crystal and as verified by size exclusion chromatography. Analysis of the dimer interface by PISA reveals that 12% of the total surface area is buried at the dimer interface for both proteins.
EcGuaD and Gud1 bind zinc within their active site
Noticeable difference density was observed at the presumed metal-binding site in both crystal structures. EcGuaD had previously been reported to bind the divalent cation zinc (Zn), and during protein production, the growth media had been supplemented with 50 μM zinc sulfate.23 Thus, the difference density was modeled as a Zn cation in the EcGuaD crystal structure. An X-ray fluorescence scan around the zinc K-edge absorption energy was measured on a Gud1 crystal at a synchrotron beamline to determine the identity of the bound metal. A large fluorescence signal was observed around 9660 eV, close to the Zn absorption K-edge of 9659 eV (Fig. S3A). Attempts to remove Zn from Gud1 by treatment with the metal chelators 1,10-phenanthroline, 2,2’-bipyridine, and ethylenediaminetetraacetic acid (EDTA) were not successful (Fig. S3B) suggesting that Zn is tightly bound and cannot be removed without denaturing the protein. Furthermore, Gud1 activity, measured by the release of ammonia with the one-to-one stoichiometric oxidation of NADH via the enzyme glutamate dehydrogenase, was not increased by supplementation with other divalent cations (Fig. S3B). The addition of certain metals, however, caused noticeable protein precipitation leading to an observed decrease in activity in some cases. These results are consistent with a tightly bound zinc cation within the Gud1 active site and zinc as the physiologically relevant metal required for catalytic activity.
Active sites of EcGuaD and Gud1.
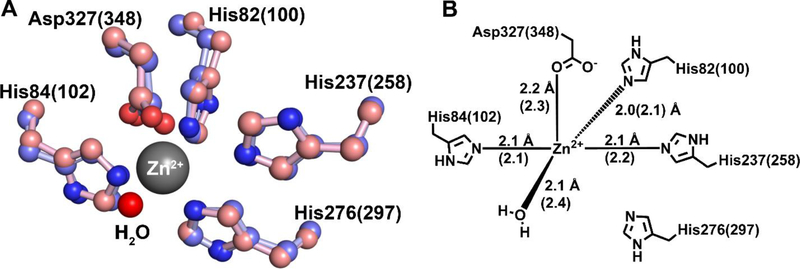
EcGuaD and Gud1 share similar active site architectures with other AHS guanine deaminase enzymes. The active site is located at the C-terminus of the (β/α)8 TIM barrel and is formed by conserved metal binding and catalytic residues on loops connecting the β-strands. Examination of the active sites of EcGuaD and Gud1 revealed a single metal ion binding site with trigonal bipyrimidal geometry. In both enzymes, Zn is directly coordinated by four protein ligands and a solvent molecule (Fig. 3). The four metal binding residues are conserved among all members of COG0402 and include two histidines from a bidentate HxH metal binding motif found at the C-terminal end of β-strand 1, a histidine residue from an HxxE motif found after β-strand 5, and an aspartate residue found at the end of β-strand 8. Specifically, the metal coordination is provided by His82, His84, His237, and Asp327 in EcGuaD, and His100, His102, His258, and Asp348 in Gud1. The metal protein ligands are clearly resolved in both crystal structures. A spherical density consistent with a water molecule was also found coordinated to the metal. The refined metal-water distances in EcGuaD and Gud1 varied from 2.1 Å – 2.5 Å, which suggests that the water coordinated to the metal is in fact a water molecule and not a hydroxide ion (–OH−).24 There is also a conserved histidine residue, His276 (His297) at the end of β-strand 6 that lies at a slightly longer distance of 4.0 Å, which is too far to coordinate the metal.
Figure 3. Metal coordination within EcGuaD and Gud1 active sites.
A superposition of the EcGuaD and Gud1 active site (A) shows the conserved residues responsible for coordination of the zinc cation (shown in ball-and-stick representation; EcGuaD has purple carbon atoms and Gud1 has pink carbon atoms; nitrogen atoms are blue and oxygen atoms are red). The zinc ion is penta-coordinated by three histidine residues, an aspartate residue, and a water molecule (shown as a red sphere). There is also a conserved histidine residue His276 (297) in the correct orientation to bind but is not close enough to coordinate the metal (4 Å). The residues are numbered according to the EcGuaD sequence, with the corresponding residues in Gud1 indicated in parenthesis. (B) shows a schematic of the metal coordination. Residues and distances corresponding to Gud1 are indicated in parenthesis.
As reported for other guanine deaminase enzymes, the water molecule that occupies the fifth coordination site is most likely the source of the hydroxide ion that participates in the catalytic reaction. Coordination of a water molecule by a zinc ion would result in a drastic lowering of its pKa value, thus favoring its deprotonation.25 The water molecule is also situated between 2.5 Å – 3.2 Å from Asp327 (Asp348) in the coordination sphere. Thus, we propose that Asp327 (Asp348) acts as the general base to abstract a proton and generate the reactive hydroxide species. Binding of the guanine substrate is hypothesized to push the water molecule closer to Zn, further accelerating its deprotonation.
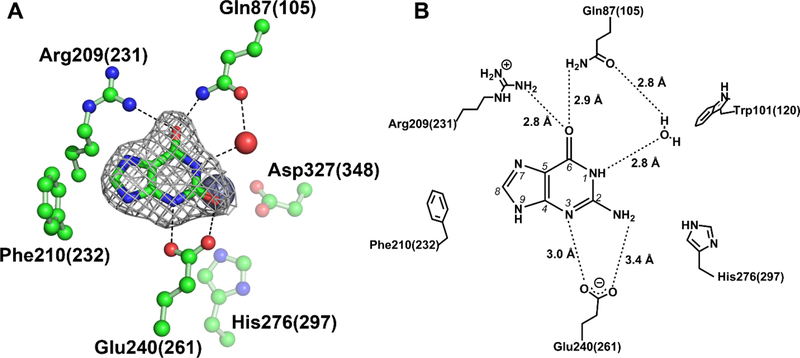
To gain further insights into substrate binding and selectivity, a crystal structure of Gud1 with xanthine bound was also determined (Fig. 4). The crystallization conditions used in this case included guanine to be co-crystallized with Gud1. However, considering the kinetics of this enzyme (see below), guanine would have been fully converted to xanthine during the time-scale of crystallization (overnight). To ensure that this was the case, we selected several crystals grown under the co-crystallization conditions, washed them in well solution lacking guanine, and then dissolved them in fresh buffer. This solution was then boiled to denature the protein and release any bound ligand. After centrifugation to remove the denatured protein, a sample was run on HPLC to confirm that the ligand bound to the crystals was, in fact, xanthine (Fig. S4). Thus, the ligand density observed in the structure was modelled as the product, xanthine. The ligand is oriented in the active site by hydrogen bond interactions with the side chains of Gln105, Arg231, and Glu261. Both Gln105 and Arg231 hydrogen bond with the carbonyl oxygen at C6, while Glu261 makes two additional hydrogen bond contacts with N3 and the carbonyl oxygen at C2 (where the amine group is removed from the substrate – see Figure 4B).
Figure 4. Guanine deaminase substrate interactions.
Cocrystallization of Gud1 with hypoxanthine ligand-bound structure (A). Electron density for xanthine is from a POLDER map generated by omitting the ligand and is contoured at 3σ. The zinc metal ion is represented by a gray sphere and a water molecule by a red sphere. Carbon atoms are colored green, oxygen atoms are red, and nitrogen atoms are blue. A schematic showing the proposed network of binding interactions for guanine is shown in (B). Note that H-bond distances are those observed in the Gud1 liganded structure with xanthine. In both (A) and (B) residues are numbered according to the EcGuaD sequence with corresponding Gud1 residues indicated in parenthesis.
Only two previous structures of AHS guanine deaminase report guanine bound, PDB entries 2OOD and 2I9U, neither of which is associated with a published analysis. Notably, the orientation of guanine is distinctly different in the two structures. In the structure reported in entry 2OOD, the carbonyl oxygen of guanine is rotated 180 degrees away from the zinc metal. Furthermore, the positioning of guanine in this structure is also markedly different to that of the other liganded guanine deaminase structure. We visually inspected the electron density map associated with PDB entry 2OOD and found the electron density to be inconsistent with the structure of guanine as shown. In addition, large negative difference peaks surrounded the molecule as fit. Based on this analysis and comparison to our structures of ligand free guanine deaminase, we believe the density peaks in the maps for entry 2OOD are indicative of water molecules and not a guanine molecule, as indicated. Our data demonstrates that our Gud1 cocrystallization experiments with guanine (Fig. S4) yield crystals that have catalytically competent enzyme and capture a product-bound form. In addition, the position of xanthine in our structure is consistent with the location of the product in the product-bound form of human guanine deaminase (2UZ9).
Substrate specificity of EcGuaD.
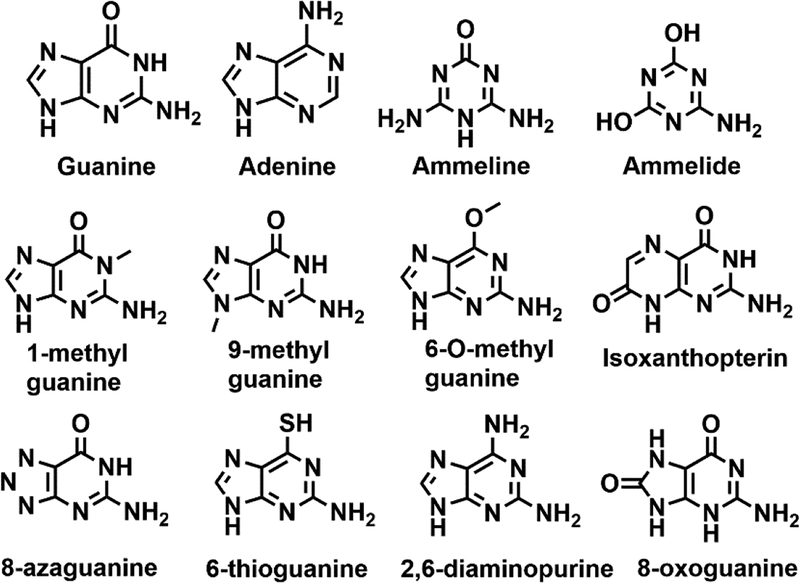
A continuous assay coupling ammonia production to NADH consumption (see Materials and Methods) was used to spectrophotometrically monitor guanine deaminase activity.19, 20 The steady-state kinetics of guanine and ammeline were determined both for Gud1 and EcGuaD (Fig. S5). In addition, EcGuaD was screened for activity against a small library of compounds to probe the molecular determinants of substrate specificity (Fig. 5). Two guanine derivatives, 1-methylguanine and 8-azaguanine were identified as EcGuaD substrates. Several other compounds, including 9-methylguanine, 6-O-methylguanine, 6-thioguanine, and 2, 6-diaminopurine, also exhibited measurable activity, but turned over too slowly (<0.001 s−1) to justify further investigation. The kinetic parameters for the EcGuaD catalyzed reaction with these substrate s are presented in Table 2.
Figure 5.
Structures of compounds tested as possible substrates.
Table 2.
Steady-state kinetic parameters for the substrates tested
| Km (mM) | kcat (s−1) | kcat/KM (M−1s−1) | Fold decrease in kcat/KM | |
|---|---|---|---|---|
| EcGuaD | ||||
| Guanine | 0.21 ± 0.04 | 2.8 ± 0.2 | 13.3 ± 0.6 | - |
| Ammeline | 1.2 ± 0.2 | 1.0 ± 0.09 | 0.83 ± 0.15 | 16 |
| 1-methylguanine | 0.93 ± 0.26 | 0.08 ± 0.01 | 0.086 ± 0.026 | 155 |
| 8-azaguanine | 1.0 ± 0.3 | 7 ± 1 | 7 ± 2.3 | 2 |
| 6-O-methylguainine | N.D.* | <0.001 | N.D. | - |
| 9-methylguanine | N.D. | <0.001 | N.D. | - |
| 2,6-diaminopurine | N.D. | <0.001 | N.D. | - |
| 8-oxoguanine | N.D. | <0.001 | N.D. | - |
| Isoxanthopterin | N.D. | <0.001 | N.D. | - |
| Gud1 | ||||
| Guanine | 0.061 ± 0.008 | 8.9 ± 0.3 | 146 ± 20 | - |
| Ammeline | ≥ 2.6 ± 1.0 | ≥ 2.8 ± 0.8 | 1.1 ± 0.5 | 134 |
ND – not determined, kcat < 0.001 s−1
Guanine was deaminated by EcGuaD and Gud1 with a kcat/Km of 13.3 M−1s−1 and 146 M−1s−1, respectively. In contrast, 8-azaguanine was deaminated at half the rate of guanine, with a kcat/Km of 7 M−1s−1 and a Km of 1.0 mM. The observed catalytic activity with 1-methylguanine was also substantially reduced compared to the native substrate with a kcat/Km of 0.086 M−1s−1. Based on analysis of existing guanine deaminase structures and our structures of EcGuaD and Gud1, 8-azaguanine is expected to have slightly altered binding within the active site compared to guanine. The presence of a double bond between N7 to a second nitrogen at N8 in 8-azaguanine would make the nitrogen at N7 less electronegative. This would decrease the ability of guanine deaminase to bind 8-azaguanine through hydrogen bonding interactions at N7 compared with guanine. Consistent with this observation, 8-azaguanine had a higher Km of 1.0 mM. Furthermore, examination of crystal structures with ligand bound suggests a sole arginine residue is responsible for binding interactions with the imidazole ring on guanine. Specifically, the guanidino side chain of arginine makes interactions with N7 and the carbonyl oxygen of guanine. This suggests small substitutions at the 7, 8, and 9 positions on the imidazole ring could be tolerated by guanine deaminase. This would explain the observed activity for 9-methylguanine. The observed activity is also consistent with the crystal structure of Valacyclovir bound to human guanine deaminase (PDB: 4AQL). Valacyclovir is analogous to guanine with the sole exception that it is functionalized at C9. In addition, analysis of the crystal structures did not reveal any residues within hydrogen bonding distance of N1 except for a water molecule, which explains why the insertion of a methyl group in 1-methylguanine allowed it to be accepted as a substrate. Conversely, substitutions at the carbonyl oxygen of guanine are not well tolerated. Replacement of the carbonyl oxygen with an amino group in 2,6-diaminoguanine or with a sulfhydryl group in 6-thioguanine resulted in little to no activity observed. The nitrogen at C2 in 2,6-diaminopurine is less electronegative than oxygen, thus disrupting normal hydrogen bonding interactions with the carbonyl oxygen of guanine. Sulfur has a larger radius than oxygen, which may preclude 6-thioguanine binding due to steric hindrance. Similarly, the presence of a methyl group on 6-O-methylguanine would also prevent binding. Addition of a carbonyl at C8 (8-oxoguanine) or increasing the size of the imidazole ring, as in isoxanthopterin, are also not tolerated by the enzyme. The proximity of Phe210 and Arg209 to C8 and N7 would most likely prevent binding of these more sterically hindered substrates.
While EcGuaD had previously been reported to exhibit secondary ammeline deaminase activity, this activity was not verified with purified enzyme nor characterized kinetically.26 EcGuaD had a Km of 1.2 mM and a kcat/Km of 0.83 M−1s−1 for ammeline (Table 2). The ability of Gud1 to deaminate ammeline was similarly tested with a Km of approximately 2.6 mM and a kcat/Km of approximately 1.1 M−1s−1 for ammeline. Note that we were unable to reach saturating concentrations of ammeline for Gud1 due to its limited solubility. The diminished catalytic activity of ammeline could be attributed to its smaller size relative to guanine. Thus, ammeline would have a reduced capacity to bind in a catalytically competent orientation, which is consistent with the observed increase in Km. Overall, our results suggest that ammeline binding is much weaker than guanine with a Km in the 1–3 mM range and a turnover number about 3–4 times lower than that of guanine (Table 2). A comparison between EcGuaD and Gud1 ammeline deaminase activity suggests that while the E. coli enzyme may have a slightly higher affinity for this guanine analog, the yeast protein turns the substrate over at least 2-fold faster. Cytosine and adenine were also tested as potential substrates and displayed no deaminase activity at all concentrations tested (up to 1 mM). The selectivity of guanine over adenine, and ammeline over ammelide (see below), point to the importance of the two hydrogen bonding interactions to N3 and the amine group at N2 of guanine by the conserved glutamate residue (Glu240 in EcGuaD and Glu261 in Gud1; Figs. 4 and S1).
Ammeline deaminase activity was previously reported for guanine deaminases of both the amidohydrolase and cytidine-deaminase like superfamilies.6, 27 We have verified that both EcGuaD and Gud1 are capable of catalyzing the deamination of ammeline. As reported for other guanine deaminase enzymes, this activity occurs at a much lower rate compared to its native substrate. EcGuaD and Gud1 displayed no noticeable activity for the deamination product of ammeline, ammelide, however. Ammeline and related s-triazines are xenobiotic compounds that cause renal toxicity in mammals and guanine deaminase could potentially be used to detect these contaminants in food sources or as a means to facilitate their bioremediation.28–30 Other members of COG0402 such as TriA (melamine and ammeline deaminase), AtzA (atrazine chlorohydrolase), and TrzA (s-triazine hydrolase), have been reported to deaminate various triazine substrates.27 These enzymes could have diverged from guanine deaminase and evolved to have greater specificities for triazine compounds.
Analysis of EcGuaD mutants
Our crystal structures of EcGuaD and Gud1 displayed virtually identical active sites to other structurally characterized AHS guanine deaminase enzymes. The positioning of xanthine within our structure of Gud1 is also consistent with existing liganded structures of guanine deaminase. Furthermore, no conformational changes were observed in xanthine bound Gud1 compared to the ligand free structure. This allowed us to compare and further examine residues important for substrate binding. Since the tertiary structure of AHS enzymes remains highly conserved, it is believed that the variable loop regions connecting the β-strands confers substrate specificity and selectivity.2
Examination of ligand bound guanine deaminase structures revealed few direct interactions between the purine base and guanine deaminase, with only a few hydrogen bonding interactions mediated by the side chains of arginine, glutamine, and glutamate residues. In our xanthine bound structure of Gud1, xanthine is coordinated by Gln105, Arg231, and Glu261 (Fig. 4). The guanidino sidechain of Arg231 forms hydrogen bonding interactions with the carbonyl oxygen and the N7 atom of xanthine. Arg231 is conserved as part of an Arg-Phe dyad in guanine deaminase enzymes, but is replaced by Cys-Ser in 8-oxoguanine deaminase and Thr-Thr in isoxanthopterin deaminase.31, 32 The replacement of Arg-Phe with more compact amino acids in 8-oxoguanine and isoxanthopterin deaminase may be responsible for the change in substrate specificity. The carbonyl oxygen on xanthine is further stabilized by a hydrogen bond with the side chain amino group of Gln105. Gln105 also makes a water-mediated hydrogen bond to N1 on xanthine. An additional interaction is found between the side chain of Glu261 and N3 of xanthine. Glu261 is likely to assist in binding and orienting guanine through interactions with the amino group at C2 before turnover. Glu261 is part of the highly conserved HxxE motif, where His258 is ligated to the metal ion, while Glu261 is proposed to be involved in the catalytic reaction. The lack of many obvious protein-ligand interactions suggests that guanine analogues could bind guanine deaminase with roughly similar affinities. The results of our kinetic characterization of various substrate analogs, however, do not support this assertion. This suggests the enzyme may utilize additional strategies for controlling substrate specificity, including via hydrophobic interactions. Two aromatic residues Phe231 and Trp120 within the active site could potentially help orient the substrate and shape the active site through hydrophobic interactions.
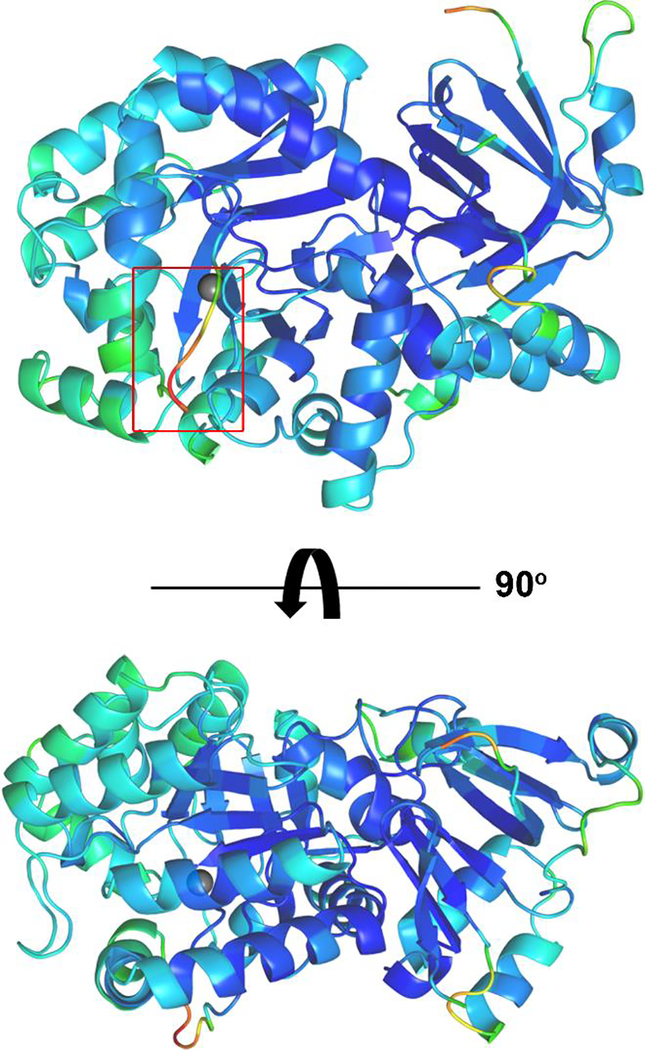
In our structure of E. coli guanine deaminase, a flexible loop near the active site exhibited increased B-factors compared to the overall structure (Fig. 6). The corresponding loop region in human guanine deaminase with Valacyclovir bound (PDB entry 4AQL) was also observed to be shifted in comparison to the corresponding structure with xanthine bound (PDB entry 2UZ9). In the Valacyclovir bound structure, the guanine base was positioned identically to that of the xanthine bound structure, with only a small displacement in the loop following β-strand 1 to accommodate the C9 valine-ester moiety.33 This led us to hypothesize that this loop could contribute to binding specificity or selectivity in EcGuaD. Notably, the homologous residue to Trp120 in EcGuaD (Trp101) is found within this flexible loop with its side chain in an alternative conformation relative to the Gud1 structure.
Figure 6. Flexible loop region adjoining EcGuaD active site.
A structure of EcGuaD, colored by B-factors (low – blue, green, yellow, orange, red – high), shows that a loop region near the active site (boxed in red) has the highest B-factors. This loop, following β-strand 1, was disordered in our P1 structure of EcGuaD and was displaced by the ligand in the corresponding region in PDB entry 4AQL of human guanine deaminase bound to Valacycolovir. The zinc metal (gray sphere) in the active site is shown for reference.
Guided by this analysis, we generated a series of EcGuaD mutants and characterized their steady-state kinetics to further investigate the substrate binding specificities of guanine deaminase enzymes. The generation of site-directed mutations in Gud1 resulted in insoluble protein under our standard expression conditions and led us to pursue mutagenesis studies with EcGuaD. An overall view of EcGuaD with the location of the mutations is shown in Figure S6. The residues coordinating the guanine base are fully conserved between Gud1 (Gln105, Arg231, and Glu261) and EcGuaD (Gln87, Arg209, and Glu240). Two of the residues responsible for orienting the purine base, Gln87 and Glu240 were selected for mutation. The aromatic residue following Arg209, Phe210 was also mutated. In addition, Leu98 and Trp101 within the flexible loop region were targeted for mutagenesis in an effort to ascertain their roles in substrate binding. Other residues targeted for mutagenesis include two metal binding ligands, His237 and Asp327, as well as a conserved residue Leu305. Ala386, which lies outside the conserved TIM barrel, was also mutated as a control. Using site-directed mutagenesis, the following point mutants were generated: Q87N, L98A, W101H, F210A, H237R, E240D, L305A, D327L, and A386S. We did not observe overexpression of the L98A mutant under our standard expression conditions and thus this point mutant was not characterized. The steady-state kinetics of all expressed mutants was tested as described previously for the wild-type enzymes.
The mutagenesis study revealed that EcGuaD does not readily tolerate point mutations, with all mutants showing significant reductions in overall rate and catalytic efficiency (Table 3, Fig. S7). Highlighting the importance of metal coordination, mutations to His237 and Asp327 resulted in a complete loss of activity. Furthermore, the D327L mutant also exhibited significantly reduced stability compared to wild-type enzyme, which suggests that alterations to the metal binding geometry are not tolerated. This suggests that the metal-binding site is relatively invariant and that EcGuaD is unlikely to bind alternative metals. In addition, mutations to Gln87 and Glu240, which are homologous to residues responsible for making hydrogen bonding interactions with xanthine in Gud1 (Gln105 and Glu261), also resulted in a complete loss of catalytic activity. The loss of activity in the Q87N mutant could be due to the inability of the shorter asparagine side chain to form hydrogen bonds with the carbonyl oxygen on the guanine base at C6 as well as a water mediated hydrogen bond to N1. The loss in hydrogen bonding interactions with guanine deaminase would severely limit the ability of guanine to bind to the enzyme. A mutation to the conserved glutamate residue in the HxxE motif, E240D also resulted in loss of any measurable catalytic activity. This glutamate residue may stabilize guanine upon initial binding within the active site, and given its proximity to N3, could also serve as a proton donor in the catalytic reaction.
Table 3.
Steady-state kinetic parameters for EcGuaD mutants
| Km (mM) | kcat (s−1) | kcat/KM (M−1s−1) | Fold decrease in kcat/KM | |
|---|---|---|---|---|
| WT | 0.21 ± 0.04 | 2.8 ± 0.2 | 13.3 ± 0.6 | - |
| Q87N | NA* | NA | NA | - |
| W101H | 0.08 ± 0.01 | 0.13 ± 0.004 | 1.5 ± 0.2 | 8.9 |
| F210A | 0.21 ± 0.06 | 0.14 ± 0.01 | 0.7 ± 0.2 | 20 |
| H237R | NA | NA | NA | - |
| E240D | NA | NA | NA | - |
| L305A | NA | NA | NA | - |
| D327L | NA | NA | NA | - |
| A386S | 0.14 ± 0.03 | 0.39 ± 0.02 | 2.8 ± 0.5 | 4.8 |
Bold – residues that coordinate metal cation or involved in catalysis
NA – no activity detectable under assay conditions
Mutations to two aromatic residues found within the active site were also generated, W101H and F210A. F210 is part of a conserved Arg-Phe dyad in guanine deaminases. The F210A mutant exhibited severe defects in catalytic efficiency with a kcat/Km of 0.67 M−1S−1, 20 times lower than wild-type enzyme. Despite the lower overall efficiency, the F210A mutant had a comparable Km to wild-type at 0.21 mM. This suggests F210 might play a role in shaping the active site for catalysis, but is likely not contributing significantly to substrate binding. The W101H mutant displayed a Km of 0.084 mM, and a kcat/Km that was 9-fold lower (1.5 M−1S−1) compared to wild-type EcGuaD. The imidazole ring on histidine is a smaller aromatic side chain than the indole ring on trypotophan, which could slightly open up the active site allowing guanine to bind more efficiently leading to the observed reduction in Km. However, the W101H mutant also resulted in reduced activity, which suggests that W101 also contributes to shaping the active site for optimal catalytic efficiency. Interestingly, both the W101H and F210A mutants were able to catalyze deamination of guanine, but had no detectable ammeline deaminase activity.
Additional mutants that were generated include L305A and A386S, with only the A386S mutant having detectable activity. The lack of activity in the L305A mutant was unexpected. L305 is conserved between the structurally characterized guanine deaminases and is located in a loop at the base of the barrel following β-strand 7. L305 could serve a gatekeeping function or could play a role in maintaining the tertiary structure of the barrel, but further investigation would be needed to make this determination. While the L305A mutant protein did express and purify as expected, we cannot rule out the possibility that the lack of activity is attributed to incorrect folding of the protein. The A386S mutant, located a large distance from the active site, was also made and tested as a control. Unexpectedly, this mutant displayed several-fold lower activity (kcat/Km of 2.8 M−1S−1) when compared to wild-type. Such long-distance effects are not without precedent and likely indicate that some degree of global protein motion is important throughout the catalytic cycle of this protein.34–37
The results of our structural analysis and site-directed mutagenesis studies of EcGuaD have identified key residues involved in substrate binding by guanine deaminase enzymes. Specifically, ligand binding in the active site is mainly mediated through hydrogen bonding interactions with a conserved arginine residue (part of a conserved Arg-Phe dyad) following β-strand 4, a glutamine residue following β-strand 1, and by the glutamate residue that is part of the conserved HxxE motif. The interaction with the glutamate residue may serve to both stabilize and optimize positioning of the substrate for catalysis. Ligand coordination is likely also assisted by additional hydrophobic interactions provided by a tryptophan and phenylalanine residue that are found within the active site. We observed that mutation of these aromatic residues led to altered catalytic activity suggesting that the hydrophobic side chains may contribute to the occlusion of water from the active site. We were also unable to fully rationalize the complex effects of mutations at L305 and A386 on the kinetic parameters. These mutations were not expected to have an effect on catalysis. Nevertheless, these results indicate that future engineering studies examining residues outside the active site could be useful to make subtle changes to activity. Given the few direct protein-ligand interactions, solvation thermodynamics involving the displacement of water in the active site by a ligand may also be critical for binding and catalysis. Of course, this conjecture would require further investigation.
Proposed catalytic mechanism of guanine deaminase
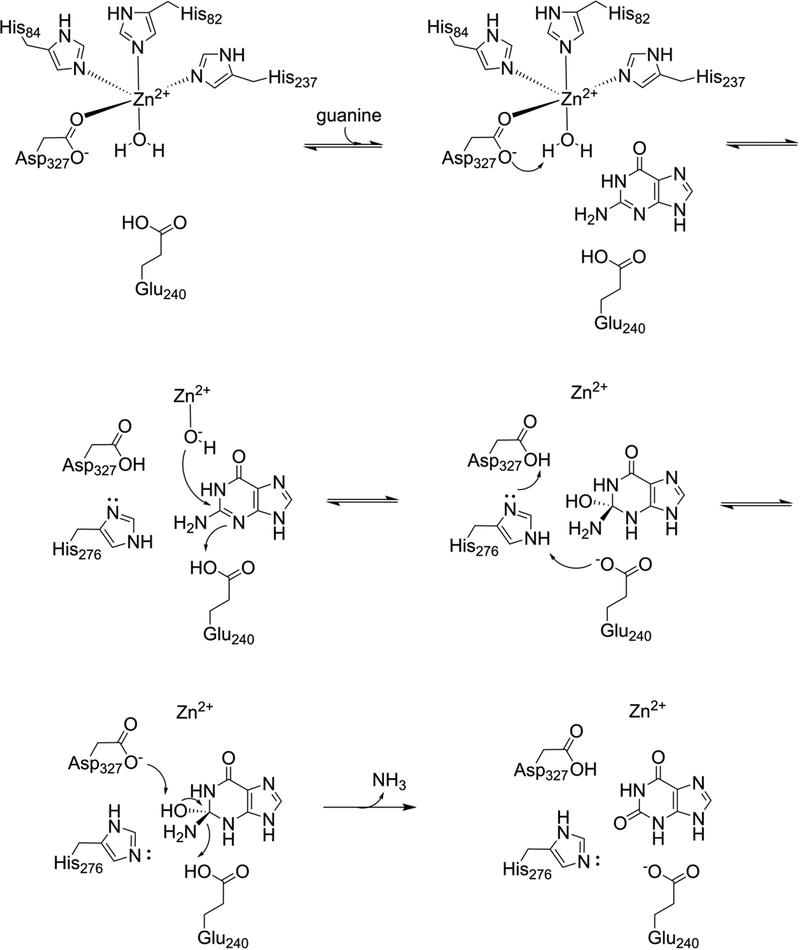
The catalytic mechanism of guanine deaminase members of the amidohydrolase superfamily has not been extensively explored, but is thought to proceed similarly to that of E. coli cytosine deaminase, which has been characterized structurally and mechanistically with some detail.4, 38, 39 Our structures clearly indicate that the metal atom is coordinated in a trigonal bipyramidal geometry by three His residues and one Asp residue (Fig. 3). Our data also indicates that the physiologically relevant metal is a zinc ion (Fig. S3). Furthermore, a fourth, conserved, histidine (H276 in EcGuaD, H297 in Gud1) is found in the correct geometry for metal coordination, but is located too far (> 4 Å) from the metal ion to be involved in its coordination. This histidine residue likely plays a role facilitating proton transfer (see below). Spherical density consistent with a water molecule was also observed in hydrogen bonding distance with the metal ion. The observed metal-water bond length (2.1 Å for EcGuaD and 2.4 Å for Gud1) suggests that, in the unliganded state, this is a water molecule and not a hydroxide ion (Fig. 3). The aspartate residue that coordinates the metal (D327 in EcGuaD, D348 in Gud1) also hydrogen bonds with this water molecule. On the basis of existing cytosine deaminase mechanistic characterization, analysis of our structures of E. coli and yeast guanine deaminase, and the results from our mutational analysis of EcGuaD, we proposed a catalytic mechanism for this class of enzymes (Fig. 7). Despite the relatively high overall sequence and structural diversity amongst the sub-families of nucleobase deaminases, the conservation in and around the active site, and supporting mechanistic studies, suggest that the catalytic mechanism is likely conserved across species.
Figure 7. Proposed mechanism of AHS guanine deaminase.
Deprotonation of the activated water molecule by Asp327 generates a hydroxide ion. Nucleophilic attack at the C2 carbon atom leads to a tetrahedral intermediate. A proton transfer from the aspartate to Glu240 is facilitated by His276. Abstraction of a proton from the hydroxyl by Asp327, and concomitant release of ammonia, yields the final product.
The position of the substrate upon binding, as indicated by our structure of Gud1 in complex with xanthine (Fig. 4) and the other available structural data (PDB: 4AQL, 2UZ9, 2I9U), is expected to push the metal-coordinated water molecule closer to the metal ion, weakening the OH bond of the already polarized water molecule to further facilitate deprotonation. Based on its position in the structures, the aspartate residue (D327 in EcGuaD, D348 in Gud1) would act as the general base to abstract a proton from the water to generate a reactive hydroxide species. The hydroxide nucleophile would then attack at C2 of the purine ring, generating a tetrahedral intermediate. Prior to the next step, a proton would be transferred between the aspartate residue and a glutamate residue (E240 in EcGuaD, E261 in Gud1). This glutamate residue is positioned within hydrogen bonding distance to N3 of xanthine in our liganded structure (Fig. 4). A similar proton transfer step has been proposed to occur in the enzyme mechanism of E. coli cytosine deaminase.4 The transfer would likely be facilitated by the conserved histidine residue (H276 in EcGuaD, H297 in Gud1) positioned equidistant between the aspartate and glutamate residues (Fig. 4). An alternative mechanism, whereby a tautomer of guanine protonated at the N1 position is the initial substrate, is not consistent with our observation that 1-methylguanine is a favorable substrate (Table 2). In the final step, deprotonation of the tetrahedral hydroxyl group by the aspartate residue is accompanied by release of the amine group as ammonia, yielding the final product of the reaction. Consistent with their critical roles in this proposed mechanism, mutations to either the aspartate residue (E240) or glutamate residue (D327) led to a complete loss of catalytic activity (Table 3).
CONCLUSION
This is the first detailed analysis of AHS guanine deaminase structure and kinetics, which furthers our understanding of the molecular determinants for substrate binding and catalysis. Consistent with existing AHS guanine deaminase structures, EcGuaD and Gud1 displayed a high degree of structural conservation and retained a TIM barrel tertiary structure. Guanine deaminase enzymes display a remarkable fidelity for their cognate nucleobase. From this work, critical features for ligand binding, position, and metal ion geometry for effective catalysis were identified. These include hydrogen-bonding interactions provided by conserved arginine, glutamine, and glutamate residues as well as hydrophobic interactions provided by conserved phenylalanine and tryptophan residues. In general, the principles observed could likely be extended to guanine deaminase enzymes that remain to be characterized. Overall, our findings also contribute to our knowledge of the molecular basis of substrate specificity by nucleobase deaminases of the amidohydrolase superfamily. It is interesting to note that eukaryotic guanine deaminase is retained from prokaryotes, whereas neither cytosine nor adenine deaminase are present in higher eukaryotes. Explicit knowledge of both the structure as well as the molecular determinants of substrate recognition and catalysis is required to facilitate protein engineering and design. This work establishes an important framework for our long term goal to rationally engineer this important class of enzymes with altered functionality and substrate preferences.
Supplementary Material
Figure S1. Sequence alignment of guanine deaminase homologues
Figure S2. Overall guanine deaminase structure with β-strand and termini labeled
Figure S3. Gud1 binds zinc.
Figure S4. Confirmation that ligand bound in co-crystallization experiment is xanthine
Figure S5. Kinetics of EcGuaD and Gud1 for guanine and ammeline
Figure S6. E. coli guanine deaminase structure showing position of mutations made in this study
Figure S7. Steady-state kinetics of E. coli guanine deaminase mutants
Table S1. Structural homologues of EcGuaD and Gud1 identified by SSM server
ACKNOWLEDGEMENTS
The authors would like to thank Anne Ostermeyer-Fay for help with cloning, Dr. Ed Luk for the kind donation of yeast cDNA, and the staff at NE-CAT and AMX for support and direction with data collection. This work was funded in part by the National Institute of General Medical Science of the National Institutes of Health (R35GM124898 to JBF). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357, and the AMX and FMX beamlines of the National Synchrotron Light Source II, a US DOE Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under contract number DE-SC0012704.
Footnotes
REFERENCES
- 1.Raushel FM (2016) Finding homes for orphan enzymes, Perspectives in Science 9, 3–7. [Google Scholar]
- 2.Holm L, and Sander C (1997) An evolutionary treasure: unification of a broad set of amidohydrolases related to urease, Proteins 28, 72–82. [PubMed] [Google Scholar]
- 3.Seibert CM, and Raushel FM (2005) Structural and catalytic diversity within the amidohydrolase superfamily, Biochemistry 44, 6383–6391. [DOI] [PubMed] [Google Scholar]
- 4.Hall RS, Fedorov AA, Xu C, Fedorov EV, Almo SC, and Raushel FM (2011) Three-dimensional structure and catalytic mechanism of cytosine deaminase, Biochemistry 50, 5077–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liaw SH, Chang YJ, Lai CT, Chang HC, and Chang GG (2004) Crystal structure of Bacillus subtilis guanine deaminase: the first domain-swapped structure in the cytidine deaminase superfamily, The Journal of biological chemistry 279, 35479–35485. [DOI] [PubMed] [Google Scholar]
- 6.Bitra A, Biswas A, and Anand R (2013) Structural basis of the substrate specificity of cytidine deaminase superfamily Guanine deaminase, Biochemistry-Us 52, 8106–8114. [DOI] [PubMed] [Google Scholar]
- 7.Tatusov RL, Koonin EV, and Lipman DJ (1997) A genomic perspective on protein families, Science (New York, N.Y.) 278, 631–637. [DOI] [PubMed] [Google Scholar]
- 8.Fan H, Hitchcock DS, Seidel RD 2nd, Hillerich B, Lin H, Almo SC, Sali A, Shoichet BK, and Raushel FM (2013) Assignment of pterin deaminase activity to an enzyme of unknown function guided by homology modeling and docking, Journal of the American Chemical Society 135, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ko T-P, Lin J-J, Hu C-Y, Hsu Y-H, Wang AH-J, and Liaw S-H (2003) Crystal Structure of Yeast Cytosine Deaminase: INSIGHTS INTO ENZYME MECHANISM AND EVOLUTION, Journal of Biological Chemistry 278, 19111–19117. [DOI] [PubMed] [Google Scholar]
- 10.Ireton GC, McDermott G, Black ME, and Stoddard BL (2002) The structure of Escherichia coli cytosine deaminase, Journal of molecular biology 315, 687–697. [DOI] [PubMed] [Google Scholar]
- 11.Kamat SS, Bagaria A, Kumaran D, Holmes-Hampton GP, Fan H, Sali A, Sauder JM, Burley SK, Lindahl PA, Swaminathan S, and Raushel FM (2011) Catalytic Mechanism and Three-Dimensional Structure of Adenine Deaminase, Biochemistry 50, 1917–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabsch W (2010) XDS, Acta Crystallogr D Biol Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans PR, and Murshudov GN (2013) How good are my data and what is the resolution?, Acta Crystallogr D Biol Crystallogr 69, 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007) Phaser crystallographic software, Journal of applied crystallography 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joosten RP, Long F, Murshudov GN, and Perrakis A (2014) The PDB_REDO server for macromolecular structure model optimization, IUCrJ 1, 213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emsley P, and Cowtan K (2004) Coot: model-building tools for molecular graphics, Acta crystallographica. Section D, Biological crystallography 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 17.Murshudov GN, Vagin AA, and Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method, Acta crystallographica. Section D, Biological crystallography 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 18.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine, Acta Crystallogr D Biol Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.French JB, Cen Y, Vrablik TL, Xu P, Allen E, Hanna-Rose W, and Sauve AA (2010) Characterization of nicotinamidases: steady state kinetic parameters, classwide inhibition by nicotinaldehydes, and catalytic mechanism, Biochemistry 49, 10421–10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muratsubaki H, Satake K, and Enomoto K (2006) Enzymatic assay of allantoin in serum using allantoinase and allantoate amidohydrolase, Anal. Biochem. 359, 161–166. [DOI] [PubMed] [Google Scholar]
- 21.Krissinel E, and Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions, Acta Crystallogr D Biol Crystallogr 60, 2256–2268. [DOI] [PubMed] [Google Scholar]
- 22.Krissinel E, and Henrick K (2007) Inference of macromolecular assemblies from crystalline state, Journal of molecular biology 372, 774–797. [DOI] [PubMed] [Google Scholar]
- 23.Maynes JT, Yuan RG, and Snyder FF (2000) Identification, expression, and characterization of Escherichia coli guanine deaminase, Journal of bacteriology 182, 4658–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christianson DW, and Cox JD (1999) Catalysis by metal-activated hydroxide in zinc and manganese metalloenzymes, Annual review of biochemistry 68, 33–57. [DOI] [PubMed] [Google Scholar]
- 25.Holm RH, Kennepohl P, and Solomon EI (1996) Structural and Functional Aspects of Metal Sites in Biology, Chemical reviews 96, 2239–2314. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch EF, and Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Vol. 3, Cold Spring Harbor Laboratory Press, Plainview, New York. [Google Scholar]
- 27.Seffernick JL, Dodge AG, Sadowsky MJ, Bumpus JA, and Wackett LP (2010) Bacterial ammeline metabolism via guanine deaminase, Journal of bacteriology 192, 1106–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almatarneh MH, Abu-Saleh AA-AA, and Elayan IA (2017) Mechanistic and spectral investigation on the deamination of ammeline and ammelide, Computational and Theoretical Chemistry 1117, 92–99. [Google Scholar]
- 29.Braekevelt E, Lau BP, Feng S, Ménard C, and Tittlemier SA (2011) Determination of melamine, ammeline, ammelide and cyanuric acid in infant formula purchased in Canada by liquid chromatography-tandem mass spectrometry, Food additives & contaminants. Part A, Chemistry, analysis, control, exposure & risk assessment 28, 698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobson RL, Motlagh S, Quijano M, Cambron RT, Baker TR, Pullen AM, Regg BT, Bigalow-Kern AS, Vennard T, Fix A, Reimschuessel R, Overmann G, Shan Y, and Daston GP (2008) Identification and characterization of toxicity of contaminants in pet food leading to an outbreak of renal toxicity in cats and dogs, Toxicological sciences : an official journal of the Society of Toxicology 106, 251–262. [DOI] [PubMed] [Google Scholar]
- 31.Hall RS, Agarwal R, Hitchcock D, Sauder JM, Burley SK, Swaminathan S, and Raushel FM (2010) Discovery and structure determination of the orphan enzyme isoxanthopterin deaminase, Biochemistry 49, 4374–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall RS, Fedorov AA, Marti-Arbona R, Fedorov EV, Kolb P, Sauder JM, Burley SK, Shoichet BK, Almo SC, and Raushel FM (2010) The hunt for 8-oxoguanine deaminase, Journal of the American Chemical Society 132, 1762–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egeblad L, Welin M, Flodin S, Gräslund S, Wang L, Balzarini J, Eriksson S, and Nordlund P (2012) Pan-Pathway Based Interaction Profiling of FDA-Approved Nucleoside and Nucleobase Analogs with Enzymes of the Human Nucleotide Metabolism, PLOS ONE 7, e37724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El Omari K, Liekens S, Bird LE, Balzarini J, and Stammers DK (2006) Mutations distal to the substrate site can affect varicella zoster virus thymidine kinase activity: implications for drug design, Mol Pharmacol 69, 1891–1896. [DOI] [PubMed] [Google Scholar]
- 35.Lee J, and Goodey NM (2011) Catalytic contributions from remote regions of enzyme structure, Chem Rev 111, 7595–7624. [DOI] [PubMed] [Google Scholar]
- 36.Oue S, Okamoto A, Yano T, and Kagamiyama H (1999) Redesigning the substrate specificity of an enzyme by cumulative effects of the mutations of non-active site residues, The Journal of biological chemistry 274, 2344–2349. [DOI] [PubMed] [Google Scholar]
- 37.Wrenbeck EE, Azouz LR, and Whitehead TA (2017) Single-mutation fitness landscapes for an enzyme on multiple substrates reveal specificity is globally encoded, Nat Commun 8, 15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hitchcock DS, Fedorov AA, Fedorov EV, Almo SC, and Raushel FM (2014) Discovery of a bacterial 5-methylcytosine deaminase, Biochemistry 53, 7426–7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manta B, Raushel FM, and Himo F (2014) Reaction Mechanism of Zinc-Dependent Cytosine Deaminase from Escherichia coli: A Quantum-Chemical Study, The Journal of Physical Chemistry B 118, 5644–5652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence alignment of guanine deaminase homologues
Figure S2. Overall guanine deaminase structure with β-strand and termini labeled
Figure S3. Gud1 binds zinc.
Figure S4. Confirmation that ligand bound in co-crystallization experiment is xanthine
Figure S5. Kinetics of EcGuaD and Gud1 for guanine and ammeline
Figure S6. E. coli guanine deaminase structure showing position of mutations made in this study
Figure S7. Steady-state kinetics of E. coli guanine deaminase mutants
Table S1. Structural homologues of EcGuaD and Gud1 identified by SSM server