Abstract
The role of dysbiosis in food allergy (FA) remains unclear. We found that dysbiotic fecal microbiota in FA infants evolved compositionally over time and failed to protect against FA in mice. Infants and mice with FA had decreased IgA and increased IgE binding to fecal bacteria, indicative of a broader breakdown of oral tolerance than hitherto appreciated. Therapy with Clostridiales species impacted by dysbiosis, either as a consortium or as monotherapy with Subdoligranulum variabile, suppressed FA in mice, as did a separate immunomodulatory Bacteroidales consortium. Bacteriotherapy induced regulatory T (Treg) cells expressing the transcription factor ROR-γt in a MyD88-dependent manner, which were deficient in FA infants and mice and ineffectively induced by their microbiota. Deletion of Myd88 or Rorc in Treg cells abrogated protection by bacteriotherapy. Thus, commensals activate a MyD88/ROR-γt pathway in nascent Treg cells to protect against FA, while dysbiosis impairs this regulatory response to promote disease.
Food allergy (FA) is a major public health concern 1. Most FA is acquired in the first years of life, indicating a critical role for early childhood exposures in disease pathogenesis. Factors impacting the gut microbiota, including method of delivery, antibiotic use and breastfeeding influence the development of atopic disease 2-6. Reduced bacterial diversity and an increased Enterobacteriaceae to Bacteroidaceae ratio in infancy have been associated with food sensitization, suggesting a role for altered gut microbiota in FA 7. Experimentally, germ-free (GF) mice cannot be orally tolerized to innocuous antigens, have reduced gut IgA and decreased IL-10-producing regulatory T (Treg) cells 8-10. Antibiotic treatment also increases food allergen sensitization 11. In contrast, colonization of GF mice with extended consortia of Clostridia species induces Treg cells 12, and protects against FA 11. Mice genetically prone to FA (Il4raF709 mice) exhibit dysbiotic fecal flora that promote FA 13. Commensals may suppress FA by producing short-chain fatty acids (SCFA), which elicit protective mucosal Treg cell responses and enhance intestinal barrier integrity 14-20.
Here, we identify an evolving dysbiosis in the gut microbiota of FA infants that impairs their capacity to protect against FA. Bacteriotherapy with culturable human-origin species from the order Clostridiales or Bacteroidales, prevents FA and suppresses established disease in Il4raF709 mice. Bacteriotherapy activates a MyD88-dependent microbial sensing pathway in nascent Treg cells that gives rise to disease-suppressing ROR-γt+ Treg cells 21,22, which are deficient in FA subjects and mice due to dysbiosis. These results identify a shared regulatory mechanism by which different commensals suppress FA, and underscore the potential for microbial therapies in treating this disorder.
Results
Patients with FA manifest early onset dynamic gut dysbiosis.
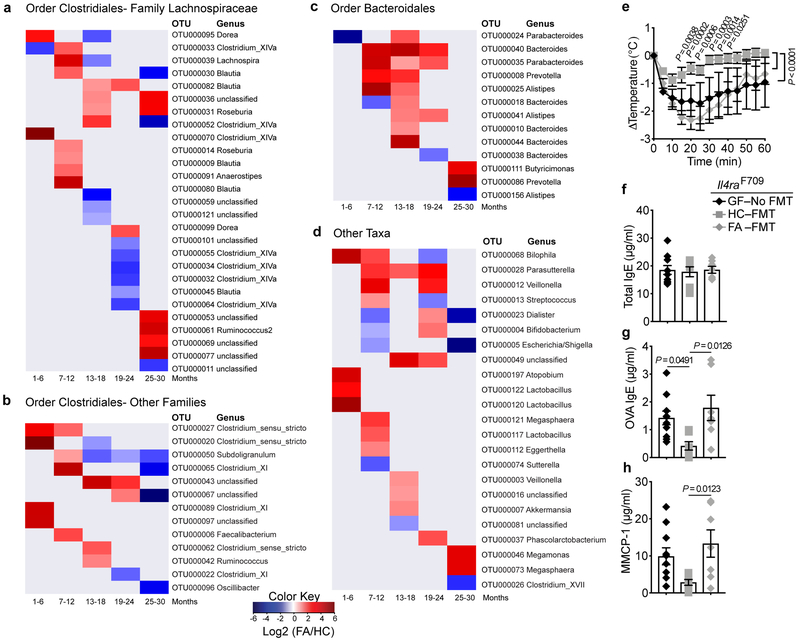
We analyzed the fecal microbiota of 56 infants with FA and 98 age matched controls recruited at 1–15 months of age and periodically sampled every 4–6 months for up to 30 months of age. Supplementary Table 1 summarizes subject demographics and Supplementary Fig. 1a summarizes the distribution of samples collected by subject age. FA versus healthy control (HC) subjects demonstrated no significant differences in the overall ecological diversity of the fecal microbiota as assessed by measures of alpha and beta diversity (Supplementary Fig. 1b,c). However, compositional differences in relative abundance among 77 Operational Taxonomic Units (OTUs) were observed in the fecal microbiome between age-stratified FA subjects and controls [False discovery rate (FDR)-adjusted p-value <0.1] (Fig. 1 and Supplementary Table 2). Differences for some taxa, such as OTU 50 [closest reference species (CRS) Subdoligranulum variabile] occurred across several age groups, while other taxa, including several Clostridiales species (clusters I, IV, XI and XIVa), showed significant differences in specific age groups. These associations in FA patients occurred even when controlling for factors including gender, mode of delivery for all age groups, and breastfeeding until 18 months of age, using multivariate statistical models. We also compared the gut microbiota of control subjects who were consuming milk products to those of FA patients who were tolerant and consuming milk but were allergic to other foods. When thus controlled for milk avoidance, most of the dysbiotic changes persisted (Supplementary Fig. 2 and Supplementary Table 3),
Fig. 1. FA infants exhibit an evolving gut dysbiosis.
(a-d) Heat map representations of log2 fold relative abundances of fecal bacterial taxa between FA and health control (HC) infants displayed across the different age groups: 1-6, 7-12, 3-18, 19-24, and 25-30 months. For detailed group description and subject characteristics, see Supplementary Figure 1 and Supplementary Table 1 and. Taxa represented included those from the order Clostridiales, family Lachnospiraceae (a), order Clostridiales, other Families (b), order Bacteroidales (c) and miscellaneous taxa (d). Blue represents higher abundance in control subjects, and red represents higher abundance in FA subjects. Taxonomic information is on the right side of the respective panel. (e) Core body temperature changes in GF Il4raF709 mice that were left uncolonized or reconstituted with FMT from HC or FA subjects, then sensitized with OVA/SEB and challenged with OVA (n=7 per group; each recipient mouse received FMT from one HC or FA subject). P values were derived by two-way ANOVA with Sidak post hoc analysis. (f,g) Total and OVA-specific serum IgE concentrations (n=7 per group, as in (e)). (h) MMCP-1 concentrations (n=7 per group, as in (e)). Results represent mean ± s.e.m. from two or three independent experiments. Each symbol represents one subject or mouse. For f-h, P values were derived by One-way ANOVA with Dunnett’s post hoc analysis.
The microbiota of FA subjects fail to protect against FA in a mouse disease model.
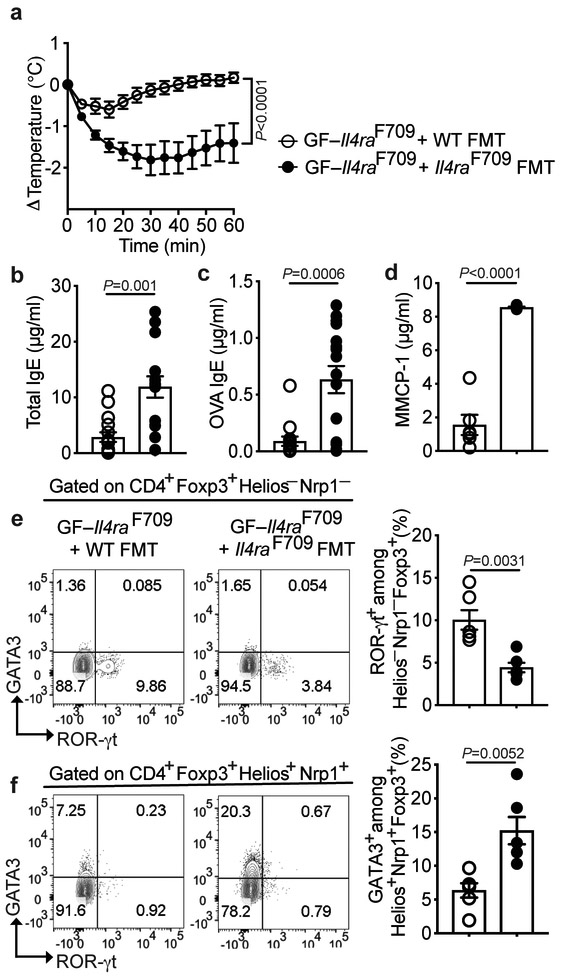
To assess the functional significance of dysbiosis in FA, adult GF Il4raF709 mice that were left either uncolonized or that received fecal microbiota transplants (FMT) from HC or FA infants, were sensitized with chicken egg ovalbumin (OVA) along with the mucosal adjuvant staphylococcal enterotoxin B (SEB) and subsequently challenged with OVA 13,23-25. GF Il4raF709 mice or those that received FMT from FA infants exhibited a rapid and sustained drop in their core body temperature, indicative of anaphylaxis, whereas mice that received donor microbiota from HC had a mild, transient drop (Fig. 1e). While the total serum IgE concentrations across the three groups were similar, induction of OVA-specific IgE was markedly decreased in mice receiving donor microbiota from HC subjects as compared to those receiving donor microbiota from FA subjects or that remained GF (Fig. 1f). Also, the increase in serum mouse mast cell protease 1 (MMCP1) concentrations post anaphylaxis was notably higher in Il4raF709 mice that were either GF or had received FMT from FA subjects as compared to those that had received FMT from healthy subjects (Fig. 1g). Thus, the capacity of the gut microbiota to protect against FA was profoundly impaired in FA as compared to HC subjects.
Il4raF709 mice also exhibit dysbiotic microbiota which, upon transfer into GF wild-type (WT) BALB/c mice, heightens their susceptibility to FA 13. We analyzed the capacity of microbiota derived from specific pathogen-free (SPF) WT mice, which are relatively resistant to FA induction, to rescue the FA phenotype of Il4raF709 mice. GF Il4raF709 mice that were reconstituted by FMT from WT mice then sensitized with OVA/SEB and challenged with OVA were protected from FA. In contrast, those reconstituted with SPF Il4raF709 mouse microbiota developed robust disease, as evidenced by a precipitous drop in core body temperature and increased serum MMCP1 concentrations post-anaphylaxis, with increased total and OVA-specific serum IgE concentrations (Extended data Fig. 1). These results confirmed that dysbiosis promotes FA in both human subjects and in mice.
Dysbiosis in FA is associated with an altered immune response to the gut microbiota.
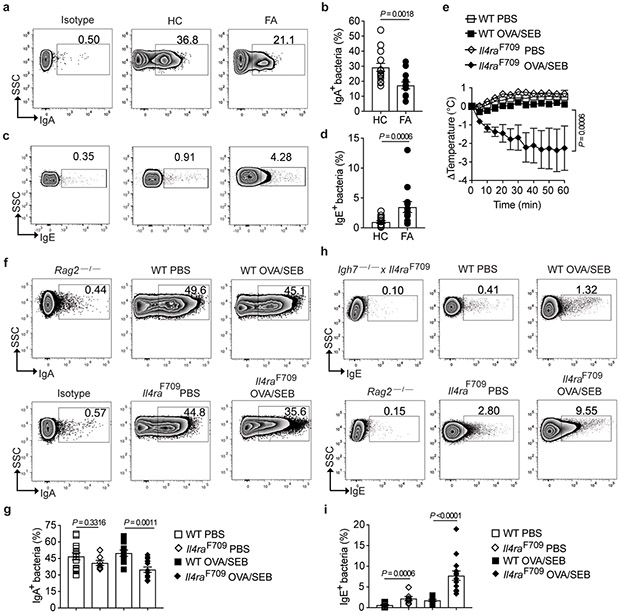
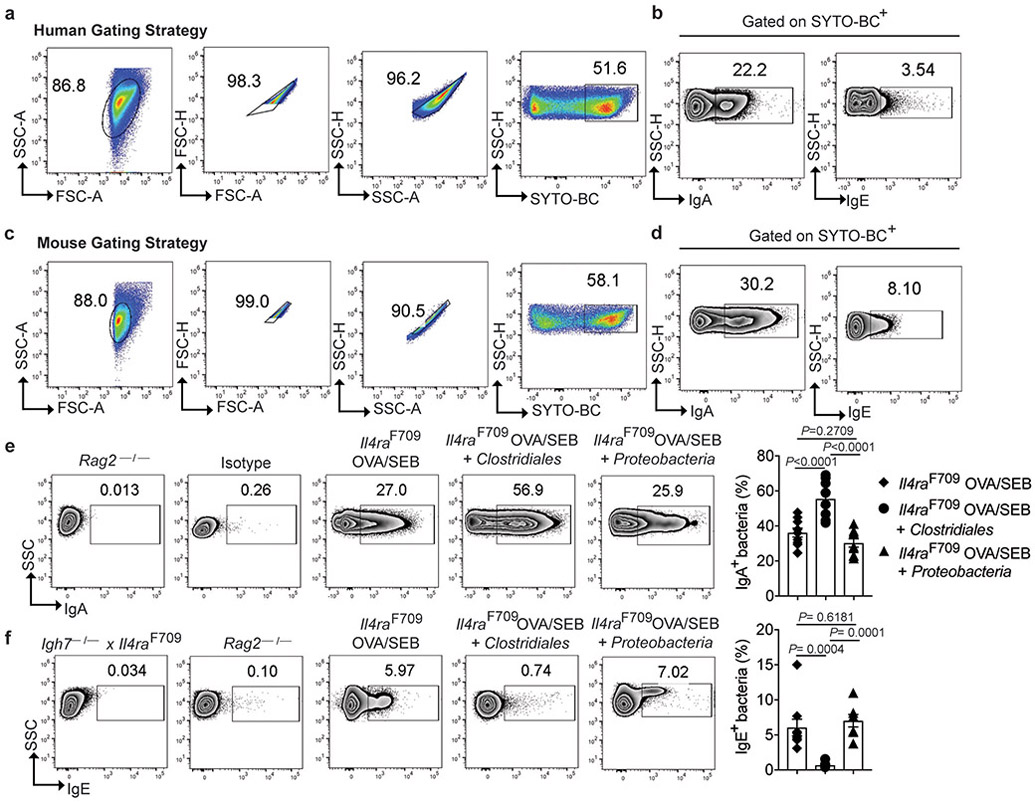
Secretory IgA (sIgA) shapes the composition of the microbiota and reinforces commensalism 26-30. We analyzed by flow cytometry the binding of sIgA to the fecal flora of FA and control subjects. The gating strategy for immunoglobulin staining of human fecal flora is demonstrated in Extended data Fig. 2a,b. FA subjects displayed decreased sIgA binding of their fecal bacteria as compared to control subjects (Fig. 2a,b). We further explored the hypothesis that FA involves dysregulated allergic responses to both food and bacteria by analyzing IgE binding to the fecal microbiota, which was found increased in FA as compared to HC subjects, indicative of an allergic response against commensals (Fig. 2c,d).
Fig. 2. Altered mucosal antibody responses to the gut microbiota in FA.
(a-d) Flow cytometric analysis and frequencies of human fecal bacteria of FA and HC subjects stained with a PE-conjugated isotype control mAb or mouse anti-human IgA (a and b) or IgE mAb (c and d). n= 15 HC and n=13 FA subjects for panel (b), and n=14 HC and n=13 FA subjects for panel (d). (e) Temperature changes in WT (n=10 mice per group) and Il4raF709 mice (n=7 per group) that have been either PBS or OVA/SEB-sensitized then challenged with OVA. (f-i) Flow cytometric analysis and frequencies of IgA (f and g) and IgE (h and i) staining of fecal bacteria of WT and Il4raF709 mice that were sensitized with PBS (n= 14 and 7 per group, respectively) or OVA/SEB (n=10 and 14 per group). Fecal pellets of Rag2−/− and Il4raF709Igh7−/− mice were used as negative controls for sIgA and IgE staining, respectively. Results represent mean ± s.e.m. from two or three independent experiments. Each symbol represents one subject or mouse. P values were derived by Student’s unpaired two tailed t test. (b,d,i), by repeat measures two-way ANOVA (e),. or by one-way analysis of variance (ANOVA) with Dunnett post hoc analysis (g).
We then analyzed the binding of sIgA and IgE to the fecal bacteria of Il4raF709 mice, following gating strategies shown in Extended data Fig. 2c,d 13,23-25,31. Il4raF709 mice and control WT mice were either sham sensitized with PBS or orally sensitized with OVA/SEB and subsequently challenged with OVA. OVA sensitized Il4raF709 but not WT control mice exhibited a rapid drop in their core body temperature, consistent with anaphylaxis (Fig. 2e). Similar to FA subjects, the fecal bacteria of OVA-sensitized Il4raF709 mice exhibited decreased sIgA binding as compared to similarly sensitized WT mice or sham sensitized Il4raF709 and WT mice (Fig. 2f,g). In addition, sensitization with OVA also resulted in an increased IgE binding to fecal bacteria of Il4raF709 mice, but not WT controls (Fig. 2h,i). These results were further confirmed by the lack of sIgA or IgE binding to fecal bacteria of Rag2-deficient mice, which do not express immunoglobulins, and the lack of IgE binding to the fecal bacteria of double mutant Igh7−/−Il4raF709 mice, which carry a targeted deletion of the IgE heavy chain gene (Fig. 2f-i) 23. Overall, these results established that dysbiosis in FA is associated with decreased sIgA responses and heightened T helper cell type 2 (Th2)/IgE responses to the commensal flora.
A defined consortium of human Clostridiales species promotes tolerance in experimental FA.
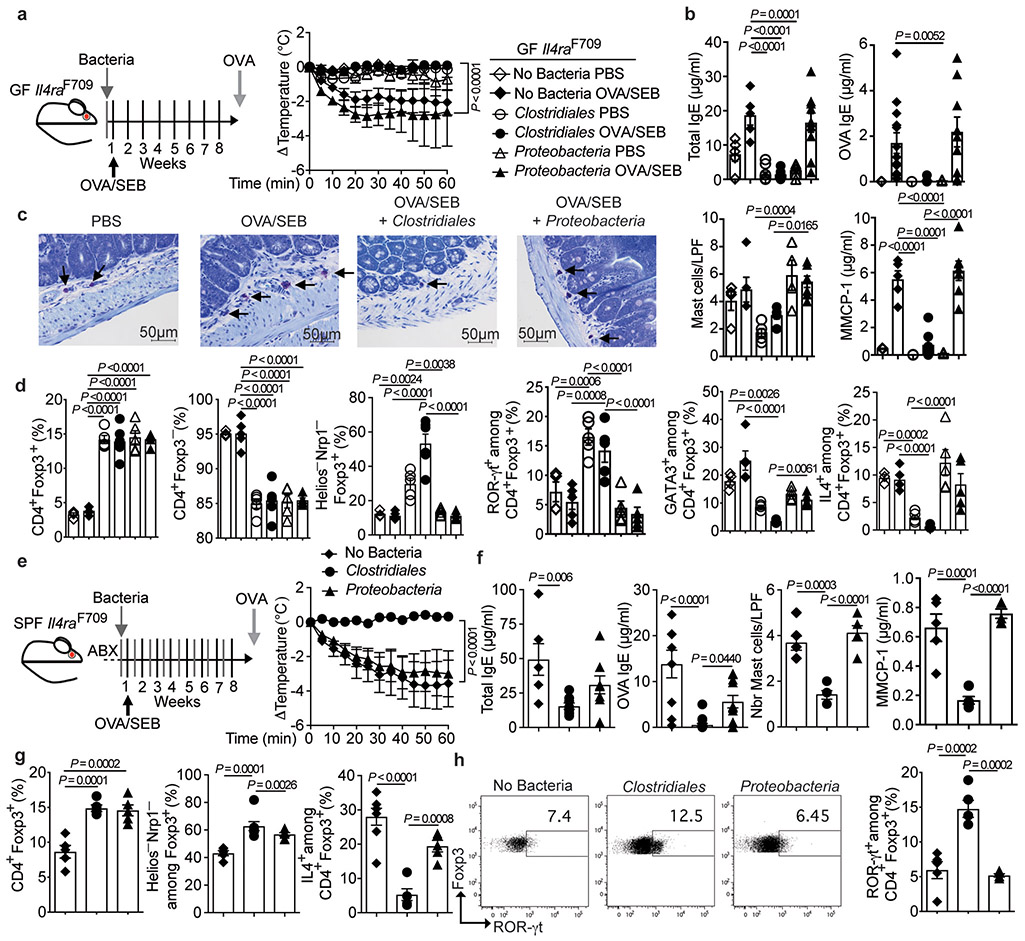
To test the hypothesis that Clostridiales taxa rendered deficient by the dysbiosis contribute to oral tolerance breakdown in FA, we examined the capacity of a defined consortium of six Clostridiales type strains, chosen as representative of Clostridiales clusters impacted by the dysbiosis in our human study, to suppress the induction of FA in Il4raF709 mice (Supplementary Table 4). Parameters driving selection of the consortium members included well-characterized genomic and metabolic profiles, prior data of in vivo effects on gut epithelium and/or immunomodulation and ease of culturability. The consortium included C. sardiniense (cluster I, e.g. OTU 20) 32, C. leptum (cluster IV, e.g. OTU 29, 50), C. hiranonis and C. bifermentans (cluster XI, e.g. OTU 22) 33, C. scindens (cluster XIVa, e.g. OTU 11) and C. ramosum (Erysipelatoclostridium ramosum) (Clostridium cluster XVIII, e.g. OTU 26) 34,35. As a negative control, we employed a consortium of species from gamma and delta Proteobacteria classes, including E. coli, P. mirabilis, K. oxytoca (Gammaproteobacteria; family Enterobacteriaceae), and B. wadsworthia (Deltaproteobacteria; family Desulfovibronaceae) (Supplementary Table 4). B. wadsworthia was increased early in life in FA subjects before declining, and E. Coli was decreased across multiple time windows (Fig. 1d and Supplementary Fig. 2d), The two other members of the Proteobacteria consortium have been implicated in gut dysbiosis associated with bowel inflammation 36.
In bacterial reconstitution studies, GF Il4raF709 mice that were either left sterile or colonized with the Proteobacteria consortium exhibited robust anaphylaxis upon OVA/SEB sensitization and OVA challenge, whereas those reconstituted with the Clostridiales consortium were fully protected (Fig. 3a). Measures of allergic sensitization and anaphylaxis, including the rise in serum concentrations of total and OVA-specific IgE, small intestinal tissue mastocytosis and the increase in serum MMCP1 concentrations post anaphylaxis, all of which were elevated in GF and Proteobacteria-supplemented mice, were inhibited by the Clostridiales consortium (Fig. 3b,c).
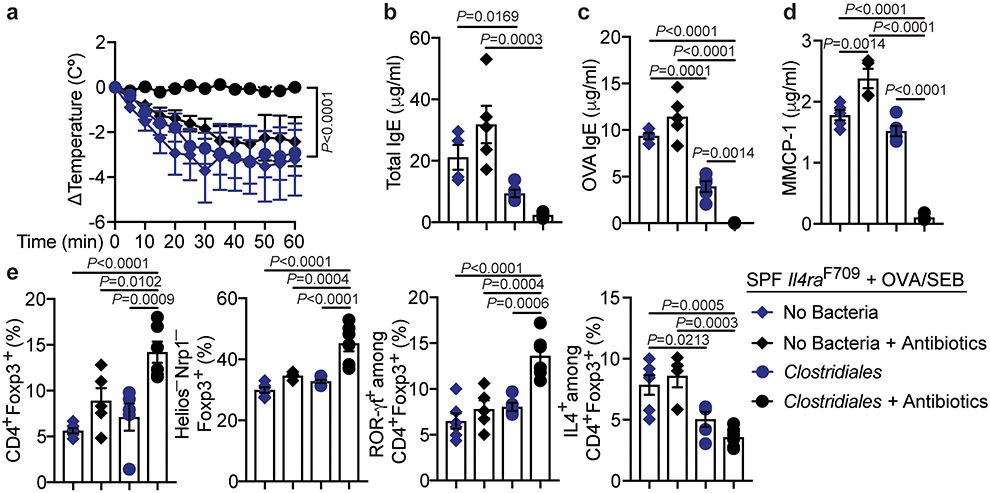
Fig. 3. A consortium of Clostridiales species prevents FA.
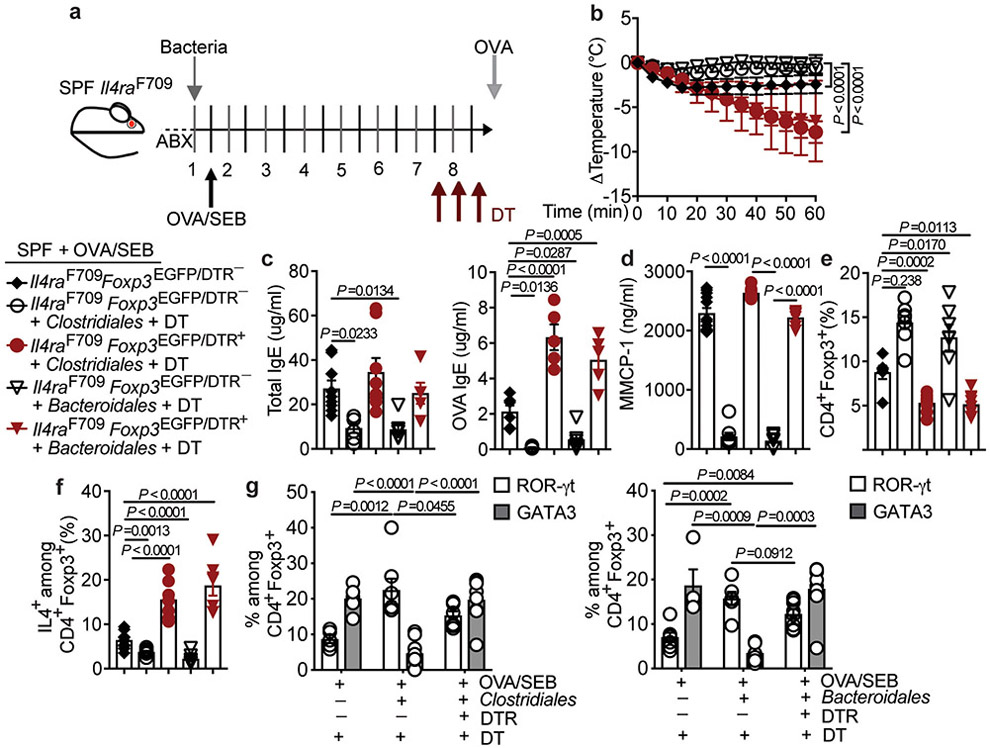
(a) Left: Experimental schema. Right: temperature changes in GF Il4raF709 mice that were colonized and sensitized as indicated then challenged with OVA. GF, PBS (n=5 mice), GF, OVA/SEB (n=10), Clostridiales, PBS (n=5), Clostridiales, OVA/SEB (n=6), Proteobacteria, PBS (n=8), Proteobacteria, OVA/SEB (n=5). (b) Total and OVA-specific IgE concentrations. GF, PBS (n=5 and 3), GF, OVA/SEB (n=5 and 13), Clostridiales, PBS (n=5 each), Clostridiales, OVA/SEB (n=6 and 7), Proteobacteria, PBS (n=5 and 4), Proteobacteria, OVA/SEB (n=10 and 9). (c) Jejunal mast cells (arrows) and counts per low powered field (LPF) and MMCP-1 concentrations. GF, PBS (n=5 per group), GF, OVA/SEB (n=5 per group), Clostridiales, PBS (n=5 per group), Clostridiales, OVA/SEB (n=5 and 7), Proteobacteria, PBS (n=4 and 5), Proteobacteria, OVA/SEB (n=7 per group). (d) Frequencies of MLN Treg and Teff cell populations (n=5 per group). (e) Left: Experimental schema; Abx: antibiotics. Right: temperature changes in OVA/SEB-sensitized and OVA-challenged Il4raF709 mice treated as indicated. No bacteria (n=6), Clostridiales (n=7), Proteobacteria, (n=6). (f) Total and OVA-specific IgE. No bacteria (n=6 and 9), Clostridiales (n=8 and 17), Proteobacteria, (n=8 and 11); mast cell counts (n=5 per group) and MMCP-1 concentrations (n=5 per group). (g) Frequencies of MLN Treg cell populations in the respectively treated mice: CD4+Foxp3+ (n=5 per group), Helios−NRP1−Foxp3+ (n=, 5, 7 and 6) and IL-4+Foxp3+ (n=6, 5 and 5). (h) Analysis of ROR-γt+Foxp3+ Treg cells (n=5 per group). Results represent mean ± s.e.m. from three independent experiments. Each symbol represents one mouse. P values were derived by repeat measures two-way ANOVA (a,e), or by one-way ANOVA with Dunnett post hoc analysis (c-d,f-h).
Whereas treatment of GF Il4raF709 mice with either the Clostridiales or Proteobacteria consortia increased the frequencies of CD4+Foxp3+ Treg cells in the mesenteric lymph nodes (MLN), only the Clostridiales consortium boosted the frequencies of iTreg cells, distinguished by their low expression of the markers Helios and neuropilin1 (Helios−Nrp1−), whose specificity is biased towards recognizing gut luminal antigens originating from food or bacteria (Fig. 3d) 37-39. Treatment also increased the frequency of ROR-γt+ Treg cells, predominantly iTreg cells with low expression of Helios (Supplementary Fig. 3) 40-43. These cells have been implicated in the control of different gut Th cell-mediated immune responses (Fig. 3d) 21,22,44. We have previously established that FA induction is associated with the reprogramming of Treg cells into Th2 cell-like cells that promote disease 25,45. Consistent with these findings, a subset of MLN Treg cells of sham and OVA-sensitized GF mice exhibited increased expression of the Th2 master transcription factor GATA3 and the Th2 cytokine IL-4. These Th2 cell-like Treg cells appeared to be thymus-derived as reflected by their Helioshigh phenotype (Supplementary Fig. 3) 40-43. A similar increase in Treg cell IL-4 expression was found in mice colonized with the Proteobacteria consortium (Fig. 3d). In contrast, the Clostridiales consortium suppressed the Th2 cell-like reprogramming of Treg cells and instead promoted the expression of ROR-γt in Treg cells independent of OVA sensitization, a gut Treg cell phenotype proposed to regulate Th2 cell responses (Fig. 3d) 21. Overall, these results established that the Clostridiales consortium could confer protection against FA in GF Il4raF709 mice independent of other bacterial species.
To determine whether the protection by the Clostridiales consortium against FA extended to mice colonized with conventional microbiota, SPF Il4raF709 mice were treated for one week with antibiotics to create a niche for the therapeutic consortia, then sensitized with OVA/SEB with bacteriotherapy administered in tandem (Fig. 3e). The Clostridiales consortium completely protected OVA/SEB sensitized Il4raF709 mice from developing anaphylaxis upon oral challenge with OVA (Fig. 3e). Total and OVA-specific serum IgE responses, gut tissue mast cell expansion and serum MMCP1 concentrations post challenge were also sharply curtailed (Fig. 3f). In contrast, mice treated with Proteobacteria were not protected. Clostridiales therapy increased Treg cells in the MLN, reflective of increased frequencies of Helios−Nrp1− cells (Fig. 3g). The Clostridiales but not the Proteobacteria suppressed the Th2 cell-like reprogramming of gut Treg cells, as evidenced by their decreased IL-4 expression, consistent with improved Treg cell function, while increasing the frequencies of the ROR-γt+ iTreg cells (Fig. 3g,h and Supplementary Fig. 3). Similar results were found in the small intestinal lamina propria lymphocytes (LPL) of Clostridiales-treated Il4raF709 mice (Supplementary Fig. 4). Of note, prior antibiotic treatment of the mice markedly improved the therapeutic efficacy of the Clostridiales consortium, suggesting that it might serve to enhance the immunomodulatory functions of the consortium by reducing the abundance of interfering bacteria (Extended data Fig. 3).
Treatment with the Clostridiales but not Proteobacteria consortium resulted in an increased sIgA response to the microbiota (Extended data Fig. 2e). Reciprocally, the Clostridiales but not Proteobacteria consortium suppressed the IgE anti-bacterial response (Extended data Fig. 2f). These findings suggested that the Clostridiales consortium normalized the aberrant mucosal immune response to the gut microbiota in FA Il4raF709 mice.
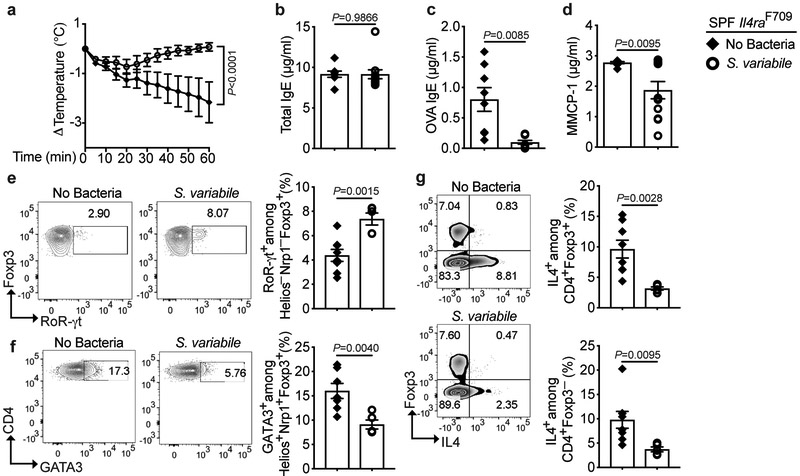
Of the Clostridial taxa affected by dysbiosis in FA, OTU 50 (CRS Subdoligranulum variabile), stood out as being decreased in FA subjects age 1 year and older, including those that were milk tolerant, suggesting that its deficiency may act as a switch for FA. Monobacterial therapy with Subdoligranulum variabile, carried out following the protocol shown in Fig. 3,e, protected SPF Il4raF709 mice from developing FA, albeit less stringently than the Clostridiales consortium, in association with the induction of ROR-γt+ iTreg cells (Extended data Fig 4). In contrast, therapy with species that were increased in FA subjects, including CRS Bilophila wadsworthia (OTU 68) and CRS Veillonela ratti (OTU 12) (Fig. 1), failed to protect Il4raF709 mice from developing FA (data not shown). Thus, the loss of key immunomodulatory bacteria may promote FA.
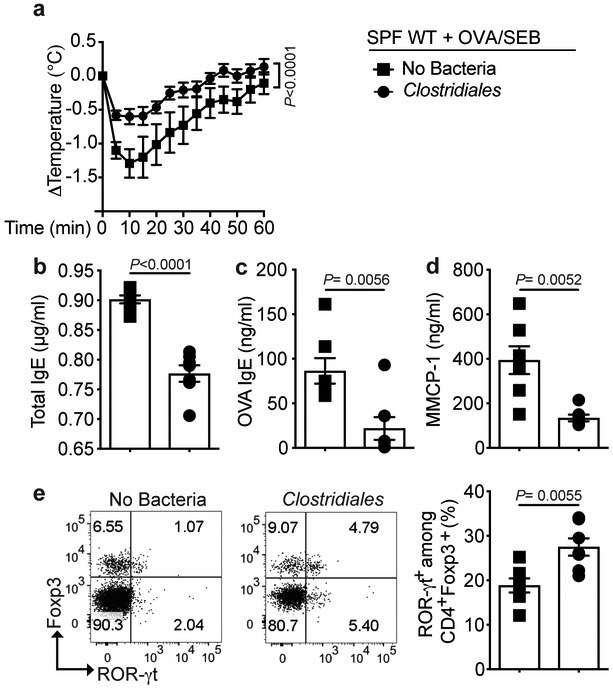
We also examined the capacity of the Clostridiales consortium to protect FA induced by epicutaneous sensitization of WT mice with OVA, a model of food sensitization in human subjects with eczema 46. Treatment with the Clostridiales consortium greatly attenuated the induction of FA via the epicutaneous sensitization route in association with the induction of ROR-γt+ iTreg cells, indicating that bacteriotherapy was protective against FA induced by different routes of allergen sensitization (Extended data Fig 5).
Promotion of oral tolerance in FA by immunomodulatory human Bacteroidales species.
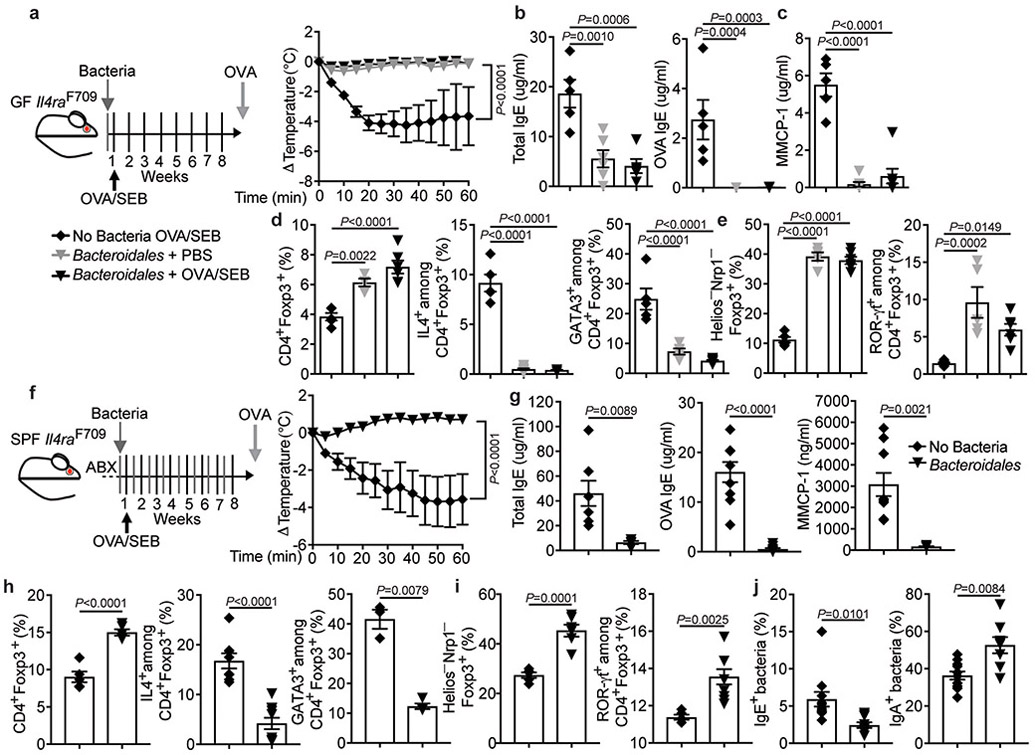
To determine if phylogenetically distinct bacteria could protect against FA, we tested another defined consortium comprised of five human-origin immunomodulatory Bacteroidales species, including B. fragilis, B. ovatus, B. vulgatus, P. melaninogenica, and P. distasonis (OTU24, CRS P. distasonis) 22,47-50. Treatment with the Bacteroidales consortium completely protected against the induction of FA in GF Il4raF709 mice upon their sensitization with OVA/SEB and challenge with OVA (Extended data Fig 6a-e). Furthermore, the Bacteroidales consortium protected SPF Il4raF709 mice from developing FA when given in tandem during the OVA/SEB sensitization (Extended data Fig 6f-i). Similar to the Clostridiales consortium, the Bacteroidales consortium increased sIgA binding and suppressed IgE binding to the fecal bacteria of treated FA Il4raF709 mice (Extended data Fig 6j).
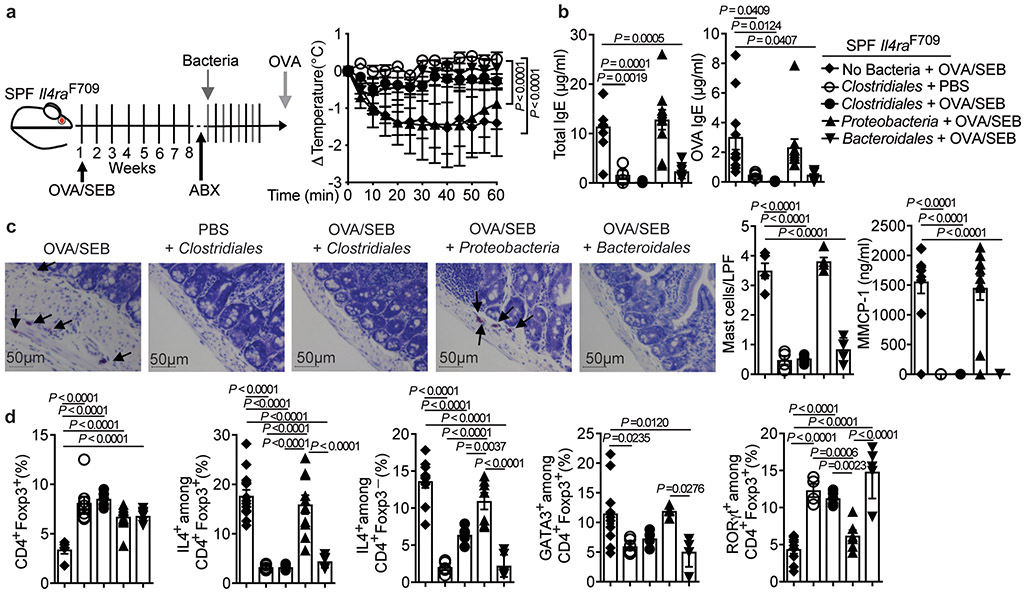
To determine whether bacteriotherapy could suppress FA once the disease was established, SPF Il4raF709 mice were sensitized with OVA/SEB, and, following a short course of antibiotics, further sensitized for an additional 4 weeks without or with bacteriotherapy with the respective consortia. The Clostridiales and Bacteroidales, but not the Proteobacteria, prevented the sensitized Il4raF709 mice from reacting to the OVA challenge (Fig. 4a). They also suppressed the total and OVA-specific serum IgE responses, the rise in serum MMCP-1 post anaphylaxis, and the mast cell expansion (Fig. 4b,c). While all bacterial consortia increased the frequencies of MLN Treg cells in this disease curative model (Fig. 4d), only the Clostridiales and Bacteroidales consortia suppressed the FA-associated Treg cell Th2 cell-like reprogramming and increased the frequency of ROR-γt+ Treg cells (Fig. 4d and Supplementary Fig. 3).
Fig. 4. Clostridiales and Bacteroidales consortia suppresses established FA.
(a) Left: experimental scheme. Right: temperature changes following OVA challenge in Il4raF709 mice sensitized and treated as follows: OVA/SEB, no bacteria (n=8 mice); PBS, Clostridiales (n=6) and OVA/SEB, Clostridiales (n=10), Bacteroidales (n=5) or Proteobacteria (n=5). (b) Total and OVA-specific IgE concentrations [OVA/SEB, no bacteria (n=10 per group); PBS, Clostridiales (n=4 and 5 per group, respectively) and OVA/SEB, Clostridiales (n=5 per group), Bacteroidales (n=7 and 5 per group) or Proteobacteria (n=10 and 11 per group). (c) Jejunal mast cells (arrows), mast cell counts per LPF [OVA/SEB, no bacteria (n=5); PBS, Clostridiales (n=4) and OVA/SEB, Clostridiales, Bacteroidales or Proteobacteria (n=5 per group)], MMCP-1 concentrations [OVA/SEB, no bacteria (n=10); PBS, Clostridiales (n=5) and OVA/SEB, Clostridiales (n=5), Bacteroidales (n=6) or Proteobacteria (n=11)]. (d) Frequencies of MLN CD4+Foxp3+ and IL-4+CD4+Foxp3+ Treg cells [OVA/SEB-, no bacteria (n=5 and 14); PBS, Clostridiales (n=8 and 5) and OVA/SEB, Clostridiales (n=5 per group), Bacteroidales (n=6 and 5) or Proteobacteria (n=12 and 10)], IL-4+CD4+Foxp3− T cells [OVA/SEB, no bacteria (n=10); PBS, Clostridiales (n=5) and OVA/SEB, Clostridiales (n=6), Bacteroidales (n=6) or Proteobacteria (n=7)], GATA3+Foxp3+ T cells [OVA/SEB, no bacteria (n=10 mice); PBS, Clostridiales (n=6) and OVA/SEB, Clostridiales (n=4), Bacteroidales (n=5) or Proteobacteria (n=4)], and ROR-γt+Foxp3+ T cells [OVA/SEB, no bacteria (n=9); PBS, Clostridiales (n=4) and OVA/SEB, Clostridiales (n=5), Bacteroidales (n=5) or Proteobacteria (n=6)]. Results represent mean ± s.e.m. from three independent experiments. Each symbol represents one mouse. P values were derived by repeat measures two-way ANOVA (a), or by one-way ANOVA with Dunnett post hoc analysis (b-d).
In vivo Treg cell depletion ablates the protective effects of bacterial therapy in FA.
We next examined the role of Treg cells in the protection against FA by the Clostridiales and Bacteroidales consortia using Il4raF709Foxp3EGFP/DTR+ mice, which express the diphtheria toxin (DT) receptor on their Treg cells, allowing for their depletion upon treatment of the mice with DT 51,52. In OVA/SEB-sensitized mice, injection of DT thrice over the last two weeks prior to challenge with OVA abrogated the protection against anaphylaxis imparted by both consortia (Extended data Fig 7a-d). In OVA/SEB-sensitized Il4raF709Foxp3EGFP/DTR+ mice treated with the respective consortia, co-treatment with DT reduced the frequencies of MLN Treg cells and rendered them Th2 cell-like with increased IL-4 production (Extended data Fig 7e,f), while reducing the expansion of ROR-γt+ Treg cells in favor of GATA3+ Treg cells (Extended data Fig 7g). These findings were recapitulated by Treg cell depletion using an anti-CD25 monoclonal antibody (mAb), which abrogated the suppression by the Clostridiales consortium of established FA in Il4raF709 mice (Supplementary Fig 5a-d). Anti-CD25 mAb treatment decreased the MLN Treg cells and suppressed the Clostridiales -induced ROR-γt+ iTreg cell skewing in favor of IL-4 and GATA3 expressing Treg cells (Supplementary Fig 5e-g).
Oral Supplementation with SCFA does not protect against FA.
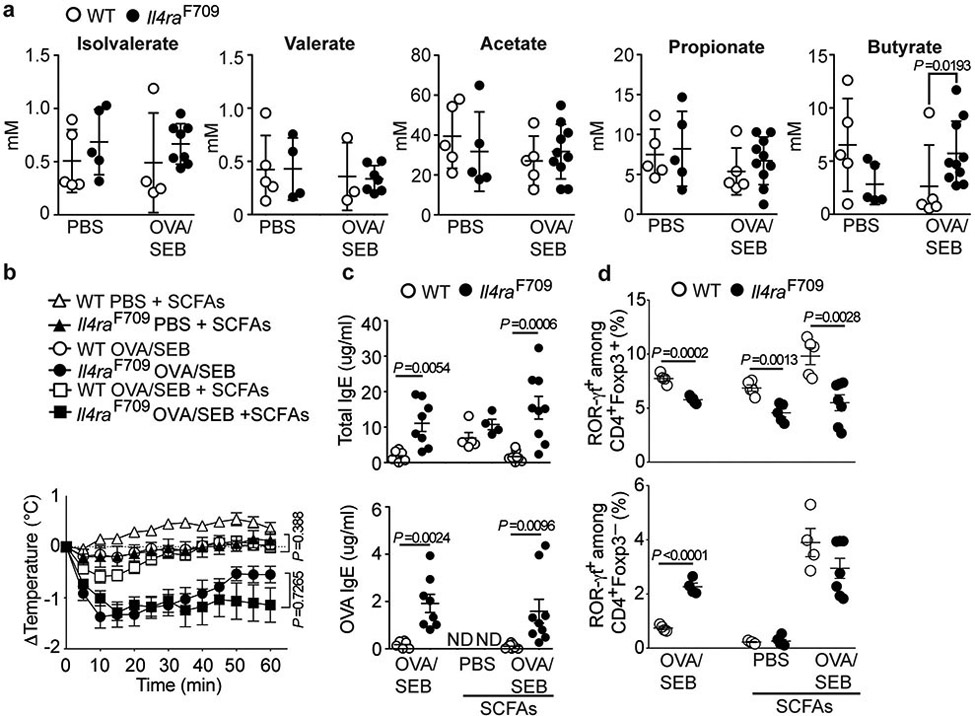
We examined the role of SCFA in the protection by the bacterial consortia against FA. WT and Il4raF709 mice had similar fecal concentrations of acetate, propionate, valerate and isovalerate, while butyrate was increased in the Il4raF709 mice (Extended data Fig 8a). Furthermore, there was no clear correlation between SCFA production by the bacterial consortia when introduced into GF Il4raF709 mice and protection against FA. All three consortia produced similar amounts of acetate. The Bacteroidales consortium selectively produced propionate but not butyrate, as previously reported 53, while butyrate was low to absent in mice reconstituted with the Clostridiales consortium, reflecting its poor production by some consortium members (Supplementary Table 5) 35.
The capacity of SCFA to protect against FA in Il4raF709 mice was tested by supplementing the drinking water with a mixture of acetate, propionate, and butyrate, given at 150 mM each, throughout the sensitization period. However, SCFA therapy failed to protect the mice against FA (Extended data Fig 8b,c). It induced increased numbers of proliferating gut Treg cells in OVA/SEB sensitized WT but not Il4raF709 mice (Supplementary Fig 6a-d). It also failed to increase the frequency of MLN ROR-γt+ Treg cells in Il4raF709 mice (Extended data Fig 8d).
We investigated the persistence of consortia species in Il4raF709 mice by real-time PCR analysis of fecal samples using species-specific primers (Supplementary Table 6). None of the Clostridial species were detected at baseline, and they were only transiently detected when individually introduced by gavage (Supplementary Fig. 7a). Similarly, they were not detected following bacteriotherapy for FA prevention (Supplementary Fig. 7b). The Bacteroidales were also not detected at baseline, but several persisted following a single gavage and also by the end of a FA prevention course, as did some Proteobacteria (Supplementary Fig. 8). Heat inactivation of the Clostridiales and Bacteroidales consortia abrogated protection against FA, indicating a requirement for bacterial viability for therapeutic efficacy (Supplementary Fig. 9 and data not shown).
Protection against FA by the commensal bacteria is dependent on ROR-γt+ Treg cells.
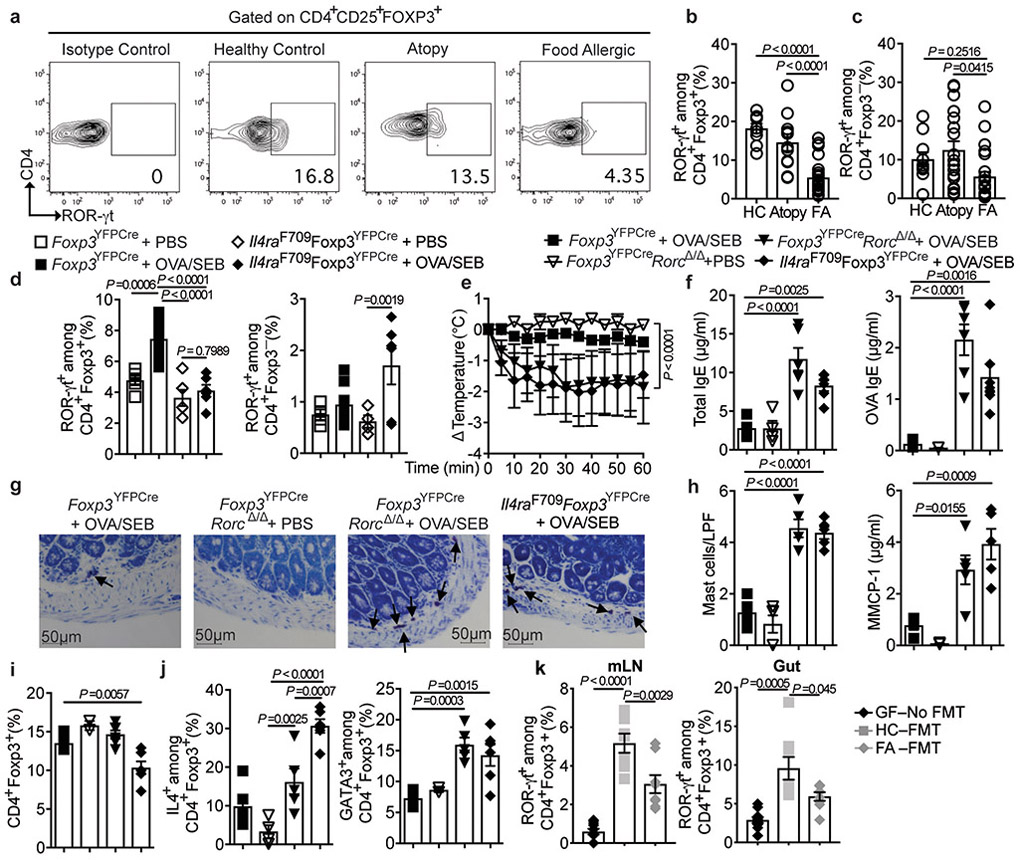
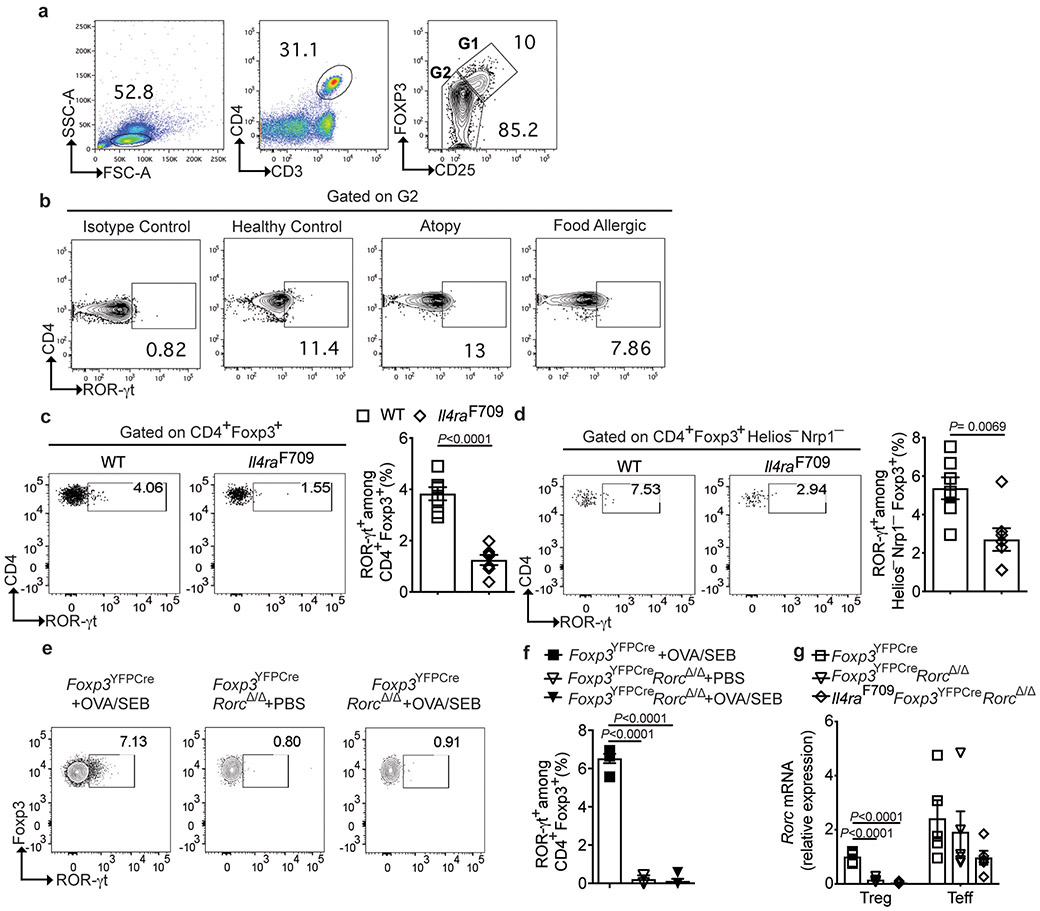
As both the Clostridiales and Bacteroidales consortia increased the frequencies and numbers of gut ROR-γt+ Treg cells in Il4raF709 mice, we examined the frequencies of circulating ROR-γt+ Treg cells in human FA subjects, and found them decreased as compared to atopic subjects without FA or to non-atopics (Fig. 5a,b and Supplementary Table 7). In contrast, the frequencies of circulating ROR-γt+ T effector (Teff) cells were not significantly different between FA and control subjects and slightly increased in atopics (Fig. 5c and Extended data Fig 9a,b). Similarly, circulating ROR-γt+ Treg cells were decreased at steady state in Il4raF709 mice (Extended data Fig 9c). OVA/SEB sensitization did not result in increased induction of ROR-γt+ Treg cells in the MLN of FA Il4raF709 mice as compared to WT controls (Fig. 5d and Extended data Fig 9c,d).
Fig. 5. ROR-γt+ Treg cell deficiency promotes FA.
(a-c) Flow cytometric analysis and frequencies of circulating ROR-γt+Foxp3+ Treg cells and ROR-γt+Foxp3− T cells in HC (n=10), atopy (n=11) and FA (n=22) subjects. (d) Frequencies of MLN ROR-γt+Foxp3+ and ROR-γt+Foxp3− T cells in the following PBS- or OVA/SEB-sensitized mouse groups: Foxp3YFPCre, PBS (n=5), Foxp3YFPCre, OVA/SEB (n=9) and Il4raF709Foxp3YFPCre, PBS or OVA/SEB (n=6 per group). (e) Temperature changes in the respective OVA-challenged mouse groups sensitized as follows: Foxp3YFPCre, OVA/SEB (n=6), Foxp3YFPCreRorcΔ/Δ PBS or OVA/SEB (n=5 per group) and Il4raF709Foxp3YFPCre OVA/SEB (n=5). (f) Total and OVA-specific IgE responses [Foxp3YFPCre OVA/SEB (n=5 per group), Foxp3YFPCreRorcΔ/Δ PBS (n=5 per group) or OVA/SEB (n=6 per group) and Il4raF709Foxp3YFPCre OVA/SEB (n=6 per group). (g) Jejunal mast cell staining (arrows), representative of two experiments. (h), mast cell numbers/LPF (n=5 mice per group) and MMCP1 concentrations [Foxp3YFPCre OVA/SEB (n=4), Foxp3YFPCreRorcΔ/Δ PBS (n=4), Foxp3YFPCreRorcΔ/Δ OVA/SEB (n=5) and Il4raF709Foxp3YFPCre OVA/SEB (n=5)]. (i) Frequencies of MLN CD4+Foxp3+ T cells [Foxp3YFPCre OVA/SEB (n=5), Foxp3YFPCreRorcΔ/Δ PBS (n=5), Foxp3YFPCreRorcΔ/Δ OVA/SEB (n=6) and Il4raF709Foxp3YFPCre OVA/SEB (n=6)]. (j), Frequencies of MLN IL-4+Foxp3+ and GATA3+Foxp3+ T cells [Foxp3YFPCre OVA/SEB (n=5), Foxp3YFPCreRorcΔ/Δ PBS (n=5), Foxp3YFPCreRorcΔ/Δ OVA/SEB (n=5) and Il4raF709Foxp3YFPCre OVA/SEB (n=6)]. (k) Frequencies of ROR-γt+Foxp3+ T cells in the MLN and small intestinal LPL of OVA/SEB-sensitized and OVA challenged GF Il4raF709 mice without (n=10) or with FMT from HC or FA subjects (n=7 per group). Results represent mean ± s.e.m. from two independent experiments. Each symbol represents one mouse or subject. P values were derived by one-way ANOVA with Dunnett post hoc analysis (b-d,f,h-k), or by repeat measures two-way ANOVA (e).
We next examined the consequences of Treg cell-specific deletion of Rorc 44, which encodes ROR-γt, in promoting FA. Treg cells of Foxp3YFPCreRorcΔ/Δ mice were profoundly deficient in ROR-γt expression as compared to those of Foxp3YFPCre mice, reflecting their specific loss of Rorc mRNA (Extended data Fig 9e-g). Foxp3YFPCreRorcΔ/Δ mice sensitized with OVA/SEB and challenged with OVA developed a vigorous anaphylactic response that was comparable to that of similarly treated Il4raF709 mice. In contrast, Foxp3YFPCre mice were resistant to FA induction (Fig. 5g,h). Also, and similar to Il4raF709 mice, Foxp3YFPCreRorcΔ/Δ mice exhibited decreased sIgA and increased IgE binding to fecal bacteria, consistent with the dysregulation of the mucosal immune response (Extended data Fig 10a-d). Treg cell-specific Rorc deletion did not affect the frequency or numbers of Treg cells in the MLN of OVA/SEB sensitized Foxp3YFPCreRorcΔ/Δ mice, but the Treg cells exhibited increased GATA3 and IL-4 expression, similar to those of Il4raF709 mice (Fig. 5f-j) 25.
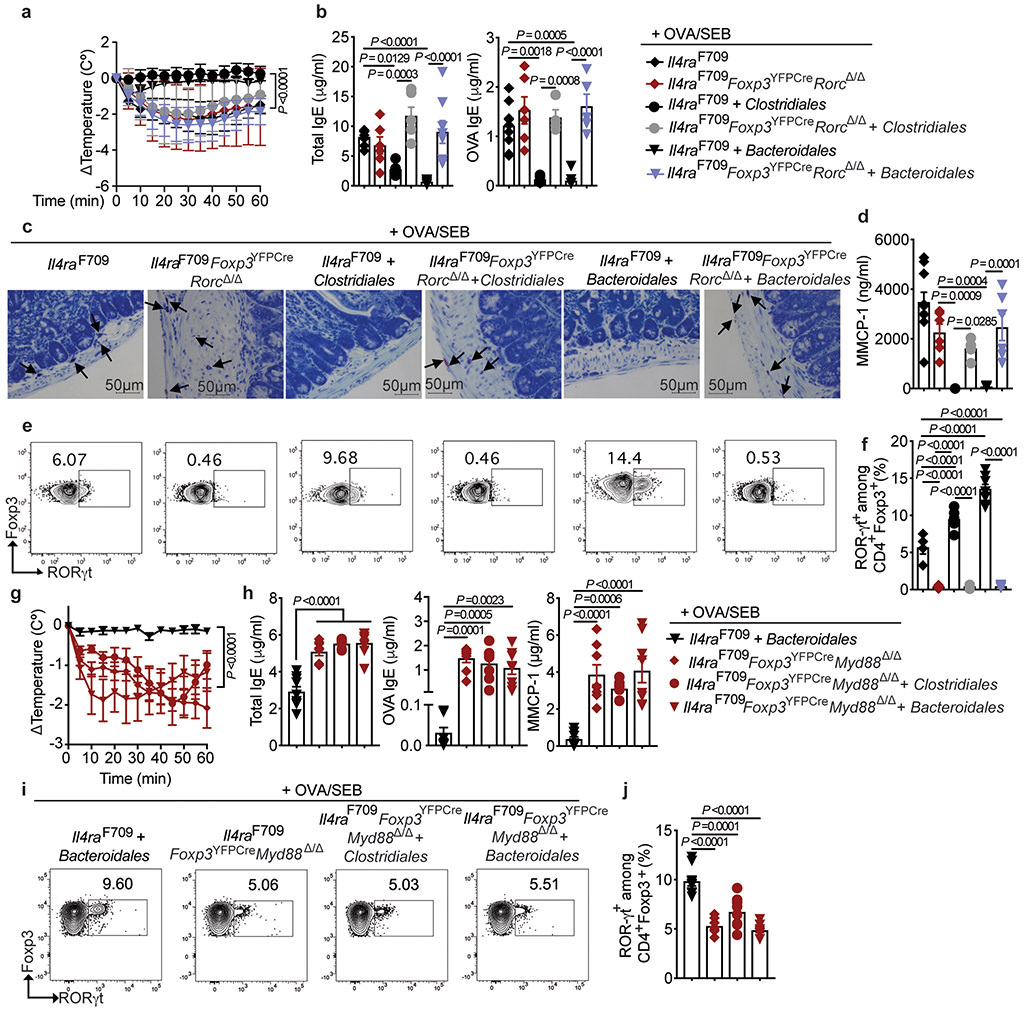
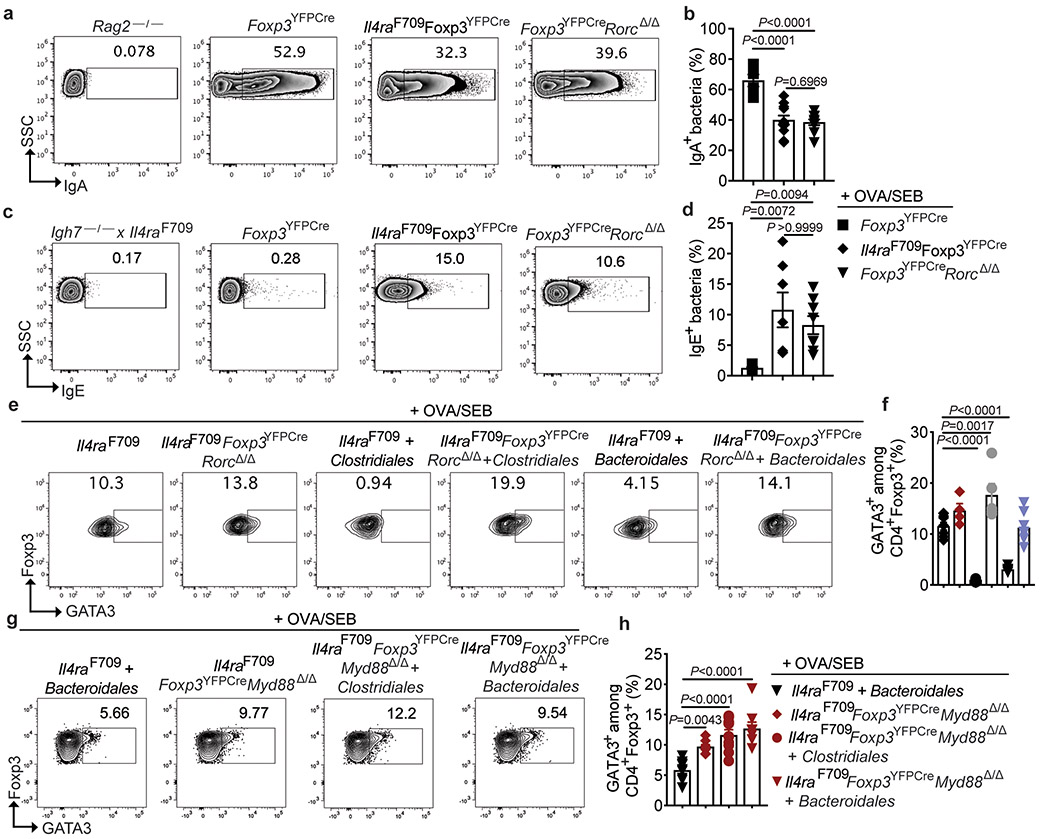
The role of ROR-γt+ Treg cells in mediating protection by the Clostridiales and Bacteroidales consortia in FA was further established by demonstrating that Treg cell-specific deletion of Rorc in Il4raF709 mice (Il4raF709Foxp3YFPCreRorcΔ/Δ), which depleted Rorc mRNA expression specifically in this cell compartment (Extended data Fig 9g), abrogated this protection (Fig. 6a-d). As expected, the Clostridiales and Bacteroidales consortia induced ROR-γt+ Treg cells in Rorc-sufficient Il4raF709Foxp3YFPCre but not Il4raF709Foxp3YFPCreRorcΔ/Δ mice (Fig. 6e,f), with the latter mice exhibiting Th2 cell-like skewing of their gut Treg cells (Extended data Fig 10e,f).
Fig. 6. Protection against FA by commensals requires ROR-γt+ Treg cells.
(a) Temperature changes in the respective OVA-challenged mouse groups sensitized as follows: Il4raF709 OVA/SEB (n=9), OVA/SEB, Clostridiales (n=5), and OVA/SEB, Bacteroidales (n=7); Il4raF709 Foxp3YFPCreRorcΔ/Δ OVA/SEB (n=8), OVA/SEB, Clostridiales (n=6) and OVA/SEB, Bacteroidales (n=10). (b) total and OVA-specific IgE [Il4raF709 OVA/SEB (n=9 and 5), OVA/SEB, Clostridiales (n=5 per group), and OVA/SEB, Bacteroidales (n=9 and 8); Il4raF709 Foxp3YFPCreRorcΔ/Δ OVA/SEB (n=6 per group), OVA/SEB, Clostridiales (n=6 and 4) and OVA/SEB, Bacteroidales (n=8 and 5)]. (c,d) Jejunal mast cells (arrows), representative of two experiments (c), and MMCP-1 concentrations (d) [Il4raF709 OVA/SEB (n=10), OVA/SEB, Clostridiales (n=5), and OVA/SEB, Bacteroidales (n=8); Il4raF709 Foxp3YFPCreRorcΔ/Δ OVA/SEB (n=6), OVA/SEB, Clostridiales (n=6) and OVA/SEB, Bacteroidales (n=6). (e,f) Analysis of MLN ROR-γt+Foxp3+ Treg cells, representative of two experiments [Il4raF709 OVA/SEB (n=8), OVA/SEB, Clostridiales (n=5), and OVA/SEB, Bacteroidales (n=9); Il4raF709 Foxp3YFPCreRorcΔ/Δ OVA/SEB (n=5), OVA/SEB, Clostridiales (n=6) and OVA/SEB, Bacteroidales (n=6). (g) Temperature changes in the following OVA/SEB-sensitized and OVA-challenged mouse groups: Il4raF709Foxp3YFPCre, Bacteroidales (n=14), and Il4raF709Foxp3YFPCreMyd88Δ/Δ mice untreated (n=8) or treated with Clostridiales (n=7) or Bacteroidales (n=8). (h) Total and OVA-specific IgE and MMCP1 concentrations [Il4raF709Foxp3YFPCre, Bacteroidales (n=9 per group), Il4raF709Foxp3YFPCreMyd88Δ/Δ mice untreated (n=8 per group) or treated with Clostridiales (n=7 per group) or Bacteroidales (n=8 per group). (I,j) Analysis of MLN ROR-γt+Foxp3+ Treg cells in the same groups in (h). Results represent mean ± s.e.m. from two independent experiments. Each symbol represents one mouse. P values were derived by one-way ANOVA with Dunnett post hoc analysis (b,d,f,h,j), or by repeat measures two-way ANOVA (a,g).
MyD88-dependent microbial sensing in nascent Treg cells directs sIgA responses to gut commensals and promotes mucosal tolerance 28. Treg cell-specific deletion of Myd88 in Il4raF709 mice (Il4raF709Foxp3YFPCreMyd88Δ/Δ) abrogated the protection by both the Clostridiales and Bacteroidales consortia against FA and also their induction of ROR-γt+ iTreg cells (Fig. 6g-j), with the gut Treg cells skewing towards a Th2 cell-like phenotype (Extended data Fig 10g,h). These results established a microbiota-responsive MyD88-ROR-γt pathway operative in nascent iTreg cells that mediates the therapeutic effects of bacteriotherapy in FA.
Discussion
Several important aspects of the role of dysbiosis in FA emerged from our studies. First, we have identified dynamic dysbiotic changes in the gut microbiota of FA infants that were pathogenic, evidenced by the failure of the microbiota of FA infants to protect against FA when introduced in GF Il4raF709 mice, whereas those of HC did. In the same model, the dysbiotic microbiota of FA-prone Il4raF709 mice were also non-protective as compared to those of WT mice, indicating an essential role for dysbiosis in FA.
Oral administration of a defined consortium of six human-origin Clostridiales species, related to taxa impacted by dysbiosis in FA infants, protected against FA and suppressed established disease in FA-prone Il4raF709 mice. The Clostridiales consortium also normalized the sIgA and suppressed the IgE responses to the gut commensals. These results are distinguished from previous studies by showing that a much smaller consortium of Clostridiales species acted to both prevent and suppress FA 11,54. Intriguingly, monobacterial therapy with Subdoligranulum variabile, a Clostridiales species found deficient in FA subjects age one year and older, was also protective, indicating that the loss of protective species is a key feature of disease pathogenesis. Critically, protection against FA was also effected by a second phylogenetically unrelated consortium of five human origin Bacteroidales species. Both consortia required an intact ROR-γt+ iTreg cell response for therapeutic efficacy. Though related taxa of Bacteroidales and Clostridales were increased in FA subjects, the inability of the microbiota of FA patients to protect GF Il4raF709 mice against FA or to effectively induce ROR-γt+ iTreg cells suggests that those taxa are poorly immunomodulatory.
Previous studies have implicated ROR-γt+ Treg cells in mediating tolerance induction by gut commensals, but differed on which immune responses they regulated 21,22,44,55. Our results indicate that the induction of ROR-γt+ iTreg cells by commensals via a Treg cell-specific MyD88-dependent pathway plays a requisite role in curtailing FA. Notably, the frequencies of ROR-γt+ iTreg cells were decreased in both human FA allergic subjects and in FA mice, consistent with their requirement in mediating oral tolerance in FA. These results provide a unifying mechanism for disease initiation in FA, involving dysbiotic gut microbiota that fail to induce protective ROR-γt+ iTreg cell populations, and they support the use of immunomodulatory bacteria to restore ROR-γt+ iTreg populations and re-establish tolerance in FA.
Finally, the extension of allergic response in FA to involve the gut microbiota indicates a broader disturbance in oral tolerance than hitherto appreciated. MyD88-dependent signaling in nascent gut Treg cells directs IgA antibody responses to gut luminal antigens including commensals and foods 28. The disruption of the MyD88-ROR-γt regulatory axis by dysbiosis in FA gave rise to decreased IgA and increased IgE responses to the gut microbiota, reproduced upon Treg cell-specific deletion of Rorc. We speculate that the presence of an anti-microbiota Th2 response may act to aggravate pathogenic immune responses to foods and may play a critical role in disease initiation, persistence and outcome.
Methods
Subjects. Subject Demographics.
Supplementary Table S1 summarizes subject demographics and Supplementary Fig. 1 summarizes the distribution of samples collected by subject age. One hundred and fifty four subjects were enrolled, 56 (36%) with food allergies to at least one of the major food allergens including milk, soy, egg, tree nuts, fish, shellfish, wheat, or peanuts, and 98 (64%) healthy controls. Seventy–eight percent of FA subjects were poly-sensitized, as defined by a positive skin test and/or specific IgE to at least 2 foods. Other FA included sesame, oat, pea, avocado, apple, grape and cantaloupe. 26 FA infants (16.8%) were diagnosed with cow’ milk allergy and were avoiding milk products at the time of stool sampling.
Healthy control subjects or subjects with FA age 1-15 months were enrolled. FA criteria included a history of allergic reactions to one or more of the major food allergens (e.g. milk, soy, egg, tree nuts, fish, shellfish, wheat, or peanuts), such as urticaria, angioedema, wheezing, diarrhea, vomiting, and moderate to severe eczema that was clearly triggered by food exposure and improving markedly after food avoidance, and a confirmatory positive food-specific skin prick test (SPT) ≥3mm compared to saline control and/or serum food-specific IgE ≥0.35. Exclusion criteria included: 1) prematurity, defined as delivery before 37 weeks of gestation, 2) recurrent or chronic infections necessitating frequent systemic (including oral) antibiotic administration, 3) history of chronic immunosuppressive therapies, 4) history of gastroenterological conditions, including non-IgE mediated colitis, eosinophilic esophagitis and food protein induced enterocolitis, allergic colitis, GE reflux or constipation necessitating medication, and 5) history of other chronic diseases, except for atopic conditions.
For studies on circulating ROR-γt+ Treg and Teff cells, demographic details on the human subjects involved, including FA, atopic but not FA and healthy controls are detailed in Supplementary Table 5. The diagnosis of FA was ascertained as detailed in the above section. Subjects who were atopic but not FA carried a diagnosis of allergic rhinitis, asthma and/or eczema, as detailed in Supplementary Table 5, while healthy control subjects did not have a history of FA or atopic diseases. All human subjects were recruited from the Boston Children’s Hospital main and satellite sites in Massachusetts. All human studies were approved by the Institutional Review Board.
Sample collection.
Parents were asked to collect stools from subjects every 4-6 months for up to 30 months of age, using a clean wood stick in Eppendorf tubes and RNA-later tubes (Thermofisher) and to freeze the sample in their home freezer immediately. Samples were collected from subjects’ homes within 1-3 weeks of the specimen collection. Overall, 60 subjects gave only one stool sample, 31 subjects gave 2 serial samples, 30 gave 3 samples, 17 gave 4 samples and 16 gave 5 samples. Each sample collection was spaced by 4-6 months. If systemic antibiotics were prescribed, sample collection was delayed for 4-6 weeks after the last dose of antibiotic administered, as guided by published studies tracking the recovery of gut microbiota following antibiotic treatment 56,57.
If a patient became tolerant to the food that they were initially allergic to (defined by being able to tolerate at least 4 grams of protein of that food), subsequent samples were not included in the analysis unless the subject had another FA confirmed by a history and positive skin and/or specific IgE. Samples from sensitized patients who lack a confirmatory history of an allergic food reaction were excluded from analysis. Parents completed an online questionnaire with each stool collection that included information regarding diet, breastfeeding, age, FA, infection, and use of antibiotics.
16S rDNA gene phylotyping.
A multiplexed amplicon library covering the 16S rDNA gene V4 region was generated from human stool sample DNA as described 58. Briefly, bacterial genomic DNA was extracted using the Mo Bio Power Fecal DNA Isolation kit (Mo Bio Laboratories) according to the manufacturer's instructions. To increase the DNA yields, the following modifications were used. An additional bead beater step using the Faster Prep FP120 (Thermo) at 6 meters/second for 1 min was used instead of vortex agitation. Incubation with buffers C2 and C3 was done for 10 min at 4°C. Starting nucleic acid concentrations were determined by a Qubit Fluromoter (Life Technologies). The amplicon library was prepared using dual-index barcodes 58. The aggregated library pool was size selected from 300-500 base pairs (bp) on a pippin prep 1.5% agarose cassette (Sage Sciences) according to the manufacturer's instructions. The concentration of the pool was measured by qPCR (Kapa Biosystems) and loaded onto the MiSeq Illumina instrument (300 bp kit) at 6-9pM with 40% phiX spike-in to compensate for low base diversity according to Illumina's standard loading protocol.
16S rDNA gene sequencing data preprocessing.
Sequencing of the 16S rDNA amplicons from the 368 human stool samples generated 14,279,132 total raw reads, with a mean of 38,802 reads per sample. Raw sequencing reads were processed using the mothur software package (v.1.35.1) 59 and custom Python scripts, which perform de-noising, quality filtering, alignment against the ARB Silva reference database of 16S rDNA gene sequences, and clustering into Operational Taxonomic Units (OTUs) at 97% identity. In total, 16387 OTUs were generated. OTUs with extremely low abundance were filtered using the following parameters: removed if mean relative abundance <0.001 across all samples or if zero reads in >80% of samples in at least one cohort.
Since the composition of the microbiota is known to change throughout childhood, we stratified subjects by age into six month intervals (1-6, 7-12, 13-18, 19-24, and 25-30 months). This interval span was chosen to provide resolution with respect to age, while ensuring sufficient numbers of subjects in each group to support meaningful comparisons. Because insufficient numbers of subjects were available for the 30-36 month group, this group was omitted from our analyses. After stratifying subjects by age groups segregated at six month intervals, 47, 53, 78, 74 and 79 OTUs were available after filtering in age group 1-6 months, 7-12 months, 13-18 months, 19-24 months, and 25-30 months, respectively.
16S rDNA gene sequencing data statistical analysis.
To assess for differences in ecological (alpha) diversity, Shannon entropy was calculated for each sample with statistical testing using the Wilcoxon rank-sum test. To assess differences in overall microbial community structure, beta-diversity was calculated using the unweighted and weighted Unifrac measures with statistical testing using the Analysis of Molecular Variance (AMOVA) method 60-62.
To statistically test for differences in the relevant abundances of fecal microbiota OTUs between control and food allergic subjects, the DESeq2 software package was employed with an analysis design depicted in Supplementary Fig. 1 63,64. Key covariates of interest (gender, mode of delivery, and breastfeeding only for younger than 18 months) were controlled for using the multi-factorial model in DESeq2. Since cow’s milk protein (CMP) intake could directly alter the microbiota, and is highly correlated with FA status, we also performed a subset analysis removing subjects without CMP intake. P-values were adjusted for multiple hypothesis testing using the method of Benjamini and Hochberg (BH) 65. OTUs reported met the following criteria: (1) adjusted p-value <=0.1; (2) absolute value of log2 fold change ≥2.
16S rDNA gene sequencing data phylogenetic analysis.
To more accurately identify the micro-organisms present in samples and their phylogenetic relationships to known species, the pplacer software package was used to perform phylogenetic placement 66. Pplacer uses a likelihood-based methodology to place short sequencing reads of 16S rDNA amplicons on a reference tree, and also generates taxonomic classifications of the short sequencing reads using a least common ancestor-based algorithm. The reference tree required for phylogenetic placement was generated using full-length or near full-length (>1,200 nt) 16S rDNA sequences of type strains from the Ribosomal Database Project (RDP) 67. In our studies, we report on the taxa that were phylogenetically placed with a like weight ratio of ≥0.8. The rest of the taxa can be seen in Supplementary Table 2. For purposes of describing OTUs in the manuscript, we refer to the closest reference species (CRS) to the phylogenetically placed consensus sequence for the OTU. While CRS does not represent an unambiguous species identification, it provides a point of reference for understanding microbiologically driven mechanisms in FA, and for deeper characterization using metagenomic or culture-based methods in future studies.
Mouse strains.
The following mice on BALB/c background have been previously described: BALB/cByJ (designated as WT mice), Rag2−/− (C.129S6(B6)-Rag2tm1Fwa), II4raF709 (C.129X1-Il4ratm3.1Tch) 68, Igh7−/−Il4raF709 69 and Foxp3EGFP/DTR+ 70. The following C57BL/6 congenic strains Rorcfl/fl (ROR-gtf/fB6(Cg)-Rorctm3Litt/J) 71,72, B6.129(Cg)-Foxp3tm4(YFP/cre)Ayr/J) (Foxp3YFPCre)73 and B6.129(Cg)-Il4raF709 69 were crossed to generate Il4raF709Foxp3YFPCre, Foxp3YFPCreRorcΔ/Δ 71, Il4raF709Foxp3YFPCreRorcΔ/Δ and Il4raF709Foxp3YFPCreMyd88Δ/Δ mice74. In all experiments, mice were matched for strain background.
Oral allergic sensitization of mice.
Mice were subjected to oral allergic sensitization with ovalbumin (OVA), mixed together with the mucosal adjuvant staphylococcal enterotoxin B (SEB) (Toxin Technology), as previously described 68,75. For antibiotic treatment, mice were treated with an antibiotic cocktail (Sigma-Aldrich) containing ampicillin 2.5 mg/ml), metronidazole (2.5 mg/ml), gentamycin (0.4 mg/ml), streptomycin (0.5 mg/ml), vancomycin (0.5 mg/ml), administered by oral gavage in a final total volume of 100 μl PBS once daily for 1 week, as indicated. For Treg cell depletion with DT, mice were injected intra-peritoneally (i.p.) DT (Sigma-Aldrich) at 250 ng/ml per injection/mouse (about 10 μg/kg), as indicated. For treatment with anti-CD25 or isotype control mAbs (BioXCell), mice were injected i.p. with the indicated antibody at 100 μg/injection/mouse, as indicated.
Percutaneous allergic sensitization of mice.
Mice were treated with antibiotics for one week, as described above. They were then sensitized through the skin with OVA/SEB as follows. The back of the mouse was shaved then tape stripped six times with Tegaderm dressings 76. OVA/SEB (at a concentration of 5 mg/ml OVA and 10 μg SEB in a final volume of 100 μl PBS were applied directly on the skin. The sensitization was repeated twice weekly for 5weeks. In subgroups of mice the Clostridiales consortium (see below) was given by gavage at 200 μl per mouse twice weekly for 5 weeks. At the end of the sensitization period, the mice are challenged with OVA in 150 mg/300 μl of PBS/mouse via gavage.
Detection of Fecal Bacteria-bound IgA and IgE by Flow Cytometry.
50mg of fecal pellet was homogenized in 1ml of sterile cold PBS and centrifuged at 40g for 10 minutes at 4°C to remove large particles. Supernatant containing the bacteria were collected, filtered through a 70μm strainer and centrifuged at 8000 g for 5 minutes to pellet the bacteria. The pellets were then washed twice with 1ml of sterile PBS and incubated on ice for 15 minutes with blocking buffer (50% fetal calf serum (FCS) in PBS for human fecal samples and 50% FCS + 10mg/ml OVA in PBS for mouse fecal samples). Samples were centrifuged at 8000g for 5 minutes and subsequently stained with 5μM SYTO-BC (eBioscience–ThermoFisher) along with either anti-mouse IgA (clone mA-6E1, eBioscience–ThermoFisher) or anti-mouse IgE (clone RME-1, Biolegend). Similarly, human fecal samples were stained with 5μM SYTO-BC along with either anti-human IgA (clone IS11-8E10, Miltenyi Biotech) or anti-human IgE (clone G7-26, BD Biosciences). Samples were then washed 3 times with 1 ml of PBS before flow cytometric analysis on a BD LSR Fortessa.
Isolation of MLN and LP lymphocytes.
MLNs were isolated and homogenized in PBS containing 2% FCS buffer. Cells were washed once PBS containing 2% FCS and used for experiments. Small intestines were dissected from mice and the fecal contents were flushed out using PBS containing 2% FCS. Payer’s patch was excised and the intestines were cut into 1cm pieces and treated with PBS containing 2% FCS, 1.5 mM DTT, and 10mM EDTA at 37 °C for 30 min with constant stirring to remove mucous and epithelial cells. The tissues were then minced and the cells were dissociated in RPMI containing collagenase (2 mg ml–1 collagenase II; Worthington), DNase I (100 μg ml–1; Sigma), 5mM MgCl2, 5mM CaCl2, 5mM HEPES, and 10% FBS with constant stirring at 37 °C for 45 min. Leukocytes were collected at the interface of a 40%/70% Percoll gradient (GE Healthcare). The cells were washed with PBS containing 2% FCS and used for experiments.
Preparation of therapeutic bacterial consortia.
Frozen stock cultures of the bacterial isolates (Supplementary Table 5) were stored at −80°C in microbank tubes (Pro-Lab Diagnostics). Obligately anaerobic species were plated from frozen stocks onto pre-reduced Brucella agar plates (BBL, Beckton Dickinson) and incubated in a Coy anaerobic chamber (Coy Labs). Facultative anaerobes were plated onto Trypticase Soy Agar (TSA) media (Remel) and incubated in the Coy chamber. All plates were incubated until visible growth was detected. Purity of materials was confirmed by Gram stain and rapid ANA (Remel) panels for anaerobes or API-20E strips (Remel) for aerotolerant facultative species.
Growth curve studies in the appropriate liquid media (Supplementary Table 4) were performed to quantitate a given biomass of organisms to the optical density (OD) measured at 600 nm. To prepare aggregate cultures, tubes containing 5 ml of the appropriate broth to support growth were inoculated. After visible growth and confirmation of culture purity, materials were then added to a larger culture volume to obtain additional biomass for aggregate mixtures. Cultures were staged based on the calculated time of the growth curves to be able to process a maximum biomass of each species on the day aggregate materials would be prepared. The bacterial consortia were prepared by normalizing the bacterial components according to OD 600 so that the OD of each component was approximately the same as the other bacteria in the cocktail. Materials were aggregated to have approximately 5 × 107 CFU/ml of each organism. After preparation of each aggregate mixture, 1ml aliquots were prepared in a Coy anaerobic chamber into cryovials, which were sealed, removed from the chamber and flash frozen in liquid nitrogen. Prior studies had identified that flash freezing by this method minimized loss of viable cells with the freeze/thaw to a ½ log-level or less 77. Frozen aliquots were stored at −80°C until use. Representative aliquots were subjected to qPCR with the probes in Supplementary Table 6 to confirm component species and relative abundance in the mixtures. Mixtures of obligate anaerobes were also plated aerobically to TSA agar media and incubated at 37°C in 5% CO2 for 72 hours to confirm absence of aerotolerant contaminants. The negative control consortium (NCC) of members of the Proteobacteria was plated to CNA sheep’s blood agar (Remel) incubated at 37°C under anaerobic conditions and at 5% CO2 under aerobic conditions for 72 hours to confirm absence of Gram positive contaminants.
Preparation of Subdoligranulum variabile therapeutic.
Microbiologic stocks of S. variabile (DSM 15176) were prepared as described. Maximal growth, ranging from 5×106-2×107 CFU/m in liquid media, was observed at 72 hours post-inoculation. For preparation of aliquots, a PRAS BHI tube (Thermo Fisher) was inoculated and incubated for 72 hours at 37°C in a Coy anaerobic chamber. Gram stain and culture to BHI media were done to confirm purity. 2.5ml of the culture was added to separate 150ml volume cultures of BHIS with hemin and vitamin K (Remel) and incubated in the anaerobic chamber for 72 hours before preparing aliquots. The strain lost substantive viability with attempts to concentrate its biomass under anaerobic conditions. Aliquots were thus prepared in a Coy chamber with nominal handling by transferring 1ml aliquots of the 72-hour culture into cryovials and sealing them in the chamber. Aliquots were then removed and snap frozen on liquid nitrogen for storage at −80°C until use. Separate aliquots were serially diluted and plated to BHI, with counting of pinpoint-sized colonies at 72hr to confirm S. variabile and at a biomass of 1.2×107 CFU/ml for administration of 2.4×106 CFU per mouse in 200 μl.
Heat killing of therapeutic consortia.
Sealed aliquots of the Clostridial or Bacteroidales consortia were placed in a heating block at 85°C for 1 hour to kill vegetative cells and spores. Control aliquots were transferred into a Coy anaerobe chamber and plated to BHI agar and broth with hemin and vitamin K to confirm killing by absence of growth. The Clostridiales consortium was also inoculated into BHI broth media + 1% maltose, hemin and vitamin K to confirm killing of C. leptum. No heat killed aliquots demonstrated any signs of growth. Heat-treated aliquots were then used in studies to assess efficacy of preparations lacking viable bacteria.
Short Chain Fatty Acid Analyses.
Samples of gut contents were kept frozen at −80°C until analysis. The samples were removed from the freezer and thawed. 500 μl of HPLC water was added to each sample and vortexed for 10 minutes and then centrifuged at 5000g for 10 minutes. 400 μl of the clear supernatant was transferred to a 2.0 ml Eppendorf tube. The pH of each sample was adjusted to 2-3 by adding 50 μl of 50% sulfuric acid. 50 μl of the internal standard (1% 2-methyl pentanoic acid solution) and 400 μl of ethyl ether anhydrous were added (Sigma-Aldrich). The tubes were mixed end over end for 10 minutes and then centrifuged at 2500g for 2 minutes. The upper ether layer was transferred to an Agilent sampling vial for analysis. 1 μl of the upper ether layer was injected into the chromatogram for analysis.
Chromatographic analysis was carried out using an Agilent 7890B system with a flame ionization detector (FID) (Agilent Technologies). A high-resolution gas chromatography capillary column 30m × 0.25 mm coated with 0.25um film thickness was used (DB-FFAP) for the volatile acids (Agilent Technologies). Nitrogen was used as the carrier gas. The oven temperature was 145°C and the FID and injection port was set to 225 °C. The injected sample volume was 1 μl and the run time for each analysis was 12 minutes. Chromatograms and data integration was carried out using the OpenLab ChemStation software (Agilent Technologies).
Standard Solutions:
A volatile acid mix containing 10 mM of acetic, propionic, isobutyric, butyric, isovaleric, valeric, isocaproic, caproic, and heptanoic acids was used (Supelco/Sigma-Aldrich). A standard stock solution containing 1% 2-methyl pentanoic acid (Sigma-Aldrich) was prepared as an internal standard control for the volatile acid extractions
Quantification of Acids:
400 μl of the standard mix was used and the extracts prepared as described for the samples except that 400 μl of ethyl ether was added. The retention times and peak heights of the acids in the standard mix were used as references for the sample unknowns. These acids were identified by their specific retention times and the concentrations determined and expressed as mM concentrations per gram of sample.
Quantitative real-time PCR for host immunological Targets.
RNA was extracted from cells using Quick-RNA MiniPrep kit (Zymo Research) according to the manufacturer protocol. Reverse transcription was performed with the SuperScript III RT-PCR system and random hexamer primers (Invitrogen) and quantitative real-time reverse transcription (RT)-PCR with Taqman® Fast Universal PCR master mix, internal house keeping gene mouse (Hprt VIC-MGB dye) and specific target gene primers for murine Rorc, as indicated (FAM Dye) (Applied Biosystems) on Step-One-Plus machine. Relative expression was normalized to Hprt and calculated as fold change compared to Foxp3YFPCre Treg cells.
Flow cytometry.
The following anti-mouse antibodies were used: CD3 (17A2), CD4 (RM4-5), IL-4 (11B11), IgE (RME-1) (Biolegend), Foxp3 (FJK-16S), GATA-3 (TWAJ), ROR-γt (BD2), Helios (22F6), NRP1 (3DS304M), IgA (mA-6E1), (eBioscience); Anti-human antibodies used in this study included CD3 (SK7), CD127 (A019D5), (Biolegend); CD4 (RPA-T4), FOXP3 (PCH101), IgE (G7-26) (eBioscience); CD25 (2A3), (BD Biosciences); IgA (IS11-8E10) (Miltenyi Biotech). Isotype control antibodies included rat IgG1 (R3-34), mouse IgG2a (G155-178) (BD Biosciences); rat IgG1 (eBRG1), mouse IgG1 (P3) (eBioscience). Cell viability dye (eFluor506) was from eBioscience. For cytokines cells were stimulated during 4 hours with PMA (50 ng/ml; Sigma-Aldrich) and ionomycin (500 ng/ml; Sigma-Aldrich) in the presence of Golgi Plug (BD Biosciences), then stained with the BD Cytofix/Cytoperm buffers (BD Biosciences) and the indicated anti-cytokine antibody. For intracellular staining of nuclear factors, the Foxp3 Transcription Factor buffer set (eBioscience) was used. Dead cells were routinely excluded from the analysis based on the staining of eFluor 506 fixable viability dye (eBioscience), and analyses were restricted to single cells using FSC-H and FSC-A signals. Stained cells were analyzed on an LSR Fortessa (BD Biosciences) and data were processed using Flowjo (Tree Star Inc.).
Table of mouse antibodies:
| Marker | Fluorochrome | clone | company | dilution | Staining |
| Foxp3 | eF450 | FJK-16S | eBioscience | 1/500 | Intracellular |
| Gata-3 | eF660 | TWAJ | eBiosicence | 1/300 | Intracellular |
| PerCP-eFluor710 | TWAJ | eBiosicence | 1/200 | Intracellular | |
| RORγt | PE | B2D | eBiosicence | 1/500 | Intracellular |
| Helios | APC | 22F6 | eBioscience | 1/300 | Intracellular |
| APC-eFluor780 | 22F6 | eBiosicence | 1/300 | Intracellular | |
| Ki67 | PE | SolA15 | eBiosicence | 1/500 | Intracellular |
| IL-4 | PE-Dazzle594 | 11B11 | Biolegend | 1/300 | Intracellular |
| CD3 | APC-Cy7 | 17A2 | Biolegend | 1/300 | Surface |
| CD4 | BV605 | RM4-5 | Biolegend | 1/500 | Surface |
| Nrp1 | PE | 3DS304M | eBioscience | 1/300 | Surface |
| PE-Cy7 | 3DS304M | eBioscience | 1/300 | Surface | |
| Viability dye | eFluor506 | Biolegend | 1/1000 | Surface |
Table of human antibodies:
| Marker | Fluorochrome | clone | company | dilution | Staining |
| CD4 | Percp-Cy5.5 | RPA-T4 | eBioscience | 2.5μl/106 cells | Surface |
| CD127 | PE-Cy7 | A019D5 | Biolegend | 1.5μl/106 cells | Surface |
| CD25 | FITC | 2A3 | BD | 20μl/106 cells | Surface |
| CD3 | APC-Cy7 | SK7 | Biolegend | 2.5μl/106 cells | Surface |
| RORγt | PE | #600380 | R&D systems | 1.5μl/106 cells | Intracellular |
| FOXP3 | APC | PCH101 | eBioscience | 3μl/106 cells | Intracellular |
Table of mouse fecal antibody staining:
| Marker | Fluorochrome | clone | company | dilution | Staining |
| IgA | PE | mA-6E1 | eBioscience | 1/2000 | Surface |
| IgE | PE | RME-1 | Biolegend | 1/200 | Surface |
| IgG1 | PE | RTK2071 | Biolegend | 1/200 | Surface |
| IgG1 | PE | eBRG1 | eBioscience | 1/2000 | Surface |
Table of human fecal antibody staining:
| Marker | Fluorochrome | clone | company | dilution | Staining |
| IgA | PE | IS11-8E10 | Miltenyi | 1/500 | Surface |
| IgE | BV605 | G7-26 | BD | 1/200 | Surface |
| IgG1 | PE | P3 | eBioscience | 1/500 | Surface |
| IgG2a | BV605 | G155-178 | BD | 1/200 | Surface |
ELISA.
Total, OVA-specific IgE and Murine mast cell protease 1 (MMCP-1) concentrations were measured in the sera of treated mice by ELISAs, as previously described 75.
Histology.
Intestinal mast cells were counted by microscopic examination of jejunal sections fixed in 10% formaldehyde and stored in ethanol 70% before staining with toluidine blue by the Harvard Rodent Histopathology Facility.
Statistical analysis.
Anaphylaxis-related Core body temperature measurements were analyzed using repeat measures 2-way ANOVA with the indicated post-test analysis. Student unpaired 2-tailed t-tests were used for 2-group comparisons. For more than 2 groups, 1-way ANOVA with the indicated post-test analysis was used. Results are presented as means and SEMs, where each point represents 1 sample. In cases in which values were spread across multiple orders of magnitude, data were log-transformed for analysis with parametric tests.
Reporting Summary.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. FMT from WT mice protects against FA in GF Il4raF709 mice.
(a) Temperature changes in GF Il4raF709 mice that were left uncolonized or reconstituted with FMT from WT or Il4raF709 mice, then sensitized with OVA/SEB and challenged with OVA (n=15 WT and 14 Il4raF709 mice). (b,c) Total and OVA-specific serum IgE (n=15 WT and 14 Il4raF709 mice). (d) MMCP-1 concentrations post OVA challenge (n=6 per group). (e,f) Analysis of ROR-γt and GATA3 expression in MLN Helios−NRP1− and Helios+NRP1+ Treg cells (n=6 per group). Each dot represents one mouse. Data represent mean ± s.e.m. from two or three independent experiments. P values were derived by repeat measures two-way ANOVA (a), or by student’s unpaired two tailed t test with Welch correction (b-f).
Extended Data Fig. 2. Analysis of IgA- and IgE-bound bacteria in fecal samples.
(a,c) Representative FACS plots showing the gating strategy for human (a) and mouse (c) fecal bacteria. Bacteria present in the feces was identified by gating on SYTO-BC+ events (right side panels). (b,d) frequencies of IgA− and IgE-bound bacteria as assessed by gating on bacteria-bound with the respective PE-labelled anti-IgA and anti-IgE antibodies, as shown in panels a and c. (e,f) Analysis of sIgA+ (e) and IgE+ (f) fecal bacteria of Il4raF709 mice sensitized with OVA/SEB without or with bacterial therapy. Fecal pellets of Rag2−/− and Igh7−/−Il4raF709 mice were used as negative controls. Each symbol in the scatter plots represents one mouse (no treatment: n=11 per group; Clostridiales: n=8 per group; Proteobacteria: n=9 and 7). Data represent mean ± s.e.m. from two independent experiments. Flow panels in (c,d) are representative of two two independent experiments. P values were derived by one-way ANOVA with Dunnett post hoc analysis.
Extended Data Fig. 3. Antibiotic therapy potentiates the therapeutic efficacy of the Clostridiales consortium in Il4raF709 mice.
(a) Temperature changes in the respective OVA/SEB-sensitized and OVA-challenged SPF Il4raF709 mouse groups treated as follows: no antibiotics (n=6), Clostridiales (n=5) and antibiotics without or with Clostridiales (n=5 per group). P values were derived by two-way ANOVA. (b,c) Total and OVA-specific IgE [no antibiotics (n=4 per group), Clostridiales (n=5 per group) and antibiotics without (n=5 per group) or with Clostridiales (n=4 per group)]. (d) MMCP-1 concentrations [no antibiotics (n=5), Clostridiales (n=5) and antibiotics without (n=4) or with Clostridiales (n=4)]. (e) Frequencies of total CD4+Foxp3+, Helios−NRP1−Foxp3+, ROR-γt+CD4+Foxp3+ and IL-4+ CD4+Foxp3+ Treg cells in the MLN of the respective mouse group [no antibiotics (n=6), Clostridiales (n=5) and antibiotics without (n=5) or with Clostridiales (n=5)]. Each dot represents one mouse. Throughout, data represent mean ± s.e.m. from two independent experiments. Unless otherwise indicated, P values were derived by one-way ANOVA with Dunnett post hoc analysis.
Extended Data Fig. 4. Bacteriotherapy with Subdoligranulum variabile protects against FA.
(a) Temperature changes in SPF Il4raF709 mice that were antibiotic-treated then sensitized with OVA/SEB while receiving no treatment (n=8) or treatment with the Subdoligranulum variabile (n=11), and thereafter challenged with OVA. P values were derived by two-way ANOVA. (b,c) Total and OVA-specific IgE (no bacteria: n=8; Subdoligranulum variabile: n=11). (d) MMCP-1 concentrations (no bacteria: n=8; Subdoligranulum variabile: n=11). (e,f) Analysis of MLN ROR-γt+ and GATA3+ cells among Helios−NRP1− and Helios+NRP1+ Foxp3+ Treg cells, respectively (no bacteria: n=8; Subdoligranulum variabile: n=5). (g) Analysis of MLN IL-4+ CD4+Foxp3+ Treg cells and IL-4+ CD4+Foxp3− Teff cells (no bacteria: n=8; Subdoligranulum variabile: n=5). Each dot represents one mouse. Throughout, data represent mean ± s.e.m. from two independent experiments. For panels b-g, P values were derived by Student’s unpaired two tailed t test with Welch correction.
Extended Data Fig. 5. Clostridiales protects against percutaneous sensitization-induced FA.
(a) Temperature changes in SPF WT BALB/c mice that were antibiotic-treated then percutaneously sensitized with OVA/SEB while receiving either no treatment (n=14) or treatment with Clostridales (n=11), and thereafter challenged with OVA. P values were derived by two-way ANOVA. (b,c) Total and OVA-specific IgE concentrations (n=7 per group). (d) MMCP-1 concentrations (n=7 per group). (e) Analysis of small intestinal LPL ROR-γt+ CD4+Foxp3+ Treg cells (n=7 per group). Each dot represents one mouse. Data represent mean ± s.e.m. from two independent experiments. For panels b-e, P values were derived by Student’s unpaired two tailed t test with Welch correction.
Extended Data Fig. 6. A Bacteroidales consortium prevents FA. A Bacteroidales consortium prevents FA.
(a) Left: Experimental schema. Right: temperature changes in GF Il4raF709 mice that were colonized and sensitized as indicated then challenged with OVA (n=5 per group). (b,c) Total and OVA-specific IgE (b) and MMCP-1 concentrations (c). GF, OVA/SEB (n=5 per group), Bacteroidales, PBS (n=6, 7 and 7), Bacteroidales, OVA/SEB (n=6, 6 and 7). (d) Frequencies of MLN CD4+Foxp3+, IL-4+Foxp3+ and GATA3+Foxp3+ T cells. GF, OVA/SEB (n=5 per group), Bacteroidales, PBS (n=4, 8 and 5), Bacteroidales, OVA/SEB (n=6, 5 and 6). (e) Frequencies of Helios−Nrp1−Foxp3+ and ROR-γt+Foxp3+ T cells. GF, OVA/SEB (n=5 and 7), Bacteroidales, PBS (n=5 per group), Bacteroidales, OVA/SEB (n=6 per group). (f) Left: Experimental Schema. Right: temperature changes in Il4raF709 mice sensitized and treated as indicated. OVA/SEB (n=6), OVA/SEB, Bacteroidales, (n=5). (g) Total and OVA-specific IgE and MMCP-1 concentrations. OVA/SEB (n=7, 9 and 9), OVA/SEB, Bacteroidales, (n=5, 10 and 5). (h,i) Frequencies of MLN CD4+Foxp3+, IL-4+Foxp3+ and GATA3+Foxp3+ (h) and Helios−Nrp1−Foxp3+ and ROR-γt+Foxp3+ T cells (i). OVA/SEB (n=5, 8 and 3, 5 and 5), OVA/SEB, Bacteroidales (n=5, 10, 4, 6 and 9). (j) IgE and IgA staining of fecal bacteria. OVA/SEB (n=11 per group), OVA/SEB, Bacteroidales (n=8 per group). Each dot represents one mouse. Data represent mean ± s.e.m. from two independent experiments. P values were derived by repeat measures two-way ANOVA (a,f), by one-way ANOVA with Dunnett post hoc analysis (b-e) or by Student’s unpaired two tailed t test (h-j).
Extended Data Fig. 7. Depletion of Treg cells abrogates protection by the microbiota.
(a) Experimental schema. (b) Temperature changes in the respective OVA/SEB-sensitized and OVA-challenged mouse groups: Il4raF709Foxp3EGFP/DTR− (n=6), Il4raF709Foxp3EGFP/DTR−+Clostridiales+DT (n=7), Il4raF709Foxp3EGFP/DTR++Clostridiales+DT (n=8), Il4raF709Foxp3EGFP/DTR−+Bacteroidales+DT (n=9), Il4raF709Foxp3EGFP/DTR++Bacteroidales+DT (n=7). (c) Total and OVA-specific IgE in the groups listed in (b) (Total IgE: n=9, 6, 8, 7 and 5; OVA-specific IgE:6, 5, 5, 5, and 5 ). (d) MMCP-1 concentrations (n=12 for Il4raF709Foxp3EGFP/DTR−, and n=8 per group for all other groups). (e,f) Frequencies of MLN CD4+Foxp3+ and IL-4+Foxp3+ T cells [Il4raF709Foxp3EGFP/DTR− (n=6 and 10), Il4raF709Foxp3EGFP/DTR−+Clostridiales+DT (n=8 and 7), Il4raF709Foxp3EGFP/DTR++Clostridiales+DT (n=8 and 7), Il4raF709Foxp3EGFP/DTR−+Bacteroidales+DT (n=8 and 7), Il4raF709Foxp3EGFP/DTR++Bacteroidales+DT (n=8 and 7)]. (g) Frequencies of ROR-γt+Foxp3+ and GATA3+Foxp3+ T cells [Il4raF709Foxp3EGFP/DTR− (n=5 per group), Il4raF709Foxp3EGFP/DTR−+Clostridiales+DT (n=7 and 9), Il4raF709Foxp3EGFP/DTR++Clostridiales+DT (n=8 per group), Il4raF709Foxp3EGFP/DTR−+Bacteroidales+DT (n=6 per group), Il4raF709Foxp3EGFP/DTR++Bacteroidales+DT (n=8 pre group)]. Each dot represents one mouse. Data represent mean ± s.e.m. from two independent experiments. P values were derived by repeat measures two-way ANOVA (b), or by one-way ANOVA with Dunnett post hoc analysis or Student’s unpaired two tailed t test (c-f).
Extended Data Fig. 8. Oral SCFA supplementation does not protect against FA.
SCFA in fecal samples of PBS or OVA/SEB-sensitized WT and Il4raF709 mice. Acetate, propionate and butyrate: WT, PBS or OVA/SEB (n=5 per group); Il4raF709, PBS (n=5 per group) or OVA/SEB (n=10 per group). Isovalerate, WT, PBS or OVA/SEB (n=5 and 4); Il4raF709, PBS or OVA/SEB (n=5 and 8). Valerate, WT, PBS or OVA/SEB (n=5 and 3); Il4raF709, PBS or OVA/SEB (n=4 and 7). (b) Temperature changes in OVA-challenged WT and Il4raF709 mice sensitized with PBS or OVA/SEB without or with SCFA supplementation. WT, PBS+SCFA (n=10), WT, OVA/SEB (n=11), WT, OVA/SEB+SCFA (n=24); Il4raF709, PBS+SCFA (n=12), Il4raF709, OVA/SEB (n=7), Il4raF709, OVA/SEB+SCFA (n=17). (c) Total and OVA-specific IgE. WT, PBS+SCFA (n=5 per group), WT, OVA/SEB (n=6 per group), WT, OVA/SEB+SCFA (n=9 per group); Il4raF709, PBS+SCFA (n=4 per group), Il4raF709, OVA/SEB (n=8 per group), Il4raF709, OVA/SEB+SCFA (n=9 per group). (d) Frequencies of MLN CD4+Foxp3+ROR-γt+ and CD4+Foxp3−ROR-γt+ T cells. WT, PBS+SCFA (n=5 per group), WT, OVA/SEB (n=4 per group), WT, OVA/SEB+SCFA (n=4 per group); Il4raF709, PBS+SCFA (n=5 per group), Il4raF709, OVA/SEB (n=5 per group), Il4raF709, OVA/SEB+SCFA (n=7 per group). Each dot represents one mouse. Data represent mean ± s.e.m. from two independent experiments. P values were derived by the Kolmogorov–Smirnov test (a), by the Student’s unpaired two tailed t test (c,d) or by two-way ANOVA (b).
Extended Data Fig. 9. Analysis of ROR-γt+ expression in human subjects and mutant mice.
(a) Gating strategy for CD4+Foxp3+ (G1) and CD4+Foxp3− T (G2) cells ex vivo. (b) Gating strategy for the expression of ROR-γt in Teff cells (G2) from FA patients, healthy controls (HC) and atopic subjects (atopy), as compared to an isotype control. (c) Flow plots and frequencies of peripheral blood CD4+Foxp3+ROR-γt+ T cells in WT and Il4raF709 mice (n=7 mice per group). (d) Flow plots and frequencies of peripheral blood CD4+Foxp3+Helios−NRP1−ROR-γt+ T cells in WT and Il4raF709 mice (n=7 mice per group). (e,f) Flow plots and frequencies of MLN CD4+Foxp3+ROR-γt+ T cells from Foxp3YFPCre mice sensitized with OVA/SEB, and Foxp3YFPCreRorcΔ/Δ either sham sensitized (PBS) or sensitized with OVA/SEB, as indicated (n= 5 mice per group). (g) Quantitative RT-PCR of Rorc gene expression in MLN CD4+Foxp3+ Treg and CD4+Foxp3− Teff cells from Foxp3YFPCre, Foxp3YFPCreRorcΔ/Δ, and Il4raF709Foxp3YFPCreRorcΔ/Δ mice. Data were normalized to the endogenous Hprt transcripts (n= 5 mice per group). Each dot represents one mouse. Results represent Means ± S.E.M. collated from 2 independent experiments. P values were derived by the Student’s unpaired two tailed t test with Welch correction (c,d), or by one-way ANOVA with Dunnett post hoc analysis (f,g).
Extended Data Fig. 10. Treg cell-specific deletion of Rorc and Myd88 impairs mucosal tolerance.
(a-d) Analysis of sIgA+ (a,b) and IgE+ (c.d) fecal bacteria in OVA/SEB-sensitized Foxp3YFPCre, Il4raF709Foxp3YFPCre and Foxp3YFPCreRorcΔ/Δ mice. Fecal pellets of Rag2−/− and Igh7−/−Il4raF709 mice were used as negative controls [Foxp3YFPCre (n=6 per group), Il4raF709Foxp3YFPCre (n=11 and 7), and Foxp3YFPCreRorcΔ/Δ mice (n=10 and 8)]. (e-f) Analysis of GATA3+Foxp3+ Treg cells in the following OVA/SEB-sensitized mice that were either untreated or treated with Clostridiales or Bacteroidales consortia: Il4raF709Foxp3YFPCre (n=9, 5 and 5), and Il4raF709Foxp3YFPCreRorcΔ/Δ (n= 4, 5 and 8). (g,h) Analysis of GATA3+Foxp3+ Treg cells in OVA/SEB-sensitized Il4raF709Foxp3YFPCre mice treated with the Bacteroidales consortium (n=9), and in OVA/SEB-sensitized Il4raF709Foxp3YFPCreMyd88Δ/Δ mice otherwise untreated or treated with the Clostridiales or Bacteroidales consortia (n= 8, 9 and 8). Each symbol represents one mouse. Results represent Means ± S.E.M. collated from 2 independent experiments. P values were derived by one-way ANOVA with Dunnett post hoc analysis (b,d,f,h).
Supplementary Material
Acknowledgements
We thank Dr. Hans Oettgen for provision of Igh7−/−Il4raF709 mice, Louis-Marie Charbonnier for critical review of the manuscript, and Mary Delaney for microbiologic support. This work was supported by NIH NIAID grants 1R56AI117983 and 1R01AI126915 (T.A.C.), NIDDK grant P30DK056338 (L.B.) the Clinical and Translational Science Center/Harvard Catalyst, the Bunning Food Allergy Fund, the Jasmine and Paul Mashikian Fund, the Massachusetts Life Sciences Center, and a Partners Healthcare Innovations Development Grant.
Footnotes
Data Availability
All data will be made available to investigators upon request. The 16S bacterial rRNA datasets generated in the course of this project have been deposited at the National Center for Biotechnology Information Sequence Read Archive (SRA) under BioProject ID: PRJNA525231.
Competing interests
L.B., G.K.G., T.A.C., R.R. and A.A.-G. are inventors on published US patent application, 15/801,811, submitted by The Brigham and Women’s Hospital, Inc. and Children’s Medical Center Corporation, that covers methods and compositions for the prevention and treatment of food allergy using microbial treatments. T.A.C., R.R., A.A.-G., and E.S.-V. have pending patent applications related to the use of probiotics in enforcing oral tolerance in food allergy (62/758,161, and, 62/823,866). L.B., G.K.G. and T.A.C. are founders of and have equity in Consortia Tx. R.R. has equity in Consortia Tx. A.A.-G. is currently an employee of, and owns shares in Seed Health Inc. The rest of the authors declare no competing interests.
References
- 1.Branum AM & Lukacs SL Food allergy among children in the United States. Pediatrics 124, 1549–1555 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Wills-Karp M, Santeliz J & Karp CL The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol 1, 69–75 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Ly NP, Litonjua A, Gold DR & Celedon JC Gut microbiota, probiotics, and vitamin D: interrelated exposures influencing allergy, asthma, and obesity? J Allergy Clin Immunol 127, 1087–1094; quiz 1095–1086 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koplin J, et al. Is caesarean delivery associated with sensitization to food allergens and IgE-mediated food allergy: a systematic review. Pediatric allergy and immunology : official publication of the European Society of Pediatric Allergy and Immunology 19, 682–687 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Bjorksten B Disease outcomes as a consequence of environmental influences on the development of the immune system. Curr Opin Allergy Clin Immunol 9, 185–189 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Rachid R & Chatila TA The role of the gut microbiota in food allergy. Curr Opin Pediatr 28, 748–753 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Azad MB, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy 45, 632–643 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Sudo N, et al. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol 159, 1739–1745 (1997). [PubMed] [Google Scholar]
- 9.Fritz JH, et al. Acquisition of a multifunctional IgA+ plasma cell phenotype in the gut. Nature 481, 199–203 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34, 794–806 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Stefka AT, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A 111, 13145–13150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noval Rivas M, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol 131, 201–212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wesemann DR & Nagler CR The Microbiome, Timing, and Barrier Function in the Context of Allergic Disease. Immunity 44, 728–738 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noval Rivas M & Chatila TA Regulatory T cells in allergic diseases. J Allergy Clin Immunol 138, 639–652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdel-Gadir A, Massoud AH & Chatila TA Antigen-specific Treg cells in immunological tolerance: implications for allergic diseases. F1000Res 7, 38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan J, et al. Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell reports 15, 2809–2824 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Ohnmacht C, et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 349, 989–993 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Sefik E, et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 349, 993–997 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burton OT, et al. Immunoglobulin E signal inhibition during allergen ingestion leads to reversal of established food allergy and induction of regulatory T cells. Immunity 41, 141–151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noval Rivas M, Burton OT, Oettgen HC & Chatila T IL-4 production by group 2 innate lymphoid cells promotes food allergy by blocking regulatory T-cell function. J Allergy Clin Immunol (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noval Rivas M, et al. Regulatory T Cell Reprogramming toward a Th2-Cell-like Lineage Impairs Oral Tolerance and Promotes Food Allergy. Immunity 42, 512–523 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fagarasan S, et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 298, 1424–1427 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Kubinak JL, et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe 17, 153–163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, et al. MyD88 Adaptor-Dependent Microbial Sensing by Regulatory T Cells Promotes Mucosal Tolerance and Enforces Commensalism. Immunity 43, 289–303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donaldson GP, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360, 795–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macpherson AJ, Yilmaz B, Limenitakis JP & Ganal-Vonarburg SC IgA Function in Relation to the Intestinal Microbiota. Annu Rev Immunol 36, 359–381 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Mathias CB, et al. IgE-mediated systemic anaphylaxis and impaired tolerance to food antigens in mice with enhanced IL-4 receptor signaling. J Allergy Clin Immunol 127, 795–805 e791–796 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalia VC, et al. Analysis of the unexplored features of rrs (16S rDNA) of the Genus Clostridium. BMC genomics 12, 18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajilic-Stojanovic M & de Vos WM The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 38, 996–1047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blander JM, Longman RS, Iliev ID, Sonnenberg GF & Artis D Regulation of inflammation by microbiota interactions with the host. Nat Immunol 18, 851–860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narushima S, et al. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut microbes 5, 333–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker AW & Lawley TD Therapeutic modulation of intestinal dysbiosis. Pharmacol Res 69, 75–86 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Kim KS, et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 351, 858–863 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russler-Germain EV, Rengarajan S & Hsieh CS Antigen-specific regulatory T-cell responses to intestinal microbiota. Mucosal Immunol (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thornton AM, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol 184, 3433–3441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss JM, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med 209, 1723–1742, S1721 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yadav M, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med 209, 1713–1722, S1711–1719 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bilate AM & Lafaille JJ Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annu Rev Immunol 30, 733–758 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Xu M, et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdel-Gadir A, et al. Oral immunotherapy with omalizumab reverses the Th2 cell-like programme of regulatory T cells and restores their function. Clin Exp Allergy 48, 825–836 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartnikas LM, et al. Epicutaneous sensitization results in IgE-dependent intestinal mast cell expansion and food-induced anaphylaxis. J Allergy Clin Immunol 131, 451–460 e451–456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kverka M, et al. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin Exp Immunol 163, 250–259 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geva-Zatorsky N, et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 168, 928–943 e911 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mangalam A, et al. Human Gut-Derived Commensal Bacteria Suppress CNS Inflammatory and Demyelinating Disease. Cell reports 20, 1269–1277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haribhai D, et al. A requisite role for induced regulatory T cells in tolerance based on expanding antigen receptor diversity. Immunity 35, 109–122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noval Rivas M, Burton OT, Oettgen HC & Chatila T IL-4 production by group 2 innate lymphoid cells promotes food allergy by blocking regulatory T-cell function. J Allergy Clin Immunol 138, 801–811 e809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rios-Covian D, Salazar N, Gueimonde M & de Los Reyes-Gavilan CG Shaping the Metabolism of Intestinal Bacteroides Population through Diet to Improve Human Health. Front Microbiol 8, 376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Britton GJ, et al. Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORgammat(+) Regulatory T Cells and Exacerbate Colitis in Mice. Immunity 50, 212–224 e214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dethlefsen L & Relman DA Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 108 Suppl 1, 4554–4561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerber GK, Onderdonk AB & Bry L Inferring dynamic signatures of microbes in complex host ecosystems. PLoS Comput Biol 8, e1002624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bucci V, et al. MDSINE: Microbial Dynamical Systems INference Engine for microbiome time-series analyses. Genome Biol 17, 121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Excoffier L, Smouse PE & Quattro JM Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lozupone C, Hamady M & Knight R UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7, 371 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lozupone C & Knight R UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71, 8228–8235 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McMurdie P & Holmes S phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PloS ONE 8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McMurdie P & Holmes S Waste not, want not: why rarefying microbiome data is inadmissible. PLoS computational biology 10, e1003531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benjamini Y & Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B 57, 289–300 (1995). [Google Scholar]
- 66.Matsen F, Kodner RB & Armbrust EV pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC bioinformatics 11, 538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cole JR, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic acids research 42, D633–642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Noval Rivas M, et al. Regulatory T Cell Reprogramming toward a Th2-Cell-like Lineage Impairs Oral Tolerance and Promotes Food Allergy. Immunity 42, 512–523 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burton OT, et al. Immunoglobulin E signal inhibition during allergen ingestion leads to reversal of established food allergy and induction of regulatory T cells. Immunity 41, 141–151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Noval Rivas M, Burton OT, Oettgen HC & Chatila T IL-4 production by group 2 innate lymphoid cells promotes food allergy by blocking regulatory T-cell function. J Allergy Clin Immunol 138, 801–811 e809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Massoud AH, et al. An asthma-associated IL4R variant exacerbates airway inflammation by promoting conversion of regulatory T cells to TH17-like cells. Nat Med 22, 1013–1022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu M, et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008). [DOI] [PubMed] [Google Scholar]
- 74.Wang S, et al. MyD88 Adaptor-Dependent Microbial Sensing by Regulatory T Cells Promotes Mucosal Tolerance and Enforces Commensalism. Immunity 43, 289–303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Noval Rivas M, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol 131, 201–212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bartnikas LM, et al. Epicutaneous sensitization results in IgE-dependent intestinal mast cell expansion and food-induced anaphylaxis. J Allergy Clin Immunol 131, 451–460 e451–456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pinciroli R, et al. Endotracheal Tubes Cleaned With a Novel Mechanism for Secretion Removal: A Randomized Controlled Clinical Study. Respir Care 61, 1431–1439 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.