Summary
The number and type of ion channels present in the membrane determines the electrophysiological function of every neuron. In many species, stereotyped output of neurons often persists for years [1], and ion channel dysregulation can change these properties to cause severe neurological disorders [2-4]. Thus, a fundamental question is how do neurons coordinate channel expression to uphold their firing patterns over long time scales [1, 5]? One major hypothesis purports that neurons homeostatically regulate their ongoing activity through mechanisms that link membrane voltage to expression relationships among ion channels [6-10]. However, experimentally establishing this feedback loop for the control of expression relationships has been a challenge: Manipulations that aim to test for voltage feedback invariably disrupt trophic signaling from synaptic transmission and neuromodulation in addition to activity [9, 11, 12]. Further, neuronal activity often relies critically on these chemicals mediators, obscuring the contribution of voltage activity of the membrane per se in forming the channel relationships that determine neuronal output [6, 13]. To resolve this, we isolated an identifiable neuron in crustaceans and then either kept this neuron silent or used a longterm voltage clamp protocol to artificially maintain activity. We found that physiological voltage activity – independent of all known forms of synaptic and neuromodulatory feedback – maintains most channel mRNA relationships, while metabotropic influences may play a relatively smaller role. Thus, ion channel relationships likely needed to maintain neuronal identity are actively and continually regulated at least in part at the level of channel mRNAs through feedback by membrane voltage.
eTOC blurb:
How do neurons coordinate channel expression to uphold their firing patterns over long time scales? Santin and Schulz found that voltage activity – independent of synaptic and neuromodulatory feedback – maintains many channel mRNA relationships.
Results & Discussion
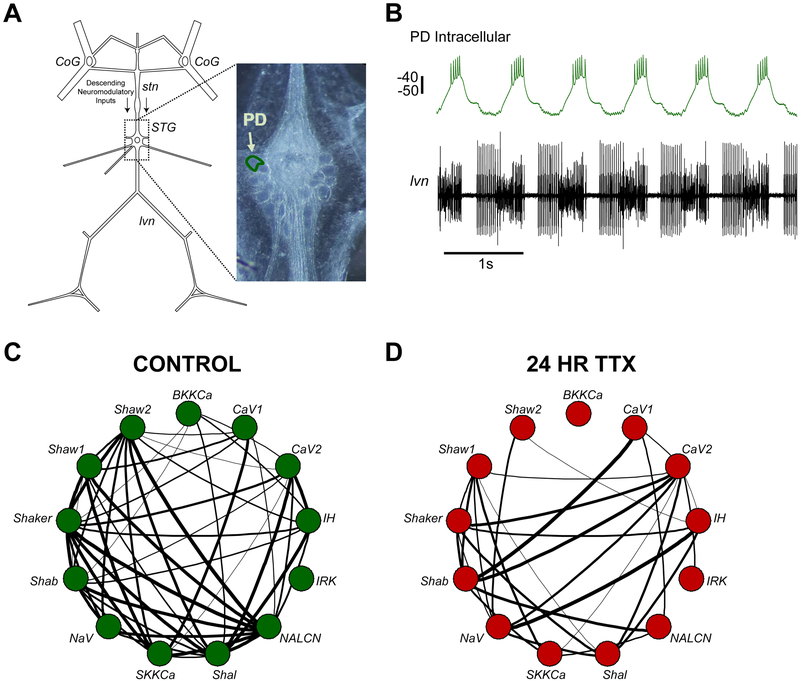
To test the role of voltage activity in generating ion channel relationships, we took advantage of the stomatogastric ganglion in Cancer borealis (STG; Figure 1A). The STG is a small motor circuit containing 26-27 neurons that are individually identifiable across animals. Each neuron type in the STG shows distinct patterns of correlated mRNA and conductance expression of voltage-gated ion channels. Thus, correlated patterns of ion channel mRNA expression are thought to be important in defining the function of each neuron [7, 11, 14-16]. We assessed correlated expression patterns of 13 ion channel genes in individually identified, hand-dissected Pyloric Dilator neurons (PD; Figure 1A-B) using single-cell quantitative PCR. We confirm that a substantial loss of correlated channel expression occurs after depriving neurons of their normal activity and chemical environment (Figure 1C-D). We set out to determine whether ongoing membrane voltage activity is the critical signal that coordinates these channel relationships.
Figure 1. Network silencing disrupts ion channel mRNA relationships in single, identified PD neurons.
(A) Schematic of the Stomatogastric Nervous System from the crab, Cancer borealis. Descending modulation from the Commissural Ganglia (CoG), whose axons travel through the stomatogastric nerve (stn), is the main neuromodulatory input to the stomatogastric ganglion (STG). The pyloric dilator (PD) neuron used in this study is outlined in the photomicrograph of the STG. (B) Example of an intracellular recording of the PD neuron (top) and the corresponding extracellular recording of the lateral ventricular nerve (Ivn) that contains PD axons. Scale bar represents 10 mV. (C) Correlation network plot used to show strong relationships between 13 ion channel mRNAs in single PD neurons (n=11; series 1 control). Two nodes connected by a line represents a pairwise correlation with an r-value greater than 0.65 (the r-value where p<0.05). Line thickness increases with r-value. (D) Correlation network plot of single PD neurons silenced with tetrodotoxin for 24 hrs (n=13).
To directly test whether physiological membrane voltage activity maintains patterns of channel expression, we recorded the ongoing activity of PD neurons in intact networks, silenced those PD neurons and the entire network with tetrodotoxin (TTX), and isolated PD cells from neuromodulatory inputs by transecting the stomatogastric nerve. With no ongoing network activity and no descending modulatory projections, these cells are without voltage, chemical synaptic, or neuromodulatory input. Immediately after isolating these cells from the network, we used two-electrode voltage clamp (TEVC) to “play back” the somatic waveform of each PD neuron (recorded previously) as a voltage-clamp protocol for 8 hr. This approach allowed us to assess the contribution of physiological membrane voltage activity while minimizing the influence of the other variables that could maintain the channel gene expression profile.
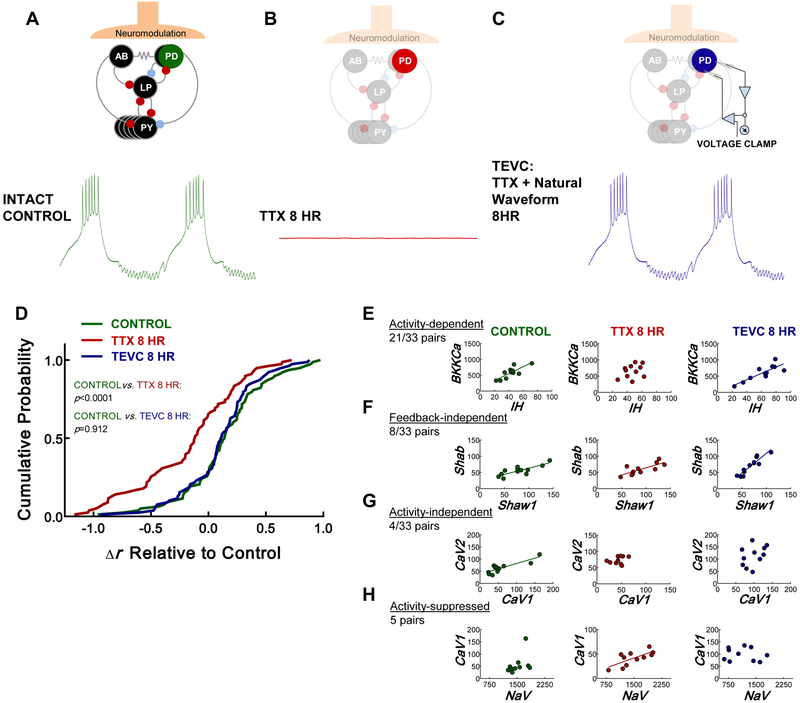
We then assessed correlated patterns of channel mRNA in three groups of PD neurons: (1) control (Figure 2A), (2) silent (isolated from inputs for 8 hr; Figure 2D), and (3) TEVC (voltage-clamped to physiological waveform while isolated from inputs for 8 hr; Figure 2C). Similar to the 24 hr TTX experiment shown in Figure 1C-D, the total strength of the correlation profile decreased relative to control after 8 hr in TTX. In contrast, when the somata of silenced PD neurons were voltage clamped to their endogenous waveforms for 8 hr, overall channel mRNA correlations were maintained (Figure 2D). Resting membrane potential did not change when measured at the end of the 8 hr TEVC protocol, indicating neurons were healthy throughout the experiment (paired t test, p = 0.46, n = 12, Figure S1).
Figure 2. Physiological voltage activity, independent of chemical input, influences coordinated ion channel expression in PD neurons.
(A-C) Diagram illustrating the experimental groups used to determine the role of voltage activity in correlating ion channel mRNA expression. (A) Activity of the PD neuron caused by normal intact synaptic inhibition, neuromodulation, and electrical coupling in the pyloric network. (B) Voltage trace from the same neuron after it has been silenced by exposing the STG to 10−7 M TTX and transecting the stn that contains the neuromodulatory fibers innervating the STG (8 hr). These preparations are devoid of activity, synaptic input, and neuromodulation. (C) The same neuron lacking endogenous input, but with its membrane potential artificially driven to its original activity pattern by a voltage clamp protocol “played back” to the cell using two-electrode voltage clamp (TEVC) for 8 hr. These preparations are devoid of neuromodulation and synaptic input, but now maintain voltage activity through TEVC. (D) Cumulative probability distribution showing that the change in Pearson’s correlation coefficient (Δr for each pairwise correlation decreases relative to the control group (green) from series 2 in silent PD neurons (TTX 8 hr, red; Kolmogorov-Smirnov test; p<0.0001; n= 12 control and n=11 silent), but not in PD neurons with physiological voltage activity maintained for 8 hr using TEVC. (TEVC 8 hr, blue; Kolmogorov-Smirnov test; p=0.912; n=12 TEVC neurons). The control distribution for this analysis was generated by subtracting the series 2 control r values from the r values in series 1. (E-H) Examples of the different ways correlations arise in the PD neuron. Left column=control, middle column=silent, and right column= TEVC. (E) Example where voltage activity determines correlation between two channel mRNAs. (F) Example where correlation does not depend on activity or chemical feedback. (G) Example where a correlation does not depend on activity, but may rely on trophic feedback. (H) Example where a correlation only appears without activity. Each point represents a single neuron. Regression lines were drawn when p<0.05 (Pearson’s test). See also Figure S1, Tables S1-S4.
When we measured mRNA levels for 13 channels, there were 78 possible pairwise relationships. We found 33 pairs of ion channels that had significantly correlated patterns of expression in the control group (Table S1-2; Pearson’s test; p<0.05). 21/33 of these relationships relied on physiological membrane voltage activity because they were found to occur regardless of whether activity was produced normally (intact networks) or artificially (via TEVC) when neurons were isolated from the network (Table S1). For example, an important relationship likely to be involved in regulating firing properties in PD neurons [11, 16], IH and BKKCa, showed significant correlations with either form of activity, but not in silenced neurons. We refer to these as bona fide activity-dependent expression relationships (Figure 2E). 8/33 relationships did not depend on voltage activity of the membrane or modulation state because they were unchanged across experimental groups (Table S2). For example, in the case of Shawl and Shab, a significant correlation existed regardless of activity and chemical input (Figure 2F). Further, 4/33 correlated relationships were likely to arise through modulatory feedback because they were eliminated in TTX and did not persist when activity was maintained artificially in TTX (Figure 2G, Table S2). We also identified 5 pairs of channel genes that only had correlated relationships when PD neurons were silenced (Table S2). Figure 2H shows one example whereby NaV and CaV1 were not correlated with normal or artificial activity, but became correlated when activity was silenced with TTX. The remaining relationships were never correlated at steady-state in control PD cells. 19 were not correlated in any condition, 8 became correlated in TTX with or without artificial activity, and 13 were correlated only when activity was controlled with TEVC (Table S3-S4). Therefore, there seems to be the possibly for complex interactions between chemical and electrical feedback to inhibit correlations that we do not yet understand.
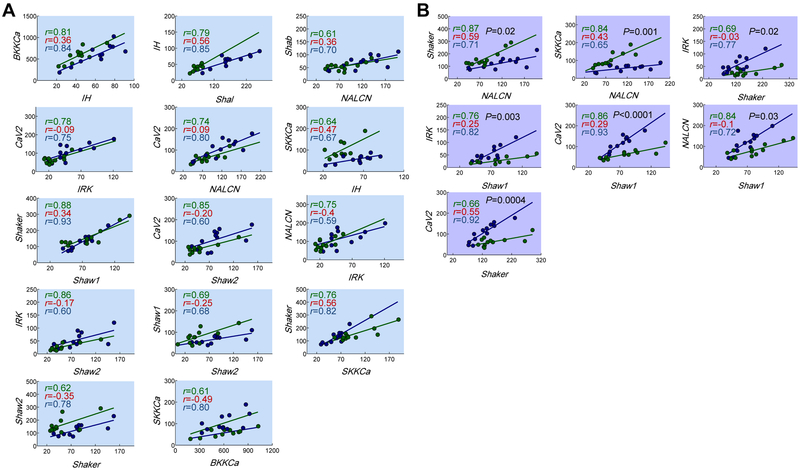
The quantitative relationship between ion channels pairs (i.e. the slope) also likely influences neuronal output, and theoretical work suggests that these relationships could arise as a result of membrane potential activity [7, 14, 17]. Is this the case in biological neurons? For 14/21 activity-dependent correlations, slopes were not statistically different between control and TEVC groups (Figure 3A, Table S1). Thus, ongoing voltage activity regulates not only the existence of correlated channel expression, but also the quantitative relationship between ion channel pairs. Seven pairs, however, showed significantly different slopes when activity was maintained in isolated neurons using TEVC (Figure 3B, Table S1). In these cases, voltage activity constrains the linear expression pattern and other factors likely fine-tune the precise quantitative relationship.
Figure 3. Physiological voltage activity maintains the quantitative relationships between most correlated ion channel pairs.
(A) 14 of the 21 bona fide voltage-dependent channel relationships have slopes that are not significantly different when activity is caused by normal network interactions (green) or TEVC (blue) [analysis of covariance (ANCOVA); p values ranged from 0.08-0.94]. Each relationship shown in (A) was significant with control (green) or the TEVC (blue), but not in isolated neurons (Pearson’s test; p<0.05). r values for silent PD neurons are presented in red text on the plots to show the loss of correlation. (B) 7/21 bona fide activity-dependent channel relationships have slopes that were significantly different when activity is caused by the voltage clamp in isolated neurons (ANCOVA; p <0.05; shown in black text on plots). Each point represents a single neuron. Regression lines were drawn when p<0.05 (Pearson’s test). See also Table S1.
Over twenty years ago, a classic series of studies suggested that ongoing voltage activity may feed back to coordinate the expression of ion channels to, in turn, regulate the activity of a neuron in a homeostatic manner [8, 18-20]. Theoretical work has since expanded on these ideas to include predictions about how activity may cause the quantitative expression relationships between ion channels to maintain neuron identity [7, 17]. Here, we present direct evidence that correlated channel expression patterns thought to define biological neuron activity are maintained by ongoing, physiological voltage activity. Testing this was the result of our ability to perform long-term voltage clamp experiments on electrically silent, chemically isolated, and individually identifiable neurons. Using this approach, our data indicate that ongoing physiological membrane activity at the soma regulates at least 64% of the steady-state channel correlation profile in PD neurons. Moreover, physiological membrane activity maintained the quantitative relationship of two-thirds of those correlated ion channel pairs. Therefore, our results support the notion that activity of the membrane is the dominant signal that maintains most of the ion channel mRNA relationships that are likely to determine the activity of PD neurons. We would be remiss to ignore the ion channel relationships that either arise or are quantitatively set by processes other than membrane voltage activity (Figure 2F, Figure 2G, Figure 3B). These results are consistent with a broad array of mechanisms that stabilize the function of neurons through metabotropic signaling or coordinated transcription and translation [12, 22]. Interestingly, Khorkova & Golowasch [12] showed that neuromodulation, and not activity, regulates ionic conductance relationships in the same PD neuron, which is, in general, consistent with the pattern of 4 relationships found in our study. Given that channel correlations seem to be important for controlling neuronal output, these cells are likely to use multiple strategies across multiples scales of organization to regulate their channel profile.
Several ion channel pairs also became correlated only when isolated from their inputs, both without (Figure 2H) or with activity (Table S3-S4). A similar response has been reported for the BKKCa-Shal relationship in the lateral pyloric neuron, LP, of the crab STG after loss of neuromodulation and activity [11]. We suggest there are two ways to interpret these results with important implications for regulating neuronal activity. First, voltage, trophic feedback, or the interaction of the two may prevent channel relationships from forming. Fixed expression ratios of channel pairs are thought to produce stereotyped activity patterns [7, 15, 17]. Thus, certain channel relationships may be actively restricted to avoid unwanted interactions from forming. The failure of such a regulation scheme has the potential to contribute to neurological disorders such as epilepsy, where changes in sodium channel activity relative to other conductances alters excitation:inhibition ratios that result in network hyperexcitability [23, 24]. Second, it has been demonstrated that some neurons can regain their ability to burst when isolated from their networks [12, 18, 21]. Therefore, the formation of new relationships in isolated neurons may represent an attempt to find a different profile of channels to compensate for the loss of activity and metabotropic signaling. The mechanisms that prevent correlations from forming have the potential to be an important way to defend the functional identify of a neuron but have received little attention thus far.
In conclusion, we show strong evidence that ongoing voltage activity is a major signal needed to maintain channel relationships thought to be important for cell-type specific neuronal output. However, interesting questions remain. First, neurons in this study have a repetitive output that carries on throughout life, much like the neurons that produce breathing in mammals [25]. Do neurons with intermittent activation histories or variable modes of output use activity-dependent feedback, or do they take advantage of activity-independent processes to regulate channel expression? Further, a large body of evidence has shown that voltage activity of the membrane regulates gene expression through Ca2+ channels and intracellular Ca2+ signaling [26, 27]. How do biological calcium sensors orchestrate the relationships of many different ion channels to produce a desired firing pattern? What are the signaling pathways involved in controlling activity-independent channel relationships? Only by addressing how these processes intersect can neuronal output be fully understood. Our study allows a line to be drawn directly from membrane voltage to steady-state mRNA relationships across ion channels. The answers to these critical remaining questions are likely to be found along this path.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
For information and requests for resources, please contact David Schulz (schulzd@missouri.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Adult male Jonah Crabs (Cancer borealis) were purchased from The Fresh Lobster Company (Gloucester, MA, USA) and kept in artificial sea water at 12°C. Crabs were anesthetized on ice for 30 min. and then were used for dissecting the stomach. The stomatogastric nervous system (STNS) was dissected from the stomach in physiological saline that contained the following salts (in mM): 440 NaCl, 11 KCl, 13 CaCl2, 26 MgCl2 and 10 HEPES buffer, pH 7.45. The STNS containing the paired commissural ganglia, the esophageal ganglion, and the stomatogastric ganglion (STG) was then pinned out in a Sylgard-coated petri dish (The Dow Chemical Company, Midland, MI, USA). The STG was desheathed using a thin stainless steel wire.
METHOD DETAILS
Electrophysiology and neuron identification:
Extracellular recordings of the pyloric rhythm produced by the STG were made by placing stainless-steal pin electrodes in a petroleum jelly well placed around the lateral ventricular nerve (Ivn). The signals were amplified using an A-M Systems Model 1700 extracellular amplifier (A-M Systems, Carlsborg, WA, USA). PD neurons were identified using intracellular recordings and standard cell identification procedures [6]. Intracellular recordings were made using 10-20 MΩ microelectrodes filled with 600 mM K2SO4 and 20 mM KCl and amplified using an Axoclamp 2A intracellular amplifier (Molecular Devices, San Jose, CA, USA). Data were acquired using a Digidata 1322A digitizer (Molecular Devices, San Jose, CA, USA).
Experimental groups:
We performed two series of experiments in this study. The first contained two treatment groups. (1) Control (n=11): these PD neurons were identified and acutely harvested from the ganglion, (2) Silent (=13): 24 hr. incubation with physiological saline containing 10−7 M tetrodotoxin with the stomatogastric nerve transected (TTX; Sigma-Aldrich, St. Louis, MO, USA). The second series of experiments had three groups of PD neurons. (1) Control (n=12): these PD neurons were identified and acutely harvested from the ganglion. This control group was distinct from the control neurons of the first series of experiments, (2) Silent (n=11): 8 hr. incubation with physiological saline containing 10−7 M TTX with the stomatogastric nerve transected, (3) TEVC (n=12): 8 hr. incubation with physiological saline containing 10−7 M TTX with the stomatogastric nerve transected, but voltage clamped to their endogenous waveform. To obtain the physiological waveform to design the voltage clamp protocol, we recorded PD activity for 5 minutes. We then selected two cycles of PD bursts and used this voltage activity as a voltage clamp protocol for two-electrode voltage clamp (TEVC). Given that PD has a highly stereotyped voltage waveform that persists over the course of days in vitro [12], and is similar to what is observed in vivo [28], we are confident that the recordings we “played back” with the voltage-clamp protocol are representative of normal PD activity. For TEVC, PD neurons were impaled with two 15-20 MΩ microelectrodes filled with 20 mM KCl + 600 mM K2SO4 and the gain was set to ~25 (×100 V/V).
Harvesting identified neurons:
After experiments, a petroleum jelly well was built around the STG. The ganglion was then exposed to ~2.5 mg/ml protease (Sigma – P6911, St. Louis, MO, USA) diluted in physiological crab saline to digest the connective tissue. After loosening of the neurons during digestion, the protease was thoroughly washed away from the ganglion. 70% ethylene glycol diluted in crab saline then replaced the saline in the well over the course of ~15-20 min. The petri dish was then placed at - 20°C for ~1-2 hrs. PD neurons were hand dissected from the ganglion using fine hand-help forceps. Each neuron was placed in 400 μl of lysis buffer (Zymo Research, Irvine, CA, USA) and stored at −80’C.
cDNA synthesis and pre-amplification of cDNA targets:
Total RNA was isolated from each neuron using the Quick-RNA MicroPrep kit (Zymo Research, Irvine, CA, USA) per the manufacturer’s instructions. Reverse transcription of total RNA was then performed using a mixture of oligo-dT and random hexamer primers (qScript cDNA Supermix; QuantaBio, Beverly, MA, USA). Half of the cDNA produced from each neuron (10 μL) was then pre-amplified using PerfeCTa PreAmp Supermix (QuantaBio, Beverly, MA, USA) according to the manufacturer’s instructions (20 μL reaction volume). This protocol utilizes a 14-cycle PCR reaction primed with a pool of target-specific primers to enrich subsequent qPCR reactions when starting sample is limited. After preamplification, cDNA samples were diluted 7.5x in nuclease-free water (150 μL final volume).
Multiplex primer and probe design:
Primer and probe sequences for the following channel targets were used in this study: Na+ leak channel (NALCN), Ca2+ voltage-gated channel auxiliary beta subunit (CACNAB), inward rectifying K+ channel (IRK), hyperpolarization-activated current (IH), small conductance Ca2+-activated K+ channel (SKKCa), delayed-rectifying K+ channel (Shawl), delayed-rectifying K+ channel (Shaw2), voltage-gated Na+ channel (NaV), A-type K+ channel (Shaker), delayed-rectifying K+ channel (Shab), L-type Ca2+ channel (CaV1), P/Q-type Ca2+ channel (CaV2), A-type K+ channel (Shal), large conductance Ca2+ activated K+ channel (BKKCa).
PD neurons are known to use acetylcholine as a neurotransmitter [29]. As a quality control measure, we measured the expression of genes associated with cholinergic and glutamatergic transmission: (1) acetylcholinesterase (AChE), (2) vesicular acetylcholine transporter (vAChT), (3) choline acetyltransferase (ChAT), and (4) vesicular glutamate transporter (vGluT). All neurons in the study had high levels of cholinergic genes and low levels of vGluT, enhancing our confidence that only PD neurons were used in this study.
Assays were designed using the RealTimeDesign™ qPCR Assay Design Software by LGC Biosearch Technologies (Teddington, UK) based on open reading frames determined previously [14] or from sequences from Cancer borealis nervous system transcriptome [30]. Forward and reverse primer pairs, probe sequences, and labeling fluorophores were as listed in Table S5.
Quantitative polymerase chain reaction (qPCR):
10 μL qPCR reactions included 2.5 μL of preamplified cDNA and were run using PerfeCTa Multiplex qPCR ToughMix per the manufacturer’s instructions (5X, QuantaBio, Beverly, MA, USA). Multiplex reactions for the for groups of genes shown in Table S5 were run in triplicate on 96-well plates using a CFX96 Touch™ Real-Time PCR Detection System from Bio-Rad (Hercules, CA, USA). Cycling conditions for qPCR reactions were as follows: 95°C for 3 min; 40 cycles of 95°C for 15 sec and 58°C for 1 min. Fluorescent mea surements were taken at the end of each cycle. Conversion of quantitation cycle (Cq) value to absolute copy number for each gene was estimated by interpolating Cq values for each gene into a standard curve of known copy number from 106 to 101 copies. We then corrected for the amount of sample used and also the 14-cycle preamplification in our final estimation.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistics were run with either Graphpad Prism 6.01 (La Jolla, CA, USA) or R [31]. For all pairwise correlations, we obtained Pearson’s correlation coefficients (r) and p values. Pairwise comparisons of channel mRNA abundance were plotted using Graphpad Prism. Comparisons of the slopes between activity-dependent relationships were carried out using an analysis of covariance in Graphpad Prism. To visualize the correlation network in PD neurons, correlation network plots in Figure 1 were made using the qgraph package in R [32]. Briefly, Pearson’s r for all pairwise correlations were drawn as lines that connected correlated genes. The thickness of the line scales proportionally with strength of the correlation. The lowest correlation (and therefore, the thinnest lines) was set at a correlation coefficient of 0.65 because in all cases, when r was ≥0.65, p was <0.05. Occasionally, individual genes had uncharacteristically low or high abundance (~ 3 qPCR cycles different from the mean Cq). To reduce errors in correlation analysis, these individual data points were omitted. Cumulative probability distributions of the 78 pairwise comparisons made in this data set were analyzed using the Kolmogorov-Smirnov test. Δr was calculated by subtracting the Pearson’s r of the series 2 control group from the r value obtained for each pairwise correlation in treatment conditions (silent – control series 2 or TEVC – control series 2). To provide a control distribution for these analyses, we determine control Δr by subtracting r values obtained from the control group in series 2 from the r values for each pairwise comparison in the experiment series 1 (control series 1 – control series 2). Therefore, a negative (left) shift in the distribution of Δr represents a reduced correlation profile. For all tests, significance was accepted when p<0.05
Supplementary Material
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Oligonucleotides | ||
| See Table S5 | Integrated DNA Technologies | |
| Chemicals | ||
| Tetrodotoxin | Alomone Labs | T-550 |
| Protease, Streptomyces griseus | Sigma-Aldrich | P6911 |
| Experimental Models: Organisms/Strains | ||
| Cancer borealis | The Fresh Lobster Company (Gloucester, MA) | https://thefreshlobstercompany.com/ |
Highlights:
The expression ratios among ion channels determine neuronal activity.
The signals that regulate these patterns remain unclear.
Ongoing membrane voltage is a major coordinator of ion channel relationships.
Membrane activity can maintain or suppress correlated ion channel mRNA levels.
Acknowledgements
We would like to thank Virginia Garcia for validating several of the qPCR assays used in this study. This work was funded by NIH grant R01 MH46742 to Eve Marder and David Schulz.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
REFERENCES
- 1.Marder E, and Goaillard J-M (2006). Variability, compensation and homeostasis in neuron and network function. Nature Reviews Neuroscience 7, 563–574. [DOI] [PubMed] [Google Scholar]
- 2.Beck H, and Yaari Y (2008). Plasticity of intrinsic neuronal properties in CNS disorders. Nature Reviews Neuroscience 9, 357–369. [DOI] [PubMed] [Google Scholar]
- 3.Staley K (2015). Molecular mechanisms of epilepsy. Nature neuroscience 18, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kourrich S, Calu DJ, and Bonci A (2015). Intrinsic plasticity: an emerging player in addiction. Nature Reviews Neuroscience 16, 173. [DOI] [PubMed] [Google Scholar]
- 5.Turrigiano G (2012). Homeostatic synaptic plasticity: local and global mechanisms for stabilizing neuronal function. Cold Spring Harbor perspectives in biology 4, a005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Temporal S, Lett KM, and Schulz DJ (2014). Activity-dependent feedback regulates correlated ion channel mRNA levels in single identified motor neurons. Current biology 24, 1899–1904. [DOI] [PubMed] [Google Scholar]
- 7.O’Leary T, Williams AH, Franci A, and Marder E (2014). Cell types, network homeostasis, and pathological compensation from a biologically plausible ion channel expression model. Neuron 82, 809–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeMasson G, Marder E, and Abbott L (1993). Activity-dependent regulation of conductances in model neurons. Science 259, 1915–1917. [DOI] [PubMed] [Google Scholar]
- 9.Desai NS, Rutherford LC, and Turrigiano GG (1999). Plasticity in the intrinsic excitability of cortical pyramidal neurons. Nature neuroscience 2, 515–520. [DOI] [PubMed] [Google Scholar]
- 10.O’Leary T, van Rossum MC, and Wyllie DJ (2010). Homeostasis of intrinsic excitability in hippocampal neurones: dynamics and mechanism of the response to chronic depolarization. The Journal of physiology 588, 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Temporal S, Desai M, Khorkova O, Varghese G, Dai A, Schulz DJ, and Golowasch J (2012). Neuromodulation independently determines correlated channel expression and conductance levels in motor neurons of the stomatogastric ganglion. Journal of Neurophysiology 107, 718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khorkova O, and Golowasch J (2007). Neuromodulators, not activity, control coordinated expression of ionic currents. Journal of Neuroscience 27, 8709–8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fong M.-f., Newman JP, Potter SM, and Wenner P (2015). Upward synaptic scaling is dependent on neurotransmission rather than spiking. Nature communications 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulz DJ, Goaillard J-M, and Marder EE (2007). Quantitative expression profiling of identified neurons reveals cell-specific constraints on highly variable levels of gene expression. Proceedings of the National Academy of Sciences 104, 13187–13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson AE, and Prinz AA (2010). Conductance ratios and cellular identity. PLoS Comput Biol 6, e1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao S, and Golowasch J (2012). Ionic current correlations underlie the global tuning of large numbers of neuronal activity attributes. Journal of Neuroscience 32, 13380–13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Leary T, Williams AH, Caplan JS, and Marder E (2013). Correlations in ion channel expression emerge from homeostatic tuning rules. Proceedings of the National Academy of Sciences 110, E2645–E2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turrigiano G, Abbott L, and Marder E (1994). Activity-dependent changes in the intrinsic properties of cultured neurons. Science 264, 974–976. [DOI] [PubMed] [Google Scholar]
- 19.Siegel M, Marder E, and Abbott L (1994). Activity-dependent current distributions in model neurons. Proceedings of the National Academy of Sciences 91, 11308–11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turrigiano G, LeMasson G, and Marder E (1995). Selective regulation of current densities underlies spontaneous changes in the activity of cultured neurons. Journal of Neuroscience 15, 3640–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haedo RJ, and Golowasch J (2006). Ionic mechanism underlying recovery of rhythmic activity in adult isolated neurons. Journal of neurophysiology 96, 1860–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacLean JN, Zhang Y, Johnson BR, and Harris-Warrick RM (2003). Activity-independent homeostasis in rhythmically active neurons. Neuron 37, 109–120. [DOI] [PubMed] [Google Scholar]
- 23.Martin MS, Dutt K, Papale LA, Dubé CM, Dutton SB, de Haan G, Shankar A, Tufik S, Meisler MH, and Baram TZ (2010). Altered function of the SCN1A voltage-gated sodium channel leads to GABAergic interneuron abnormalities. Journal of Biological Chemistry, jbc. M109. 078568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank HY, Mantegazza M, Westenbroek RE, Robbins CA, Kalume F, Burton KA, Spain WJ, McKnight GS, Scheuer T, and Catterall WA (2006). Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nature neuroscience 9, 1142. [DOI] [PubMed] [Google Scholar]
- 25.Del Negro CA, Funk GD, and Feldman JL (2018). Breathing matters. Nat Rev Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ibata K, Sun Q, and Turrigiano GG (2008). Rapid synaptic scaling induced by changes in postsynaptic firing. Neuron 57, 819–826. [DOI] [PubMed] [Google Scholar]
- 27.Li B, Tadross MR, and Tsien RW (2016). Sequential ionic and conformational signaling by calcium channels drives neuronal gene expression. Science 351, 863–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rezer E, and Moulins M (1983). Expression of the crustacean pyloric pattern generator in the intact animal. Journal of Comparative Physiology A: Neuroethology, Sensory, Neural, and Behavioral Physiology 153, 17–28. [Google Scholar]
- 29.Marder E (1976). Cholinergic motor neurones in the stomatogastric system of the lobster. The Journal of physiology 257, 63–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Northcutt AJ, Lett KM, Garcia VB, Diester CM, Lane BJ, Marder E, and Schulz DJ (2016). Deep sequencing of transcriptomes from the nervous systems of two decapod crustaceans to characterize genes important for neural circuit function and modulation. BMC genomics 17, 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Team, R.C.D. (2014). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. [Google Scholar]
- 32.Epskamp S, Cramer AO, Waldorp LJ, Schmittmann VD, and Borsboom D (2012). qgraph: Network visualizations of relationships in psychometric data. Journal of Statistical Software 48, 1–18. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.