Abstract
Background:
Most eukaryotic protein-coding genes exhibit alternative cleavage and polyadenylation (APA), resulting in mRNA isoforms with different 3′ untranslated regions (3′ UTRs). Studies have shown that brain cells tend to express long 3′ UTR isoforms using distal cleavage and polyadenylation sites (PASs).
Methods:
Using our recently developed, comprehensive PAS database PolyA_DB, we developed an efficient method to examine APA, named Significance Analysis of Alternative Polyadenylation using RNA-seq (SAAP-RS). We applied this method to study APA in brain cells and neurogenesis.
Results:
We found that neurons globally express longer 3′ UTRs than other cell types in brain, and microglia and endothelial cells express substantially shorter 3′ UTRs. We show that the 3′ UTR diversity across brain cells can be corroborated with single cell sequencing data. Further analysis of APA regulation of 3′ UTRs during differentiation of embryonic stem cells into neurons indicates that a large fraction of the APA events regulated in neurogenesis are similarly modulated in myogenesis, but to a much greater extent.
Conclusion:
Together, our data delineate APA profiles in different brain cells and indicate that APA regulation in neurogenesis is largely an augmented process taking place in other types of cell differentiation.
Keywords: alternative polyadenylation, brain cells, RNA-seq, scRNA-seq
Author summary:
Most eukaryotic protein-coding genes express isoforms with different 3′ UTR lengths. Studies have shown that transcripts expressed in brain tend to have longer 3′ UTRs compared to other tissues. We have developed an efficient computational method to analyze 3′ UTR isoforms using RNA-seq data. We show that neurons have the longest 3′ UTRs among all brain cell types and 3′ UTRs are the shortest in microglia and endothelial cells. This finding is also supported by single cell sequencing data. We further show that 3′ UTRs lengthen in neurogenesis, similar to that in myogenesis. However, 3′ UTR lengthening is much potent in differentiating neurons.
INTRODUCTION
Cleavage and polyadenylation (C/P) is an essential step for 3′ end maturation of almost all eukaryotic mRNAs [1]. The C/P site, also known as the polyA site or PAS, is defined by surrounding sequence motifs [2], which are recognized by the C/P machinery [3]. Over 70% of mammalian genes display alternative cleavage and polyadenylation (APA), resulting in mRNA isoforms with different 3′ ends [4,5]. Most APA sites are located in the 3′ untranslated region (3′ UTR) of mRNAs, leading to isoforms with different 3′ UTR lengths [4]. Differential expression of APA isoforms has been shown in different cells and tissue types [6,7]. For example, genes in brain express longer 3′ UTRs as compared to other tissues [7,8], whereas transcripts in testis have short 3′ UTRs [9]. In addition, APA is dynamically and globally regulated in a number of biological conditions, such as cell proliferation and differentiation, development, cancer, and neuronal activation [10–14].
Because 3′ UTRs harbor regulatory elements for aspects of mRNA metabolism, such as nuclear export, stability, translational efficiency, and subcellular localization [1,15,16], APA is believed to play an important role in post-transcriptional control of gene expression in brain. Isoforms with long 3′ UTRs in brain have been shown to have functional impacts on the nervous system. For example, dysregulation of APA of MeCP2 has been implicated in intellectual disability and neuropsychiatric diseases [17,18]. Another example is the gene encoding brain derived neuropathic factor (BDNF), which has two 3′ UTR isoforms. While the short 3′ UTR isoform is restricted to soma, the long isoform is enriched in dendrites [19,20]. Despite some conflicting data [21], it is generally believed that long 3′ UTR isoforms in brain cells are localized differently than short 3′ UTR isoforms [22,23].
A number of 3′ end sequencing methods have been developed in recent years, allowing specific interrogation of APA isoforms [4,5,24–27]. However, the vast amount of RNA-seq data available in the public domain offers a treasure trove for mining APA profiles. Several bioinformatic methods have been developed to examine APA using RNA-seq data, falling into two categories. One group of methods examines APA using annotated PASs [28,29]; and other group predicts PASs based on difference in RNA-seq read coverage before and after a PAS [30–33].
Here, using RNA-seq data and our recently created database PolyA_DB 3 with comprehensive PAS annotations, we examine APA in different cell types of mouse brain. We use RNA-seq data from bulk samples as well as from single cells. Comparison of APA regulation in neurogenesis with that in myogenesis indicates general similarities but different extents of 3′ UTR APA.
RESULTS
Analysis of APA using RNA-seq data and annotated PASs
We recently comprehensively cataloged PASs in human, mouse, and rat genomes using a large number of samples from diverse biological conditions [34]. We reasoned that combining well annotated PASs with RNA-seq data could offer an efficient approach to study APA. To this end, we developed a method, named Significance Analysis of Alternative Polyadenylation using RNA-seq (SAAP-RS). As illustrated in Figure 1A, for each interrogated PAS in a 3′ UTR, we calculated RNA-seq read counts in its upstream (UP) and downstream (DN) sequences in the 3′ UTR, followed by a statistical test to derive a P-value for significance of difference in relative isoform expression between samples (see Methods for detail). For the statistical test, we used either the Fisher’s exact test when there was no replicate or the DEXSeq method [35] when there were replicates to obtain data dispersion.
Figure 1. APA analysis using RNA-seq data.
(A) Schematic of the SAAP-RS strategy. RNA-seq reads are divided into upstream (UP) and downstream (DN) region groups relative to the reference PAS obtained from the PolyA_DB database. A statistical test, such as the Fisher’s exact test or DEXSeq, is used to compare the read distribution in UP and DN regions between two sample groups. As indicated, the relative expression difference (RED) value for each reference PAS is the difference in log2(DN/UP) between two sample groups (test vs. reference). (B) 3′ UTR APA analysis of 3′ READS data from brain and testis. Each dot represents a gene with two 3′ UTR isoforms, named proximal PAS (x-axis) and distal PAS (y-axis). Blue (91) and red (1,291) dots represent genes with significantly shortened and lengthened 3′ UTRs in brain vs. testis, respectively (P <0.05, DEXSeq, n = 2). Ratio of number of red dots to blue dots is indicated. (C)Box plots showing RED values based on different methods to choose a reference PAS for APA analysis. Median RED values are indicated. Highest RPM and Highest PSE PASs are those with the highest expression levels of all samples in the PolyA_DB database and with the highest percent of samples with expression across samples, respectively. (D) An example gene Hspa4l shown in USCS genome browser. Conserved and non-conserved PASs annotated in the PolyA_DB are shown in different colors as indicated. Selected PASs for different PAS selection methods are highlighted in red. (E) Scatter plots comparing RED values from 3′ READS and those from RNA-seq data. Different reference PAS selection methods are shown. Pearson correlation coefficient (r) is shown.
To test SAAP-RS, we selected an RNA-seq dataset for mouse brain and testis, which our lab previously generated [4]. Because we also processed the same RNA samples by 3′ region extraction and deep sequencing (3′ READS), a specialized method which generates reads at the 3′ ends of poly(A)+ transcripts [4], we could directly compare results from the two sequencing methods.
As we previously reported [4], a substantial global APA bias was detected between brain and testis with the 3′ READS data, based on comparison of top two most abundant 3′ UTR isoforms of each gene. The number of genes expressing the long 3′ UTR isoform to a higher level in brain than in testis was significantly greater than those with the opposite trend by 14-fold (1,291 vs. 91, Figure 1B). To examine the extent of 3′ UTR size difference for each gene between samples, we calculated a relative expression difference (RED) value, reflecting difference in log2(distal PAS/proximal PAS) between two samples. Log2(distal PAS/proximal PAS) was based on expression levels (reads per million, RPM) of distal and proximal PAS isoforms. The median RED between brain and testis was 1.75 using the3′ READS data (Figure 1C).
To examine APA with RNA-seq data, we considered several options in choosing a reference PAS to gauge 3′ UTR length changes, including the first conserved PAS, the PAS with the highest expression levels (highest RPM) based on all samples used in PolyA_DB [34], the PAS with the widest expression breadth based on the percent of samples with expression (PSE) value in PolyA_DB [34], and the most significantly regulated PAS based on SAAP-RS P-values (see Methods for detail). As shown in Figure 1C, all four methods gave rise to a positive median RED value for brain vs. testis, consistent with the 3′ READS result. None of the RED values, however, were as high as that from the 3′ READS analysis, indicating lower sensitivity to detect 3′ UTR length changes using RNA-seq data as compared to 3′ end sequencing. However, this is expected, because RNA-seq data could not resolve individual PAS isoforms and, hence, reads in upstream and downstream regions of a reference PAS could come from multiple APA isoforms. By contrast, 3′ READS data are specific for individual PASs, providing sharper differences between isoforms. We found that using the most significant PAS as a reference gave a higher RED value (1.07) than other methods (0.78–0.79) (Figure 1C). An example gene, Hspa4l, is shown in Figure 1D, with reference PASs used by the four methods indicated.
We next directly compared RED values obtained from 3′ READS with those from RNA-seq data for individual genes (Figure 1E). The method using the first conserved PAS as a reference showed a better correlation (r = 0.33, Pearson correlation, Figure 1E) than other methods (r = 0.28–0.29, Figure 1E). Therefore, we conclude that RNA-seq data coupled with annotated PASs can be effectively used to examine APA changes despite lower sensitivity than data from 3′ end sequencing.
Neurons globally express longer 3′ UTRs than other cell types in mouse brain
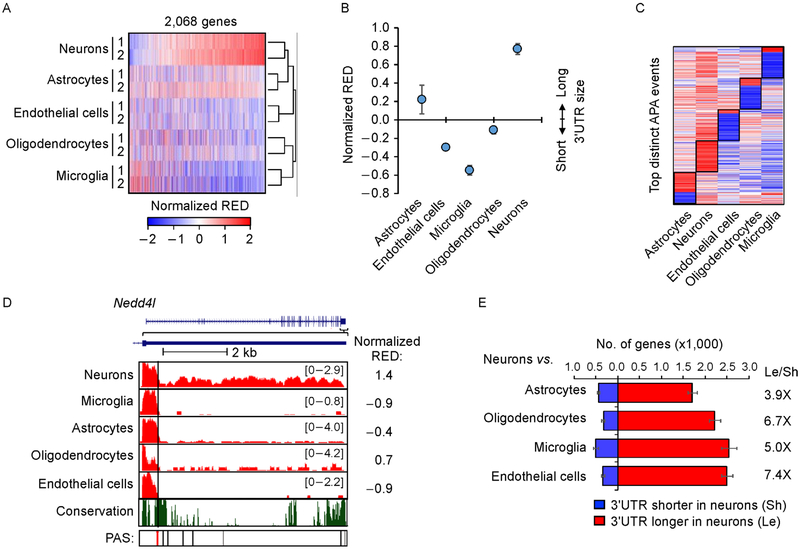
Previous studies indicated longer 3′ UTRs in brain than other tissues [7,8]. However, how different cell types in brain differ in APA is unclear. With SAAP-RS, we next set out to examine APA profiles in different cell types of brain using an RNA-seq dataset generated by Zhang et al. [36]. With this data, different types of cells in mouse cerebral cortex were isolated through immunopanning and fluorescence-activated cell sorting (FACS) [36]. The cell types include astrocytes, neurons, oligodendrocytes, endothelial cells, and microglia. To compare APA profiles across these cell types, we first calculated a normalized RED value for each gene using the first conserved 3′ UTR APA site as a reference. Only genes with expression in all cell types were used (see Methods for detail). Heatmap and clustering analyses indicate that the APA profile is distinct among different brain cells (Figure 2A). Neurons displayed the longest 3′ UTRs overall (median RED = 0.77, Figure 2B), followed by astrocytes (median RED = 0.2) and oligodendrocytes (median RED= − 0.1). Endothelial cells and microglia had shorter 3′ UTRs globally, with median RED= − 0.30 and − 0.54, respectively.
Figure 2. APA differences among brain cells.
(A) Heat map of APA profiles of different brain cells. Normalized RED values by standardization across samples are shown in the heat map. RED is based on comparison of each sample with the mean of all samples. The first conserved PAS was used as the reference PAS for RED calculation. A total of 2,068 genes are shown. Samples are clustered using Pearson correlation as a metric and genes are sorted by the mean RED values of neurons. (B) Median normalized RED values of each cell type. Two replicates for each cell type were averaged for plotting. Error bars are standard deviation of two replicates. (C) Heatmap of top distinct APA events of different cell types. Distinct APA events were selected by Wilcoxon test comparing RED values of one cell type with those of other cell types (see Methods for detail). (D) An example gene Nedd4l shown in UCSC genome browser, which showed longest 3′ UTRs in neurons. The reference PAS is indicated by a red line. Other conserved (black) and non-conserved (gray) PASs also indicated. The conservation track is based on seven mammalian species (rat, human, orangutan, horse, dog, chicken, and opossum). (E) Bar graph showing the number of genes with significantly shortened (blue) or lengthened (red) 3′ UTRs (P<0.05, DEXSeq) in neurons vs. another cell type. The most significant 3′ UTR PAS was used as the reference for analysis. The ratio of number of 3′ UTR lengthened genes to number of 3′ UTR shortened genes for each cell type is indicated. Error bars are standard deviation obtained by bootstrapping (see Methods for detail).
We next identified top 50 APA events that were most distinct in each cell type as compared to other types, using gene RED values (see Methods for detail). Consistent with the global analysis result, all distinct APA events in neurons showed longer 3′ UTRs than other cell types (Figure 2C). Astrocytes also showed longer 3′ UTRs in most of the events. By contrast, distinct APA events in endothelial cells, microglia and oligodendrocytes corresponded largely to shorter 3′ UTRs (Figure 2C). An example gene, Nedd4l, is shown in Figure 2D, which encodes an E3 ubiquitin-protein ligase involved in regulation of several molecules and pathways, such as EGFR, WNT signaling pathway and ion channels [37,38]. While neurons had abundant RNA-seq reads in the downstream region of the reference PAS, other cells had much fewer reads in the region (Figure 2D). Some example marker genes for other cell types are shown in Supplementary Figure S1A.
To further corroborate the first conserved PAS-based result, we next carried out pair-wise comparisons between neurons and cells of another type (Figure 2E), requiring expression of gene only in the two comparing cell types. We used the most significant PAS out of all possible PASs as a reference to gauge 3′ UTR length differences. Consistent with the all cell comparison result using the first conserved PAS, neurons showed longer 3′ UTRs than other cell types (Figure 2E). The ratio of genes with longer 3′UTRs in neurons to genes with the opposite trend was greater than three in all comparisons (Figure 2E). Taken together, our data indicate that neurons globally express longer 3′ UTRs than other cell types in brain.
Some neuron-enriched genes appear to express longer 3′ UTRs in non-neuronal cells
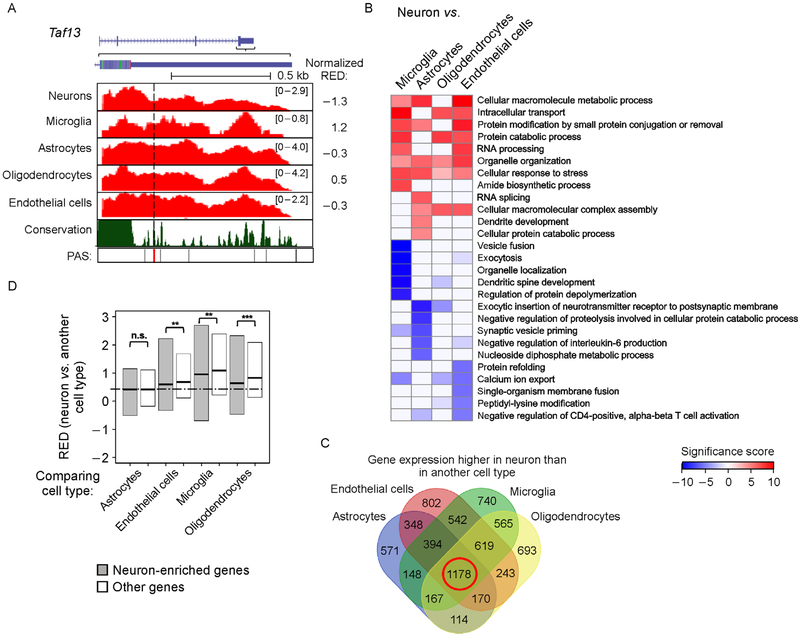
We also noticed from all sample comparisons (Figure 2A) and pair-wise comparisons (Figure 2E) that some genes showed shorter 3′ UTRs in neurons than in other cells (blue genes in Figure 2A and 2E). An example gene Taf13, encoding transcription initiation factor TFIID subunit 13, is shown in Figure 3A, which had a lower RED value in neurons than in other cell types. Venn diagram analysis indicated that genes with longer 3′ UTRs in non-neuronal cells were more likely to have restricted expression in certain cell types. For example, whereas 637 genes showed longer 3′ UTRs in neurons than all other cell types (Supplementary Figure S1B), only 12 genes had shorter 3′ UTRs in neurons than all other cell types (Supplementary Figure S1C).
Figure 3. Some neuron-enriched genes have longer 3′ UTRs in non-neuronal cells.
(A) An example gene Taf13 shown in UCSC genome browser, which displayed longer 3′ UTRs in other cell types as compared to neurons. See Figure 2D for figure format. (B) Top biological processes GO terms enriched for genes with different 3′ UTR lengths in neurons vs. another cell type. Top five GO terms were selected for each comparison shown in Figure 2E. Significance score (SS) was calculated by (−log10P)*s, where P is based on the Fisher’s exact test indicating the significance of association between a GO term and genes with longer or shorter 3′ UTRs. s = 1 for association with genes with longer 3′ UTRs in neurons and s = −1 for association with genes with shorter 3′ UTRs in neurons. GO terms are sorted based on the most significant term across all comparisons. SS values are shown in a heatmap. (C) Venn diagram showing significantly upregulated genes in neurons as compared to four other cell types (FDR<0.05, DESeq; fold change>2). The 1,178 genes overlapping among all four comparisons are defined as neuron-enriched genes. (D) Box plot showing RED values of neurons vs. another cell type for neuron-enriched genes and other genes. Significance of difference between neuron-enriched genes and other genes is indicated (* P<0.01, ** P<0.001 and *** P<1 × 10−5, Wilcoxon test).
We next carried out Gene Ontology (GO) analysis of the genes with different 3′ UTR lengths in neurons vs. other cell types (Figure 3B). Interestingly, the top GO terms for genes with longer 3′ UTRs in neurons were related to basic cellular functions, such as “cellular macromolecular metabolic process” and “intracellular transport”, as well as RNA metabolism processes, such as “RNA processing”. A few GO terms related to protein degradation and modification, such as “protein catabolic process”, “protein modification by small protein conjugation or removal” and “cellular protein catabolic process” were also enriched. By contrast, GO terms enriched for genes with longer 3′ UTRs in other cell types varied between different comparisons and some were related to neuronal features (Figure 3B). The most significant GO terms were those associated with longer 3′ UTRs in microglia, including “vesicle fusion”, “exocytosis”, “organelle localization”, “dendritic spine development”, etc.
Next, we specifically examined neuron-enriched genes. Using the same RNA-seq data, we identified a total of 1,178 genes that had significantly higher expression levels in neurons as compared to other cell types (FDR < 0.05, DESeq analysis, fold change>2, Figure 3C). Interestingly, these genes, named “neuron-enriched genes”, displayed lesser lengthened 3′ UTRs in neurons as compared to other cell types (Figure 3D). Some examples are shown in Supplementary Figure S1D. Taken together, our results indicate that some neuron-enriched genes tend to show longer 3′ UTRs in non-neuronal cells where their expression levels are low.
Single cell RNA-seq data corroborate bulk RNA sample results
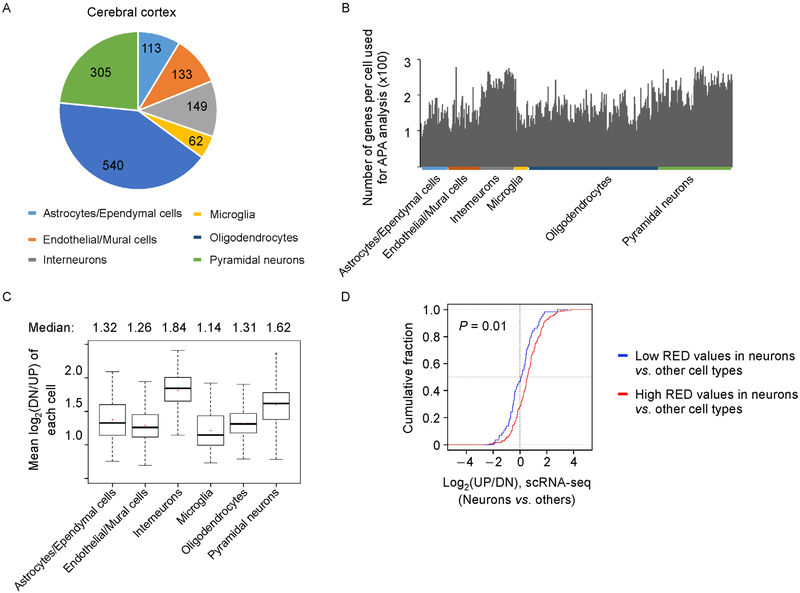
To corroborate our findings based on RNA-seq data with RNA from bulk samples, we resorted to single cell RNA-seq (scRNA-seq) data generated by Zeisel et al. [39], where single cells from mouse cerebral cortex and hippocampus were analyzed. For cerebral cortex, which is the same region used by Zhang et al. [36], the authors identified 113 astrocytes/ependymal cells, 133 endothelial/mural cells, 149 interneurons, 62 microglial cells, 540 oligodendrocytes and 305 pyramidal neurons (Figure 4A). We first identified genes that had reads in 3′ UTRs and then calculated log2(DN/UP) using the first conserved PAS as the reference for APA analysis (see Methods for detail). Because of the shallow sequencing depth of scRNA-seq, we were able to examine only 100–300 genes in each cell (Figure 4B). As shown in Figure 4C, interneurons and pyramidal neurons had the longest 3′ UTRs (median RED = 1.84 and 1.62, respectively) compared to other cell types (median RED = 1.32 or lower, Figure 4C). Again, microglia showed the shortest 3 ′ UTRs among all cell types (Figure 4C).
Figure 4. Analysis of APA events using single cell RNA-seq data.
(A) A pie chart showing number of cells for each cell type in cerebral cortex used in the analysis [39]. (B) Bar graph showing number of genes of each cell used for APA analysis. Genes that have at least 1 read in either UP or DN region are shown. Cell types are indicated at the bottom with colored bars. (C) Box plot showing mean log2(DN/UP) of all single cells of a cell type. (D) Cumulative distribution function (CDF) curves showing log2(DN/UP) values of genes based on neurons vs. other cells. Two gene groups are shown. Gene with high RED (red) or low RED (blue) values are those selected from bulk RNA-seq analysis shown in Figure 2A. P-value (Kolmogorov-Smirnov (K-S) test) indicating difference between two gene groups is shown.
We next applied the same method to examine APA in hippocampal samples, which included 81 astrocytes/ependymal cells, 33 endothelial/mural cells, 126 interneurons, 14 microglial cells, 121 oligodendrocytes and 936 pyramidal neurons (Supplementary Figure S2A and S2B). Again, we observed longer 3′ UTRs in interneurons and pyramidal neurons than in other cell types (Supplementary Figure S2C). Note the data of microglial cells were not conclusive due to their small sample size (only 14 cells) and, hence, high data variation (Figure S2C).
We next wanted to compare bulk RNA-seq results with scRNA-seq results. To this end, we selected top and bottom 25% of genes with respect to RED values in neurons (heatmap in Figure 2A), and examined their respective log2(DN/UP) values in the scRNA-seq data. As shown in Figure 4D, genes with high RED values in neurons with bulk RNA samples also showed significantly higher log2(DN/UP) values in neurons based on the scRNA-seq data, as compared to genes with low RED values (P = 0.01, Kolmogorov-Smirnov (K-S) test). In summary, our scRNA-seq analysis supports the conclusion that neurons in general have longer 3′ UTRs than other brain cells, and different genes vary their 3′ UTR lengths to different degrees between neurons and other cell types.
3′ UTR lengthening in neurogenesis
Genes display 3′ UTR lengthening during cell differentiation and development [4,10]. We next wanted to address to what extent the 3′UTR length differences between neurons and other cell types are attributable to APA regulation during neurogenesis. To this end, we analyzed RNA-seq datasets from two studies that involved differentiation of mouse embryonic stem cells (mESCs) into terminally differentiated neurons [40,41].
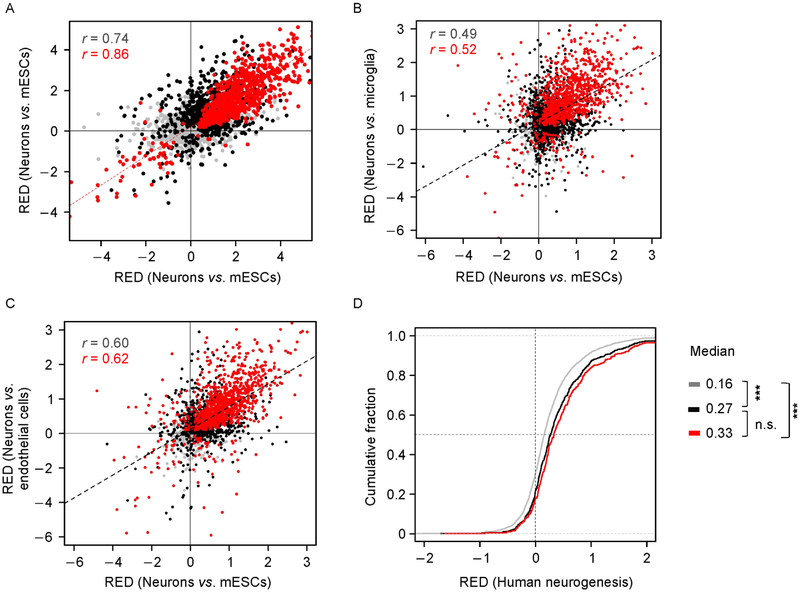
Using SAAP-RS, we observed overall 3′ UTR lengthening during neurogenesis with both data sets (median RED = 0.56 and 0.44 for Ref. [41] and Ref. [40], respectively) and a high correlation between these two data sets (r = 0.74 for all genes and r = 0.86 for significantly regulated genes in both studies, Figure 5A).
Figure 5. APA in neurogenesis.
(A) Scatter plot comparing RED values during neurogenesis (differentiated neurons vs. mESC) from Ref. [41] (x-axis, n = 1) and from Ref. [40] (y-axis, n = 2). Significantly regulated genes (P <0.01, Fisher’s Exact Test) in only one of the data sets are shown in black (2,028), and those significant in both data sets are in red (1,101). Pearson correlation coefficient (r) for each gene group is indicated. (B and C) Comparison of APA events in neurogenesis with those in neurons vs. microglia (B) or in neurons vs. endothelial cells (C). Neurogenesis RED values are based on averaged REDs from two different studies. Significantly regulated genes in only one of the comparisons are shown in black and those significant in both are in red (P<0.01, Fisher’s Exact Test). Pearson correlation coefficient (r) is indicated. (D) Comparison of APA regulation in mouse neurogenesis vs. human neurogenesis. Gene groups (indicated by colors) are those from (A). Median values and P-values for differences between groups (K-S test) are indicated ( *** P<1 × 10−7).
To address whether long 3′ UTRs in mature neurons are established in neurogenesis, we compared RED values in neurogenesis with that in neuron vs. other brain cells (Figure 5B and 5C). We observed overall high correlations between neurogenesis and neuron vs. microglia cells (r = 0.52, Pearson correlation) or neurons vs. endothelial cells (r = 0.62, Pearson correlation).
GO analysis indicated that the genes with lengthened 3′ UTRs during neurogenesis tended to be enriched for several biological processes, such as “peptide metabolic process”, “RNA processing” and “cellular macromolecular complex assembly”, and some cellular components, such as “intracellular part”, “intracellular ribonucleoprotein complex”, “mitochondrial part” (Supplementary Figure S3A). Interestingly, similar GO terms were also enriched for genes with longer 3′ UTRs in neurons as compared to other brain cells (Supplementary Figure 3B).
To examine how 3′ UTR lengthening in neurogenesis is conserved between mouse and human, we analyzed RNA-seq data from Ref. [42], in which induced pluripotent stem cells (IPSCs) were differentiated into mature neurons [42], and from Blair et al., in which human ESCs (hESCs) were differentiated into mature neurons [43]. Both data sets showed significant 3′ UTR lengthening (Figure S3B) and were well correlated (r = 0.49 for all genes, and 0.70 for significantly regulated genes in both studies). In addition, using orthologous genes, we found that the gene set with the most significant 3′ UTR lengthening in murine neurogenesis also displayed the greatest 3′ UTR lengthening in human neurogenesis (Figure 5D), supporting conservation of 3′ UTR lengthening in neurogenesis between the two species.
Similarity in APA regulation between neurogenesis and myogenesis
We previously showed that 3′ UTRs generally lengthen in myogenesis, which recapitulates APA regulation in embryonic development [4]. We next wanted to examine how 3′ UTR lengthening in neurogenesis is related to that in myogenesis. To this end, we first analyzed APA of C2C12 differentiation data sets from two different studies (Supplementary Figure S4A) [44,45]. Overall, myogenesis displayed 3′ UTR lengthening to a lesser extent than neurogenesis (median RED = 0.13 vs. 0.47). However, a modest positive correlation between these two processes could be discerned (r = 0.40 for all genes and r = 0.44 for significantly regulated genes in both, Figure 6B), indicating APA regulation in myogenesis is related to that in neurogenesis. We identified 347 genes that were commonly lengthened in both neurogenesis and myogenesis. Interestingly, GO analysis showed that these consistently regulated genes were enriched for several GO terms, including “RNA processing”, “translation” and “mitochondrial ribosome” (Table 1). An example gene Pdk1 encoding pyruvate dehydrogenase kinase 1 is shown in Figure 6C, which displayed 3′ UTR lengthening in both neurogenesis and myogenesis (Figure 6C). This gene is involved in many biological events from cancer to Alzheimer’s disease [46–48]. A previous study showed that Pdk1 deficiency in mouse brain caused abnormalities such as microcephaly and neuronal hypertrophy [49].
Figure 6. APA regulation in neurogenesis vs. myogenesis.
(A) CDF curves showing RED values for myogenesis (black) and neurogenesis (red). RED values are based on the first conserved 3′UTR PAS as the reference for APA analysis. Each curve was based on two different studies. (B) Scatter plot comparing 3′UTR APA in neurogenesis vs. myogenesis. Neurogenesis and myogenesis RED values are based on two different studies each. Significantly regulated genes in only one process are shown in black and those significant in both are shown in red (P <0.01, Fisher’s Exact Test). Pearson correlation coefficient (r)is indicated. (C) An example gene, Pdk1, with lengthened 3′ UTRs in both neurogenesis and myogenesis. The reference PAS used for analysis is indicated by a red arrow. RPM ranges (in brackets) are indicated. RED and P-values (Fisher’s Exact Test) for both comparisons are indicated. (D) Relationship between aUTR size and APA regulation. Gene are divided into 5 bins based on distance between reference PAS and last PAS (aUTR size). Median RED for each bin is shown in the plot. (E) Violin plot showing log2 fold change of 87 C/P factor genes expressed during both neurogenesis and myogenesis. Median values and P-value indicating difference in gene expression changes between neurogenesis and myogenesis (Wilcoxon test) are indicated.
Table 1.
Top biological process and cellular component GO terms enriched for genes with lengthened 3′ UTRs in both neurogenesis and myogenesis
| Category | GO term | P |
|---|---|---|
| Biological process | RNA processing | 6.2E-06 |
| Ribosome biogenesis | 6.2E-05 | |
| Regulation of ubiquitin-protein transferase activity | 2.6E-04 | |
| Translation | 1.2E-03 | |
| Inner mitochondrial membrane organization | 1.9E-03 | |
| Negative regulation of organelle assembly | 2.0E-03 | |
| Viral budding | 4.4E-03 | |
| Establishment of protein localization to mitochondrial membrane | 4.4E-03 | |
| Outer mitochondrial membrane organization | 5.2E-03 | |
| Ribonucleoprotein complex subunit organization | 7.0E-03 | |
| Cellular component | Macromolecular complex | 1.5E-05 |
| Mitochondrial ribosome | 7.1E-04 | |
| Nucleolus | 1.4E-03 | |
| Membrane protein complex | 3.3E-03 | |
| Catalytic step 2 spliceosome | 4.3E-03 |
Genes (347 in total) correspond those red dots in Figure 6B. P-values are based on the Fisher’s exact test.
Consistent with our previous study [50,51], we found that the extent of 3′ UTR lengthening in myogenesis correlates with aUTR size (Figure 6D). Interestingly, a similar trend could be discerned with neurogenesis (Figure 6D), but with a much greater extent of 3′ UTR lengthening.
Previous studies indicated that downregulation of C/P factors leads to 3′ UTR lengthening in cell differentiation [10,50]. We thus wanted to know how C/P factor expression was regulated in neurogenesis. Interestingly, we observed marked downregulation of C/P factor transcripts during neurogenesis, the extent of which was greater than that during myogenesis (median log2 fold change: − 0.7 and − 0.38 for neurogenesis and myogenesis, respectively; P = 4.4 ×10−5, Wilconxon test, Figure 6E). We also observed a moderate correlation of C/P factor expression changes between neurogenesis and myogenesis (r = 0.36 and= 0.51 for all C/P factors and core C/P factors, respectively, Supplementary Figure S4B). Taken together, our data indicate that a common set of APA events are regulated in both myogenesis and neurogenesis. The latter shows augmented 3′ UTR lengthening compared to the former, plausibly due to lower C/P activities in neurons.
DISCUSSION
In this study we developed a method, named SAAP-RS, to examine APA using RNA-seq data combined with comprehensively annotated PAS database, PolyA_DB. We show that using the first conserved PAS as a reference offers an efficient approach to examine global APA profiles across multiple samples, whereas each PAS can be individually examined when only two samples are compared. When replicates are available, the DEXSeq method can be readily used to obtain data dispersion and false discovery rate. Because no de novo prediction of PAS is needed, our method is computationally lightweight and is well suited for large scale mining of APA profiles. While we focused on 3′ UTR APA sites in this study, APA events in introns [52] could also be analyzed with minor changes.
We applied SAAP-RS to studying APA in brain cells and neurogenesis. Using the widely used brain cell RNA-seq data from Ref. [36], we defined 3′ UTR APA profiles in different brain cell types. We show that the APA profile can be used to distinguish brain cell types, similar to using gene expression levels, and neurons express longest 3′ UTRs among all brain cells. However, intriguingly, genes with basic cellular functions appear to express long 3′ UTR isoforms to a greater extent than other genes in neurons, including “RNA processing”, “macromolecular complex”, “translation”, etc. By contrast, some neuron-enriched genes were found to express long 3′ UTR isoforms to a greater extent in non-neuronal cells. It remains a possibility that RNA stability may be involved in shaping the APA pattern for neuron-specific genes. For example, long 3′ UTR isoforms are more rapidly degraded in neurons where the gene expression level is high and are more stable in non-neuronal cells where the gene expression level is low. However, a more parsimonious explanation is that APA site choice is coupled with transcriptional gene regulation for neuron-specific genes. That is proximal PASs are preferred when genes are expressed at high levels whereas distal PASs are preferentially used when gene expression levels are low. This coupling mechanism was previously suggested to entail more efficient recruitment of the 3′ end processing machinery when transcription is activated [53,54]. Future studies will need to address why the coupling seems obvious in the neuronal system but not so in other systems [25].
Recent studies of 3′ UTR isoforms in hESC-derived neurons and in the Drosophila nervous system indicate that long 3′ UTR isoforms in general have repressed translation [43,55]. Our data indicate that neuron-specific genes in fact do not show the greatest 3′ UTR lengthening in neurons. Therefore, these genes, through expression of short 3′ UTR isoforms, can avoid translational repression, leading to enhanced protein production in neurons. This needs to be examined in the future.
We found that while neurogenesis exhibits much stronger 3′ UTR lengthening than myogenesis, 3′ UTR APA regulations in these two processes are generally correlated. Therefore, with respective to 3′ UTR length control, neurogenesis appears to be an augmented differentiation process that takes place in other cell types. Future studies need to address whether this augmentation is due to greater PAS usage control in neurons, for example, through transcriptional pausing [56], or to long 3′ UTR stabilization, for example, through repressed mRNA decay [57].
In this study, we used scRNA-seq data to corroborate our findings based on bulk RNA samples. While the low read coverage of scRNA-seq data made it infeasible to examine APA of individual genes, the cell-based global patterns were consistent with those of regular RNA-seq. As expected, our analysis indicated that a small number of cells would result in high data variability. Therefore, future studies using a large number of cells would be critical to further unravel APA dynamics in single cells.
METHODS
Datasets and data processing
Bulk RNA-seq data for different cell types in cerebral cortex of mouse brain [36] and single cell RNA-seq data generated by Zeisel et al. [39] were downloaded from European Nucleotide Archive (ENA). 3′ READS and RNA-seq datasets for brain and testis samples were previously generated in our lab [4]. Neurogenesis RNA-seq data [40,41] and myogenesis data [44,45] were downloaded from the gene omnibus expression (GEO) database. 3′ READS data were analyzed as previously described [4]. Briefly, reads were mapped to the mouse genome (mm9) using bowtie2 [58] and reads with more than two unaligned 5′ Ts were considered as PAS reads. PASs within 24 nucleotides from one another were clustered together. RNA-seq data were aligned to the mouse genome with STAR [59] using default settings. Raw bam files were further processed using R packages: RSamtools for processing bam files, GenomicAlignments for counting reads, and GenomicFeatures for defining genomic regions.
APA analysis using RNA-seq data
Mouse PAS locations were downloaded from PolyA_DB 3 (http://polya-db.org/v3/). Information about conservation, percent of samples with expression (PSE) and mean reads per million (RPM) for each PAS was obtained from the database. For each reference PAS, upstream RNA-seq reads until the stop codon were used as upstream reads (UP) and those to the last PAS were used as downstream reads (DN). We used DEXSeq [35] to examine APA difference when there were replicates. The Fisher’s exact test was used when there were no replicates. Significant events were defined as P < 0.05. Standard deviation was obtained by sampling data with a bootstrapping method for 20 times, as described previously [51]. Relative expression difference (RED) was calculated as difference in log2(DN/UP) between two samples, where DN and UP are reads in DN and UP regions, respectively. We required that the read density (read number/length) of the DN region to be lower than that of the UP region. For RNA-seq data without strand information, we filtered out genes that overlapped with downstream antisense genes using our strand-specific RNA-seq data from brain and testis. A sense/antisense ratio was calculated for the aUTR region of each gene using reads on sense and antisense strands. Genes with a sense/antisense ratio greater than 10 were selected for further analysis. Sample clustering was performed with the R heatmap package using Pearson correlation. Each row was normalized by standardization (minus mean and divided by standard deviation). Marker genes were selected by comparing normalized RED values of each gene with those of other cell types by the Wilcoxon test.
Gene expression analysis
Differential gene expression analysis was performed with DESeq [60]. Significant events were defined as FDR < 0.05 and fold change > 2. Only CDS reads were used for gene expression analysis to avoid confounding issues with APA analysis.
Single cell RNA-seq (scRNA-seq) analysis
scRNA-seq reads were mapped to genes similar to bulk RNA-seq data. We required DN or UP regions to have at least 1 read. We calculated log2(DN/UP) for each gene. A pseudo count of 1 was used to avoid infinity values. Log2(DN/UP) was averaged for each cell. For gene set-based analysis, we required both DN and UP regions of each gene to have at least 1 read. Log2(DN/UP) values of a gene from different cells were normalized.
Data access
All data are accessible through the Gene Expression Omnibus (GEO) database, including brain cell bulk RNA-seq data (GSE52564); single cell RNA-seq data (GSE60361); neurogenesis data from Ref. [41] (GSE25533), Ref. [40] (GSE33252), Ref. [42]. (GSE60548) and Ref. [43] (GSE100007); and C2C12 differentiation data from Ref. [45] (GSE94560) and Ref. [44] (GSE84279).
Supplementary Material
ACKNOWLEDGMENTS
We thank members of Bin Tian lab for helpful discussions. This work was supported by grants from NIH (Nos. R01 GM084089 and R21 NS097992) and a grant from the Rutgers Brain Health Institute.
Footnotes
SUPPLEMENTARY MATERIALS
The supplementary materials can be found online with this article at https://doi.org/10.1007/s40484-018-0148-3.
COMPLIANCE WITH ETHICS GUIDELINES
The authors Aysegul Guvenek and Bin Tian declare that they have no conflict of interests.
This article does not contain any studies with human or animal subjects performed by any of the authors.
REFERENCES
- 1.Tian B and Manley JL (2017) Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol, 18, 18–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tian B and Graber JH (2012) Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip. Rev. RNA, 3, 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Y and Manley JL (2015) The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev, 29, 889–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G and Tian B (2013) Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat. Methods, 10, 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM and Babak T (2012) A quantitative atlas of polyadenylation in five mammals. Genome Res, 22, 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP and Burge CB (2008) Alternative isoform regulation in human tissue transcriptomes. Nature, 456, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Lee JY and Tian B (2005) Biased alternative polyadenylation in human tissues. Genome Biol, 6, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miura P, Shenker S, Andreu-Agullo C, Westholm JO and Lai EC (2013) Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome Res, 23, 812–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Park JY, Zheng D, Hoque M, Yehia G and Tian B (2016) Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol, 14, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji Z, Lee JY, Pan Z, Jiang B and Tian B (2009) Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. USA, 106, 7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayr C and Bartel DP (2009) Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell, 138, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandberg R, Neilson JR, Sarma A, Sharp PA and Burge CB (2008) Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science, 320,1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontes MM, Guvenek A, Kawaguchi R, Zheng D, Huang A, Ho VM, Chen PB, Liu X, O’Dell TJ, Coppola G, et al. (2017) Activity-dependent regulation of alternative cleavage and polyadenylation during hippocampal long-term potentiation. Sci. Rep, 7, 17377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM and Greenberg ME (2008) Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron, 60, 1022–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutz CS and Moreira A (2011) Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip Rev. RNA, 2, 22–31 [DOI] [PubMed] [Google Scholar]
- 16.Mayr C (2016) Evolution and biological roles of alternative 3′ UTRs. Trends Cell Biol, 26, 227–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gennarino VA, Alcott CE, Chen CA, Chaudhury A, Gillentine MA, Rosenfeld JA, Parikh S, Wheless JW, Roeder ER, Horovitz DD, et al. (2015) NUDT21-spanning CNVs lead to neuropsychiatric disease and altered MeCP2 abundance via alternative polyadenylation. Elife, 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han K, Gennarino VA, Lee Y, Pang K, Hashimoto-Torii K, Choufani S, Raju CS, Oldham MC, Weksberg R, Rakic P, et al. (2013) Human-specific regulation of MeCP2 levels in fetal brains by microRNA miR-483-5p. Genes Dev, 27, 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, et al. (2008) Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell, 134, 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau AG, Irier HA, Gu J, Tian D, Ku L, Liu G, Xia M, Fritsch B, Zheng JQ, Dingledine R, et al. (2010) Distinct 3′ UTRs differentially regulate activity-dependent translation of brain-derived neurotrophic factor (BDNF). Proc. Natl. Acad. Sci. USA, 107, 15945–15950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taliaferro JM, Vidaki M, Oliveira R, Olson S, Zhan L, Saxena T, Wang ET, Graveley BR, Gertler FB, Swanson MS, et al. (2016) Distal alternative last exons localize mRNAs to neural projections. Mol. Cell, 61, 821–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andreassi C and Riccio A (2009) To localize or not to localize: mRNA fate is in 3′ UTR ends. Trends Cell Biol, 19, 465–474 [DOI] [PubMed] [Google Scholar]
- 23.Tushev G, Glock C, Heumuller M, Biever A, Jovanovic M and Schuman EM (2018) Alternative 3′ UTRs modify the localization, regulatory potential, stability, and plasticity of mRNAs in neuronal compartments. Neuron, 98, 495–511e6 [DOI] [PubMed] [Google Scholar]
- 24.Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kühn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K, et al. (2012) The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell, 149, 538–553 [DOI] [PubMed] [Google Scholar]
- 25.Lianoglou S, Garg V, Yang JL, Leslie CS and Mayr C (2013) Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev, 27, 2380–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ and Shi Y (2011) Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA, 17, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng D, Liu X and Tian B (2016) 3′ READS +, a sensitive and accurate method for 3′ end sequencing of polyadenylated RNA. RNA, 22, 1631–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grassi E, Mariella E, Lembo A, Molineris I and Provero P (2016) Roar: detecting alternative polyadenylation with standard mRNA sequencing libraries. BMC Bioinformatics, 17, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katz Y, Wang ET, Airoldi EM and Burge CB (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods, 7, 1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Z and Teeling EC (2017) ExUTR: a novel pipeline for large-scale prediction of 3′-UTR sequences from NGS data. BMC Genomics, 18, 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim M, You BH and Nam JW (2015) Global estimation of the 3′ untranslated region landscape using RNA sequencing. Methods, 83, 111–117 [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Wei Z and Li H (2014) A change-point model for identifying 3′ UTR switching by next-generation RNA sequencing. Bioinformatics, 30, 2162–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ and Li W (2014) Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun, 5, 5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R, Nambiar R, Zheng D and Tian B (2018) PolyA_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res, 46, D315– D319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anders S, Reyes A and Huber W (2012) Detecting differential usage of exons from RNA-seq data. Genome Res, 22, 2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci, 34, 11929–11947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao S, Alarcón C, Sapkota G, Rahman S, Chen PY, Goerner N, Macias MJ, Erdjument-Bromage H, Tempst P and Massagué J (2009) Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol. Cell, 36, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rotin D and Kumar S (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol, 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 39.Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. (2015) Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science, 347, 1138–1142 [DOI] [PubMed] [Google Scholar]
- 40.Lienert F, Mohn F, Tiwari VK, Baubec T, Roloff TC, Gaidatzis D, Stadler MB and Schübeler D (2011) Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells. PLoS Genet, 7, e1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiwari VK, Stadler MB, Wirbelauer C, Paro R, Schubeler D and Beisel C (2011) A chromatin-modifying function of JNK during stem cell differentiation. Nat. Genet, 44, 94–100 [DOI] [PubMed] [Google Scholar]
- 42.Busskamp V, Lewis NE, Guye P, Ng AH, Shipman SL, Byrne SM, Sanjana NE, Mum J, Li Y, Li S, et al. (2014) Rapid neurogenesis through transcriptional activation in human stem cells. Mol. Syst. Biol, 10, 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blair JD, Hockemeyer D, Doudna JA, Bateup HS and Floor SN (2017) Widespread translational remodeling during human neuronal differentiation. Cell Rep, 21, 2005–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doynova MD, Markworth JF, Cameron-Smith D, Vickers MH and O’Sullivan JM (2017) Linkages between changes in the 3D organization of the genome and transcription during myotube differentiation in vitro. Skelet Muscle, 7, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamed M, Khilji S, Dixon K, Blais A, Ioshikhes I, Chen J and Li Q (2017) Insights into interplay between rexinoid signaling and myogenic regulatory factor-associated chromatin state in myogenic differentiation. Nucleic Acids Res, 45, 11236–11248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newington JT, Rappon T, Albers S, Wong DY, Rylett RJ and Cumming RC (2012) Overexpression of pyruvate dehydrogenase kinase 1 and lactate dehydrogenase A in nerve cells confers resistance to amyloid β and other toxins by decreasing mitochondrial respiration and reactive oxygen species production. J. Biol. Chem, 287, 37245–37258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wigfield SM, Winter SC, Giatromanolaki A, Taylor J, Koukourakis ML and Harris AL (2008) PDK-1 regulates lactate production in hypoxia and is associated with poor prognosis in head and neck squamous cancer. Br. J. Cancer, 98, 1975–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma X, Li C, Sun L, Huang D, Li T, He X, Wu G, Yang Z, Zhong X, Song L, et al. (2014) Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat Commun, 5, 5212. [DOI] [PubMed] [Google Scholar]
- 49.Chalhoub N, Zhu G, Zhu X and Baker SJ (2009) Cell type specificity of PI3K signaling in Pdk1- and Pten-deficient brains. Genes Dev, 23, 1619–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ji Z and Tian B (2009) Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE, 4, e8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W, You B, Hoque M, Zheng D, Luo W, Ji Z, Park JY, Gunderson SI, Kalsotra A, Manley JL, et al. (2015) Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet, 11, e1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian B, Pan Z and Lee JY (2007) Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res, 17, 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M and Manley JL (2011) Transcriptional activators enhance polyadenylation of mRNA precursors. Mol. Cell, 41, 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji Z, Luo W, Li W, Hoque M, Pan Z, Zhao Y and Tian B (2011) Transcriptional activity regulates alternative cleavage and polyadenylation. Mol. Syst. Biol, 7, 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hilgers V, Perry MW, Hendrix D, Stark A, Levine M and Haley B (2011) Neural-specific elongation of 3′ UTRs during Drosophila development. Proc. Natl. Acad. Sci. USA, 108, 15864–15869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oktaba K, Zhang W, Lotz TS, Jun DJ, Lemke SB, Ng SP, Esposito E, Levine M and Hilgers V (2015) ELAV links paused Pol II to alternative polyadenylation in the Drosophila nervous system. Mol. Cell, 57, 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dai W, Li W, Hoque M, Li Z, Tian B and Makeyev EV (2015) A post-transcriptional mechanism pacing expression of neural genes with precursor cell differentiation status. Nat Commun, 6, 7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B and Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders S and Huber W (2010) Differential expression analysis for sequence count data. Genome Biol, 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.