Abstract
Alternative splicing (AS) is a major post-transcriptional mechanism to enhance the diversity of proteome in response to environmental signals. Among the numerous external signals perceived by plants, light is the most crucial one. Plants utilize complex photoreceptor signaling networks to sense different light conditions and adjust their growth and development accordingly. Although light-mediated gene expression has been widely investigated, little is known regarding the mechanism of light affecting AS to modulate mRNA at the post-transcriptional level. In this minireview, we summarize current progresses on how light affects AS, and how sensory photoreceptors and retrograde signaling pathways may coordinately regulate AS of pre-mRNAs. In addition, we also discuss the possibility that AS of the mRNAs encoding photoreceptors may be involved in feedback control of AS. We hypothesize that light regulation of the expression and activity of splicing factors would be a major mechanism of light-mediated AS. The combination of genetic study and high-throughput analyses of AS and splicing complexes in response to light is likely to further advance our understanding of the molecular mechanisms underlying light control of AS and plant development.
INTRODUCTION
In eukaryotes, introns usually need to be spliced out to form mature mRNAs in the nucleus (1). The splicing machinery utilizes different splice sites to produce two or more isoforms at the same locus (2,3). The discovery of alternative splicing (AS) is a key milestone for understanding the post-transcriptional regulation (4–7). Early studies based on EST and full-length cDNA demonstrate that AS presents a larger proportion of multi-intron genes in plants (8). Recent genomewide studies based on next-generation sequencing confirm that a large number of genes are regulated by AS, including more than 60% of the intron-containing genes in both Arabidopsis and Glycine max (9–11), approximately 33% of the annotated genes in Oryza sativa (12), and at least 56% of the genes in Zea mays (12,13).
Alternative splicing can be classified into four major types: intron retention, exon skipping, alternative donor and acceptor sites (9,14). Intron retention, as the most prevalent type of AS in plants, often produces splicing isoforms containing premature termination codon (PTC) that can be targeted by nonsense-mediated mRNA decay (NMD) to regulate the transcriptional level of functional transcription (8–10,15,16). Furthermore, splicing isoforms with PTC can also produce truncated proteins to regulate the full-length functional proteins (17–20). Other types of AS including exon skipping, alternative donor and acceptor sites produce various proteins with distinct subcellular localization, stability and binding properties by in-frame deletion or addition of alternative domains (3,14,21,22).
Alternative splicing enhances the adaptability of plants in response to environmental alterations (23). Emerging studies have revealed that AS plays an essential role in light response by regulating core-clock genes, including RVE8, JMJD5, LHY, TIC and CKB3 (24–26). Meanwhile, light quality and quantity can regulate AS of numerous plant genes (24,27,28). These findings enhance our understanding of how plants adapt themselves to track light oscillations via AS-mediated regulation. However, the mechanisms of light regulation of AS are still not fully understood. Here, we summarize the latest findings of light-mediated AS regulation and propose the possible mechanisms of AS regulation in response to light.
Transcriptome-wide regulation of AS in response to light
Due to technical limitations, only a subset of AS regulated by light has been revealed by earlier studies (29,30). For example, the first study reports nine genes with differential spliced pattern upon light treatment, including carboxypeptidase, tyrosyl tRNA synthetase transcripts and two serine/arginine-rich (SR) genes using the AS RT-PCR panel (31). Another study shows that splice forms from ten genes displays opposite splicing patterns in the light- vs. dark-treated samples using whole-genome oligonucleotide array (32). Resent development of high-throughput sequencing technologies allows genomewide survey of splicing isoforms (33,34). Three genomewide transcriptome studies using RNA-seq reveal that light affects AS of hundreds to thousands of genes (24,27,28). In Arabidopsis, at least 7% of protein-coding genes exhibit AS pattern mediated by phytochrome A (phyA) and phyB, two major molecular species of red/far-red-sensing phytochromes (27). Genomewide study by Wu et al. (28) in the moss Physcomitrella patens shows 8.4% and 8.9% of AS events in response to red and blue light, respectively. Most recently, a large-scale transcriptome profiling in Arabidopsis estimates different AS patterns in 382 genes upon 2-h pulse of white light in the middle of the night (24). Alternatively, as the most frequent AS events, intron retentions are mainly induced by phytochromes (24,28). These observations suggest that mRNA splicing is directly involved in the photomorphogenic response in plants (Fig. 1). Taking advantage of current whole-transcriptome sequencing strategies, more light-mediated splicing isoforms will be discovered in the future.
Figure 1.
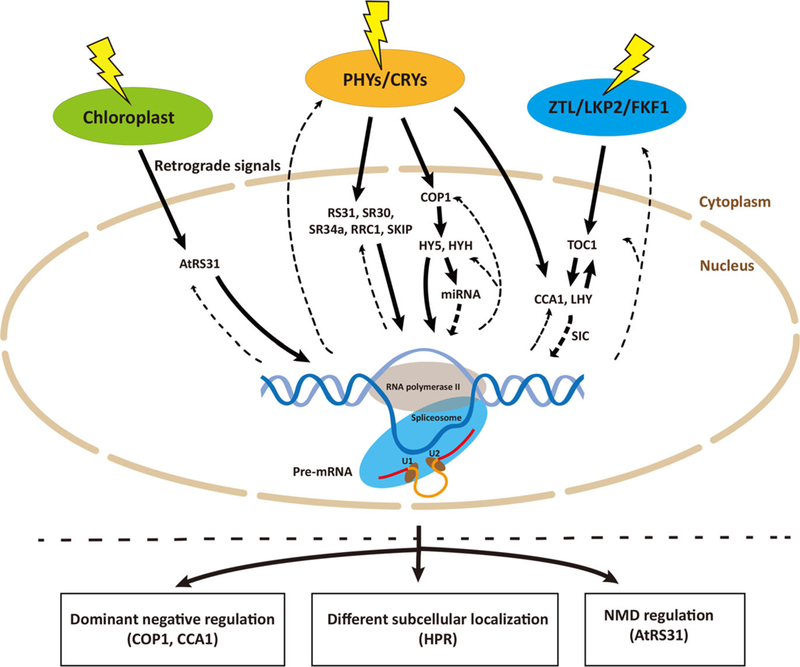
Model of the regulation of light-mediated AS. Light regulates AS widely via three primary types of sensory photoreceptor pathways (phytochromes, cryptochromes and ZTL/LKP2/FKF1) or retrograde signals from chloroplast. Splicing factors, such as RS31, SR30, SR34a, RRC1 and SKIP, are involved in light-mediated AS regulation, whereas these splicing factors are also under the regulation of AS. Intriguingly, AS of PHYA, CRY2, ZTL and other genes involved in light signaling pathways, such as COP1, HYH, CCA1 and TOC1, might form a feedback loop in response to light. Solid arrow illustrates genes involved in light signaling pathway to regulate AS. Thick broken arrow represents weak indirect evidence that genes involved in light signaling pathway to regulate AS. Dash arrow indicates genes that are under the regulation of AS. Light-mediated AS has function on dominant negative regulation, different subcellular localization or taking part in NMD pathway.
AS mediated by sensory photoreceptors and retrograde signaling pathway
A couple of photosensory receptors, including five phytochromes (phyA–phyE), UV-A/blue light-absorbing receptors such as two phototropins (phot1, phot2), two cryptochromes (cry1, cry2) and three Zeitlupe proteins (ZTL, FKF1 and LKP2), and the UV-B photoreceptors (UVR8) have been reported (35,36). Phytochrome-dependent change in AS was firstly reported in Arabidopsis and demonstrated that 15% of splicing-related genes possess phytochrome-mediated AS after exposing to red light for 1 h (27). Furthermore, phytochrome-deficient mutants in Physco-mitrella patens cannot response to AS regulation upon light treatment, supporting the involvement of phytochromes in splicing regulation (28). Collectively, these studies suggest that the signaling from sensory photoreceptors, at least phytochromes, is the primary pathway in the light-regulated AS, although the involvement of other photoreceptor in the AS regulation has not been experimentally proved. In contrast, another study reports that light-triggered AS in AtRS31 is independent of phytochrome and cryptochrome pathways (37). Further study demonstrates that white light-induced splicing alteration in SR30 is not affected in phyAB, phyABCDE and cry1cry2 mutants, suggesting white light may regulate the activity of splicing factors via photosynthetic process (24). In addition, retrograde signals from the chloroplast are also involved in the regulation of AS events (37). These observations support the view that light regulation of AS is not only dependent on photosensory photoreceptors, but also mediated by the photosynthetic process or retrograde signals from the chloroplast (Fig. 1).
Circadian clock genes regulated by AS in response to light
In the central loop of circadian clock, TIMING OF CAB EXPRESSION 1 (TOC1) and CIRCADIAN CLOCK-ASSOCIATED1 (CCA1)/LATE ELONGATED HYPOCOTYL (LHY) show reciprocal suppression (38,39). ZTL, together with CCA1 and LHY, can suppress the expression of TOC1 (40,41). AS is an important mechanism for the regulation of circadian clock-related genes, including ZTL, CCA1, LHY and TOC1 in Arabidopsis (17,42). AS of core-clock genes, such as LHY, RVE8, JMJD5, TIC and CKB3, is reported to be affected by light (24). It is noteworthy that AS and circadian clock-related genes can be mutually regulated (17,43,44). Clock-related genes, for example, AtGRP7 and its paralog AtGRP8 can reciprocally control AS of their transcripts (15,44,45), while splicing factors, such as SKIP and STIPL1, can regulate the circadian clock in Arabidopsis (46,47). Furthermore, more recent studies also pointed out that protein arginine methyltransferase 5 (PRMT5) can regulate the AS of core-clock gene PSEUDO-RESPONSE REGULATOR9 (PRR9) in Arabidopsis (48–50), and the expression of PRMT5 is clock-regulated (51). These studies strongly indicate that the circadian clock is tightly associated with the regulation of AS.
Mechanism of light regulation of AS
Alternative splicing patterns are determined by both cis-splicing regulatory elements and trans-acting splicing factor (52). Serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucle-oproteins (hnRNPs) are two well-known trans-splicing factors that either facilitate or restrain spliceosome assembly in response to environmental cues (53–56), and their expression or activity is regulated by light (28,57,58). Five splicing factors (RS31, SR30, SR34a, SR34b and U2AF65a) have been reported to participate in the phytochrome-mediated AS regulation (27,37). AS regulation of AtRS31 is particularly essential for the response to changing level of light conditions in Arabidopsis (37). Additionally, AtSR45a, another SR-coding gene, suppresses the splicing efficiency by facilitating to form a bridge between 5′ and 3′ splice sites in response to high light stress (59). Given the widespread AS events affected by light are enriched in encoding mRNA processing (24) and splicing factors are significantly enriched among phytochrome-regulated genes in Arabidopsis (27), we posit that light-mediated AS is mostly regulated through the modulation of AS of RNA-processing genes them-selves (Fig. 1). Most photoreceptors, including phytochromes, cryptochromes, ZTL/LKP2/FKF1 and UVR8, are located in the nucleus (60–63), which may directly interact with the spliceo-somes. It will be intriguing to investigate whether the components of photoreceptors can interact with the above splicing factors directly.
RRC1 (reduced red-light responses in cry1cry2 background 1) encodes a potential splicing factor that regulates the AS of several SR genes in response to red light, such as RS31 and SR34b (64). Splicing-defective mutant of rrc1 is insensitive to red light in the phyB-mediated responses (64). However, another splicing factor mutant SKIP is hypersensitive to both red and blue light (46). Therefore, these inverse effects of splicing factors will prompt us to systematically determine the quantitative regulation of AS in response to light.
Aberrant photomorphogenic phenotypes are observed in defects of transcription factors (TFs), including B-box zinc finger (e.g. BBX22), basic helix–loop–helix (e.g. PIF3) and basic region/leucine zipper motif (e.g. HYH, HY5) TFs (65). In addition to splicing factors, TFs also affect AS by influencing the transcription elongation rate of RNA polymerase II (52,66,67). ChIP-seq peaks of CCA1 include binding peaks near the promoter of several splicing-related genes, which suggests that CCA1 might regulate AS indirectly by regulating splicing-related genes, such as U11/U12–65K, GRP7 and GRP8 (68). SIC regulates pre-mRNA metabolism and regulates the AS of circadian clock transcripts, including LHY, CCA1, ELE3 and PRR7 (69). The sic-associated clock impairment phenotypes required the presence of CCA1 and LHY (69). However, the linkage among light-responsive TFs and the subsequent AS pattern is still not fully understood (Fig. 1). It is possible that light-regulated splicing factors, as well as transcription factors, act in coordination to regulate AS patterns in response to light (Fig. 1).
Regulation of AS is achieved by dynamic reciprocity between trans-splicing factor and cis-regulatory elements to define exon/intron boundaries and produce accurate splicing variants (70). There is no enriched motif around splicing sites from light-mediated AS events based on RNA-seq data (24), which might distinguish both positive and negative influences from different light-regulated splicing factors. Thus, it would be informative to analyze the cis-acting regulatory elements individually to identify both positive and negative regulatory elements. Cross-linking and immunoprecipitation, followed by high-throughput sequencing (CLIP-seq), are the most appropriate techniques to capture really binding motif of a single RNA-binding protein in plant (71). New breakthroughs are anticipated by combination of CLIP-seq with genetic overexpression or knockouts of splicing factors to clarify the molecular mechanism of specific splicing factors that are highly regulated by light.
AS of photoreceptors and clock-related genes
As shown in Table 1, AS plays a key role in regulating photo-morphogenesis and clock-related genes. For example, PHYA in Arabidopsis was annotated to include intron retention and produce an N-terminal truncated protein according to the TAIR10 annotations (72). In tomato, the PHYA gene is also regulated by AS which lend credence to the idea that it is a conserved mechanism (73). In addition to phytochromes, CRY2 has one alternative accept site in exon 2 and produces different 5′ UTR according to the annotation in TAIR10 (72) and ASIP database (8). AS variants on 5′ UTR of CRY2 may affect their mRNA stability or translatability. As mentioned above, the light regulation of genomewide AS switch is dependent, at least in part, on photosensory photoreceptors (27). On the other hand, these photosensory photoreceptor genes undergo the AS regulation; thus, it can form a feedback loop in response to light (Fig. 1). In addition to phytochromes and cryptochromes photoreceptors, other regulators of photomorphogenesis and circadian clocks such as COP1, HYH, HY5, LHY and CCA1 also undergo AS. It has been reported that COP1 produces different AS isoforms (74). Overexpression of short isoform of COP1 with truncated WD-40 repeat shows short hypocotyl and developed cotyledons in the dark (74), implying that AS of COP1 may be relevant to the regulation of its physiological activities. Phytochromes and cryptochromes regulate light responses primarily through the transcriptional regulation of a great number of genes. Photoactivated phytochromes and cryptochromes inactivate COP1/SPA E3 ligase complex to regulate the abundance of two bZIP transcription factors, HY5 and HYH, which are the positive regulators in photomorphogenesis (75,76). Comparing with full-length HYH, the truncated HYH generated from the product of AS regulation, which lacks the COP1-interaction domain, is less susceptible to COP1-mediated degradation (77). HY5 also includes intron retention and alternative donor in the coding region according to AtRTD2 annotation (34). The splicing isoforms of LHY can be recognized by NMD pathway (78). For another example, CCA1β encoding truncated proteins interfere with the activity of functional protein CCA1α by competitively forming functional heterodimers (19).
Table 1.
Photomorphogenesis-related gene and circadian clock genes regulated by AS.
| Gene | Locus | AS type | AS region | Light-regulated | Reference |
|---|---|---|---|---|---|
| PHYA | AT1G09570 | IR; AltD | CDS | phy-regulated | (27) |
| CRY2 | AT1G04400 | AltA | 5′-UTR | phy-regulated | (27,34) |
| ZTL | AT5G57360 | IR | CDS | Unknown | (72) |
| COP1 | AT2G32950 | AltA | CDS | phy-regulated | (27,74) |
| HY5 | AT5G11260 | IR; AltD | CDS | 2-h white light | (24,34) |
| HYH | AT3G17609 | IR; AltA | CDS | phy-regulated | (27,77) |
| LHY | AT1G01060 | IR; ES | 5′-UTR/CDS | phy-regulated/2-h white light | (24,27) |
| CCA1 | AT2G46830 | IR; AltD | CDS | phy-regulated | (27,79) |
| TOC1 | AT5G61380 | IR | CDS | Unknown | (34) |
| RVE8 | AT3G09600 | AltA | CDS | phy-regulated/2-h white light | (24,27) |
| JMJD5 | AT3G20810 | IR | CDS | phy-regulated/2-h white light | (24,27) |
| TIC | AT3G22380 | IR | CDS | phy-regulated/2-h white light | (24,27) |
| CKB3 | AT3G60250 | IR; AltA | CDS | phy-regulated/2-h white light | (24,27) |
| PIF6 | AT3G62090 | IR | CDS | Unknown | (93) |
The function of AS alteration in response to light
The correlation between light and AS has been explored in a genomewide scale by RNA-seq. However, investigation of biological function or specific splicing variants is time-consuming because there is no high-throughput experimental method to identify whether isoforms possess specific function in vivo. Even so, distinct functions of splicing isoforms induced by light have been identified in some cases (Fig. 1). For example, phytochromes induce splicing alteration of SPA3 and produce a truncated protein lacking in WD40 repeats (27), which is still able to interact with COP1 and form COP1/SPA complex but fails to bind to DAMAGE DNA-BINDING PROTEIN 1 (DDB1) to make up a multimeric E3 ubiquitin ligase (27). Another example is CCA1, which produces CCA1β lacking the MYB domain. It works as a regulator by competitively inhibiting the DNA binding of functional CCA1α by forming CCA1α-CCA1β heterodimers (19,79). It was also reported that splicing isoforms (mRNA3) of AtRS31, which possess PTC, showed increased expression in dark (37). However, the full-length splicing isoforms (mRNA1) encoding functional protein present decreased expression (37). Finally, light-mediated AS produces proteins with different subcellular localization. For example, light signaling induces the expression of HPR2 located in the cytosol rather than HPR1 located in leaf peroxisomes (80). Given that AS is the major contributor to protein diversity, it is necessary to explore the function of splicing isoforms in a large scale. It will be interesting to investigate the photomorphogenic changes by overexpressing the light-induced isoforms to interrogate the functional information.
CONCLUSIONS AND PERSPECTIVES
As shown above, light regulates AS mainly via photoreceptors or retrograde signals from chloroplast, while photoreceptors themselves are also under the regulation of AS to form a feedback loop. The regulation of light-mediated AS is mostly through the regulation of AS of splicing factors themselves. Light-mediated AS mainly confers its functional effects on dominant negative regulation, different subcellular localization or taking part in NMD pathway.
Recent observation reveals that mRNAs and protein abundances present a negative correlation upon illumination, suggesting diverse post-transcriptional regulation takes place in light response (81). In addition to AS, alternative polyadenylation (APA) and small RNAs, including miRNAs and siRNAs, are another post-transcriptional regulators in plants (82,83). MiRNAs are involved in photomorphogenesis according to studies on HY5 (84), HEN1 (85) and AGO1 (86). The ago 1 mutant displays light hypersensitive phenotype, suggesting that miRNAs act as negative regulators of photomorphogenesis (86). The mutation in HY5 causes aberrant light-mediated phenotypes, which might result from the direct binding on the upstream of eight miRNA genes and subsequently affect the expression of miRNA target genes (84). Several animal studies confirm that miRNA regulates AS by targeting splicing regulator, such as miR-124 targeting PTBP1 (87), miR-133 targeting nPTB (88) and miR-222 targeting Rmb24 (89). In Arabidopsis, SERRATE plays roles in both pri-miRNA processing and mRNA splicing (90) and miRNAs can target AS region (91); thus, further evidences in future will extend our understanding of the interplay between miRNA and AS (Fig. 1).
Like AS, APA is also a widespread post-transcriptional regulator to generate transcript diversity in plants by changing the coding region or the length of the 3′-UTR (71,92). Recent study shows that CCA1 has direct binding peak on the promoter region of poly(A) binding protein 6 (68). However, the role of APA in response to light has never been reported. Does APA take part in light-regulated development, and if so, does it have dynamic interplay with either miRNA or AS? It may be expected that, taking full account of the coordinated influences above, most common post-transcriptional regulation will fully dissect the molecular mechanisms underlying photomorphogenic responses in plants.
Acknowledgements—
The authors thank Dr. Marcelo J. Yanovsky and Yoshito Oka for critical reading of the manuscript. This work was supported by the National Key Research and Development Program of China (2016YFD0600106) and the National Natural Science Foundation of China Grant (Grant No. 31570674 to L.G.).
AUTHOR BIOGRAPHIES
 Hangxiao Zhang graduated with a B.Sc. in Biotechnology from Shandong University followed by a Ph.D. in Genomics from Beijing Institute of Genomics, Chinese Academy of Sciences, under the direction of Prof. Jun Yu. After a postdoctoral fellowship in the laboratory of Prof. Haibing Wang at the Institute of Zoology, Chinese Academy of Sciences, she joined Fujian Agriculture and Forestry University as an assistant researcher in 2016. Her present research focuses on the regulation of alternative splicing in plant.
Hangxiao Zhang graduated with a B.Sc. in Biotechnology from Shandong University followed by a Ph.D. in Genomics from Beijing Institute of Genomics, Chinese Academy of Sciences, under the direction of Prof. Jun Yu. After a postdoctoral fellowship in the laboratory of Prof. Haibing Wang at the Institute of Zoology, Chinese Academy of Sciences, she joined Fujian Agriculture and Forestry University as an assistant researcher in 2016. Her present research focuses on the regulation of alternative splicing in plant.
 Chentao Lin is currently a Professor at the Department of Molecular, Cell & Developmental Biology, University of California, Los Angeles. He went to the United States in 1985 after getting his B.Sc. in Agronomy at South China College of Tropical Crops and obtained an M.Sc. at Iowa State University. After receiving his Ph.D. degree in Genetics at Michigan State University, he became a NIH postdoctoral fellow at the University of Pennsylvania. Now, he is engaged in using various approaches, including molecular genetics, biochemistry and high-throughput sequencing, to further investigate how cryptochromes act in Arabidopsis.
Chentao Lin is currently a Professor at the Department of Molecular, Cell & Developmental Biology, University of California, Los Angeles. He went to the United States in 1985 after getting his B.Sc. in Agronomy at South China College of Tropical Crops and obtained an M.Sc. at Iowa State University. After receiving his Ph.D. degree in Genetics at Michigan State University, he became a NIH postdoctoral fellow at the University of Pennsylvania. Now, he is engaged in using various approaches, including molecular genetics, biochemistry and high-throughput sequencing, to further investigate how cryptochromes act in Arabidopsis.
 Lianfeng Gu obtained his Ph.D. degree from the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences in 2012 under the supervision of Prof. Xiao-feng Cao. During his postdoctoral training at the same institute, he spent a period of time at the University of California, Riverside, as a visiting scholar. He is currently a Professor at Fujian Agriculture and Forestry University after having returned to China in 2015. His scientific interests include investigation of the interplay between different post-transcriptional regulations in plants and developing bioinformatics methods and computational tools to analyze large-scale datasets.
Lianfeng Gu obtained his Ph.D. degree from the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences in 2012 under the supervision of Prof. Xiao-feng Cao. During his postdoctoral training at the same institute, he spent a period of time at the University of California, Riverside, as a visiting scholar. He is currently a Professor at Fujian Agriculture and Forestry University after having returned to China in 2015. His scientific interests include investigation of the interplay between different post-transcriptional regulations in plants and developing bioinformatics methods and computational tools to analyze large-scale datasets.
Footnotes
This article is part of the special issue highlighting Dr. Aziz Sancar’s outstanding contributions to various aspects of the repair of DNA photodamage in honor of his recent Nobel Prize in Chemistry.
REFERENCES
- 1.Moore MJ and Proudfoot NJ (2009) Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700. [DOI] [PubMed] [Google Scholar]
- 2.Blencowe BJ (2006) Alternative splicing: new insights from global analyses. Cell 126, 37–41. [DOI] [PubMed] [Google Scholar]
- 3.Wang Z and Burge CB (2008) Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14, 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert W (1978) Why genes in pieces? Nature 271, 501. [DOI] [PubMed] [Google Scholar]
- 5.Gupta S, Zink D, Korn B, Vingron M and Haas SA (2004) Genome wide identification and classification of alternative splicing based on EST data. Bioinformatics 20, 2579–2585. [DOI] [PubMed] [Google Scholar]
- 6.Nagasaki H, Arita M, Nishizawa T, Suwa M and Gotoh O (2006) Automated classification of alternative splicing and transcriptional initiation and construction of visual database of classified patterns. Bioinformatics 22, 1211–1216. [DOI] [PubMed] [Google Scholar]
- 7.Gu L and Guo R (2007) Genome-wide detection and analysis of alternative splicing for nucleotide binding site-leucine-rich repeats sequences in rice. J. Genet. Genomics 34, 247–257. [DOI] [PubMed] [Google Scholar]
- 8.Wang BB and Brendel V (2006) Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA 103, 7175–7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK and Mockler TC (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20, 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marquez Y, Brown JW, Simpson C, Barta A and Kalyna M (2012) Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22, 1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen Y, Zhou Z, Wang Z, Li W, Fang C, Wu M, Ma Y, Liu T, Kong L-A and Peng D-L (2014) Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26, 996–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X, Chen L, Tian W, Tao Y, Kristiansen K, Zhang X, Li S, Yang H, Wang J and Wang J (2010) Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res. 20, 646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, Reidel EJ, Turgeon R, Liu P, Sun Q, Nelson T and Brutnell TP (2010) The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42, 1060–1067. [DOI] [PubMed] [Google Scholar]
- 14.Reddy AS (2007) Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 58, 267–294. [DOI] [PubMed] [Google Scholar]
- 15.Schoning JC, Streitner C, Meyer IM, Gao Y and Staiger D (2008) Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 36, 6977–6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spellman R, Llorian M and Smith CW (2007) Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell 27, 420–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui Z, Xu Q and Wang X (2014) Regulation of the circadian clock through pre-mRNA splicing in Arabidopsis. J. Exp. Bot. 65, 1973–1980. [DOI] [PubMed] [Google Scholar]
- 18.Dinesh-Kumar S and Baker BJ (2000) Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 97, 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo PJ, Park M-J, Lim M-H, Kim S-G, Lee M, Baldwin IT and Park C-M (2012) A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24, 2427–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X-C and Gassmann W (2007) Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol. 145, 1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim E, Magen A and Ast G (2007) Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 35, 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbazuk WB, Fu Y and McGinnis KM (2008) Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 18, 1381–1392. [DOI] [PubMed] [Google Scholar]
- 23.Syed NH, Kalyna M, Marquez Y, Barta A and Brown JW (2012) Alternative splicing in plants-coming of age. Trends Plant Sci. 17, 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mancini E, Sanchez SE, Romanowski A, Schlaen RG, San-chez-Lamas M, Cerdan PD and Yanovsky MJ (2016) Acute effects of light on alternative splicing in light-grown plants. Photochem. Photobiol 92, 126–133. [DOI] [PubMed] [Google Scholar]
- 25.Millar AJ (2004) Input signals to the plant circadian clock. J. Exp. Bot. 55, 277–283. [DOI] [PubMed] [Google Scholar]
- 26.Fankhauser C and Staiger D (2002) Photoreceptors in Arabidopsis thaliana: light perception, signal transduction and entrainment of the endogenous clock. Planta 216, 1–16. [DOI] [PubMed] [Google Scholar]
- 27.Shikata H, Hanada K, Ushijima T, Nakashima M, Suzuki Y and Matsushita T (2014) Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl. Acad. Sci. USA 111, 18781–18786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu HP, Su YS, Chen HC, Chen YR, Wu CC, Lin WD and Tu SL (2014) Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol. 15, R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee C and Roy M (2004) Analysis of alternative splicing with microarrays: successes and challenges. Genome Biol. 5, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandenbroucke II, Vandesompele J, Paepe AD and Messiaen L (2001) Quantification of splice variants using real-time PCR. Nucleic Acids Res. 29, E68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson CG, Fuller J, Maronova M, Kalyna M, Davidson D, McNicol J, Barta A and Brown JW (2008) Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant J. 53, 1035–1048. [DOI] [PubMed] [Google Scholar]
- 32.Jung K-H, Bartley LE, Cao P, Canlas PE and Ronald PC (2009) Analysis of alternatively spliced rice transcripts using microarray data. Rice 2, 44–55. [Google Scholar]
- 33.Moore MJ and Silver PA (2008) Global analysis of mRNA splicing. RNA 14, 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang R, Calixto CP, Tzioutziou NA, James AB, Simpson CG, Guo W, Marquez Y, Kalyna M, Patro R, Eyras E, Barta A, Nimmo HG and Brown JW (2015) AtRTD - a comprehensive reference transcript dataset resource for accurate quantification of transcript-specific expression in Arabidopsis thaliana. New Phytol 208, 96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kami C, Lorrain S, Hornitschek P and Fankhauser C (2010) Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91, 29–66. [DOI] [PubMed] [Google Scholar]
- 36.Jenkins GI (2014) The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 26, 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrillo E, Godoy Herz MA, Fuchs A, Reifer D, Fuller J, Yanovsky MJ, Simpson C, Brown JW, Barta A, Kalyna M and Kornblihtt AR (2014) A chloroplast retrograde signal regulates nuclear alternative splicing. Science 344, 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alabadýì D, Oyama T, Yanovsky MJ, Harmon FG, Más P and Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. [DOI] [PubMed] [Google Scholar]
- 39.Huang W, Pérez-García P, Pokhilko A, Millar A, Antoshech-kin I, Riechmann JL and Mas P (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336, 75–79. [DOI] [PubMed] [Google Scholar]
- 40.Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Hen-riques R, Pruneda-Paz JL, Chua NH, Tobin EM, Kay SA and Imaizumi T (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22, 606–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Más P, Kim W-Y, Somers DE and Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426, 567–570. [DOI] [PubMed] [Google Scholar]
- 42.Wang X and Ma L (2013) Unraveling the circadian clock in Arabidopsis. Plant Signal Behav. 8, e23014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hazen SP, Naef F, Quisel T, Gendron JM, Chen H, Ecker JR, Borevitz JO and Kay SA (2009) Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol. 10, R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staiger D and Green R (2011) RNA-based regulation in the plant circadian clock. Trends Plant Sci. 16, 517–523. [DOI] [PubMed] [Google Scholar]
- 45.Streitner C, Hennig L, Korneli C and Staiger D (2010) Global transcript profiling of transgenic plants constitutively overexpressing the RNA-binding protein AtGRP7. BMC Plant Biol. 10, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Wu F, Xie Q, Wang H, Wang Y, Yue Y, Gahura O, Ma S, Liu L, Cao Y, Jiao Y, Puta F, McClung CR, Xu X and Ma L (2012) SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24, 3278–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones MA, Williams BA, McNicol J, Simpson CG, Brown JW and Harmer SL (2012) Mutation of Arabidopsis spliceosomal time-keeper locus1 causes circadian clock defects. Plant Cell 24, 4066–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng X, Gu L, Liu C, Lu T, Lu F, Lu Z, Cui P, Pei Y, Wang B and Hu S (2010) Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 107, 19114–19119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, Cuevas JC, Herz MAG, Depetris-Chauvin A and Simpson CG (2010) A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468, 112–116. [DOI] [PubMed] [Google Scholar]
- 50.Deng X, Lu T, Wang L, Gu L, Sun J, Kong X, Liu C and Cao X (2016) Recruitment of the NineTeen Complex to the activated spliceosome requires AtPRMT5. Proc. Natl. Acad. Sci. USA 113, 5447–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong S, Song H-R, Lutz K, Kerstetter RA, Michael TP and McClung CR (2010) Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 107, 21211–21216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naftelberg S, Schor IE, Ast G and Kornblihtt AR (2015) Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 84, 165–198. [DOI] [PubMed] [Google Scholar]
- 53.Lopato S, Mayeda A, Krainer AR and Barta A (1996) Pre-mRNA splicing in plants: characterization of Ser/Arg splicing factors. Proc. Natl. Acad. Sci. USA 93, 3074–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reddy AS and Shad Ali G (2011) Plant serine/arginine-rich proteins: roles in precursor messenger RNA splicing, plant development, and stress responses. Wiley Interdiscip. Rev. RNA 2, 875–889. [DOI] [PubMed] [Google Scholar]
- 55.Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T and Chabot B (2007) hnRNP proteins and splicing control. Adv. Exp. Med. Biol. 623, 123–147. [DOI] [PubMed] [Google Scholar]
- 56.Wachter A, Ruhl C and Stauffer E (2012) The role of Polypyrimidine tract-binding proteins and other hnRNP proteins in plant splicing regulation. Front. Plant Sci. 3, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin F, Zhang Y and Jiang MY (2009) Alternative splicing and differential expression of two transcripts of nicotine adenine dinucleotide phosphate oxidase B gene from Zea mays. J. Integr. Plant Biol. 51, 287–298. [DOI] [PubMed] [Google Scholar]
- 58.Yoshimura K, Mori T, Yokoyama K, Koike Y, Tanabe N, Sato N, Takahashi H, Maruta T and Shigeoka S (2011) Identification of alternative splicing events regulated by an Arabidopsis serine/arginine-like protein, atSR45a, in response to high-light stress using a tiling array. Plant Cell Physiol. 52, 1786–1805. [DOI] [PubMed] [Google Scholar]
- 59.Tanabe N, Kimura A, Yoshimura K and Shigeoka S (2009) Plant-specific SR-related protein atSR45a interacts with spliceosomal proteins in plant nucleus. Plant Mol. Biol. 70, 241–252. [DOI] [PubMed] [Google Scholar]
- 60.Wu G and Spalding EP (2007) Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 104, 18813–18818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J and Lin C (2007) Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19, 3146–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zoltowski BD and Imaizumi T (2014) Structure and function of the ZTL/FKF1/LKP2 group proteins in Arabidopsis. Enzymes 35, 213–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Franklin KA and Quail PH (2010) Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shikata H, Shibata M, Ushijima T, Nakashima M, Kong SG, Matsuoka K, Lin C and Matsushita T (2012) The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction. Plant J. 70, 727–738. [DOI] [PubMed] [Google Scholar]
- 65.Wu S-H (2014) Gene expression regulation in photomorphogenesis from the perspective of the central dogma. Annu. Rev. Plant Biol. 65, 311–333. [DOI] [PubMed] [Google Scholar]
- 66.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D and Kornblihtt AR (2003) A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12, 525–532. [DOI] [PubMed] [Google Scholar]
- 67.Dujardin G, Lafaille C, de la Mata M, Marasco LE, Muñoz MJ, Le Jossic-Corcos C, Corcos L and Kornblihtt AR (2014) How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell 54, 683–690. [DOI] [PubMed] [Google Scholar]
- 68.Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR and Kay SA (2015) Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 112, E4802–E4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marshall CM, Tartaglio V, Duarte M and Harmon FG (2016) The Arabidopsis sickle Mutant Exhibits Altered Circadian Clock Responses to Cool Temperatures and Temperature-Dependent Alternative Splicing. Plant Cell 28, 2560–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cartegni L, Chew SL and Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3, 285–298. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y, Gu L, Hou Y, Wang L, Deng X, Hang R, Chen D, Zhang X, Zhang Y, Liu C and Cao X (2015) Integrative genome-wide analysis reveals HLP1, a novel RNA-binding protein, regulates plant flowering by targeting alternative polyadenylation. Cell Res. 25, 864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasid-haran R, Muller R, Dreher K, Alexander DL and Garcia-Hernan-dez M (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lazarova GI, Kerckhoffs LH, Brandstadter J, Matsui M, Kendrick RE, Cordonnier-Pratt MM and Pratt LH (1998) Molecular analysis of PHYA in wild-type and phytochrome A-deficient mutants of tomato. Plant J 14, 653–662. [DOI] [PubMed] [Google Scholar]
- 74.Zhou D-X, Kim Y-J, Li Y-F, Carol P and Mache R (1998) COP1b, an isoform of COP1 generated by alternative splicing, has a negative effect on COP1 function in regulating light-dependent seedling development in Arabidopsis. Mol. Gen. Genet. 257, 387–391. [DOI] [PubMed] [Google Scholar]
- 75.Nagy F and Schafer E (2002) Phytochromes control photomorpho-genesis by differentially regulated, interacting signaling pathways in higher plants. Annu. Rev. Plant Biol. 53, 329–355. [DOI] [PubMed] [Google Scholar]
- 76.Holm M, Ma LG, Qu LJ and Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. GenesDev. 16, 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sibout R, Sukumar P, Hettiarachchi C, Holm M, Muday GK and Hardtke CS (2006) Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet. 2, e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, Herzyk P, Brown JW and Nimmo HG (2012) Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24, 961–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Park M-J, Seo PJ and Park C-M (2012) CCA1 alternative splicing as a way of linking the circadian clock to temperature response in Arabidopsis. Plant Signal. Behav. 7, 1194–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mano S, Hayashi M and Nishimura M (1999) Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J. 17, 309–320. [DOI] [PubMed] [Google Scholar]
- 81.Liang C, Cheng S, Zhang Y, Sun Y, Fernie AR, Kang K, Panagiotou G, Lo C and Lim BL (2016) Transcriptomic, proteomic and metabolic changes in Arabidopsis thaliana leaves after the onset of illumination. BMC Plant Biol. 16, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He L and Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531. [DOI] [PubMed] [Google Scholar]
- 83.Di Giammartino DC, Nishida K and Manley JL (2011) Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang H, He H, Wang X, Wang X, Yang X, Li L and Deng XW (2011) Genome-wide mapping of the HY5-mediated genenet-works in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65, 346–358. [DOI] [PubMed] [Google Scholar]
- 85.Tsai H-L, Li Y-H, Hsieh W-P, Lin M-C, Ahn JH and Wu S-H (2014) HUA ENHANCER1 is involved in posttranscriptional regulation of positive and negative regulators in Arabidopsis photo-morphogenesis. Plant Cell 26, 2858–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sorin C, Bussell JD, Camus I, Ljung K, Kowalczyk M, Geiss G, McKhann H, Garcion C, Vaucheret H and Sandberg G (2005) Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 17, 1343–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Makeyev EV, Zhang J, Carrasco MA and Maniatis T (2007) The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boutz PL, Chawla G, Stoilov P and Black DL (2007) Micro-RNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 21, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cardinali B, Cappella M, Provenzano C, Garcia-Manteiga J, Lazarevic D, Cittaro D, Martelli F and Falcone G (2016) MicroRNA-222 regulates muscle alternative splicing through Rbm24 during differentiation of skeletal muscle cells. Cell Death Dis. 7, e2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G and Weigel D (2008) Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105, 8795–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang X, Zhang H and Li L (2012) Alternative mRNA processing increases the complexity of microRNA-based gene regulation in Arabidopsis. Plant J. 70, 421–431. [DOI] [PubMed] [Google Scholar]
- 92.Wu X, Liu M, Downie B, Liang C, Ji G, Li QQ and Hunt AG (2011) Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. USA 108, 12533–12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Penfield S, Josse E-M and Halliday KJ (2010) A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol. Biol. 73, 89–95. [DOI] [PubMed] [Google Scholar]


