Summary
The mandibular or first pharyngeal arch forms the upper and lower jaws in all gnathostomes. A gene regulatory network that defines ventral, intermediate, and dorsal domains along the dorsal-ventral (D-V) axis of the arch has emerged from studies in zebrafish and mice, but the temporal dynamics of this process remain unclear. To define cell fate trajectories in the arches we have performed quantitative gene expression analyses of D-V patterning genes in pharyngeal arch primordia in zebrafish and mice. Using NanoString technology to measure transcript numbers per cell directly we show that, in many cases, genes expressed in similar D-V domains and induced by similar signals vary dramatically in their temporal profiles. This suggests that cellular responses to D-V patterning signals are likely shaped by the baseline kinetics of target gene expression. Furthermore, similarities in the temporal dynamics of genes that occupy distinct pathways suggest novel shared modes of regulation. Incorporating these gene expression kinetics into our computational models for the mandibular arch improves the accuracy of patterning, and facilitates temporal comparisons between species. These data suggest that the magnitude and timing of target gene expression help diversify responses to patterning signals during craniofacial development.
Keywords: fate specification, neural crest, organogenesis, patterning, skeletal
1 |. INTRODUCTION
The development of a mobile jaw is a defining feature of gnathostomes (jawed vertebrates), and a key factor in the success of this clade. The formation of jaws is linked to another vertebrate innovation—neural crest (NC) cells. These cells delaminate from the lateral edge of the neural plate and migrate throughout the embryo, where they form a variety of tissues, such as neurons, glia, and pigment cells, as well as cartilage and bone in the cranial region, including the jaw. Skeletogenic NC cells migrate ventrally in streams to surround the pharynx in pouch-like structures called pharyngeal arches, up to seven of which are arranged bilaterally along the anteroposterior (A-P) axis. The jaw arises from the first arch, which is subdivided into dorsal (proximal, maxillary, and upper jaw) and ventral (distal, mandibular, and lower jaw) portions in zebrafish embryos, and a critical step in jaw development is patterning this arch into dorsalventral (D-V) domains. How gene functions regulate formation of these different jaw components remains unclear.
A D-V arch gene regulatory network (GRN) has emerged from studies in zebrafish and mice (reviewed in Clouthier, Garcia, & Schilling, 2010; Medeiros & Crump, 2012; Schilling & Le Pabic, 2014). At least three domains form along the D-V axis of the embryonic mandibular arch—dorsal, intermediate, and ventral—defined by the expression of distinct sets of transcription factors. Dlx1/2 are expressed in the whole arch. Expression of Hand family transcription factors marks the ventral-most arch, with successively more dorsal domains marked by overlapping regions of expression of Dlx3/4 and Dlx5/6, followed by the most dorsal region which only expresses Dlx1/2. These transcription factors are induced by the activities of signaling molecules from the ventral arch ectoderm, including Bmp2/4 and Endothelin-1 (Edn1), and Notch from the dorsal arch. Initially both Edn1 and Bmp promote ventral/intermediate gene expression (e.g., Hand2 and Dlx3/4/5/6), but later Edn1 function becomes more restricted to the maintenance of intermediate genes. Edn1 acts as a permissive morphogen, required for the initiation of a mandibular arch-specific gene expression cascade that includes Hand and Dlx genes (Clouthier et al., 2000, 2010; Miller, Schilling, Lee, Parker, & Kimmel, 2000; Nair, Li, Cornell, & Schilling, 2007). This occurs, at least in part, through the negative regulation of Nr2f family members, which repress expression of genes associated with ventral mandibular development (Barske et al., 2018). Absence of Edn1 results in a change in D-V identity, with the resulting structures resembling more maxillary-like structures. Homeotic transformations along the D-V axis are also observed in Dlx5/6 and Hand2 mutant embryos (Barron et al., 20ll; Beverdam et al., 2002; Depew, Lufkin, & Rubenstein, 2002; Miller, Schilling, Lee, Parker, & Kimmel, 2003; Talbot, Johnson, & Kimmel, 2010; Walker, Miller, Coffin Talbot, Stock, & Kimmel, 2006; Yanagisawa, Clouthier, Richardson, Charité, & Olson, 2003). In contrast, overexpression of Edn1 in the dorsal arch (zebrafish) or maxillary prominence (mouse) or loss of Nr2f leads to the upregulation of ventral gene expression and subsequent homeotic transformation of maxillary into more mandibular-like structures (Alexander et al., 2011; Barske et al., 2018; Tavares & Clouthier, 2015; Zuniga, Rippen, Alexander, Schilling, & Crump, 2011). Bmps also act as morphogens during mandibular arch development, inducing Hand2 expression, at least in zebrafish (Alexander et al., 2011; Zuniga et al., 2011). Loss of Bmp signaling disrupts lower jaw development but does not appear to result in homeosis (Bonilla-Claudio et al., 2012; Liu et al., 2005a, 2005b; Tucker, Khamis, & Sharpe, l998a; Tucker, Matthews, & Sharpe, 1998b; Vincentz et al., 2016).
In addition to the effects of different levels of Bmp and Edn1 signaling on arch patterning, there are differences in the timing of their activities. In mice Ednra signaling is required between E8.25 and E9.5, corresponding to when NC cells arrive into the mandibular arch (Ruest & Clouthier, 2009). After completion of NC cell migration, Ednra signaling is dispensable for NC patterning, indicating that Edn1’s role is in the initial establishment of NC cell identity (Ruest & Clouthier, 2009). Similarly, injection of Edn1 into the zebrafish arch near the end of NC migration rescues facial development, again arguing that Edn1 acts in a narrow early window of NC cell post-migratory development around 20–28 hr post-fertilization (hpf) (Miller et al., 2000). Bmp also regulates signaling primarily during the early postmi- gratory phase in zebrafish (17–24 hpf) (Alexander et al., 2011; Zuniga et al., 2011). These general aspects of timing are largely conserved between fish and mammals, despite their evolutionary distance and differences in spatiotemporal developmental scale. Recently, our mathematical modeling of D-V patterning has shown that the order in which patterning genes are activated by combinatorial Bmp and Edn1 signaling is important for proper patterning; expression of intermediate factors (such as dlx3b) prior to the ventral factor hand2 leads to the most precise boundary formation in the face of signaling and gene expression noise (Meinecke et al., 20l8).
Many developmental contexts involve integration of multiple morphogens (Briscoe & Small, 2015; Rogers & Schier, 2011; Sagner & Briscoe, 2017). Examples include the interaction of Bmp and Shh signaling in D-V neural tube patterning (Liem, Jessell, & Briscoe, 2000; Zagorski et al., 20l7) and the interaction of Wnt, retinoic acid (RA) and Fgf signaling in A-P body axis patterning (McGrew, Hoppler, & Moon, l997; White, Nie, Lander, & Schilling, 2007). Measurements of morphogen diffusion and immediate downstream target genes, combined with computational modeling, have clarified which of several alternative models of signaling are applicable, such as a source-sink model for early embryonic patterning in zebrafish by the Bmp pathway (Pomreinke et al., 20l7; Zinski et al., 20l7). Precise measurements of target gene expression, such as those in the Nodal signaling pathway, have also suggested that in some morphogen systems the kinetics of expression are more important than more classical threshold models for proper patterning (Dubrulle et al., 2015).
Gene expression in the pharyngeal arches has previously been studied in both zebrafish (Askary et al., 2017; Barske et al., 2016) and mouse (Bonilla-Claudio et al., 2012; Brunskill et al., 2014; Feng et al., 2009; Hooper et al., 2017) using high-throughput microarray or RNA-seq techniques, at multiple time points and in various mutant conditions. While these methods excel at gene discovery, elucidating broad trends across hundreds or thousands of genes, they rely on enzymatic reverse-transcription and PCR amplification of transcripts, which are inherently non-linear in terms of signal produced relative to amount of transcript input. In contrast, the NanoString technique uses direct labeling of transcripts with barcoded probes to measure gene expression levels (Geiss et al., 2008). This approach maintains a high degree of linearity, and has been shown to be at least as good and frequently better at linearity and reproducibility than amplification-based approaches, while being more robust to degradation of RNA (Geiss et al., 2008; Malkov et al., 2009; Neubert et al., 2016; Omolo et al., 2016; Raman et al., 2018; Reis et al., 2011; Richard et al., 2014; Speranza et al., 2017; Veldman-Jones et al., 2015). These advantages of NanoString measurements have also made them particularly useful for computational analysis and modeling of expression patterns (Dubrulle etal., 2015).
We previously used NanoString to examine temporal expression patterns of a small number of D-V patterning genes at selected time points in order to model arch patterning computationally (Meinecke et al., 2018). To investigate expression kinetics of genes in the D-V patterning GRN more thoroughly, we have now quantified expression of over 50 genes in embryonic arch primordia (including known Bmp and Edn1 targets) at several more time points in embryonic zebrafish and mice. Our measurements show unexpected differences in the temporal patterns of expression of related genes with similar spatial patterns of expression. Computational clustering of zebrafish arch gene expression according to these temporal patterns also reveals previously unexpected co-regulation. Moreover, we extend our spatiotemporal computational model of arch patterning (Meinecke et al., 2018) to individual genes and match their temporal profiles. The modeling results indicate that a decoupling between genes that pattern the same domains and between genes that behave similarly over time improves robustness to stochastic fluctuations. In addition, aligning gene expression profiles between zebrafish and mouse facilitates better stage-matching between these model systems during craniofacial development.
2 |. MATERIALS AND METHODS
2.1 |. Zebrafish lines
We used fli1a:GFP (Lawson & Weinstein, 2002) and sox10:dsRed (Das & Crump, 2012) embryos (double-transgenics kindly provided by the Crump lab at the University of Southern California). As simultaneous processing of multiple samples is important to reduce batch effects in expression quantification, embryos were raised at combinations of 25°C or 28.5°C to ensure desired age at the time of dissociation.
2.2 |. Zebrafish live imaging
fli1a:GFP;sox10:dsRed embryos were mounted in 0.5% low-melt agarose immersed in embryo medium containing tricaine and PTU. Imaging was done on a Nikon C1 confocal with a heated stage held at 28.5°C.
2.3 |. Zebrafish arch dissociation and sorting
Transgenic zebrafish embryos were dissociated at 20, 22, 24, 26, 30,36, 40, and 48 hpf using a combination of mechanical disruption and trypsin/collagenase P treatment (as described in Barske et al., 2016). Dissociated cells were sorted for double-positive GFP and dsRed signal using a BD FACSAria II sorter. Sorted cells were resuspended in Qiagen Buffer RLT and submitted for NanoString processing. No further RNA extraction or quality control steps are required for NanoString analysis of dissociated cells in suspension. For each time point, three samples were submitted, consisting of several pooled embryos (from ~10 for older embryos to 30+ for the youngest).
2.4 |. Zebrafish hybridization chain reaction in situ hybridization
Hybridization chain reaction (HCR) was performed as described (Choi, Beck, & Pierce, 2014). About 13 hpf AB zebrafish embryos were fixed overnight at 4°C in 4% paraformaldehyde. Following the HCR protocol. They were mounted in low-melt agarose and imaged on a Nikon Eclipse Ti confocal microscope. The dlx2a probe was ordered from Molecular Instruments (Los Angeles, CA, USA) using the accession number NM_131311.2.
2.5 |. Mouse arch dissection and sorting
Wild-type mouse embryos from the 129S6 strain were collected at 18–20 somites (E9.25), 23–24 somites (E9.5), 27–28 somites (E9.75), and 31–33 somites (E10.0) (staging based on somite counts at Facebase.org). First arch mandibular processes from individual embryos (consisting of all tissue layers) were collected into RNALater. Arches were collected from three embryos for each time point. RNA was then isolated using the RNA Mini kit (Qiagen), with tissue dissociation first accomplished using a QiaShredder column. RNA quality was confirmed using a Bioanalyzer (Agilent) and then processed for NanoString analysis.
2.6 |. Selection of genes for NanoString codesets
NanoString is a candidate-based approach. Zebrafish and mouse genes were selected through a literature search that was up-to-date in 2015, when the codesets were ordered to generate probes. Briefly, we selected genes known to be involved in arch patterning, their known interactors, and several genes for which their homologs in the other species (i.e., zebrafish or mouse) were known to be involved in these processes. The full list of zebrafish and mouse genes examined is listed in Supporting Information File 1.
2.7 |. Data preprocessing and normalization
All raw NanoString counts in both zebrafish and mouse were normalized to the geometric mean of the same four housekeeping genes: actb2, cdc42, stau2, and ubb. Only genes expressed at least 2 standard deviations above artificial negative control levels were chosen for further analysis.
2.8 |. Zebrafish temporal correlation map
A 2nd-order polynomial fit for normalized expression counts was computed using the least-squares method in Microsoft Excel, in order to smooth the effects of small changes in expression. We examined seven time intervals bounded by eight time points (20, 22, 24, 26, 30, 36, 40, and 48 hpf). For each interval, each gene was scored as having increasing or decreasing expression. Thus, each gene had a seven-bit temporal signature. Each gene was then compared with every other gene to test if, at each time interval, their change in expression was similar or different. For each similar change the score for the gene pair was incremented by 1, and for each dissimilar change the score was decremented by 1. Gene pairs that were perfectly matched (+7) were placed in the same group.
2.9 |. Spatiotemporal modeling in the zebrafish mandibular arch
The spatiotemporal computational model was derived from our published computational model (Meinecke et al., 2018). To improve the model we extended the GRN from dorsal, ventral and intermediate gene groups to having multiple genes patterning the same domain, with dlx3b and dlx5a as intermediate genes and jag1b and hey1 as dorsal genes. The ventral gene group was represented solely by hand2. The GRN was modeled by five stochastic differential equations (Meinecke et al., 2018). To compute gene expression in the whole arch gene expression levels across all cells were integrated and normalized with the total arch area at each time point. Parameters in the GRN were then chosen such that this normalized gene expression qualitatively agreed with the NanoString measurements between 22 hpf and 5 hpf (Figure 4;Supporting Information Figure S1; Supporting Information File 2).
FIGURE 4.
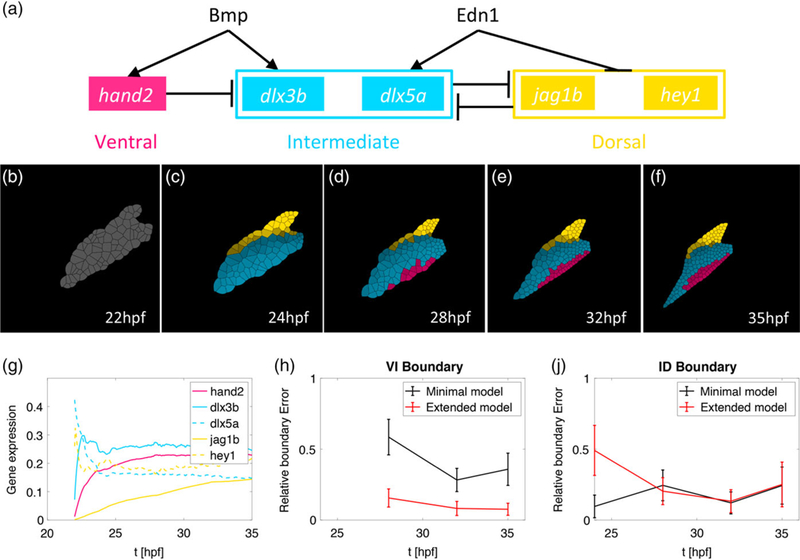
Computational modeling reveals that multiple genes with distinct temporal expression patterns stabilize V-I domain boundary formation. (a) The gene regulatory network (GRN) where individual genes specify the ventral (pink), intermediate (blue), and dorsal (yellow) identities. This model is an extension of the original model introduced in Meinecke et al., 2018 (b-f) simulation results of the spatial gene expression profiles incorporating measured temporal trajectories. The hey1 profile particularly drives dorsal domain establishment to earlier time points. (g) The time-dependent gene expression profiles in the simulation match those measured in vivo in zebrafish (Figure 3) for the time interval 22–35 hpf. (h, j) The y-axis depicts the normalized error of the boundary position in the computational simulations compared to the position observed by quantifying hand2 and dlx5 transgene expression in Meinecke et al., 2018. The solid line represents the mean value over 100 stochastic simulations and the error bars one standard deviation. (h) Comparison with the original model reveals that the parallel patterning of the three spatial domains with different genes with distinct temporal expression profiles stabilizes the accuracy of positioning the V-I boundary. (j) at the I-D boundary, initially higher error due to the early expression of the dorsal gene hey1 resolves to similar error levels at later time points
2.10 |. Mouse temporal correlation map
Similar to the zebrafish data, a second-order polynomial fit was computed for the mouse data. We examined three time intervals bounded by four time points (E9.25, E9.5, E9.75, and E10.0). For each interval, each gene was scored as having increasing or decreasing expression. Due to the fewer time points for mouse, each gene had a three-bit temporal signature. Each gene was compared with every other gene at each time interval to score their temporal expression patterns. Gene pairs that were perfectly matched (+3) were placed in the same group.
2.11 |. Gene selection for interspecies correspondence
Mouse genes were generally less variable in expression than zebrafish genes over the time periods examined. In order to capture the greatest possible temporal variation between gene expression, we selected those mouse genes expressed in the arch that had fish orthologs expressed in the same arch compartment, and that had at least a 1.5-fold change in expression over the time course. We then selected the fish orthologs of these genes for pair-wise analysis.
2.12 |. Aligning interspecies temporal data using cross-correlation
In order to estimate correspondence between developmental stages in fish and mouse, we first carried out a general comparison of developmental rates in arches in these species. We first attempted to infer the relative developmental rates using patterning gene expression data alone, by testing fish: mouse developmental rates ranging from 1:1 to 4:1 (i.e., mouse developing four times slower than fish). However, given the infrequent time point measurements and the relatively low signal-to-noise ratio in these data, no clear consensus emerged regarding a relative developmental rate that maximized the cross-correlation between arch patterning genes. Thus, we could not infer this parameter from the gene expression data alone. Instead, we used physical developmental landmarks to establish an initial relative rate difference between these species. We chose the initial arrival of cranial neural crest cells (NCCs) in the arches as our initial time point (the earliest time at which patterning can commence) and the beginning of chondrogenesis as the final time point (the latest time by which patterning must be established). In fish, NCCs arrive in the arch at around 13–14 hpf (Supporting Information Figure S2 and Meinecke et al., 2018), and in mouse by E8.5 (Abe, Ruest, et al., 2002; Jacques-Fricke, Roffers-Agarwal, & Gammill, 2012; Ruest & Clouthier, 2009). In fish, chondrogenesis initiates at around 53–54 hpf (Barske et al., 2016; Dale & Topczewski, 2011; Le Pabic, Ng, & Schilling, 2014; Schilling & Kimmel, 1997), and in mouse at E12.5 (Wood, Ashhurst, Corbett, & Thorogood, 1991; Wright et al., 1995). Thus, a period of 40 hr in fish development corresponds to a period of 96 hr in mouse, and fish arch development thus proceeds at about 2.4× the rate of mouse arch development. While these measurements establish the window and overall rate at which arches develop, they do not on their own define how the temporal dynamics of patterning compare between the two species.
We then followed two independent strategies for establishing correspondence between the orthologous fish-mouse gene pairs. The first strategy was a manual alignment strategy, wherein the expression trajectories for both species were graphed on the same x-axis, which is hpf in zebrafish. While the zebrafish expression trajectory was placed directly on the shared axis, the first mouse time point, E9.25, was initially placed at 20 hpf, matching the first zebrafish time point, while the last mouse time point, E10.0, was placed according to the 2.4× developmental rate, with the 18 hr elapsed in mouse development from E9.25 to E10.0 equating to 18×2.4 = 43.2 hr on the shared axis, and thus E10.0 initially being placed at 63.2 hpf. The entire trajectory of the mouse gene was then shifted forward (positive value) or backward (negative value) until a subjectively best-possible alignment was achieved. Each gene pair thus was given a value leading to the best-possible alignment, and the average of all these values provided the best-possible global adjustment, γman. This global adjustment was then applied to the starting hpf values to calculate the global correspondence, such that E9.25 = 20+γman, and E10.0 = 63.2+γman. The adjustment was also applied to the mouse trajectory to display the global correspondence applied to each gene pair.
We also undertook an unbiased computational approach for comparing stages and developmental rates between zebrafish and mouse. Given two time series described by u(τ) and v(τ), the cross-correlation between them is
where tl is the lag time between u and v (Papoulis, 1962). If the trajectory is sampled only at a few points in time, we estimate the cross-correlation using a discrete sum:
where (τ1, τ2, …, τn) are the time points at which u and v are sampled. Here, we want to find the best alignment between the developmental trajectories of the zebrafish and the mouse, hence we want to find the lag time (or delay time, td) that maximizes the cross-correlation. That is,
which can be found by repeatedly aligning subsections of the trajectory. Due to the sampling rate and inevitable gaps between time points at which gene expression in measured, the cross-correlation curve may not always be well-resolved. In such cases there can be multiple peaks, and we visually inspect the alignment for each such peak to assess correspondence.
In order to assess the trajectory alignment by cross-correlation, we used cross-validation, and calculated the cross-correlation for a subset (~80%) of the genes, to test how close this value was to the measured cross-correlation of the remaining 20% (Hastie, Tibshirani, & Friedman, 2009). We randomly sampled 9/11 variable genes multiple times to perform cross-validation. However, as discussed above, regarding the determination of relative developmental rates, due to the sampling frequency and the relatively low signal-to-noise ratio, it was difficult to detect clear peaks for the cross-correlation and thus compare across subsets.
3. |. RESULTS
3.1 |. Temporal expression profiles of zebrafish arch patterning genes
Embryonic pharyngeal arches are bilateral, segmentally reiterated structures located ventrally in the head, posterior to the eye, which undergo rapid changes in size, shape and position during development (Figure 1a). As no single transgenic marker uniquely labels NC-derived pharyngeal arch cells in zebrafish, we used fli1a:GFP;sox10:dsRed double-transgenic embryos (Figure 1a)(Barske et al., 2016). fli1a:GFP is expressed in blood vessels, migratory angioblasts, hematopoietic cells, arch NCCs and developing cartilage (Lawson & Weinstein, 2002). sox10:dsRed is expressed in most NCCs and the otic vesicle (Das & Crump, 2012). Sorting with both transgenes (i.e., by gating on both red and green channels) allowed us to isolate arch NCCs with minimal contamination from other populations. Confocal time-lapsed images of these double-transgenics revealed the complexity of arch morphogenesis between 20–48 hpf (Figure 1b). As arches are not patterned simultaneously but instead in an anterior-to-posterior sequence we measured the volumes of different arches as a proxy for the number of cells contained in each arch (Figure 1c) in order to account for variability in gene expression measurements caused by heterogeneity in developmental stage between arches. At 24 hpf, arch 2 accounts for about 70% of the total arch volume, with arch2 accounting for 25% and arch 3 5%. At later time points, the measured volume of arch 1 decreases due to compaction and medial movement of the arch, which obscures medial cells from imaging. As there are no reported differences between cell division and death rates between the arches, we expect the relative proportions of cell numbers in the arches to remain roughly constant. As the majority of cells come from the first arch, and the vast majority from the first two arches, we expect general temporal heterogeneity to have a relatively small contribution to noise in our measurements. Differences in spatial expression between the first two arches have been reported for several genes (Barske et al., 2016; Talbot et al., 2010; Zuniga et al., 2011), and we expect this heterogeneity to have a somewhat larger contribution to measurement noise. FAC-sorting of NC cells from double-transgenic embryos at eight-time points between 20–48 hpf revealed that combined NCC number in all arches per embryo greatly increases over this period (Figure 1d).
FIGURE 1.
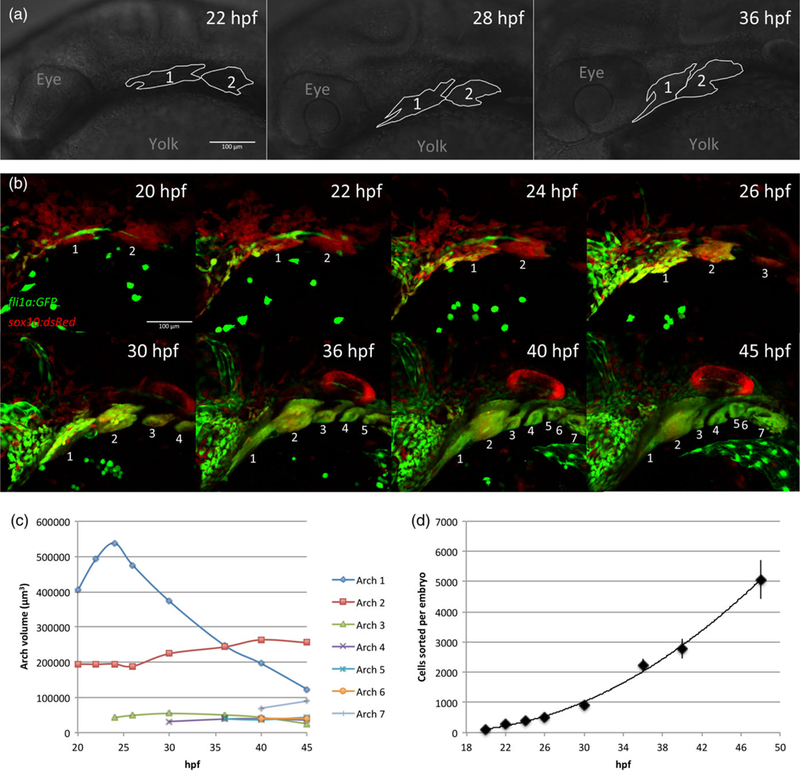
Embryonic growth and morphogenesis of zebrafish pharyngeal arches. (a) Outlines of brightfield images indicating positions of pharyngeal arches 1 and 2 in zebrafish embryos at 22, 28, and 36 hr postfertilization (hpf ). Anterior is left and dorsal is up. (b) Maximum projections of live confocal images of a fli1a:GFP;sox10:dsRed embryo, lateral views, anterior left. Pharyngeal arches are numbered. Cells that co-express both transgenes (yellow) are neural crest cells, which were FAC sorted at the time points shown and processed for NanoString measurements. (c) Quantification of the volume of pharyngeal arches from the images in (a). Measured volume of arch 1 decreases due to compaction and medial movement. At 24 hpf, before significant compaction/migration, arch 1 makes up 69.4% of total arch volume, with arch 2 making up 25.2% and arch 3 5.5%. (d). Average number of fli1a:GFP;sox10:dsRed co-expressing cells sorted per embryo. Three replicates per time point, error bars indicate standard deviation
To assess gene expression dynamics in zebrafish, we used NanoString technology to quantify levels of expression per cell of 55 genes in fli1a:GFP;sox10:dsRed FAC-sorted cells at eight-time points corresponding to Figure 1 between 20 hpf and 48 hpf (Figure 2 and Supporting Information Figure S3). Genes were categorized based on their D-V localization (whole arch, ventral, ventral/intermediate, dorsal, or other) as well as their known regulation by key D-V signaling pathways that pattern the arches, including Edn1, Bmp, Wnt, and Jag/Notch (Supporting Information File 3)(Alexander et al., 2011, 2014; Miller et al., 2000; Talbot et al., 2010; Zuniga et al., 2010, 2011). Intriguingly, genes within each category showed divergent temporal expression patterns in many cases. For example, among genes expressed throughout the arch, some close relatives such as ednraa and ednrab showed opposite trends of increasing or decreasing levels, respectively (Nair et al., 2007). Others, such as ventral/intermediate genes dlx3b/4b and dlx5a/6a, which occupy bigenic clusters (Ellies et al., 1997; Talbot et al., 2010) increased in expression at dramatically different rates. Other examples of highly dynamic genes included the Edn1 effector mef2ca and the direct Wnt target mycn, whose expression declined rapidly similar to ednrab. We note that for many key patterning genes (ventral: hand2; ventral/intermediate: dlx4b/5a/6a; dorsal: hey1, jag1b) expression peaked around 36 hpf, perhaps reflecting the completion of D-V patterning and onset of differentiation programs (as suggested by the concomitant increase in sox9 expression). A consistent theme in these data is that even closely related genes have different responses to the same morphogen inputs.
FIGURE 2.
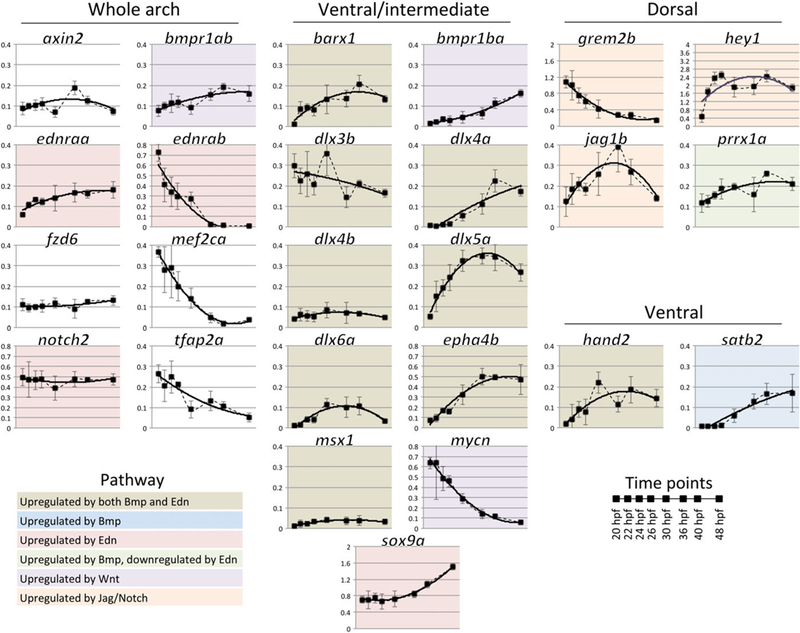
Temporal expression profiles of selected zebrafish arch patterning genes. Transcript counts per cell from NanoString measurements, normalized to a set of housekeeping genes, are shown for the most dynamic or critical genes at eight time points between 20 and 48 hpf. Three biological replicates, error bars are standard deviation. Dotted lines smoothly connect data points, while solid lines are second-degree polynomial fits. Genes are clustered according to their spatial expression in the arches, and colors indicate their regulation by arch patterning pathways (based on the literature). Most but not all genes have the same scale of expression
In addition to these qualitative comparisons, we also conducted a systematic quantitative analysis of arch gene expression. Temporal patterns of gene expression were categorized as increasing or decreasing at seven time intervals (between each of the eight time points measured from 20 hpf to 48 hpf) and used to construct a heat map of all 55 genes analyzed (Figure 3). This revealed nine groups with very similar trajectory profiles within each group (groups A-I), and three individual genes with unique profiles (groups J-L) (Figure 3). Within each group, genes followed similar trends of expression kinetics even when the scale of change in expression was different. For example, in group A expression increases at every interval while in group B expression initially increases but falls at the last two intervals. Thus, genes with subtle changes were grouped along with genes with more dramatic changes as long as they followed the same up-or-down pattern. The two largest groups were group A (13 genes, whose expression kept increasing over all time points), and group B (12 genes, whose expression peaked between 36 hpf and 40 hpf). Several key patterning genes fell into groups B, D, and E, all of which have trajectories that peak midway through the time series. In addition to these, several other genes in these groups are not known to be expressed in NC-derived arch ectomesenchyme cells (marked by O), but their temporal patterns suggest that they may be regulated similarly to NC patterning genes. As we used sorted NC cells for our analysis, we could be detecting low levels of expression that were not previously detected by other (i.e., in situ hybridization) methods, or, despite sorting on two channels, our samples may be contaminated by small numbers of non-NCCs. Group A members are known to be regulated by several different signaling pathways, as indicated by their color-coding (Figure 3), and the monotonically increasing expression pattern of this group suggests that—unlike key patterning genes—group A genes play roles both in patterning and in later developmental stages and differentiation. Alternatively, these genes may be basally expressed at high levels, and the increase seen marks accumulation of transcript as the tissue continues to grow. Group C consists of genes whose expression decreases monotonically. This most likely reflects their early roles in regulation of the migratory NC (tfap2a, Knight et al., 2003, 2; Knight, Javidan, Nelson, Zhang, & Schilling, 2004, 2; Li & Cornell, 2007), or in initial D-V patterning specification (dlx3b, Meinecke et al., 2018). Expression of group C genes may drop either due to repression or degradation.
FIGURE 3.
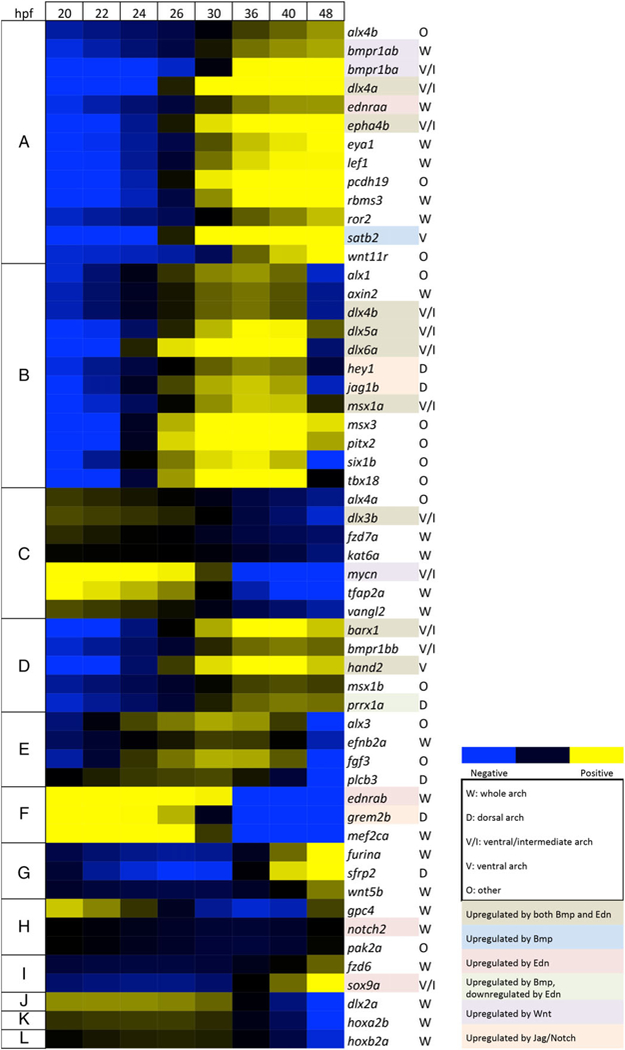
Temporal heat map of zebrafish arch patterning genes. Heat map for 55 zebrafish genes scored based on their expression trajectories (increasing or decreasing expression) at each of seven time intervals (20–22, 22–24, 24–26, 26–30, 30–36, 36–40, and 40–48 hpf). Nine groups (A-1) had similar temporal expression trajectories, either expressed early or late. Three genes expressed early (J-L) showed unique temporal patterns. The heat map is based on log2 fold-changes from the geometric mean for each gene, with coloring based on 10th and 90th percentile fold-change values for the whole data set
3.2 |. Temporal order of gene expression modulates robustness of patterning
We previously performed NanoString measurements for our published computational model for the mandibular arch (Meinecke et al., 2018), but only described the results qualitatively. In our model, domains were patterned by abstracted “ventral”, “intermediate”, or “dorsal” genes, without incorporating actual gene expression trajectories. Here, we extend this computational model with ventral identity represented by hand2, intermediate identity by dlx3b and dlx5a, and dorsal identity by jag1b and hey1 (Figure 4). The time course of the two-dimensional simulation (Figure 4b–f) reveals that incorporating hey1 leads to faster dorsal patterning, compared to the previous model in which dorsal cells remained unpatterned for longer (Meinecke et al., 2018). The normalized gene expression over the time period simulated (20–35 hpf) qualitatively agrees with our NanoString measurements, (Figure 4g). Our previous model also showed that gene expression and signaling noise leads to errors in boundary position and we find that incorporating multiple genes per domain with distinct temporal expression profiles significantly reduces error in formation of the ventral-intermediate domain boundary (Figure 4h). In contrast, error in formation of the intermediate-dorsal domain boundary initially increases, due to the earlier establishment of the dorsal domain in this extended model. At later time points the error converges to similar levels as our previous model.
3.3 |. Temporal expression profiles of mouse arch patterning genes and interspecies comparisons
To assess gene expression dynamics in mice, arches were dissected and RNA prepared for NanoString analysis at E9.25, E9.5, E9.75, and E10.0. These were gross dissections, which included ectoderm, mesoderm, and endoderm—however, the majority of the tissue consisted of NCC-derived mesenchyme. Similar to our analysis in zebrafish, we measured the expression of 65 genes, which were categorized by spatial expression and known regulatory pathways (Figure 5 and Supporting Information Figure S4). As with zebrafish, many genes with similar spatial localization or occupying similar signaling pathways showed distinct temporal patterns of expression. Notably, in contrast to zebra-fish, the temporal patterns of the intermediate/ventral Dlx genes analyzed (e.g., Dlx3 and Dlx5) were similar to one another. Expression was generally less dynamic in the mouse than in zebrafish over the time period examined, but we did note a few patterning genes that underwent significant changes in expression. Transcript levels of the ventral factors Hand1, Hand2, and Satb2 increased, as did Cited1, a transcriptional co-activator which is regulated by Hand factors in cardiac development (McFadden et al., 2005). Meanwhile expression of the ventral morphogen Edn1 and the dorsal factors Hey1 and Jag1 both decreased markedly. We also quantified the temporal patterns of expression of mouse genes and constructed a heat map, revealing five gene groups with similar trajectories (groups I-V) and one gene with a unique profile (group VI) (Figure 6), which we named with Roman numerals to avoid confusion with the zebrafish groups. The largest groups were group I with 26 genes (with a consistent trajectory of increased expression) and group II with 15 genes (with a consistent trajectory of decreased expression). Several ventral and ventral/intermediate factors such as Hand1/2 and Msx1 were in group I. Group II includes the dorsal markers Jag1 and Hey1, and Tfap2a and Sox10, markers of migrating NC cells. Similar to zebrafish, some genes with similar spatial patterns fell into different groups, for example, Dlx5 in group I and Dlx6 in group V, or Msx1 in group I and Msx2 in group III. However, since many mouse genes generally had a smaller dynamic range of expression than their zebrafish counterparts, minor differences in expression trajectory could lead to differential categorization for genes that have qualitatively similar expression. Interestingly, unlike with their zebrafish orthologs, mouse Dlx3 and Dlx4 grouped together in group III.
FIGURE 5.
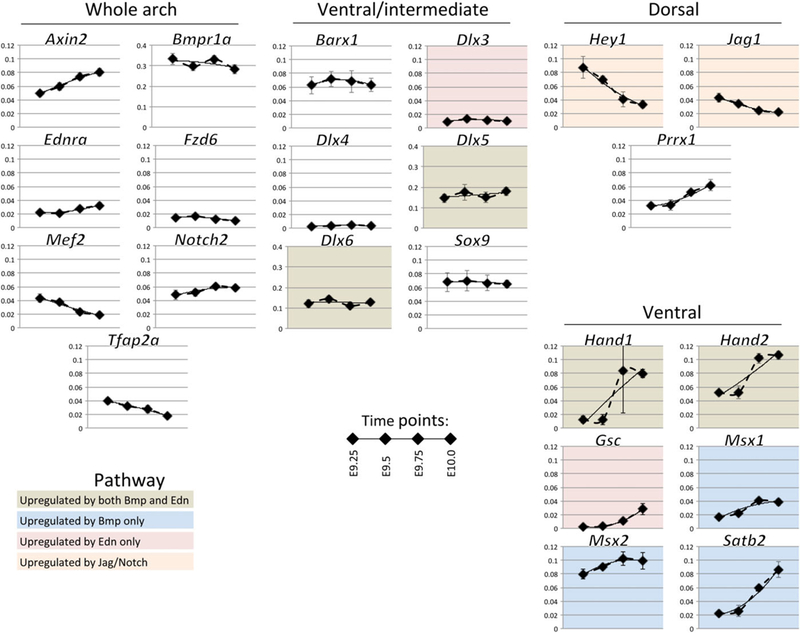
Temporal expression profiles of selected mouse arch patterning genes. Transcript counts per cell from NanoString measurements, normalized to a set of housekeeping genes are shown for the most dynamic or critical genes at four time points between E9.25 and E10.0, three biological replicates, error bars are standard deviation. Dotted lines smoothly connect data points, while solid lines are second-degree polynomial fits. Genes are clustered according to their spatial expression in the arches, and colors indicate their regulation by arch patterning pathways (based on the literature). Most but not all genes have the same scale of expression
FIGURE 6.
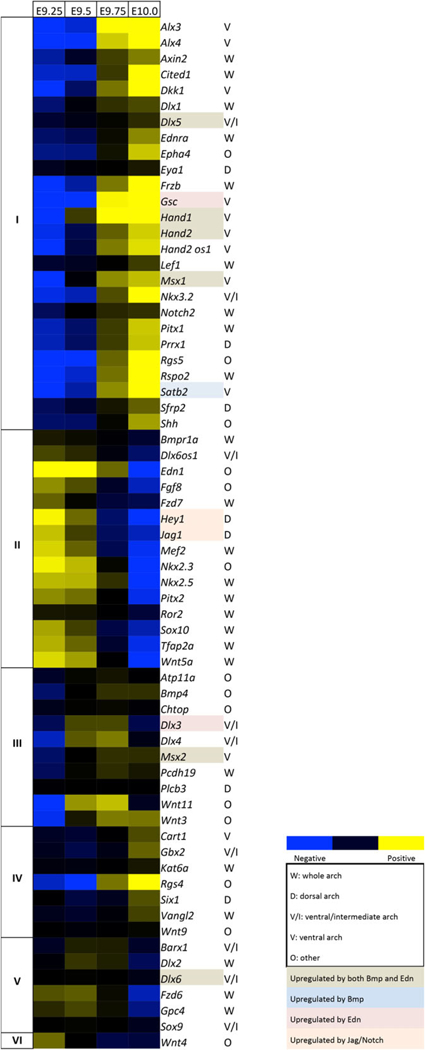
Temporal heat map of mouse arch patterning genes. Heat map for 65 mouse genes scored based on their expression trajectories at each of three time intervals (E9.25–E9.5, E9.5–E9.75, and E9.75–E10.0). Five-gene groups (I–V) had similar temporal expression trajectories, either expressed early or late. 1 (VI) had a unique trajectory. The heat map is based on log2 fold-changes from the geometric mean for each gene, with coloring based on 10th and 90th percentile fold-change values for the whole data set
Due to the smaller number of time points in the mouse data, it was difficult to compare temporally correlated gene groups between zebra-fish and mouse directly. Instead, temporal trajectories of pairs of putative homologous genes were compared to gain insight into relative developmental trajectories between species. We selected mouse genes that were expressed in NCC-derived arch mesenchyme cells, with at least 1.5-fold change in expression over time, that had zebrafish orthologs expressed in the same compartment. This gave us 11 homologous gene pairs (axin2/Axin2, dlx4b/Dlx4, ednraa/Ednra, fzd6/Fzd6, hand2/Hand2, heyl/Heyl, jaglb/Jagl, mef2ca/Mef2, prrxla/Prrxl, satb2/Satb2, and tfap2a/Tfap2a) (Figure 7). For each pair, we used an independent manual or computational approach to calculate gene-pair correspondences, which were averaged over 11 pairs to establish the global correspondence (individual gene-pair alignments in Supporting Information File 4). Our manual approach calculated a global correspondence of E9.25 = 17.5 hpf/E10.0 = 60.7 hpf (Supporting Information Figure S5). Our computational approach calculated a global correspondence of E9.25 = 15.3 hpf/E10.0 = 58.1 hpf (Figure 7). Individual gene pairs were also aligned (Supporting Information File 4). Each of these independent approaches resulted in generally comparable results, differing by less than 5% over the time period (40–50 hr). We can thus predict with confidence that the overall interspecies range of arch patterning occurs within the window: E9.25≈15–18 hpf, and E10.0≈58–61 hpf. As D-V patterning in the zebrafish arches must be complete before initiation of chondrogenesis at 53–54 hpf, thissuggests that D-V patterning in the mouse arch is largely established at least 2 days before initiation of chondrogenesis at E12.5.
FIGURE 7.
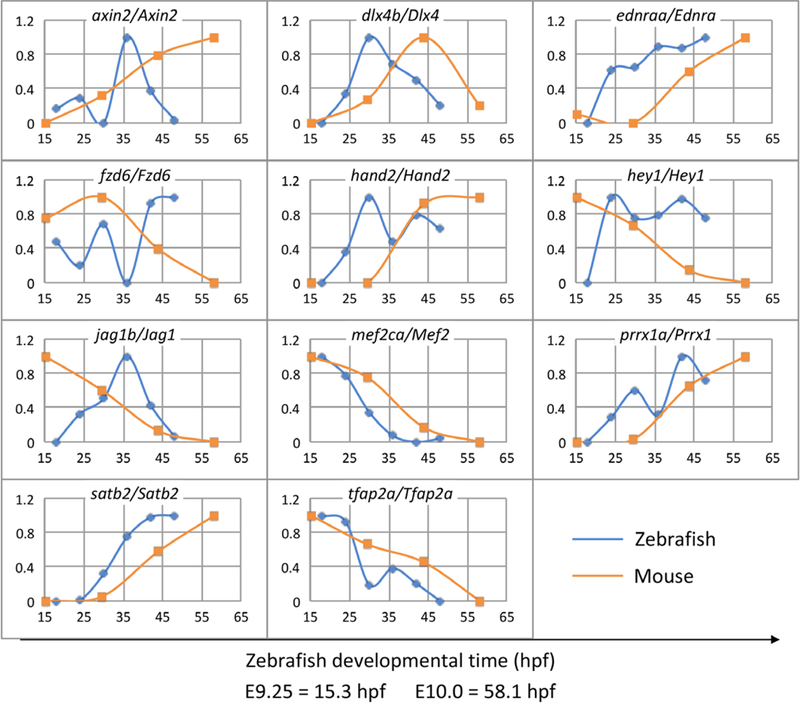
Computational correspondence between timing of zebrafish and mouse arch patterning gene expression. Algorithmic alignment of 11 genes with highly dynamic temporal expression profiles in the arches. Global alignment is: E9.25 = 15.3 hpf/E10.0 = 58.1 hpf. The x-axis shows development time in hpf units on a common scale. The y-axis shows normalized gene expression levels. Expression levels are normalized to 1 for maximum and 0 for minimum
4. |. DISCUSSION
Bmp and Edn1 morphogen gradients are thought to induce distinct gene expression patterns along the D-V axis of the mandibular arch. Target genes of these and other signals that are expressed in similar regions, such the ventral/intermediate dlx3b/4a/4b/5a/6a genes or the dorsal hey1 and jag1b genes, are expected to have similar expression trajectories (i.e., up or downregulation over time) if based simply on the momentary concentration of their controlling morphogens. However, our data do not support this threshold “snapshot” model of morphogen control (Rogers & Schier, 2011). Similarly, recent work quantifying Nodal target gene induction in the early zebrafish (blastula-stage) embryo has shown that, instead of being controlled by threshold, ratchet or time-integrative modes of morphogen signaling, the primary determinant of gene expression patterns is the maximal transcription rate of each gene (Dubrulle et al., 2015). We propose that a similar principle may apply to many D-V patterning genes in the arch GRN.
As evidence for this idea, we find that even paralogs that share cis-regulatory elements can have varying responses to the same upstream signals. The dlx genes are the most dramatic example. dlx3b/4a/4b/5a/6a are all controlled by the same morphogens and are expressed in similar ventral/intermediate regions of the arch. They are not expressed, however, in completely identical spatial regions, with dlx5a/6a expression extending further dorsally and ventrally than dlx3b/4a/4b (Talbot et al., 2010). Additionally, here and in other recent work (Meinecke et al., 2018) we have shown that their temporal expression differs too, with dlx3b high early and then decreasing, dlx4b/5a/6a peaking at ~36 hpf but each changing at very different rates. dlx4a expression is also initiated late and then gradually increases. As the dlx3b/4b and dlx5a/6a pairs are located in bigenic clusters (Ellies et al., 1997), with largely common cis-regulatory regions, we suggest that these differences in spatiotemporal expression could reflect differences in transcription rates due to varying promoters. Maximal transcription rate is determined more by highly proximal promoters than by more distal enhancer regions (Krumm, Hickey, & Groudine, 1995). Dissection of the roles of proximal and distal cis-regulatory regions in temporal regulation of gene expression will require alteration of small sequences while leaving the rest of the cis-regulatory apparatus unchanged (e.g., via CRISPR/Cas9-mediated homologous recombination).
Certain factors such as the Edn1 effector mef2ca are known to regulate members of bigenic clusters such as dlx3b/4b differently, inhibiting dlx3b expression while promoting dlx4b expression (Miller et al., 2007). Further examination of Mef2ca binding sites will reveal whether this disparity arises due to differences in transcription factor activity near the transcriptional start sites or by more distal sequences. However, as dlx3b expression declines at the same time as mef2ca expression in our measurements, this regulatory interaction alone is insufficient to explain the observed dlx3b trajectory. In addition, incorporating these different expression kinetics into our computational model improves the robustness of patterning, for example, V-I domain boundary formation improves in our extended model, which we hypothesize is due mainly to the decoupling of dlx3b and dlx5a expression. Future studies will examine how decoupling spatial and temporal expression improves responses to noise.
Common temporal features of gene expression also hint at unexpected intersections between morphogens and pathways within the arch D-V patterning GRN. For example, mef2ca expression in zebrafish declines rapidly similar to ednrab, perhaps reflecting a late repression of both genes by Edn1 signaling, while earlier mef2ca expression at 20 hpf does not require edn1 (Miller et al., 2007). Wnt signaling positively regulates Edn1 signaling in the arches (Alexander et al., 2014), and the direct Wnt target mycn also declines similar to ednrab and mef2ca, hinting at further points of convergence between these pathways. We also note the gradually declining expression of grem2b: a secreted dorsal Bmp signaling inhibitor, which is upregulated by both Edn1 and Jag/Notch, and downregulated by Bmp signaling (Zuniga et al., 2011). These results add a new level of temporal complexity to our computational models for the zebrafish D-V arch GRN (Clouthier et al., 2010; Meinecke et al., 2018) similar to models in other patterning systems (Pomreinke et al., 2017; Sagner & Briscoe, 2017; Zinski et al., 2017) particularly for processes involving multiple signals in concert (Zagorski et al., 2017). Testing how combinatorial signals lead to such temporal expression kinetics will require experimental manipulation of morphogen gradients at specific times and locations such as with heat-shock inducible transgenic constructs (Alexander et al., 2011; Zuniga et al., 2011). Testing their spatial correlates will require high-resolution measurements of expression in situ with techniques such as hybridization chain reaction (Choi et al., 2018).
Although patterning gene expression is less dynamic in the mouse arch over the period examined compared to the fish arch, we do note some informative patterns. An increase in Hand1/2 expression at late time points might reflect their roles in the dentition (Abe, Tamamura, et al., 2002; Barbosa et al., 2007; Barnes et al., 2010; Catón & Tucker, 2009; Ruest et al., 2003). Although zebrafish only have a single hand2 gene and lack oral teeth, other teleosts such as medaka and pufferfish possess oral teeth and have both handl and hand2 (Vincentz, Barnes, & Firulli, 2011), consistent with this idea. We note that the decrease in Edn1 expression coincides with a reduced requirement for Edn1 signaling at E9.5 (Ruest & Clouthier, 2009), suggesting a general reduction in Edn1 signaling after this stage. In both zebrafish and mouse, expression of the dorsal genes Jag1 and Hey1 decrease at later time points, hinting that Jag/Notch signaling may participate in establishment, but not maintenance, of the dorsal domain.
Other groups have undertaken similar transcriptomic approaches to study craniofacial development in zebrafish and mouse (Askary et al., 2017; Barske et al., 2016; Brunskill et al., 2014). In zebrafish, RNA-seq approaches focused on the arches at 36 hpf in a variety of mutant conditions with cells sorted from specific arch regions have revealed several co-regulated gene clusters (Askary et al., 2017). These researchers performed a meta-analysis based on both wild-type and mutant datasets and clustered all ventral-intermediate dlx genes together in cluster 3 (dlx3b, dlx4a, dlx4b, dlx5a, and dlx6a), along with msxla, and placed jaglb separately in cluster 1 (Askary et al., 2017). In contrast, our temporal clustering places dlx4b, dlx5a, dlx6a, msxla, and jaglb together in group B, with dlx3b in group C and dlx4a in group A. These and other differences likely reflect the absence of granular temporal data in such previous studies. Similarly, in mouse, a microarray/RNA-seq approach examined microdissected regions of wild-type arches at multiple time points yielding gene expression clusters (Brunskill et al., 2014). In that case Dlx1/2/5/6 clustered together, while Dlx1/5 clustered in group I and Dlx2/6 in group V and there are numerous other differences from our results (e.g., Barx1, Msx2, Hand2, Axin2, Prrx1, and Ednra), which we would argue reflect, at least in part, the absence of precise temporal data.
Finally, our comparison of the dynamics of zebrafish and mouse patterning gene expression points to a temporal correspondence of E9.25 mice with ~15–18 hpf zebrafish, and of E10.0 mice with ~58–61 hpf zebrafish. These similarities in gene expression trajectories occur despite dramatic differences in size and shape between mouse and fish arches. For example, the maxillary portion of the mandibular arch is much less prominent in zebrafish than in mammalian embryos, but these data suggest that an equivalent domain with similar expression profile is patterned during this period. More detailed measurements of cell numbers and division rates in the mouse arch, along with computational modeling of patterning in this system, will provide further insight into the coordination of temporal dynamics of gene expression and growth during craniofacial patterning.
Supplementary Material
Acknowledgments
Funding information
Division of Mathematical Sciences, Grant/Award Numbers: 1562176,1763272; National Institute of Dental and Craniofacial Research, Grant/Award Numbers: 5R01DE023050–05, 5R01DE013828–17; National Institute of General Medical Sciences, Grant/Award Number: 5R01GM107264–04; Simons Foundation, Grant/Award Number: 594598
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article
REFERENCES
- Abe M, Ruest L-B, & Clouthier DE (2002). Fate of cranial neural crest cells during craniofacial development in endothelin - A receptor-deficient mice. The International Journal of Developmental Biology, 51, 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe M, Tamamura Y, Yamagishi H, Maeda T, Kato J, Tabata MJ, … Kurisu K (2002). Tooth-type specific expression of dHAND/Hand2: Possible involvement in murine lower incisor morphogenesis. Cell and Tissue Research, 310, 201–212. [DOI] [PubMed] [Google Scholar]
- Alexander C, Zuniga E, Blitz IL, Wada N, Pabic PL, Javidan Y, … Schilling TF (2011). Combinatorial roles for BMPs and endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development, 138, 5135–5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C, Piloto S, Le Pabic P, & Schilling TF (2014). Wnt signaling interacts with bmp and Edn1 to regulate dorsal-ventral patterning and growth of the craniofacial skeleton. PLoS Genetics, 10, e1004479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askary A, Xu P, Barske L, Bay M, Bump P, Balczerski B, … Crump JG (2017). Genome-wide analysis of facial skeletal regionalization in zebrafish. Development, 144, 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AC, Funato N, Chapman S, McKee MD, Richardson JA, Olson EN Yanagisawa H. (2007). Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Developmental Biology, 310, 154–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes R. m. , Firulli B. a. , Conway SJ, Vincentz JW, & Firulli AB (2010). Analysis of the Hand1 cell lineage reveals novel contributions to cardiovascular, neural crest, extra-embryonic, and lateral mesoderm derivatives. Developmental Dynamics, 239, 3086–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron F, Woods C, Kuhn K, Bishop J, Howard MJ, & Clouthier DE (2011). Downregulation of Dlx5 and Dlx6 expression by Hand2 is essential for initiation of tongue morphogenesis. Development, 138, 2249–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barske L, Askary A, Zuniga E, Balczerski B, Bump P, Nichols JT, … Crump JG (2016). Competition between jagged-notch and Endothe- lin1 signaling selectively restricts cartilage formation in the zebrafish upper face. PLoS Genetics, 12, e1005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barske L, Rataud P, Behizad K, Del Rio L, Cox SG, & Crump JG (2018). Essential role of Nr2f nuclear receptors in patterning the vertebrate upper jaw. Developmental Cell, 44, 337–347.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverdam A, Merlo G. r. , Paleari L, Mantero S, Genova F, Barbieri O, … Levi G (2002). Jaw transformation with gain of symmetry after Dlx5/Dlx6 inactivation: Mirror of the past? Genesis, 34, 221–227. [DOI] [PubMed] [Google Scholar]
- Bonilla-Claudio M, Wang J, Bai Y, Klysik E, Selever J, & Martin JF (2012). Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development, 139, 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, & Small S (2015). Morphogen rules: Design principles of gradient-mediated embryo patterning. Development, 142, 3996–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Potter AS, Distasio A, Dexheimer P, Plassard A, Aronow BJ, … Potter SS (2014). A gene expression atlas of early craniofacial development. Developmental Biology, 391, 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catón J, & Tucker AS (2009). Current knowledge of tooth development: Patterning and mineralization of the murine dentition. Journal of Anatomy, 214, 502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Beck VA, & Pierce NA (2014). Next-generation in situ hybridization chain reaction: Higher gain, lower cost, greater durability. ACS Nano, 8, 4284–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, … Pierce NA (2018). Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. bioRxiv 285213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, & Yanagisawa M (2000). Signaling pathways crucial for craniofacial development revealed by endothelin-a receptor-deficient mice. Developmental Biology, 217, 10–24. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Garcia E, & Schilling TF (2010). Regulation of facial morphogenesis by endothelin signaling: Insights from mice and fish. American Journal of Medical Genetics. Part A, 152A, 2962–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale RM, & Topczewski J (2011). Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Developmental Biology, 357, 518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, & Crump JG (2012). Bmps and Id2a act upstream of Twistl to restrict Ectomesenchyme potential of the cranial neural crest. PLoS Genetics, 8, e1002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew MJ, Lufkin T, & Rubenstein JLR (2002). Specification of jaw subdivisions by Dlx genes. Science, 298, 381–385. [DOI] [PubMed] [Google Scholar]
- Dubrulle J, Jordan BM, Akhmetova L, Farrell JA, Kim S-H, Solnica- Krezel L, … Schier AF (2015). Response to nodal morphogen gradient is determined by the kinetics of target gene induction. eLife, 4, e05042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellies DL, Stock DW, Hatch G, Giroux G, Weiss KM, & Ekker M (1997). Relationship between the genomic organization and the over-lapping embryonic expression patterns of the ZebrafishdlxGenes. Genomics, 45, 580–590. [DOI] [PubMed] [Google Scholar]
- Feng W, Leach SM, Tipney H, Phang T, Geraci M, Spritz RA, … Williams T (2009). Spatial and temporal analysis of gene expression during growth and fusion of the mouse facial prominences. PLoS One, 4, e8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, … Dimitrov K (2008). Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature Biotechnology, 26, 317–325. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani RJ, & Friedman JH (2009). The elements of statistical learning:Data mining, inference, and prediction. New York: Springer-Verlag. [Google Scholar]
- Hooper JE, Feng W, Li H, Leach SM, Phang T, Siska C, … Williams T (2017). Systems biology of facial development: Contributions of ectoderm and mesenchyme. Developmental Biology, 426, 97–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques-Fricke BT, Roffers-Agarwal J, & Gammill LS (2012). DNA methyltransferase 3b is dispensable for mouse neural crest development. PLoS One, 7, e47794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight RD, Nair S, Nelson SS, Afshar A, Javidan Y, Geisler R, … Schilling TF (2003). Lockjaw encodes a zebrafish tfap2a required for early neural crest. Development, 130, 5755–5768. [DOI] [PubMed] [Google Scholar]
- Knight RD, Javidan Y, Nelson S, Zhang T, & Schilling T (2004). Skeletal and pigment cell defects in the lockjaw mutant reveal multiple roles for zebrafish tfap2a in neural crest development. Developmental Dynamics, 229, 87–98. [DOI] [PubMed] [Google Scholar]
- Krumm A, Hickey LB, & Groudine M (1995). Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes & Development, 9, 559–572. [DOI] [PubMed] [Google Scholar]
- Lawson ND, & Weinstein BM (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Developmental Biology, 248, 307–318. [DOI] [PubMed] [Google Scholar]
- Le Pabic P, Ng C, & Schilling TF (2014). Fat-Dachsous signaling coordinates cartilage differentiation and polarity during craniofacial development. PLoS Genetics, 10, e1004726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, & Cornell RA (2007). Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Developmental Biology, 304, 338–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem KF, Jessell TM, & Briscoe J (2000). Regulation of the neural patterning activity of sonic hedgehog by secreted BMP inhibitors expressed by notochord and somites. Development, 127, 4855–4866. [DOI] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, … Martin JF (2005a). Distinct functions for Bmp signaling in lip and palate fusion in mice. Development, 132, 1453–1461. [DOI] [PubMed] [Google Scholar]
- Liu W, Selever J, Murali D, Sun X, Brugger SM, Ma L, … Martin JF (2005b). Threshold-specific requirements for Bmp4 in mandibular development. Developmental Biology, 283, 282–293. [DOI] [PubMed] [Google Scholar]
- Malkov VA, Serikawa KA, Balantac N, Watters J, Geiss G, Mashadi-Hossein A, & Fare T (2009). Multiplexed measurements of gene signatures in different analytes using the Nanostring nCounter™ assay system. BMC Research Notes, 2, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, & Olson EN (2005). The Handl and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development, 132, 189–201. [DOI] [PubMed] [Google Scholar]
- McGrew LL, Hoppler S, & Moon RT (1997). Wnt and FGF pathways cooperatively pattern anteroposterior neural ectoderm in Xenopus. Mechanisms of Development, 69, 105–114. [DOI] [PubMed] [Google Scholar]
- Medeiros DM, & Crump JG (2012). New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Developmental Biology, 371, 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinecke L, Sharma PP, Du H, Zhang L, Nie Q, & Schilling TF (2018). Modeling craniofacial development reveals spatiotemporal constraints on robust patterning of the mandibular arch. PLoS Computational Biology, 14, e1006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K, Parker J, & Kimmel CB (2000). Sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch. Development, 127, 3815–3828. [DOI] [PubMed] [Google Scholar]
- Miller CT, Yelon D, Stainier DYR, & Kimmel CB (2003). Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development, 130, 1353–1365. [DOI] [PubMed] [Google Scholar]
- Miller CT, Swartz ME, Khuu PA, Walker MB, Eberhart JK, & Kimmel CB (2007). mef2ca is required in cranial neural crest to effect Endothelin1 signaling in zebrafish. Developmental Biology, 308, 144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Li W, Cornell R, & Schilling TF (2007). Requirements for endothelin type-A receptors and Endothelin-1 signaling in the facial ectoderm for the patterning of skeletogenic neural crest cells in zebra- fish. Development, 134, 335–345. [DOI] [PubMed] [Google Scholar]
- Neubert NJ, Soneson C, Barras D, Baumgaertner P, Rimoldi D, Delorenzi M, … Speiser DE (2016). A well-controlled experimental system to study interactions of cytotoxic T lymphocytes with tumor cells. Frontiers in Immunology, 7, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omolo B, Yang M, Lo FY, Schell MJ, Austin S, Howard K, … Yeatman TJ (2016). Adaptation of a RAS pathway activation signature from FF to FFPE tissues in colorectal cancer. BMC Medical Genomics, 9, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoulis A (1962). The Fourier integral and its applications. New York: McGraw-Hill. [Google Scholar]
- Pomreinke AP, Soh GH, Rogers KW, Bergmann JK, Bläßle AJ, & Müller P (2017). Dynamics of BMP signaling and distribution during zebrafish dorsal-ventral patterning. eLife, 6, e25861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman AT, Pohodich AE, Wan Y-W, Yalamanchili HK, Lowry WE, Zoghbi HY, ‥ Liu Z (2018). Apparent bias toward long gene misregulation in MeCP2 syndromes disappears after controlling for baseline variations. Nature Communications, 9, 3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis PP, Waldron L, Goswami RS, Xu W, Xuan Y, Perez-Ordonez B, … Kamel-Reid S (2011). mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnology, 11, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard AC, Lyons PA, Peters JE, Biasci D, Flint SM, Lee JC, … Smith KG (2014). Comparison of gene expression microarray data with count-based RNA measurements informs microarray interpretation. BMC Genomics, 15, 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KW, & Schier AF (2011). Morphogen gradients: From generation to interpretation. Annual Review of Cell and Developmental Biology, 27, 377–407. [DOI] [PubMed] [Google Scholar]
- Ruest L-B, & Clouthier DE (2009). Elucidating timing and function of endothelin - A receptor signaling during craniofacial development using neural crest cell-specific gene deletion and receptor antagonism. Developmental Biology, 328, 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest L-B, Dager M, Yanagisawa H, Charité J, Hammer RE, Olson EN, … Clouthier DE (2003). Dhand-cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Developmental Biology, 257, 263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagner A, & Briscoe J (2017). Morphogen interpretation: Concentration, time, competence, and signaling dynamics. Wiley Interdisciplinary Reviews: Developmental Biology, 6(4), e271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling TF, & Kimmel CB (1997). Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development, 124, 2945–2960. [DOI] [PubMed] [Google Scholar]
- Schilling TF, & Le Pabic P (2014). Chapter 7 - Neural crest cells in craniofacial skeletal development In Trainor PA (Ed.), Neural Crest Cells (pp. 127–151). Boston, MA: Academic Press. [Google Scholar]
- Speranza E, Altamura LA, Kulcsar K, Bixler SL, Rossi CA, Schoepp RJ, … Connor JH (2017). Comparison of transcriptomic platforms for analysis of whole blood from Ebola-infected cynomolgus macaques. Scientific Reports, 7, 14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JC, Johnson SL, & Kimmel CB (2010). hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development, 137, 2507–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares ALP, & Clouthier DE (2015). Cre recombinase-regulated Endothelin1 transgenic mouse lines: Novel tools for analysis of embryonic and adult disorders. Developmental Biology, 400, 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS, Khamis AA, & Sharpe PT (1998a). Interactions between Bmp-4 and Msx-1 act to restrict gene expression to odontogenic mesenchyme. Developmental Dynamics, 212, 533–539. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Matthews KL, & Sharpe PT (1998b). Transformation of tooth type induced by inhibition of BMP signaling. Science, 282, 1136–1138. [DOI] [PubMed] [Google Scholar]
- Veldman-Jones MH, Brant R, Rooney C, Geh C, Emery H, Harbron CG, … Marshall G (2015). Evaluating robustness and sensitivity of the NanoString technologies nCounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Research, 75, 2587–2593. [DOI] [PubMed] [Google Scholar]
- Vincentz JW, Barnes RM, & Firulli AB (2011). Hand factors as regulators of cardiac morphogenesis and implications for congenital heart defects. Birth Defects Research Part A: Clinical and Molecular Teratology, 91, 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, Casasnovas JJ, Barnes RM, Que J, Clouthier DE, Wang J, Firulli AB (2016). Exclusion of Dlx5/6 expression from the distal-most mandibular arches enables BMP-mediated specification of the distal cap. Proceedings of the National Academy of Sciences of the United States of America, 113, 7563–7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MB, Miller CT, Coffin Talbot J, Stock DW, & Kimmel CB (2006). Zebrafish furin mutants reveal intricacies in regulating Endothelin1 signaling in craniofacial patterning. Developmental Biology, 295, 194–205. [DOI] [PubMed] [Google Scholar]
- White RJ, Nie Q, Lander AD, & Schilling TF (2007). Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebra- fish embryo. PLoS Biology, 5, e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Ashhurst DE, Corbett A, & Thorogood P (1991). The transient expression of type II collagen at tissue interfaces during mammalian craniofacial development. Development, 111, 955–968. [DOI] [PubMed] [Google Scholar]
- Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, … Koopman P (1995). The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nature Genetics, 9, 15–20. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Clouthier DE, Richardson JA, Charité J, & Olson EN (2003). Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development, 130, 1069–1078. [DOI] [PubMed] [Google Scholar]
- Zagorski M, Tabata Y, Brandenberg N, Lutolf MP, Tkačik G, Bollenbach T Kicheva, A. (2017). Decoding of position in the developing neural tube from antiparallel morphogen gradients. Science, 356, 1379–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinski J, Bu Y, Wang X, Dou W, Umulis D, & Mullins MC (2017). Systems biology derived source-sink mechanism of BMP gradient formation. eLife, 6, e22199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga E, Stellabotte F, & Crump JG (2010). Jagged-notch signaling ensures dorsal skeletal identity in the vertebrate face. Development, 137, 1843–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga E, Rippen M, Alexander C, Schilling TF, & Crump JG (2011). Gremlin 2 regulates distinct roles of BMP and endothelin 1 signaling in dorsoventral patterning of the facial skeleton. Development, 138, 5147–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


