Abstract
The Angiotensin II Receptor Blocker (ARB) Telmisartan reduces inflammation through Angiotensin II AT1 receptor blockade and peroxisome proliferator-activated receptor gamma (PPARγ) activation. However, in a mouse microglia-like BV2 cell line, imitating primary microglia responses with high fidelity and devoid of AT1 receptor gene expression or PPARγ activation, Telmisartan reduced gene expression of pro-injury factors, enhanced that of anti-inflammatory genes, and prevented LPS-induced increase in inflammatory markers. Using global gene expression profiling and pathways analysis, we revealed that Telmisartan normalized the expression of hundreds of genes upregulated by LPS and linked with inflammation, apoptosis and neurodegenerative disorders, while downregulating the expression of genes associated with oncological, neurodegenerative and viral diseases. The PPARγ full agonist Pioglitazone had no neuroprotective effects. Surprisingly, the PPARγ antagonists GW9662 and T0070907 were neuroprotective and enhanced Telmisartan effects. GW9226 alone significantly reduced LPS toxic effects and enhanced Telmisartan neuroprotection, including downregulation of pro-inflammatory TLR2 gene expression. Telmisartan and GW9662 effects on LPS injury negatively correlated with pro-inflammatory factors and upstream regulators, including TLR2, and positively with known neuroprotective factors and upstream regulators. Gene Set Enrichment Analysis (GSEA) of the Telmisartan and GW9662 data revealed negative correlations with sets of genes associated with neurodegenerative and metabolic disorders and toxic treatments in cultured systems, while demonstrating positive correlations with gene sets associated with neuroprotection and kinase inhibition. Our results strongly suggest that novel neuroprotective effects of Telmisartan and GW9662, beyond AT1 receptor blockade or PPARγ activation, include downregulation of the TLR2 signaling pathway, findings that may have translational relevance.
Keywords: Angiotensin receptor blockers; Inflammation; Neuroprotection, microglia; PPARγ; TLR2
Introduction
Dysregulated and excessive inflammation is a significant factor in the initial stages and development of many brain diseases, where resident microglia play fundamental roles [1–3]. Microglia have key roles to maintain homeostasis, support brain repair and remodeling when their anti-inflammatory M2 phenotype predominates [3]. Conversely, the microglia pro-inflammatory Ml phenotype is a major player and contributor to neurotoxicity, with excessive production of inflammatory cytokines that are considered important participants in inflammatory, traumatic and degenerative brain disorders [4, 5].
Excessive Angiotensin II activity is one important injury factor contributing to the development of brain inflammation [6–10]. Angiotensin II stimulates two receptor types, AT1 and AT2. Pathological effects on inflammation and neurotoxicity were identified as the consequence of brain AT1 receptor activation [6–10]. Consequently, treatment with selective Angiotensin II AT1 Receptor Blockers (ARBs) reduces inflammation, cell injury and apoptosis, demonstrated in neuronal, cerebrovascular endothelial, primary microglia and astrocyte cultures, and after oral administration in many rodent models representing inflammatory, traumatic and neurodegenerative brain disorders [8–15].
We wished to better understand the ARB protective effects using microglia cultures. Lipopolysaccharide (LPS)-induced injury is a representative damaging, pro-inflammatory factor in microglia [16]. ARBs reduce the M1 pro-inflammatory phenotype while stimulating the M2 anti-inflammatory phenotype, documented in cultured primary rat microglia for Candesartan [13] and in cultures of primary mouse microglia and microglia-like BV2 cells for Telmisartan [17].
We selected Telmisartan, the most effective ARB in neuronal cultures [14] with the widest pleiotropic pharmacological profile, blocking AT1 receptors and stimulating the anti-inflammatory, pro-metabolic peroxisome proliferator-activated receptor gamma (PPARγ) [14, 17–19].
To reveal the relative role of AT1 receptor blockade and PPARγ activation in Telmisartan neuroprotection, we analyzed the effects of the full PPARγ agonist Pioglitazone [20] and two PPARγ antagonists, GW9662 and T0070907 [21, 22]. We selected immortalized mouse microglia-like BV2 cell line cultures, injured in vitro by exposure to the inflammatory factor LPS. BV2 cells are frequently used as a suitable model for in vitro studies on microglia and models of brain inflammation [17, 23–27]. Upon LPS exposure, BV2 cells mimic primary microglia responses with high fidelity [23–27]. We performed global gene expression analysis of selected experiments and confirmed the expression of several important pathways and key genes by qPCR.
Materials and Methods
BV2 Cell Culture
BV2 cells were obtained from William Rebeck, Ph.D. Department of Neurosciences, Georgetown University Medical Center, and a mouse Short Tandem Repeat (STR) profile for genotyping and interspecies contamination test was generated (IDEXX, Columbia, MO) (Supplemental Table 1).
BV2 cells were cultured in DMEM (1X) Dulbecco’s Modified Eagle Medium [+] 4.5 g/l D-Glucose [+] L-Glutamine [−] Sodium Pyruvate, with addition of 5% penicillin/streptomycin (Gibco lot# 3304c238), 10% heat inactivated Fetal Bovine Serum (FBS), at 37 °C in an atmosphere of 5% CO2. When reached 80% confluence, 400,000 cells per well were seeded in 6-well plates for further experiments.
Mouse Frontal Cortex
Three individual samples of frontal cortex from C57BL/6J mice (Jackson Laboratories, Farmington, CT), were supplied by Sonia Villapol, Ph.D., Department of Neuroscience, Georgetown University Medical Center (protocol number 2016–1263, approved by the Georgetown University Animal Use and Care Committee (ACUC) and conducted following the NRC guide to the Care and Use of Laboratory animals. These mice were from one of our previous experiments [10]. They had been subjected to traumatic brain injury and treated with vehicle. We demonstrated that traumatic brain injury did not increase AT1 receptor gene expression in the cerebral cortex [10].
Experimental Design and Randomization to Confirm Microarray Results Using qPCR
Dimethylsulfoxide (DMSO), Telmisartan, Valsartan, Pioglitazone, GW9662, T0070907 and LPS were from Sigma- Aldrich (St. Louis, MO). All drugs were used at 10 μM, diluted in 1.5% DMSO, and LPS at 100 ng/ml, diluted in water. All treatments consisted of three individual independent samples per group, each sample analyzed in triplicates, and included 1.5% DMSO (Sigma-Aldrich, St. Louis, MO). Vehicle-treated groups received 1.5% DMSO for 3 h. Groups treated only with drugs (Telmisartan, Pioglitazone, GW9662 and T0070907) received the drugs for 3 h. Groups treated only with LPS, received vehicle for 2 h, followed by LPS for 1 h. Groups treated with drugs + LPS received drugs for 2 h followed by LPS for 1 h. All experiments were conducted for 3 h. The experiments were terminated by discarding the medium and treating each well with 350 μl of lysis buffer RLT from RNeasy Mini Kits (Qiagen, Valencia, CA). Researchers performing the experiments were blinded to the protocols with a third party concealing the treatments with individually coded vials.
Separate experiments were conducted to test the effects of:
Telmisartan, LPS and Telmisartan + LPS;
Pioglitazone and Pioglitazone + LPS;
Valsartan, LPS and Valsartan + LPS
GW9662, GW9662 + LPS, and GW9662 + LPS + Telmisartan;
T0070907, T0070907 + Telmisartan, and T0070907 + LPS + Telmisartan.
Quantitative PCR
To compare the relative gene expression of AT1 receptor and PPARγ, total RNA was extracted from three different BV2 cell stocks and three different mouse frontal cortex samples using 1 ml TRIzol reagent (Invitrogen, Carlsbad, CA) followed by purification using the RNeasy Mini Kit (Qiagen, Valencia, CA), according to manufacturer’s instructions. To test microarray results on selective gene expression, total RNA was extracted from cultured BV2 cells as described above. Synthesis of complementary DNA (cDNA) was performed with 0.6 μg of total RNA and Super-Script III first-Strand Synthesis Kit (Invitrogen, Carlsbad, CA). The remaining reagents for RNA isolation and reverse transcription were from Invitrogen. Quantitative real-time PCR reactions were performed using an Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA) with PowerUp™ SYBR® Green Master Mix (Thermo Fisher). qPCR was performed in a 10 μl reaction mixture containing 8 μl SYBR Green PCR Master Mix, 2 μl cDNA and 0.3 μmol/l of each primer for specific target (Supplemental Table 2). Amplification conditions consisted of 1 denaturation/activation cycle at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 60 s. Serial dilutions of cDNA from the same source as samples were used to obtain a standard curve. Individual targets for each sample were quantified by determining the cycle threshold and comparison with the standard curve (ΔΔCt method). The relative amount of the target mRNA was normalized with the housekeeping gene GAPDH.
Statistical Analysis for qPCR Samples
Data in Fig. 1 were expressed as fold-change relative to CTX after correction for GAPDH expression and were analyzed by one-way ANOVA followed by Duncan test. Multiple group comparisons for data obtained for all other qPCR experiments were performed by ANOVA followed by Newman-Keuls post-test. Statistical significance was determined using GraphPad Prism 5 Software (GraphPad Software, San Diego, CA, USA). In all cases, data are expressed as means ± SEM and considered statistically significant given a probability value of ≤0.05.
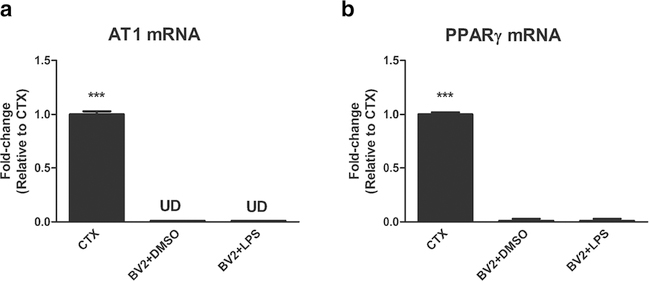
Fig. 1.
Expression of AT1 and PPARγ genes in mouse cortex, BV2 cells treated with DMSO and BV2 cells treated with DMSO + LPS. In contrast to the mouse frontal cortex (CTX) BV2 cells do not express the AT1 receptor gene, whether treated with LPS or not (A). PPARg gene expression is extremely low in BV2 cells when compared to mouse frontal cortex and does not change whether treated with LPS or not. Results are means of three samples analyzed independently. Data are expressed as fold-change relative to CTX after correction for GAPDH expression and were analyzed by one way ANOVA followed by Duncan test. ***p < 0.0001 vs all others
Gene Expression Analysis
Total RNA was extracted from groups of cultured BV2 cells; each group consisted of three independent experiments, with investigators blinded to the protocol. Experiment # 1 consisted on separated groups treated with vehicle (DMSO, 1.5%), Telmisartan, LPS, and Telmisartan + LPS. Experiment # 2 consisted on separate groups treated with vehicle, Telmisartan, GW9662, LPS, Telmisartan + LPS, GW9662 + LPS, and GW9662 + Telmisartan + LPS. Treatment times were identical to those used for qPCR experiments: Telmisartan and/or GW9662 were administered 2 h before LPS, and the experiments terminated 1 h after LPS administration. Standard procedures for labeling, hybridization, washing, staining and scanning were as per manufacturer’s recommendation (Affymetrix, Santa Clara, CA) and as described in detail [28]. The RNAwas purified using a RiboPure Kit (Ambion, Austin, TX, USA) according to manufacturer’s protocol. The quality and quantity of RNA were ensured using the Bioanalyzer (Agilent, Santa Clara, CA) and NanoDrop (Thermo Scientific, Waltham, MA), respectively. All analyses were performed using Partek Genomics Suite (Chesterfield, MO). Data were considered statistically significant at a p value < 0.05 and included with a cutoff of 1.2-fold-change. For gene expression analysis and microarray data mining and dataset description, we used Ingenuity pathway analysis (IPA) http://www.ingenuity.com. To identify whether sets of genes may have an association with known functional pathways or disease phenotypes, we performed gene set enrichment analysis (GSEA) http://software.broadinstitute.org/gsea/ [28, 29].
The raw data has been submitted to Gene Expression Omnibus (GEO) under accession numbers GSE108669 and GSE108670.
Results and Discussion
Expression of AT1, AT1A, AT1B, AT2 Receptor and PPARγ Genes in BV2 Cells
Angiotensin II activates two receptor types, AT1 and AT2 receptors. While in humans only one AT1 is expressed, in rodents AT1 receptors are expressed as two different receptor subtypes, the AT1A and AT1B receptors. These receptor subtypes have 98% homology in their coding regions [30] and for this reason both receptor subtypes are inhibited by ARBs. The subtypes may only be distinguished by their gene expression, using primers directed to untranslated regions that are not homologous for AT1A and AT1B [30]. On the other hand the gene expression of both subtypes may be simultaneously detected by using primers directed to their common coding region [30]. We used primers directed to the common AT1A and AT1B coding regions to demonstrate expression of these receptor subtypes simultaneously (Supplemental Table 2).
In contrast with the clear expression of AT1 receptor gene in the mouse frontal cortex, AT1 receptor genes were not expressed in our BV2 cell line (Fig. 1a). Microarray analysis confirmed these results, revealing that genes for all Angiotensin II receptors (Agtr1 encoding the AT1 receptor type, Agtr1a encoding the AT1A receptor subtype, Agtr1b encoding the AT1B receptor subtype, and Agtr2, encoding the AT2 receptor type) were not significantly expressed in any of the groups tested (Supplemental Table 3).
The expression of AT1 and AT2 receptors in primary microglia and BV2 cell lines has been controversial. AT1 gene expression was expressed in BV2 cell lines of different origin than the one used in our experiments and in unstimulated primary rat microglia [31, 32]. Conversely, AT1A, AT1B and AT2 gene expression in isolated cortical mouse microglia was not higher than background noise [33]. We did not find AT1 or AT2 receptor gene expression in a BV2 cell line of different origin [17] and in the human macrophage cell line HTP-1 or in human circulating monocytes [34]. Analysis of a complete transcriptome conclusively demonstrated that AT1A, AT1B and AT2 genes are not expressed in human or mouse microglia [35].
The neuroprotective effects of Telmisartan in BV2 cells lacking AT1 receptor expression described here agree with prior observations demonstrating that in some systems, ARBs, including Telmisartan, can be neuroprotective beyond AT1 receptor blockade [36–38].
It has been reported that the AT1 receptor gene, not present in unstimulated primary microglia, was expressed only after 6 h of LPS injury [31]. We have not detected AT1 gene expression in our BV2 cell line after 1 h of LPS injury (Fig. 1a) and these results were confirmed in our microarray analysis (Supplemental Table 3).
The function of AT2 receptors in the brain is controversial, has not been clarified, and most of the evidence indicates that they do not play a significative role in Angiotensin II-induced brain toxicity [8, 9, 13, 39]. AT2 receptors are not present in mouse or human microglia [33, 35] or in BV2 cells [17] and were not expressed in our microarray analysis (Supplemental Table 3).
There is clear evidence of PPARγ gene expression in primary microglia and in several BV2 cell lines, and its stimulation is a major protective factor [2, 14, 35, 40–42]. It was also established that part of Telmisartan neuroprotective effects are the consequence of PPARγ activation [12, 14–18, 43]. However, there are also some previous indications that in some systems, the mechanisms of Telmisartan neuroprotection may not only be beyond AT1 receptor blockade but also unrelated to PPARγ activation [22, 37].
We could only detect very low PPARγ gene expression (Pparg) in our BV2 cell line (Fig. 1b) and none in our microarray analysis (Supplemental Table 3). To test whether PPARγ could be activated in our system, we tested the effect of the PPARγ agonist pioglitazone and the PPARγ agonists GW9662 and T0070907. We found that activation of PPARγ with pioglitazone was not neuroprotective (Fig. 2). The PPARγ antagonists GW9662 (Fig. 3) and T0070907 (Supplemental Fig. 1) were neuroprotective and enhanced, rather than reduced Telmisartan neuroprotection, and there is evidence that T0070907 utilizes mechanisms beyond PPARγ activation [21]. In addition, Valsartan, an ARB without direct PPARγ stimulation [36, 43, 44] and in some systems acting beyond AT1 blockade [36], significantly reduced LPS activation of pro-inflammatory factors (Supplemental Fig. 2), showing that the neuroprotective mechanisms of Telmisartan neuroprotection in our system are not unique to this ARB.
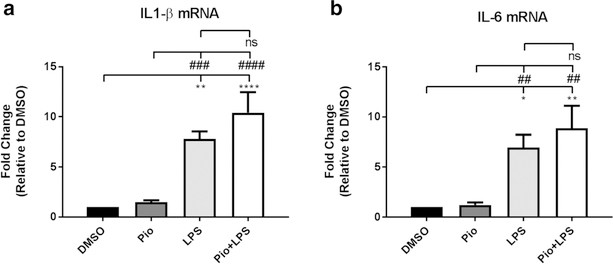
Fig. 2.
The PPARγ agonist Pioglitazone does not reduce LPS-induced increase in IL-1β and IL-6 gene expression. a Pretreatment with the PPARγ full agonist Pioglitazone (Pio) 10 μM for 2 h does not decrease the enhanced IL-1β gene expression produced after 1 h of exposure to LPS (100 ng/ml). ANOVA F (3, 16) = 18.07, p < 0.0001. b Pretreatment with the PPARγ full agonist Pioglitazone (Pio) 10 μM for 2 h does not decrease the enhanced IL-6 gene expression produced after 1 h of exposure to LPS (100 ng/ml). ANOVA F (3, 16) = 9.578, p = 0.0007. Results are means ± SEM for three to five groups analyzed independently. Data were analyzed by one-way ANOVA with Newman-Keuls to correct for multiple comparisons. ****p < 0.0001, **p < 0.01, *p<0.05 compared to DMSO; ####p < 0.0001, ###p < 0.001, ##p<0.01 compared to Pio; ns (not significant)
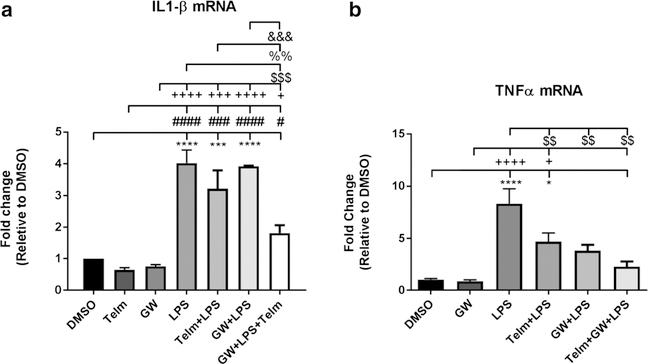
Fig. 3.
The PPARγ antagonist GW9662 enhances the Telmisartan-induced reduction of LPS-induced increase in IL-1β gene expression and eliminates the LPS-induced increase in TNFα gene expression. A Exposure to the PPARγ antagonist GW9662 (GW) 10 μM of for 2 h potentiates the effect of Telmisartan to reduce the increase in IL-1β gene expression produced after 1 h of exposure to LPS (100 ng/ml). ANOVAF (6, 14) = 27.95, p < 0.0001. B Exposure to the PPARγ antagonist GW9662 (GW) 10 μM or Telmisartan (Telm) 10 μM alone for 2 h eliminates the increase in TNFα gene expression produced after 1 h of exposure to LPS (100 ng/ml). ANOVA F (5, 16) = 13.19, p < 0.0001 Results are means ± SEM for three to five groups analyzed independently. Data were analyzed by one-way ANOVA with Newman- Keuls to correct for multiple comparisons. ****p < 0.0001, ***p < 0.001, *p < 0.05 compared to DMSO; ####p < 0.0001, ###p < 0.001, #p < 0.05 compared to Telm; ++++p < 0.0001, +++p<0.001, +p < 0.05 compared to GW; $$$p < 0.001, $$p<0.01 compared to LPS; %%p<0.01 compared to LPS + Telm; &&&p < 0.001 compared to GW+ LPS
In addition, we found that the expression of a number of PPARγ target genes (CD16, CD36, TAF12, CDKN3, MAGOH, GAPDH, STAT1, STAT6, CEBPB) [34, 45] and Supplemental Table 3) was not altered in our microarray analysis (Supplemental Table 3) and that the gene expression of ABCG1, significantly downregulated by Telmisartan in our array analysis (Supplemental Table 3) has been reported to be upregulated by PPARγ activation [14].
From the above we conclude that Telmisartan neuroprotection in our BV2 cells was unrelated to AT1 receptor blockade or PPARγ activation and that our BV2 cell line is an excellent model to characterize additional, novel mechanisms of Telmisartan neuroprotection from LPS.
We propose that apparently contradictory results obtained in different laboratories and including not only AT1 but also PPARγ receptor gene expression may be the result of alterations occurring in different BV2 cell lines over time. The BV2 cell line has been first produced in 1990 [24, 46, 47] and it may now be obtained from diverse sources around the World [17, 41, 42]. Cell lines may not remain as homogeneous clonal cells sharing a similar phenotype over time, and some established cell lines give rise to heterogenous progeny [48]. It is possible that phenotype changes in BV2 cell over time may explain apparently contradictory results. Unfortunately, the different BV2 cell lines from different laboratories have not been fully characterized and compared with each other. To facilitate replication and explain potential future discrepancies with our data, we performed, for the first time, a Cell Check including an STR profile of our BV2 cell line (Supplemental Table 1).
Administration of Telmisartan Alone to Uninjured BV2 Cells Is Neuroprotective, Downregulating Pro-injury and Upregulating Protective Gene Expression
Using global gene analysis, we discovered that when administered to cultured BV2 cells not injured by LPS, as compared with vehicle-treated samples, Telmisartan altered the expression of 492 genes (Supplemental Table 3), including down-regulation of Ccrl2, Dusp2, Dusp5, Csf1, and Bcl11b expression (Supplemental Table 4). These genes have been demonstrated by others to markedly reduce inflammation [49–52]. Other downregulated genes included 11 miRs, such as miR-874 and miR-574 (Supplemental Table 4). These miRs have been previously reported to be involved in multiple functions as well as promoting stroke [53], neuronal vulnerability to injury [54] and cognitive impairment in pre-clinical models of Alzheimer’s disease [55]. We found that Cib1 was also downregulated (Supplemental Table 4). Cib1 encodes CIB1, that activates kinase oncogenic pathways and pathological cardiovascular hypertrophy [56].
Conversely, Telmisartan upregulated numerous histone genes such as Hist1h, Hist2h and Hist4h (Supplemental Table 4). It has already been reported that these histone genes limit inflammation, [57]. Ang4 was also found upregulated (Supplemental Table 4) and it has been demonstrated that Ang4 encodes an angiogenin with microbicidal activity involved in innate immunity [58]. Slc25a51 and Usp17le (Dub3) were also upregulated by Telmisartan (Supplemental Table 4). Slc25a51 is known to protect metabolism [59], and Usp17le (Dub3), encodes a deubiquitinating enzyme regulating multiple cellular processes [60].
The Ingenuity Pathway Analysis (IPA) confirmed the protective effect of Telmisartan. The IPA’s diseases and functions analysis with largest numbers of network molecules included activation, movement, and migration of vascular endothelial cells, inflammatory response and immunological disease (Supplemental Table 4). The upstream regulator analysis showed negative correlations with many inflammatory factors, such as LPS, IL-1β, TNF-α, TGF-beta 1, TLR4 and TLR7 (Table 1 and Supplemental Table 4). There were additional negative correlations with many known inflammation transcription regulators, although these correlations were not significant. Networks with largest numbers of molecules include cell death and survival, inflammatory response and many aspects of cell biology and cancer (Supplemental Table 4).
Table 1.
IPA up-stream regulator’s z-score comparisons in BV2 cells treated with LPS, LPS + Telmisartan and LPS + Telmisartan + GW9662. The genes differentially expressed between LPS/DMSO, between LPS/LPS + Telmisartan and between LPS + Telmisartan/LPS + Telmisartan+GW9662 were put in Ingenuity Pathway Analysis program upstream regulators to identify the transcriptional regulator genes/drugs that may be responsible for most of the differentially expressed genes in each comparison. The z-score values infer the activation (positive number) or inhibition (negative number) states of predicted transcriptional regulators. The list of all the common upstream regulators in the 3 comparisons were put in a table and sorted based on the LPS comparison z-score. Except for NUPR1, Forskolin and ADRB, all the others upstream regulators show a complete reversal of z-score when comparing LPS to LPS + Telmisartan or to LPS + Telmisartan+GW9662. On the other hand, the z-score of LPS + Telmisartan+GW9662 is in the same direction and always greater than the one with only LPS + Telmisartan
| Upstream regulator | LPS/DMSO z-score | LPS/LPS + Telmisartan z-score | LPS + Telmisartan/GW + LPS + Telmisartan z-score |
|---|---|---|---|
| TNF | 8.83 | −5.442 | −3.088 |
| CSF2 | 5.848 | −4.438 | −2.793 |
| MYD88 | 5.816 | −2.641 | −2.818 |
| TLR4 | 5.47 | −3.434 | −3.991 |
| IL2 | 5.461 | −2.31 | −3.305 |
| TLR3 | 5.329 | −2.393 | −3.928 |
| TGFB1 | 5.226 | −3.039 | −3.051 |
| TICAM1 | 5.216 | −3.376 | −3.451 |
| TLR9 | 4.999 | −2.008 | −3.484 |
| TLR7 | 4.58 | −2.377 | −3.533 |
| TNF (family) | 4.522 | −2.96 | −2.929 |
| ERK1/2 | 4.43 | −1.094 | −2.174 |
| APP | 4.416 | −2.077 | −2.852 |
| IL18 | 4.416 | −2.04 | −2.928 |
| TLR2 | 4.416 | −1.518 | −2.575 |
| IL6 | 4.226 | −1.548 | −2.39 |
| STAT1 | 3.923 | −2.422 | −2.909 |
| E. coli lipopolysaccharide | 3.9 | −2.092 | −2.79 |
| JUN | 3.745 | −0.827 | −1.696 |
| MAPK14 | 3.655 | −1.614 | −2.13 |
| MAP3K7 | 3.518 | −1.607 | −2.938 |
| IL7 | 3.514 | −1.443 | −1.709 |
| NUPR1 | 3.182 | −2 | 3.464 |
| TNFRSF1A | 3.056 | −1.745 | −0.102 |
| CEBPB | 3.017 | −0.606 | −1.978 |
| OSM | 2.791 | −0.829 | −2.107 |
| IL4 | 2.684 | −0.532 | −2.528 |
| IL33 | 2.584 | −1.367 | −2.404 |
| ERBB2 | 2.255 | −0.528 | −2.184 |
| IL21 | 2.001 | −0.149 | −1.709 |
| TNFRSF1B | 1.893 | −1.095 | −2.4 |
| forskolin | 1.783 | 0.487 | −0.373 |
| cycloheximide | 1.589 | −0.829 | −1.937 |
| PPARG | −1.202 | 1.49 | 1.54 |
| dexamethasone | −1.238 | 0.368 | 1.597 |
| MYC | −1.505 | 0.532 | 1.297 |
| troglitazone | −1.559 | 1.749 | 2.759 |
| CD28 | −1.581 | 0.761 | 1.493 |
| vorinostat | −1.736 | 0.067 | 0.522 |
| NFE2L2 | −1.741 | 1.734 | 3.905 |
| APOE | −1.823 | 0.211 | 1.452 |
| ADRB | −2.119 | 0.508 | −1.941 |
| caffeic acid phenethyl ester | −2.224 | 1.474 | 1.727 |
| GW3965 | −2.276 | 0.584 | 1.275 |
| 15-deoxy-delta-12,14 -PGJ 2 | −2.402 | 2.268 | 2.863 |
| rosiglitazone | −2.452 | 1.119 | 2.136 |
| PTGER4 | −2.454 | 1.577 | 3.058 |
| TNFAIP3 | −2.498 | 1.776 | 2.213 |
| ZBTB16 | −2.508 | 1.094 | 1.732 |
| wortmannin | −2.588 | 0.601 | 1.356 |
| resveratrol | −2.723 | 2.403 | 3.252 |
| SOCS1 | −2.809 | 0.303 | 2.771 |
| VIP | −3.061 | 2.022 | 2.365 |
| NR3C1 | −3.279 | 2.413 | 2.511 |
| PD98059 | −4.49 | 1.259 | 1.622 |
| CD3 | −4.56 | 2.209 | 3.158 |
| U0126 | −4.679 | 1.214 | 1.91 |
| LY294002 | −5.419 | 2.264 | 1.992 |
| SB203580 | −5.623 | 1.207 | 1.486 |
Geneset enrichment analysis revealed that genes upregulated by Telmisartan positively correlated with those upregulated in two aged mouse strains undergoing calories restriction diets (Supplemental Table 4). Calories restriction was associated with prolongation of life, favor lipid metabolism and protect from renal disease [61] (GSE75569). These effects were reported to be like those of ARB administration, and it was suggested that Telmisartan administration and calories restriction share common protective and anti-aging mechanisms [62].
In addition, GSEA demonstrated a strong correlation of genes downregulated by Telmisartan that were reported to be upregulated in a mouse model of neuronal ceroid lipofuscinoses (NCL), a severe monogenic neurodegenerative disease of childhood with widespread neuronal loss, demyelination, astrocytosis and microglial activation and without effective therapy [63] (GSE37643) (Supplemental Table 4). Whether Telmisartan administration may ameliorate NCL has not been yet considered.
The potential benefits of Telmisartan administration to uninjured cells support the proposal that this compound may prevent or delay brain injury when administered to populations vulnerable to brain diseases with strong inflammatory components, such as neurodegenerative disorders [8–10].
LPS, when Administered Alone, Upregulated Expression of Pro-inflammatory and Downregulated that of Protective Genes Associated to Multiple Mechanisms of Injury and Brain Disorders
Differential gene expression comparing results from LPS-treated BV2 cells with those of vehicle-treated cells yielded over 979 annotated transcripts significantly upregulated (534) or downregulated (445) by LPS (Supplemental Tables 3 and 5).
The response of the BV2 cells to LPS-induced injury was like that demonstrated in primary microglia [64], indicating that our BV2 cell culture was a reliable substitute for primary microglia cultures.
We confirmed and expanded the previously reported major upregulation of several genes by LPS in our BV2 cell cultures (Supplemental Tables 3 and 5). These genes included several pro-inflammatory cytokines such as IL1-β and IL-6, NF-κB and the TNF superfamily. The LPS-induced increase of IL1-β, IL-6, TNFα and NF-κB was confirmed by qPCR (Fig. 4). In addition, we found that the miR-155, miR-221, Cxc110, and Ccrl2 genes were also upregulated by LPS (Supplemental Tables 3 and 5). All the genes mentioned have been defined as major players in inflammation, and their upregulation by LPS was previously reported [65]. They include the pro-inflammatory cytokines IL1-β [66], NF-κB [67], the TNF superfamily [68], NO and ROS production, miR-155 and miR-221 [69], Cxc110 [70] and Ccrl2 [49].
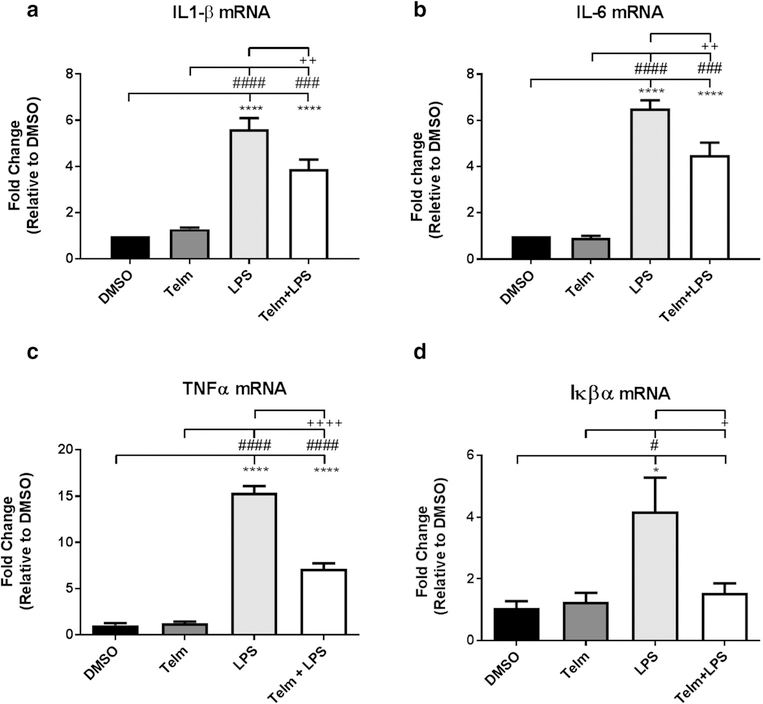
Fig. 4.
Telmisartan significantly reduces LPS-induced IL-β, IL-6, TNFα and Iκβα gene expression. Pretreatment with Telmisartan 10 μM (Telm) for 2 h significantly reduces the increase in a IL-1β gene expression produced after 1 h of exposure to LPS (100 ng/ml). ANOVA F (3, 14) = 37.9 p < 0.0001. b IL-6 gene expression produced after 1 h of exposure to LPS (100 ng/ml). ANOVA F (3, 10) = 52.9 p < 0.0001. c TNFα gene expression produced after 1 h of exposure to LPS (100 ng/ml). ANOVA F (3, 8) = 196.4 p<0.0001. d Iκβα gene expression produced after 1 h of exposure to LPS (100 ng/ml) F (3, 8) = 6.114 p = 0.0182. Results are means ± SEM for three to five groups analyzed independently. Data are expressed as fold-change relative to DMSO and were analyzed by one-way ANOVA followed by Newman-Keuls test for multiple comparisons. ****p < 0.0001, *p < 0.05 compared to DMSO; ####p < 0.0001, ###p < 0.001, #p < 0.05 compared to Telm; ++p < 0.01, +p < 0.05 compared to LPS
Several kinases and the olfactory receptors Olfr887 and Olfr97 were also upregulated (Supplemental Tables 3 and 5). Both the kinases and the olfactory receptors Olfr887 and Olfr97 have been previously associated with neurodegenerative disorders [71, 72].
Conversely, LPS downregulated genes including Histh3b and Hist1h4c, 8 miRs, including miR-129, 28 snoRNAs, including Snora64, Snora20, Snord57 and Snord69, and Sirt7 (Supplemental Tables 3 and 5). Histh3b and Hist1h4c were reported to reduce inflammation [73]. Downregulation of miR-129 and snoRNAs has been reported to be associated with stress, oncological, neurodegenerative and viral diseases [74, 75]. Sirt7 is a member of the sirtuin family proposed to attenuate aging [76].
The list of functionally/biologically annotated only genes (564 transcripts) was submitted to IPA analysis, confirming multiple injury mechanisms stimulated by LPS (Supplemental Table 5). As expected, and with great statistical significance, we identified major canonical pathways of neuroinflammation: IL-6, IL-1, NF-κB, TLR2, NO, ROS, glucocorticoid receptors, TREM1 and p38MAPK signaling. Diseases and functions prominently included inflammatory response (Supplemental Table 5). These major canonical pathways have been previously linked not only with inflammation, but also with Alzheimer’s disease [77], atherosclerosis [78] and cancer, with a key role of protein kinase pathways [17, 79]. Consequently, analysis of diseases and functions (Supplemental Table 5) revealed a predominance of inflammation, and multiple cell functions including apoptosis and proliferation of blood cells [80].
Upstream regulator analysis of these LPS differentially expressed genes expanded our previous observations, including positive correlations with the well-known inflammatory cytokines (IL-1β, IL-6), IFNγ, TGF-beta 1, LPS, TLRs (TLR2, 3, 4, 7 and 9) and components of the TLR signaling pathway CD14, MYD88), NF-κB complex, LCN2, a biomarker for inflammatory and metabolic and neurodegenerative disorders [81, 82] and many kinases linking inflammation and cancer [83], including JNK, MAPK7, MAPK8, ERK, ERK1/2 and Pl3K (Table 1, Supplemental Table 5). Conversely, upstream regulator analysis revealed several kinase inhibitors with known neuroprotective properties, such as SB203580 [84], PD98059, LY294002, [85], U0126 [86] and GW3965 [87] (Table 1, Supplemental Table 5).
Network analysis comprised top diseases and functions: infectious diseases, cell cycle, death and survival, cell to cell signaling, cardiovascular and neurological disease, and lipid metabolism (Supplemental Table 5). These diseases and functions have been previously shown to significantly contribute to LPS-induced senescence in BV2 cells [88].
This information not only confirms the established multiple pro-inflammatory effects of LPS, but also reveals its participation in additional widespread mechanisms of injury. For example, the association with molecular mechanisms of cancer is not surprising, since inflammation has been earlier characterized as a major factor in the development and progression of malignancies [89, 90]. In turn, the association with the role of macrophages, fibroblasts and endothelial cells, supports the earlier report of LPS-induced direct injury to the neurovascular unit and the blood-brain barrier [91].
Telmisartan Normalized Expression of Multiple Pro-injury Genes Upregulated by LPS, and that of Protective Genes Downregulated by LPS
Differential gene expression comparing results from Telmisartan pretreatment followed by exposure to LPS injury for 1 h yielded over 572 well annotated transcripts significantly upregulated (335) or downregulated (237) by LPS (Supplemental Tables 3 and 6).
When followed by exposure to LPS, Telmisartan pretreatment completely reversed 145 of the 534 genes upregulated by LPS when administered alone, and the remaining genes showed a partial downregulation or normalization not reaching statistical significance (Supplemental Tables 3 and 6).
The genes upregulated by LPS and downregulated by Telmisartan include the pro-inflammatory cytokines IL1-β and IL-6, NFκB and its activator miR221, miR155, the TNF superfamily, TLR2, Adamts1 and miR129–2 (Supplemental Tables 3 and 6). These genes, including NFκB and its activator miR221 [92], miR155 [93], the TNF superfamily [94], TLR2 [95], Adamts1 [96] and miR129–2 [74] have been previously reported to be strongly associated with inflammation.
In addition, Telmisartan normalizes LPS-induced upregulation of miR-146, several olfactory receptors, ABCA1, BCB2-like 11 and ptgs2 (Supplemental Tables 3 and 6). miR-146, [97], the olfactory receptors [98] and ABCA1 [99] are genes identified by other groups to promote neurodegenerative disorders; BCB2-like 11 has been associated to apoptosis [100] and ptgs2 with diabetes [101].
Only two genes, Olfr700 and Dynap, remained upregulated after Telmisartan treatment of LPS-injured BV2 cells (Supplemental Table 3).
The effect of Telmisartan on gene expression of inflammatory factors upregulated by LPS was confirmed by qPCR analysis; Telmisartan significantly reduces IL-1β, IL-6, TNF-α and Iκβα. gene expression enhanced by LPS (Fig. 4a, b, c, d). Another ARB, Valsartan, not directly stimulating PPARγ [43, 44] similarly reduced LPS-induced increase in IL-1β and IL-6 gene expression indicating that reduction of LPS effects is not restricted to the ARB Telmisartan (Supplemental Fig. 1).
Conversely, Telmisartan pretreatment significantly reversed the expression of 141 (fold change above 1.2) and normalized 262 (fold change between 1.19 and 1.00) of the 445 genes downregulated by LPS. The remaining 42 genes have a fold change between −1.0 and − 1.19 not reaching significance (Supplemental Table 3). Only one gene (Fv1) remained downregulated after Telmisartan pre-treatment, but with a p value of 0.089 (Supplemental Table 3).
Telmisartan upregulated many genes that were downregulated by LPS, including multiple snoRNAs, Eid2b, Hist1h4m and Hist1h2bh (Supplemental Table 3). These genes have been demonstrated to be protective, including multiple snoRNAs [75], the interacting inhibitor of differentiation Eid2b, protecting against oxidative stress, repressing glucocorticoid-dependent transcription [102], endogenous suppressor of TGF-β signaling [103] and Hist1h4m and Hist1h2bh inhibiting inflammation and vascular apoptosis [57, 104]. Only one gene, the antiretroviral restriction factor Fv1 [105] remained still downregulated after Telmisartan is administered before LPS (Supplemental Table 3).
Taken together, these results indicate that Telmisartan exerts multiple protective mechanisms, not only by downregulating gene expression enhanced by LPS and associated with cell injury and brain diseases, but also by normalizing protective genes downregulated by LPS (Supplemental Table 6).
Consequently, IPA analysis revealed canonical pathways previously identified to be related to neuroinflammation, NF-kB, HMGB1 [106], glucocorticoid receptor, and many networks involving the immune response and infection, cellular function, movement, signaling, maintenance, assembly, organization, death and survival, development of neurological diseases [107] and a cancer signaling network [108] (Supplemental Table 6).
In turn, the IPA examination revealed that Telmisartan administration involved many upstream transcription regulators associated with LPS injury (Supplemental Table 6). As expected, the influence of Telmisartan administration prior to LPS treatment shows a negative z-score (or inhibition) for pro-inflammatory, cellular stress, neurodegenerative and metabolic disorders associated genes that are up-regulated by LPS and downregulated by Telmisartan such as those encoding IL-1β, TNF-α, TLR3-TRIF (TICAM1)-TRAF-INF-β and the TLR3-TRIF (TICAM1)-NF-κB pathways, the TICAM1 pathway including HMGB1, TLR4, TLR2, MyD88, and several MAP kinases; ERK1/2, Map3k7, P38 MAPK (Supplemental Table 6).
Conversely, several up-stream regulators with positive z-score were found for genes downregulated by LPS and upregulated by Telmisartan, such as Smad7, Tnfaip3, ATF4, RARA, ZFP36, the kinase inhibitors U0126, SB203580, LY294002, SP600125 and PD98059, and compounds such as N-acetyl-L-cysteine, simvastatin, trichostatin A and thapsigargin (Table 1, Supplemental Table 6). These up-stream regulators have been previously reported to be protective, including Smad7 [109], Tnfaip3 [110], ATF4 [111], RARA [112], ZFP36 [113], genes encoding the PI3K complex, U0126 [114] (GSE6675), SB203580 [84], LY294002 [115], SP600125 [116], PD98059 [117], N-acetyl-L-cysteine [118], simvastatin [119], trichostatin A [120, 121] and thapsigargin [122].
Networks with the highest number of molecules include cell death, survival and organization, lipid and carbohydrate metabolism and cancer (Supplemental Table 6).
To see whether the genes regulated by Telmisartan when administered alone overlap with genes regulated by Telmisartan followed by LPS treatment, we looked up the 622 genes upregulated by Telmisartan administered before LPS injury. Off these 622 genes upregulated by Telmisartan+LPS, only 15 genes are also upregulated by Telmisartan only (Supplemental Table 3). On the other hand, there are only 14 genes downregulated by Telmisartan alone out of the 399 genes downregulated by Telmisartan+ LPS. (Supplemental Table 3). This means that the effect of Telmisartan on uninjured BV2 cells, although associated with neuroprotection, is vastly different from the neuroprotection that Telmisartan offers when the BV2 cells are injured with LPS.
GW9662, a PPARγ Antagonist, Protected from LPS-Induced Injury and Enhanced Telmisartan Neuroprotective Effects
To further clarify PPARγ participation on Telmisartan neuroprotective effects, we treated BV2 cells, in separate experiments, with the PPARγ antagonist GW9662, administered alone, added prior to LPS, and together with Telmisartan, prior to LPS injury. We expected that GW9662 administration would significantly reduce Telmisartan protective effects. Surprisingly, we found the opposite response, a significant increase in Telmisartan protective effects, such as a reduction in the LPS-induced increase in TNFα gene expression, confirmed by qPCR analysis (Fig. 3a and b).
The finding that GW9662 exerts protective effects unrelated to PPARγ antagonism is supported by previous reports of GW9662 inhibition of tumor growth and promotion of the anticancer effects of the PPARγ agonist rosiglitazone, independently of PPARγ activation [123]. In another study, GW9662 antagonism of PPARγ was not complete, since it only partially antagonized rosiglitazone neuroprotection from NMDA-induced neurotoxicity in cultured hippocampal slices [124]. Moreover, the report of GW9662 neuroprotection in our BV2 cells is not unique, since we found similar neuroprotective effects of another PPARγ antagonist, T0070907 (Supplemental Fig. 2A and 2B).
Treatment of BV2 cells with the PPARγ antagonist GW9662 and Telmisartan followed by LPS showed 211 downregulated and 135 upregulated genes compared to those expressed in BV2 cells treated only with Telmisartan and LPS (Supplemental Tables 3 and 7).
Gene analysis expanded our findings, demonstrating a negative correlation between GW9662 effects and inflammatory genes, drugs and toxins inducing inflammation. When we compared the gene expression resulting from the addition of GW9662 to BV2 cells pretreated with Telmisartan and injured by LPS, we found significant further downregulation of genes previously associated with inflammatory, autoimmune, neurodegenerative, microvascular and metabolic disorders, including 25 genes encoding histones, that are also the most downregulated genes in our study, Edn1 and various chemokines such as Ccl7, Ccl10 and Ccl2. Histones participate in tumor progression and activate TLR receptors and the NLRP3 inflammosome, contributing to cerebrovascular injury [125, 126]. Edn1, encoding the vasoconstrictor endothelin, was proposed as an important factor in the cerebrovascular dysfunction in Alzheimer’s disease [127]. Ccl2 was reported to participate in a several neurological disorders and autoimmune disease [128].
These findings were confirmed by qPCR analysis. GW9662 significantly enhanced the Telmisartan-mediated reduction of LPS-induced increase in IL-1β (Fig. 3a) and reduced the increase in TNF-α gene expression produced by LPS (Fig. 3b). TLR2 was significantly upregulated by LPS (+1.44), down by LPS + Telimsartan (−1.18) and still down by LPS + Telmisartan+GW9662 (−1.13) (Supplemental Table 3).
Conversely, addition of GW9662 to BV2 cells pretreated with Telmisartan followed by LPS upregulated Plin2, Hmox1, and Srxn1 (Supplemental Table 3). Plin2 [129], Hmox1 [130] and Srxn1 [131] have been previously demonstrated to be neuroprotective, to reduce inflammation and to offer beneficial effects on metabolism.
Canonical pathways with larger numbers of associated genes include DNA methylation and transcriptional regulation/repression signaling, including multiple histone genes, inflammation, and oxidative stress response (Supplemental Table 7). Networks with the largest number of molecules include post-translational modifications, cardiovascular disease and cell death and survival, signaling, interaction and development (Supplemental Table 7).
At the disease and function level, the GW9662 treatment of BV2 cells treated with both Telmisartan and LPS included a decrease in inflammatory response, cell-to-cell signaling/interaction, cellular movement and macrophage activation. Several functions associated with cardiovascular disease and diabetes were also statistically significant. Interestingly, organism survival showed the highest number of upregulated genes at the disease level (Supplemental Table 7).
The IPA analysis of upstream regulators for the genes differentially expressed between BV2 cell treated with Telmisartan and LPs, and BV2 cells treated with Telmisartan and GW9662 followed by LPS revealed negative correlations with IL-1β and TNF-α, NF-κB, several TLRs, INF, poly rl:rC, enterotoxinB, cardiotoxin and APP (Table 1 and Supplemental Table 7). These factors were demonstrated to induce inflammation and autoimmune diseases normally activated by LPS, including the pro-inflammatory cytokines IL-1β and TNF-α, NF-κB, the TLRs, INF and the interferon-inducible gene poly rl:rC [132], and enterotoxin B [133]. Cardiotoxin was found to exhibit anti-neoplastic properties [134] and APP is a multifunctional protein associated with Alzheimer’s disease [135]. Additionally, there was a negative correlation with ERK1/2 and p38MAPK pathways, including MEK (MAP2K1) and with Raf1 (MAP3K) which acts upstream of MEK and ERK (Table 1 and Supplemental Table 7),
Of interest is the TGF-beta 1 pathway, as the most down-regulated pathway after LPS and TNFα, with over 90 genes upregulated by LPS, 69 of them are downregulated by Telmisartan and 32 by GW9662 (Supplemental Table 7). TGF-beta 1 was reported to increase microglial p38 MAPK and AKT phosphorylation [136] that is impaired by SB203580 and LY294002, two kinase inhibitors [84, 137] that we found to have positive correlation with GW9662 (Supplemental Table 7).
Conversely, we found that genes upregulated by GW9662 are positively correlated with upstream regulators such as genes, drugs, statins and other neuroprotective compounds (Table 1 and Supplemental Table 7). There was a positive correlation with factors previously reported to protect from oxidative stress, inflammation and age-related disorders such as cancer, metabolic and cardiovascular diseases. These include the transcription regulator NFE2l2 [138, 139] the nuclear receptor NR3C1 [140], several kinase inhibitors including LY294002, PD98059, SB203580 and U0126, statins such as fluvastatin, cerivastatin, atorvastatin and simvastatin, resveratrol [141] and curcumin [142, 143]. When comparing the IPA’s upstream regulator’s z-score activation/inhibition for LPS, LPS + Telmisartan and LPS + Telmisartan + GW9662, we found a striking reversal action of Telmisartan over LPS and that reversal is even enhanced with the addition of GW9662 (Table 1).
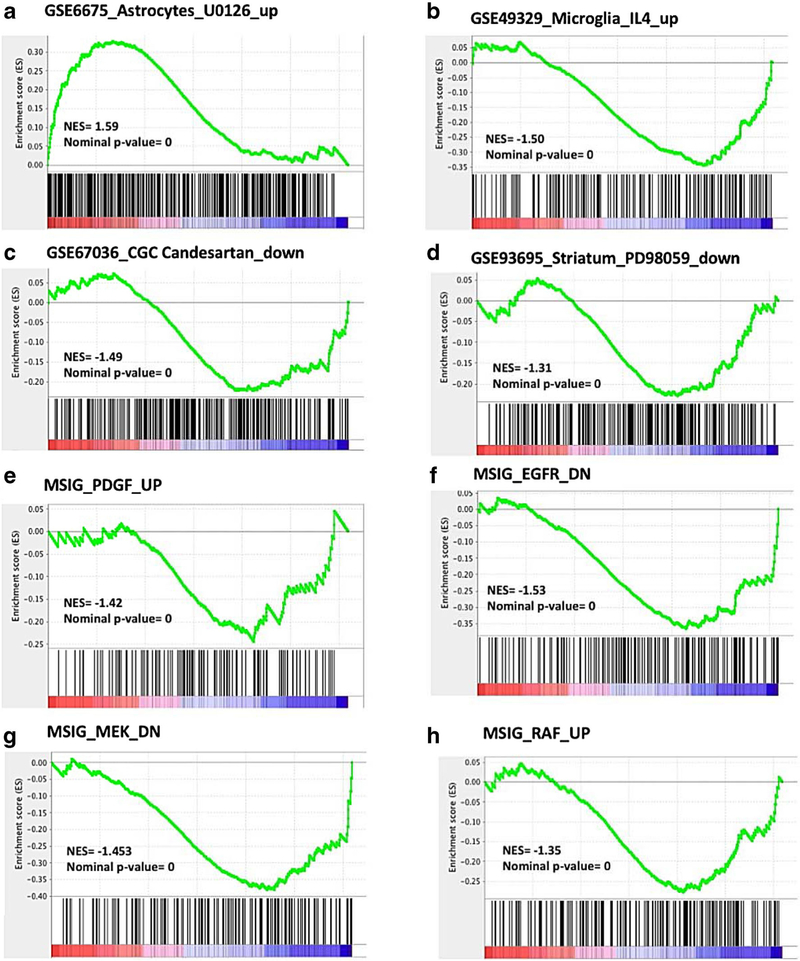
Using GSEA, we compared the results obtained after treating the BV2 cell culture with the PPARγ antagonist GW9662 in the presence of Telmisartan and LPS with published genesets. We found several remarkable positive correlations between GW9662 effects with those reported for inhibitors of the MAP kinase pathways. These include a positive correlation with genes reported to be expressed in astrocytes activated with FGF2 and then treated with the MEK inhibitor U0126 (Supplemental Table 8), [114] (GSE6675) a compound that reduces apoptosis and protects from ischemia [144, 145]. Supplemental Table 8 reveals that GW9662 and U0126 share over 100 genes associated with neuroprotection and reduction of inflammation (Fig. 5a and Supplemental Table 8). Conversely, the effects of GW9662 were negatively correlated with upregulated genes in mouse primary microglia cells treated with IL-4 [146] (GSE49329) (Fig. 5b) and with genes upregulated with excitotoxic glutamate concentrations and down regulated by Candesartan in rat primary cerebellar neurons treated with glutamate [14] (GSE67036), (Fig. 5c). There was a positive correlation between GW9662 effects with the antidyskinetic effect of PD98059 in a pre-clinical model of Parkinson disease [147] (GSE93695), (Fig. 5d and Supplemental Table 8). These GW9662 analyses demonstrated also correlations with the effects of the kinase inhibitors PD98059, SB203580 and LY294002 as revealed by our IPA analysis (Supplemental Table 8).
Fig. 5.
Gene Set Enrichment Analysis (GSEA) of BV2 cells treated with Telmisartan + LPS versus Telmisartan + LPS + GW9662. Gene signatures (vertical bars) from GEO (NCBI) and MSIG (Broad Institute) were overlaid on the ranked list of genes from our microarray data (red and blue bar). In mouse astrocytes, genes activated by FGF2 and up regulated by U0126 correlate with genes upregulated by GW9662 (Fig. 4a) [114]. Genes up regulated by IL-4 in murine microglia are negatively correlated with genes up-regulated by GW9662 (Fig. 4b, GSE49329, [146]). Genes upregulated by glutamate and down regulated by Candesartan in rat cerebellar cortical neurons correlate with genes downregulated by GW9662 (Fig. 4c, GSE67036 [14]). In the striatum of dyskinetic rats treated with L-DOPA, genes down regulated by the anti-dyskinesia MEK inhibitor (PD98059) are also down-regulated by GW9662 (Fig. 4d, GSE93695 [147]). Genes up-regulated by PDGF in the neuroblastoma cell line SH-SY5Y pre-treated by the MEK inhibitor U0126 are negatively correlated with genes up-regulated by GW9662 (Fig. 4e [117, 144]). In the MCF7 breast cancer cell line stably overexpressing ligand-activable EGFR, genes down-regulated by EGFR are also down-regulated by GW9662 (Fig. 4f [148]). In the MCF7 breast cancer cell line stably overexpressing constitutively active MAP2K1 (MEK), genes down-regulated by MEK are also down-regulated by GW9662 (Fig. 4g [148]). In the MCF7 breast cancer cell line stably overexpressing constitutively active RAF1, genes upregulated by RAF1 are downregulated by GW9662 (Fig. 4h GSE3542 [148])
GW9662 effects negatively correlated with a number of gene signatures indicating cellular injury, such as the neuroblastoma cell line SH-SY5Y treated with PDGF and pretreated with the ERK inhibitors U0126 [117] (GSE7403) (Fig. 5e), and another negative correlation with a gene signature of upregulated genes in MCF-7 cell lines stably overexpressing growth factor signaling, constitutively active EGFR [148] (Fig. 5f and Supplemental Table 8), MEK (MAP2K1) [148] (Fig. 5g and Supplemental Table 8) or Raf1 [148] (Fig. 5h and Supplemental Table 8).
In addition to our findings using GW9662, we found additional evidence of the paradoxical effect of PPARγ inhibition of LPS-induced alterations in gene expression. Administration of another PPARγ antagonist, T0070907, enhanced the Telmisartan reduction of LPS-induced gene expression of inflammatory cytokines; T0070907, administered alone, abolished the IL-1β gene expression enhanced by LPS and significantly reduced that of IL-6 (Supplemental Fig. 2A and 2B). Furthermore, PPARγ activation with the full agonist Pioglitazone did not alter the LPS-induced increase in IL-1β and IL-6 gene expression (Fig. 5).
Hypothesis
In our BV2 mouse cell model, we have found that LPS significantly upregulated, and that Telmisartan and GW9662 significantly downregulated TLR2 gene expression (Supplemental Table 3). Mouse microglia strongly expresses most of TLRs [3] and LPS upregulates TLR2 and TLR4 gene expression in microglia [149]. LPS was positively, and Telmisartan and GW9662 were negatively, correlated with several TLR upstream regulators including TLR2, and with several components of the TLR signaling pathways (Table 1). This makes downregulation of the TLR2 pro-inflammatory gene and signaling pathway a candidate for a novel neuroprotective mechanism for Telmisartan and GW9662 in microglia, beyond AT1 receptors or PPARγ.
The association of AT1 receptor blockade and TLR down-regulation is well established in the literature. AT1 receptor stimulation enhances TLR gene expression in microglia and Telmisartan and several other ARBs reduce gene expression of several TLRs in many in vitro and in vivo models [13, 36, 150–157]. In addition, ARBs downregulate gene expression of many members of the TLR down-stream pathways, including CD14 [13], MyD88 [158, 159], IRAK1 and TRAF6 [160] and MMP-2 activation [153]. Whether the novel mechanism postulated here has a role in other conditions remain to be determined.
Conclusions
We revealed Telmisartan neuroprotection in a culture of microglia-like BV2 cells with a response to LPS like that of primary microglia, and not expressing AT1A receptors or PPARγ genes.
This indicated that in our system Telmisartan effects were the consequence of novel neuroprotective mechanisms, beyond its canonical AT1 receptor blockade and PPARγ activation.
Unexpectedly, in our system, administration of PPARγ inhibitors significantly decreased LPS-induced injury and enhanced, rather than reduced, Telmisartan neuroprotective effects.
Telmisartan and the PPARγ inhibitor GW9662 protective effects are widespread, including normalization of the expression of many pro-injury genes upregulated and that of many protective genes downregulated by LPS.
IPA analysis uncovered many associated pathways, diseases and functions and upstream regulators reducing cell toxicity and promoting protection.
GSEA analysis revealed multiple gene sets associated with several disease phenotypes, validating IPA analysis and homing in on common pathways of major clinical interest. They included not only a reduction of inflammation and a positive correlation with neuroprotective kinase inhibitors, but also indicate possible protection against cardiovascular, metabolic and neurodegenerative disorders and malignancies, where inflammation plays determinant roles.
Our results may have translational significance. Telmisartan neuroprotection in uninjured cells supports the hypothesis that this compound may have a role in preventing or delaying neurodegenerative and age-related disorders. The strong association of Telmisartan effects with that of kinase inhibitors is promising, since this class is increasingly considered for the treatment of many disorders. The protective effects of GW9662 indicate that novel derivatives of this compound with translational value may be developed. In addition, future drug development may result in Telmisartan and/or GW9662 derivatives with enhanced downregulating properties on the TLR signaling pathways.
The present study is not without limitations. Our results have been obtained using a microglia cell line, and whether they may be replicated in primary microglia remains to be determined. This study has been restricted to determine changes in gene expression without analysis of protein expression or direct cellular effects. The molecular mechanisms involved in the Telmisartan and GW9662 downregulation of the TLR signaling pathways have not been determined.
Supplementary Material
Acknowledgements
AGE was supported by the National Human Genome Research Institute Intramural Program, National Institutes of Health.
YR was a research technician at the Department of Pharmacology and Physiology, Georgetown University Medical Center.
SA was supported by a scholarship from the government of the Kingdom of Saudi Arabia and by the Master in Physiology Program at Georgetown University Medical Center.
EW was a PhD student, Department of Pharmacology and Physiology, Georgetown University Medical Center.
JMS was supported by a GX4002-705 grant from Partners in Research, Georgetown University Medical Center.
Footnotes
Compliance with Ethical Standards
Conflict of Interest The authors declare that they have no conflict of interest.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s12035-018-1300-9) contains supplementary material, which is available to authorized users.
References
- 1.Mamik MK, Power C (2017) Inflammasomes in neurological diseases: emerging pathogenic and therapeutic concepts. Brain 140: 2273–2285. 10.1093/brain/awx133 [DOI] [PubMed] [Google Scholar]
- 2.Villapol S (2018) Roles of peroxisome proliferator-activated receptor gamma on brain and peripheral inflammation. Cell Mol Neurobiol 38:121–132. 10.1007/s10571-017-0554-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kabba JA, Xu Y, Christian H, Ruan W, Chenai K, Xiang Y, Zhang L, Saavedra JM et al. (2018) Microglia: housekeeper of the central nervous system. Cell Mol Neurobiol 38:53–71. 10.1007/s10571-017-0504-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Bernhardi R, Eugenín-von Bernhardi L, Eugenín J (2015) Microglial cell dysregulation in brain aging and neurodegeneration. Front Aging Neurosci 7:124 10.3389/fnagi.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salter MW, Stevens B (2017) Microglia emerge as central players in brain disease. Nat Med 23:1018–1027. 10.1038/nm.4397 [DOI] [PubMed] [Google Scholar]
- 6.Saavedra JM, Sánchez-Lemus E, Benicky J (2011) Blockade of brain angiotensin II AT1 receptors ameliorates stress, anxiety, brain inflammation and ischemia: Therapeutic implications. Psychoneuroendocrinology 36:1–18. 10.1016/j.psyneuen.2010.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarrott B, Williams SJ (2016) Chronic brain inflammation: the neurochemical basis for drugs to reduce inflammation. Neurochem Res 41:523–533. 10.1007/s11064-015-1661-7 [DOI] [PubMed] [Google Scholar]
- 8.Saavedra JM (2017) Beneficial effects of angiotensin II receptor blockers in brain disorders. Pharmacol Res 125(Pt A):91–103. 10.1016/j.phrs.2017.06.017 [DOI] [PubMed] [Google Scholar]
- 9.Saavedra JM (2012) Angiotensin II AT(1) receptor blockers as treatments for inflammatory brain disorders. Clin Sci (Lond) 123:567–590. 10.1042/CS20120078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.10.Villapol S, Yaszemski AK, Logan TT, Sánchez-Lemus E, Saavedra JM, Symes AJ. (2012) Candesartan, an angiotensin II AT1-receptor blocker and PPAR-γ agonist, reduces lesion volume and improves motor and memory function after traumatic brain injury in mice. Neuropsychopharmacology 37:2817–2829. 10.1038/npp.2012.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danielyan L, Lourhmati A, Verleysdonk S, Kabisch D, Proksch B, Thiess U, Umbreen S, Schmidt B et al. (2007) Angiotensin receptor type 1 blockade in astroglia decreases hypoxia-induced cell damage and TNF alpha release. Neurochem Res 32:1489–1498. 10.1007/s11064-007-9337-6 [DOI] [PubMed] [Google Scholar]
- 12.Danielyan L, Klein R, Hanson LR, Buadze M, Schwab M, Gleiter CH, Frey WH (2010) Protective effects of intranasal losartan in the APP/PS1 transgenic mouse model of Alzheimer disease. Rejuvenation Res 13:195–201. 10.1089/rej.2009.0944 [DOI] [PubMed] [Google Scholar]
- 13.Benicky J, Sánchez-Lemus E, Honda M, Pang T, Orecna M, Wang J, Leng Y, Chuang DM et al. (2011) Angiotensin II AT1 receptor blockade ameliorates brain inflammation. Neuropsychopharmacology 36:857–870. 10.1038/npp.2010.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Pang T, Hafko R, Benicky J, Sanchez-Lemus E, Saavedra JM (2014a) Telmisartan ameliorates glutamate-induced neurotoxicity: roles of AT(1) receptor blockade and PPARγ activation. Neuropharmacology 79:249–261. 10.1016/j.neuropharm.2013.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villapol S, Balarezo MG, Affram K, Saavedra JM, Symes AJ (2015) Neurorestoration after traumatic brain injury through angiotensin II receptor blockage. Brain 138(Pt 11):3299–3315. 10.1093/brain/awv172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lund S, Christensen KV, Hedtjarn M, Mortensen AL, Hagberg H, Falsig J, Hasseldam H, Schrattenholz A et al. (2006) The dynamics of the LPS triggered inflammatory response of murine microglia under different culture and in vivo conditions. J Neuroimmunol 180:71–87. 10.1016/j.jneuroim.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Xu Y, Wang Y, Wang Y, He L, Jiang Z, Huang Z, Liao H et al. (2015) Telmisartan prevention of LPS-induced microglia activation involves M2 microglia polarization via CaMKKβ-dependent AMPK activation. Brain Behav Immun 50:298–313. 10.1016/j.bbi.2015.07.015 [DOI] [PubMed] [Google Scholar]
- 18.Benson SC, Pershadsingh HA, Ho CI, Chittiboyina A, Desai P, Pravenec M, Qi N, Wang J et al. (2004) Identification of telmisartan as a unique angiotensin II receptor antagonist with selective PPARgamma-modulating activity. Hypertension 43: 993–1002 [DOI] [PubMed] [Google Scholar]
- 19.Destro M, Cagnoni F, Dognini GP, Galimberti V, Taietti C, Cavalleri C, Galli E (2011) Telmisartan: just an antihypertensive agent? A literature review. Expert Opin Pharmacother 12:2719–2735. 10.1517/14656566.2011.632367 [DOI] [PubMed] [Google Scholar]
- 20.Gillies PS, Dunn CJ (2000) Pioglitazone. Drugs 60:333–343 [DOI] [PubMed] [Google Scholar]
- 21.Zaytseva YY, Wallis NK, Southard RC, Kilgore MW (2011) The PPARgamma antagonist T0070907 suppresses breast cancer cell proliferation and motility via both PPARgamma-dependent and - independent mechanisms. Anticancer Res 31:813–823 [PubMed] [Google Scholar]
- 22.Chen HY, Xu Z, Chen LF, Wang W, Fang Q, Yan XW (2012) Valsartan and telmisartan abrogate angiotensin II-induced down-regulation of ABCA1 expression via AT1 receptor, rather than AT2 receptor or PPARγ activation. J Cardiovasc Pharmacol 59: 570–575. 10.1097/FJC.0b013e31824fc5e3 [DOI] [PubMed] [Google Scholar]
- 23.Horvath R, McMenemy N, Alkaitis M, DeLeo J (2008) Differential migration, LPS-induced cytokine, chemokine, and NO expression in immortalized BV-2 and HAPI cell lines and primary microglia cultures. J Neurochem 107:557–569. 10.1111/j.1471-4159.2008.05633.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henn A, Lund S, Hedtjarn M, Schrattenholz A, Pörzgen P, Leist M (2009) The suitability of BV2 cells as alternative model system for primary microglia cultures or for animal experiments examining brain inflammation. ALTEX 26:83–94. 10.14573/altex.2009.2.83 [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Wang H, Guo H, Kang L, Gao X, Hu L (2011) Neuroprotection of Scutellarin is mediated by inhibition of microglial inflammatory activation. Neuroscience 185:150–160. 10.1016/j.neuroscience.2011.04.005 [DOI] [PubMed] [Google Scholar]
- 26.Pinteaux E, Parker LC, Rothwell NJ, Luheshi GN (2002) Expression of interleukin-1 receptors and their role in interleukin-1 actions in murine microglial cells. J Neurochem 83:754–763. 10.1046/j.1471-4159.2002.01184.x [DOI] [PubMed] [Google Scholar]
- 27.Moon DO, Park SY, Lee KJ, Heo MS, Kim KC, Kim MO, Lee JD, Choi YH et al. (2007) Bee venom and melittin reduce proinflammatory mediators in lipopolysaccharide-stimulated BV2 microglia. Int Immunopharmacol 7:1092–10101. 10.1016/j.intimp.2007.04.005 [DOI] [PubMed] [Google Scholar]
- 28.Elkahloun AG, Hafko R, Saavedra JM (2016) An integrative genome-wide transcriptome reveals that candesartan is neuroprotective and a candidate therapeutic for Alzheimer’s disease. Alzheimers Res Ther 8:5 10.1186/s13195-015-0167-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jöhren O, Saavedra JM (1996) Expression of AT1A and AT1B angiotensin II receptor messenger RNA in forebrain of 2-wk-old rats. Am J Phys 271(1 Pt 1):E104–E112. 10.1152/ajpendo.1996.271.1.E104 [DOI] [PubMed] [Google Scholar]
- 31.Miyoshi M, Miyano K, Moriyama N, Taniguchi M, Watanabe T (2008) Angiotensin type 1 receptor antagonist inhibits lipopolysaccharide-induced stimulation of rat microglial cells by suppressing nuclear factor kappaB and activator protein-1 activation. Eur J Neurosci 27:343–351. 10.1111/j.1460-9568.2007.06014.x [DOI] [PubMed] [Google Scholar]
- 32.Li JJ, Lu J, Kaur C, Sivakumar V, Wu CY, Ling EA (2009) Expression of angiotensin II and its receptors in the normal and hypoxic amoeboid microglial cells and murine BV-2 cells. Neuroscience 158:1488–1499. 10.1016/j.neuroscience.2008.11.046 [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P et al. (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. 10.1523/JNEUROSCI.1860-14.2014 Supplemental materials at http://web.stanford.edu/group/barres_lab/brain_rnaseq.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pang T, Benicky J, Wang J, Orecna M, Sanchez-Lemus E, Saavedra JM (2012a) Telmisartan ameliorates lipopolysaccharide-induced innate immune response through peroxisome proliferator-activated receptor-γ activation in human monocytes. J Hypertens 30:87–96. 10.1097/HJH.0b013e32834dde5f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, Jaeger BN, O’Connor C, Fitzpatrick C, Pasillas MP, Pena M, Adair A, Gonda DD, Levy ML, Ransohoff RM, Gage FH, Glass CK (2017) An environment-dependent transcriptional network specifies human microglia identity. Science 356(6344). 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ha YM, Park EJ, Kang YJ, Park SW, Kim HJ, Chang KC (2014) Valsartan independent of AT1 receptor inhibits tissue factor, TLR-2 and −4 expression by regulation of Egr-1 through activation of AMPK in diabetic conditions. J Cell Mol Med 18:2031–2043. 10.1111/jcmm.12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto K, Ohishi M, Ho C, Kurtz TW, Rakugi H (2009) Telmisartan-induced inhibition of vascular cell proliferation beyond angiotensin receptor blockade and peroxisome proliferator-activated receptor-gamma activation. Hypertension 54:1353–1359. 10.1161/HYPERTENSIONAHA.109.138750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Huang Y, Xu Y, Ruan W, Wang H, Zhang Y, Saavedra JM, Zhang L et al. (2018) A dual AMPK/Nrf2 activator reduces brain inflammation after stroke by enhancing microglia M2 polarization. Antioxid Redox Signal 28:141–163. 10.1089/ars.2017.7003 [DOI] [PubMed] [Google Scholar]
- 39.Leong DS, Terrón JA, Falcón-Neri A, Armando I, Ito T, Jöhren O, Tonelli LH, Hoe KL et al. (2002) Restraint stress modulates brain, pituitary and adrenal expression of angiotensin II AT(1A), AT(1B) and AT(2) receptors. Neuroendocrinology 75:227–240. 10.1159/000054714 [DOI] [PubMed] [Google Scholar]
- 40.Kitamura Y, Taniguchi T, Kimura H, Nomura Y, Gebicke-Haerter PJ (2000) Interleukin-4-inhibited mRNA expression in mixed rat glial and in isolated microglial cultures. J Neuroimmunol 106:95–104 [DOI] [PubMed] [Google Scholar]
- 41.Choi MJ, Lee EJ, Park JS, Kim SN, Park EM, Kim HS (2017) Anti-inflammatory mechanism of galangin in lipopolysaccharide-stimulated microglia: Critical role of PPAR-γ signaling pathway. Biochem Pharmacol 144:120–131. 10.1016/j.bcp.2017.07.021 [DOI] [PubMed] [Google Scholar]
- 42.Han Q, Yuan Q, Meng X, Huo J, Bao Y, Xie G (2017) 6-Shogaol attenuates LPS-induced inflammation in BV2 microglia cells by activating PPAR-γ. Oncotarget 8:42001–42006. 10.18632/oncotarget.16719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erbe DV, Gartrell K, Zhang YL, Suri V, Kirincich SJ, Will S, Perreault M, Wang S et al. (2006) Molecular activation of PPARgamma by angiotensin II type 1-receptor antagonists. Vasc Pharmacol 45:154–162. 10.1016/j.vph.2006.05.002 [DOI] [PubMed] [Google Scholar]
- 44.Iwashita M, Nakatsu Y, Sakoda H, Fujishiro M, Kushiyama A, Fukushima T, Kumamoto S, Shinjo T et al. (2013) Valsartan restores inflammatory response by macrophages in adipose and hepatic tissues of LPS-infused mice. Adipocyte 2:28–32. 10.4161/adip.21837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cekanova M, Yuan JS, Li X, Kim K, Baek SJ (2008) Gene alterations by peroxisome proliferator-activated receptor gamma agonists in human colorectal cancer cells. Int J Oncol 32:809–819 [PMC free article] [PubMed] [Google Scholar]
- 46.Blasi E, Barluzzi R, Bocchini V, Mazzolla R, Bistoni F (1990) Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. J Neuroimmunol 27:229–237 [DOI] [PubMed] [Google Scholar]
- 47.Romano P, Manniello A, Aresu O, Armento M, Cesaro M, Parodi B (2009) Cell line data base: structure and recent improvements towards molecular authentication of human cell lines. Nucleic Acids Res 37:D925–D932. 10.1093/nar/gkn730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato S, Rancourt A, Sato Y, Satoh MS (2016) Single-cell lineage tracking analysis reveals that an established cell line comprises putative cancer stem cells and their heterogeneous progeny. Sci Rep 6:23328 10.1038/srep23328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salvi V, Sozio F, Sozzani S, Del Prete A (2017) Role of atypical chemokine receptors in microglial activation and polarization. Front Aging Neurosci 9:148 10.3389/fnagi.2017.00148 eCollection 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ham JE, Oh EK, Kim DH, Choi SH (2015) Differential expression profiles and roles of inducible DUSPs and ERK1/2-specific constitutive DUSP6 and DUSP7 in microglia. Biochem Biophys Res Commun 467:254–260. 10.1016/j.bbrc.2015.09.180 [DOI] [PubMed] [Google Scholar]
- 51.De I, Nikodemova M, Steffen MD, Sokn E, Maklakova VI, Watters JJ, Collier LS (2014) CSF1 overexpression has pleiotropic effects on microglia in vivo. Glia 62:1955–1967. 10.1002/glia.22717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desplats P, Dumaop W, Smith D, Adame A, Everall I, Letendre S, Ellis R, Cherner M et al. (2013) Molecular and pathologic insights from latent HIV-1 infection in the human brain. Neurology 80: 1415–1423. 10.1212/WNL.0b013e31828c2e9e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khoshnam SE, Winlow W, Farbood Y, Moghaddam HF, Farzaneh M (2017) Emerging roles of microRNAs in ischemic stroke: as possible therapeutic agents. J Stroke 19:166–187. 10.5853/jos.2016.01368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Truettner JS, Motti D, Dietrich WD (2013) MicroRNA overexpression increases cortical neuronal vulnerability to injury. Brain Res 1533:122–130. 10.1016/j.brainres.2013.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li F, Wei G, Bai Y, Li Y, Huang F, Lin J, Hou Q, Deng R et al. (2015a) MicroRNA-574 is involved in cognitive impairment in 5-month-old APP/PS1 mice through regulation of neuritin. Brain Res 1627:177–188. 10.1016/j.brainres.2015.09.022 [DOI] [PubMed] [Google Scholar]
- 56.Leisner TM, Freeman TC, Black JL, Parise LV (2016) CIB1: a small protein with big ambitions. FASEB J 30:2640–2650. 10.1096/fj.201500073R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pisetsky DS (2013) Immune activation by histones: plusses and minuses in inflammation. Eur J Immunol 43:3163–3166. 10.1002/eji.201344175 [DOI] [PubMed] [Google Scholar]
- 58.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI (2003) Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol 4:269–273. 10.1038/ni888 [DOI] [PubMed] [Google Scholar]
- 59.Palmieri F (2013) The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol Asp Med 34: 465–484. 10.1016/j.mam.2012.05.005 [DOI] [PubMed] [Google Scholar]
- 60.Pereg Y, Liu BY, O’Rourke KM, Sagolla M, Dey A, Komuves L, French DM, Dixit VM (2010) Ubiquitin hydrolase Dub3 promotes oncogenic transformation by stabilizing Cdc25A. Nat Cell Biol 12:400–406. 10.1038/ncb2041 [DOI] [PubMed] [Google Scholar]
- 61.Barger JL, Vann JM, Cray NL, Pugh TD, Mastaloudis A, Hester SN, Wood SM, Newton MA et al. (2017) Identification of tissue-specific transcriptional markers of caloric restriction in the mouse and their use to evaluate caloric restriction mimetics. Aging Cell 16:750–760. 10.1111/acel.12608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Cavanagh EM, Inserra F, Ferder L (2015) Angiotensin II blockade: how its molecular targets may signal to mitochondria and slow aging. Coincidences with calorie restriction and mTOR inhibition. Am J Physiol Heart Circ Physiol 309:H15–H44. 10.1152/ajpheart.00459.2014 [DOI] [PubMed] [Google Scholar]
- 63.Blom T, Schmiedt M-L, Wong AM, Kyttala A, Soronen J, Jauhiainen M, Tyynelä J, Cooper JD et al. (2013) Exacerbated neuronal ceroid lipofuscinosis phenotype in Cln1/5 double-knockout mice. Dis Model Mech 6:342–357. 10.1242/dmm.010140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orihuela R, McPherson CA, Harry GJ (2016) Microglial M1/M2 polarization and metabolic states. Br J Pharmacol 173:649–665. 10.1111/bph.13139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Christian R, Raetz H, Whitfield C (2002) Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. 10.1146/annurev.biochem.71.110601.135414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taniguchi K, Karin M (2014) IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin Immunol 26:54–74. 10.1016/j.smim.2014.01.001 [DOI] [PubMed] [Google Scholar]
- 67.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G et al. (2004) Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science 306: 704–708. 10.1126/science.1099962 [DOI] [PubMed] [Google Scholar]
- 68.Croft M, Duan W, Choi H, Eun SY, Madireddi S, Mehta A (2012) TNF superfamily in inflammatory disease: translating basic insights. Trends Immunol 33:144–152. 10.1016/j.it.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marques-Rocha JL, Samblas M, Milagro FI, Bressan J, Martínez JA, Marti A (2015) Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J 29:3595–3611. 10.1096/fj.14-260323 [DOI] [PubMed] [Google Scholar]
- 70.Clarner T, Janssen K, Nellessen L, Stangel M, Skripuletz T, Krauspe B, Hess FM, Denecke B et al. (2015) CXCL10 triggers early microglial activation in the cuprizone model. J Immunol 194:3400–3413. 10.4049/jimmunol.1401459 [DOI] [PubMed] [Google Scholar]
- 71.Daulatzai MA (2015) Olfactory dysfunction: its early temporal relationship and neural correlates in the pathogenesis of Alzheimer’s disease. J Neural Transm (Vienna) 122:1475–1497. 10.1007/s00702-015-1404-6 [DOI] [PubMed] [Google Scholar]
- 72.Ferrer I, Garcia-Esparcia P, Carmona M, Carro E, Aronica E, Kovacs GG, Grison A, Gustincich S (2016) Olfactory receptors in non-chemosensory organs: the nervous system in health and disease. Front Aging Neurosci 8:163 10.3389/fnagi.2016.00163 eCollection 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Han JA, Kim JY, Kim JI (2014) Analysis of gene expression in cyclooxygenase-2-overexpressed human osteosarcoma cell lines. Genomics Inform 12:247–253. 10.5808/GI.2014.12.4.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ampofo E, Schmitt BM, Menger MD, Laschke MW (2017) The regulatory mechanisms of NG2/CSPG4 expression. Cell Mol Biol Lett 22:4 10.1186/s11658-017-0035-3. eCollection 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stepanov GA, Filippova JA, Komissarov AB, Kuligina EV, Richter VA, Semenov DV (2015) Regulatory role of small nucleolar RNAs in human diseases. Biomed Res Int 2015:206849 10.1155/2015/206849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim W, Kim JE (2013) SIRT7 an emerging sirtuin: deciphering newer roles. J Physiol Pharmacol 64:531–534 [PubMed] [Google Scholar]
- 77.Jiang T, Zhang YD, Gao Q, Ou Z, Gong PY, Shi JQ, Wu L, Zhou JS (2018) TREM2 ameliorates neuronal tau pathology through suppression of microglial inflammatory response. Inflammation. 10.1007/s10753-018-0735-5 [DOI] [PubMed] [Google Scholar]
- 78.Wang J, Si Y, Wu C, Sun L, Ma Y, Ge A, Li B (2012) Lipopolysaccharide promotes lipid accumulation in human adventitial fibroblasts via TLR4-NF-κB pathway. Lipids Health Dis 11:139 10.1186/1476-511X-11-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bode JG, Ehlting C, Haussinger D (2012) The macrophage response towards LPS and its control through the p38(MAPK)-STAT3 axis. Cell Signal 24:1185–1194. 10.1016/j.cellsig.2012.01.018 [DOI] [PubMed] [Google Scholar]
- 80.Poujol F, Monneret G, Pachot A, Textoris J, Venet F (2015) Altered T lymphocyte proliferation upon lipopolysaccharide challenge ex vivo. PLoS One 10:e0144375 10.1371/journal.pone.0144375 eCollection 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abella V, Scotece M, Conde J, Gómez R, Lois A, Pino J, Gómez-Reino JJ, Lago F et al. (2015) The potential of lipocalin-2/NGAL as biomarker for inflammatory and metabolic diseases. Biomarkers 20: 565–571. 10.3109/1354750X.2015.1123354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khalil M, Renner A, Langkammer C, Enzinger C, Ropele S, Stojakovic T, Scharnagl H, Bachmaier G et al. (2016) Cerebrospinal fluid lipocalin 2 in patients with clinically isolated syndromes and early multiple sclerosis. Mult Scler 22:1560–1568. 10.1177/1352458515624560 [DOI] [PubMed] [Google Scholar]
- 83.Bhatelia K, Singh K, Singh R (2014) TLRs: linking inflammation and breast cancer. Cell Signal 26:2350–2357. 10.1016/j.cellsig.2014.07.035 [DOI] [PubMed] [Google Scholar]
- 84.Cuenda A, Rousseau S (2007) p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta 1773:1358–1375. 10.1016/j.bbamcr.2007.03.010 [DOI] [PubMed] [Google Scholar]
- 85.Ueda K, Nakahara T, Akanuma K, Mori A, Sakamoto K, Ishii K (2013) Differential effects of LY294002 and wortmannin on neurons and vascular endothelial cells in the rat retina. Pharmacol Rep 65:854–862 [DOI] [PubMed] [Google Scholar]
- 86.Ong Q, Guo S, Zhang K, Cui B (2015) U0126 protects cells against oxidative stress independent of its function as a MEK inhibitor. ACS Chem Neurosci 6:130–137. 10.1021/cn500288n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sandoval-Hernández AG, Buitrago L, Moreno H, Cardona-Gómez GP, Arboleda G (2015) Role of liver X receptor in AD pathophysiology. PLoS One 10(12):e0145467 10.1371/journal.pone.0145467 eCollection 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu HM, Zhao YM, Luo XG, Feng Y, Ren Y, Shang H, He ZY, Luo XM et al. (2012) Repeated lipopolysaccharide stimulation induces cellular senescence in BV2 cells. Neuroimmunomodulation 19: 131–136. 10.1159/000330254 [DOI] [PubMed] [Google Scholar]
- 89.Atsumi T, Singh R, Sabharwal L, Bando H, Meng J, Arima Y, Yamada M, Harada M et al. (2014) Inflammation amplifier, a new paradigm in cancer biology Cancer Res 74:8–14. 10.1158/0008-5472.CAN-13-2322 [DOI] [PubMed] [Google Scholar]
- 90.Diakos CI, Charles KA, McMillan DC, Clarke SJ (2014) Cancer-related inflammation and treatment effectiveness. Lancet Oncol 15:e493–e503. 10.1016/S1470-2045(14)70263-3 [DOI] [PubMed] [Google Scholar]
- 91.Banks WA, Gray AM, Erickson MA, Salameh TS, Damodarasamy M, Sheibani N, Meabon JS, Wing EE et al. (2015) Lipopolysaccharide-induced blood-brain barrier disruption: roles of cyclooxygenase, oxidative stress, neuroinflammation, and elements of the neurovascular unit. J Neuroinflammation 12:223 10.1186/s12974-015-0434-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao D, Zhuang N, Ding Y, Kang Y, Shi L (2016) MiR-221 activates the NF-κB pathway by targeting A20. Biochem Biophys Res Commun 472:11–18. 10.1016/j.bbrc.2015.11.009 [DOI] [PubMed] [Google Scholar]
- 93.Huang Y, Chen J, Zhou Y, Tang S, Li J, Yu X, Mo Y, Wu Y et al. (2016) Circulating miR155 expression level is positive with blood pressure parameters: potential markers of target-organ damage. Clin Exp Hypertens 38:331–336. 10.3109/10641963.2015.1116551 [DOI] [PubMed] [Google Scholar]
- 94.Guo S, Messmer-Blust AF, Wu J, Song X, Philbrick MJ, Shie J-L, Rana JS, Li J (2014) Role of A20 in cIAP-2 protection against tumor necrosis factor α (TNF-α)-mediated apoptosis in endothelial cells. Int J Mol Sci 15:3816–3833. 10.3390/ijms15033816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Raetz CR, Whitfield C (2002) Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. 10.1146/annurev.biochem.71.110601.135414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lemarchant S, Pruvost M, Montaner J, Emery E, Vivien D, Kanninen K, Koistinaho J (2013) ADAMTS proteoglycanases in the physiological and pathological central nervous system. J Neuroinflammation 10:133 10.1186/1742-2094-10-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang G, Huang Y, Wang LL, Zhang YF, Xu J, Zhou Y, Lourenco GF, Zhang B et al. (2016) MicroRNA-146a suppresses ROCK1 allowing hyperphosphorylation of tau in Alzheimer’s disease. Sci Rep 6:26697 10.1038/srep26697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ansoleaga B, Garcia-Esparcia P, Llorens F, Moreno J, Aso E, Ferrer I (2013) Dysregulation of brain olfactory and taste receptors in AD, PSP and CJD, and AD-related model. Neuroscience 248: 369–382. 10.1016/j.neuroscience.2013.06.034 [DOI] [PubMed] [Google Scholar]
- 99.Lupton MK, Proitsi P, Lin K, Hamilton G, Daniilidou M, Tsolaki M, Powell JF (2014) The role of ABCA1 gene sequence variants on risk of Alzheimer’s disease. J Alzheimers Dis 38:897–906. 10.3233/JAD-131121 [DOI] [PubMed] [Google Scholar]
- 100.Sabirzhanov B, Stoica BA, Zhao Z, Loane DJ, Wu J, Dorsey SG, Faden AI (2016) miR-711 upregulation induces neuronal cell death after traumatic brain injury. Cell Death Differ 23:654–668. 10.1038/cdd.2015.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Garrigan E, Belkin NS, Seydel F, Han Z, Carter J, McDuffie M, Morel L, Peck AB et al. (2015) Csf2 and Ptgs2 epigenetic dysregulation in diabetes-prone Bicongenic B6.NODC11bxC1tb mice. Genet Epigenet 7:5–17. 10.4137/GEG.S29696 eCollection 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bensellam M, Montgomery MK, Luzuriaga J, Chan JY, Laybutt DR (2015) Inhibitor of differentiation proteins protect against oxidative stress by regulating the antioxidant-mitochondrial response in mouse beta cells. Diabetologia 58:758–770. 10.1007/s00125-015-3503-1 [DOI] [PubMed] [Google Scholar]
- 103.Lee MS, Kim B, Oh GT, Kim YJ (2013) OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat Immunol 14:346–355. 10.1038/ni.2535 Erratum in: Nat Immunol. 2013;14:355. Nat Immunol. 2013;14:877 [DOI] [PubMed] [Google Scholar]
- 104.Miao Y, Cui L, Chen Z, Zhang L (2016) Gene expression profiling of DMU-212-induced apoptosis and anti-angiogenesis in vascular endothelial cells. Pharm Biol 54:660–666. 10.3109/13880209.2015.1071414 [DOI] [PubMed] [Google Scholar]
- 105.Nair S, Rein A (2014) Antiretroviral restriction factors in mice. Virus Res 193:130–134. https://doi.Org/10.1016/j.virusres.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, Frazier A, Yang H et al. (1999) HMG-1 as a late mediator of endotoxin lethality in mice. Science 285:248–251. 10.1126/science.285.5425.248 [DOI] [PubMed] [Google Scholar]
- 107.Bao XM, He Q, Wang Y, Huang ZH, Yuan ZQ (2017) The roles and mechanisms of the Hippo/YAP signaling pathway in the nervous system. Yi Chuan 39:630–641. 10.16288/j.yczz.17-069 [DOI] [PubMed] [Google Scholar]
- 108.Harvey KF, Zhang X, Thomas DM (2013) The hippo pathway and human cancer. Nat Rev Cancer 13:246–257. 10.1038/nrc3458 [DOI] [PubMed] [Google Scholar]
- 109.Patel RK, Prasad N, Kuwar R, Haldar D, Abdul-Muneer PM (2017) Transforming growth factor-beta 1 signaling regulates neuroinflammation and apoptosis in mild traumatic brain injury. Brain Behav Immun 64:244–258. 10.1016/j.bbi.2017.04.012 [DOI] [PubMed] [Google Scholar]
- 110.Heyninck K, De Valck D, Vanden Berghe W, Van Criekinge W, Contreras R, Fiers W, Haegeman G, Beyaert R (1999) The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J Cell Biol 145:1471–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rutkowski DT, Kaufman RJ (2003) All roads lead to ATF4. Dev Cell 4:442–444. 10.1016/S1534-5807(03)00100-X [DOI] [PubMed] [Google Scholar]
- 112.Guleria RS, Choudhary R, Tanaka T, Baker KM, Pan J (2011) Retinoic acid receptor-mediated signaling protects cardiomyocytes from induced apoptosis: Role of the renin-angiotensin system. J Cell Physiol 226:1292–1307. 10.1002/jcp.22457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang H, Taylor WR, Joseph G, Caracciolo V, Gonzales DM, Sidell N, Seli E, Blackshear PJ et al. (2013) mRNA-binding protein ZFP36 is expressed in atherosclerotic lesions and reduces inflammation in aortic endothelial cells. Arterioscler Thromb Vasc Biol 33:1212–1220. 10.1161/ATVBAHA.113.301496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Heffron DS, Mandell JW (2005) Opposing roles of ERK and p38 MAP kinases in FGF2-induced astroglial process extension. Mol Cell Neurosci 28:779–790. 10.1016/j.mcn.2004.12.010 [DOI] [PubMed] [Google Scholar]
- 115.Jayasooriya RG, Lee KT, Kang CH, Dilshara MG, Lee HJ, Choi YH, Choi IW, Kim GY (2014) Isobutyrylshikonin inhibits lipopolysaccharide-induced nitric oxide and prostaglandin E2 production in BV2 microglial cells by suppressing the PI3K/Akt-mediated nuclear transcription factor-κB pathway. Nutr Res 34: 1111–1119. 10.1016/j.nutres.2014.10.002 [DOI] [PubMed] [Google Scholar]
- 116.Pang T, Wang J, Benicky J, Sánchez-Lemus E, Saavedra JM (2012b) Telmisartan directly ameliorates the neuronal inflammatory response to IL-1β partly through the JNK/c-Jun and NADPH oxidase pathways. J Neuroinflammation 102 10.1186/1742-2094-9-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Antipova AA, Stockwell BR, Golub TR (2008) Gene expression-based screening for inhibitors of PDGFR signaling. Genome Biol 9:R47 10.1186/gb-2008-9-3-r47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Berk M, Malhi GS, Gray LJ, Dean OM (2013) The promise of N-acetylcysteine in neuropsychiatry. Trends Pharmacol Sci 34:167–177. 10.1016/j.tips.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 119.Zhang F, Sun D, Chen J, Guan N, Huo X, XiH (2015) Simvastatin attenuates angiotensin II-induced inflammation and oxidative stress in human mesangial cells. Mol Med Rep 11:1246–1251. 10.3892/mmr.2014.2871 [DOI] [PubMed] [Google Scholar]
- 120.Yang W, Chauhan A, Mehta S, Mehta P, Gu F, Chauhan V (2014) Trichostatin A increases the levels of plasma gelsolin and amyloid beta-protein in a transgenic mouse model of Alzheimer’s disease. Life Sci 99:31–36. 10.1016/j.lfs.2014.01.064 [DOI] [PubMed] [Google Scholar]
- 121.De Souza C, Chatterji BP (2015) HDAC inhibitors as novel anticancer therapeutics. Recent Pat Anticancer Drug Discov 10:145–162. 10.2174/1574892810666150317144511 [DOI] [PubMed] [Google Scholar]
- 122.Doan NT, Paulsen ES, Sehgal P, MØller JV, Nissen P, Denmeade SR, Isaacs JT, Dionne CA (2015) Targeting thapsigargin towards tumors. Steroids 97:2–7. 10.1016/j.steroids.2014.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Seargent JM, Yates EA, Gill JH (2004) GW9662, a potent antagonist of PPARgamma, inhibits growth of breast tumour cells and promotes the anticancer effects of the PPARgamma agonist rosiglitazone, independently of PPARgamma activation. Br J Pharmacol 143:933–937. 10.1038/sj.bjp.0705973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wong SB, Cheng SJ, Hung WC, Lee WT, Min MY (2015) Rosiglitazone suppresses in vitro seizures in hippocampal slice by inhibiting presynaptic glutamate release in a model of temporal lobe epilepsy. PLoS One 10(12):e0144806 10.1371/journal.pone.0144806 eCollection 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Allam R, Kumar SV, Darisipudi MN, Anders HJ (2014) Extracellular histones in tissue injury and inflammation. J Mol Med (Berl) 92:465–472. 10.1007/s00109-014-1148-z [DOI] [PubMed] [Google Scholar]
- 126.Vardabasso C, Hasson D, Ratnakumar K, Chung CY, Duarte LF, Bernstein E (2014) Histone variants: emerging players in cancer biology. Cell Mol Life Sci 71:379–404. 10.1007/s00018-013-1343-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Love S, Miners JS (2016) Cerebrovascular disease in ageing and Alzheimer’s disease. Acta Neuropathol 131:645–658. 10.1007/s00401-015-1522-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.O’Connor T, Borsig L, Heikenwalder M (2015) CCL2-CCR2 signaling in disease pathogenesis. Endocr Metab Immune Disord Drug Targets 15:105–118. 10.2174/1871530315666150316120920 [DOI] [PubMed] [Google Scholar]
- 129.Cho KA, Kang PB (2015) PLIN2 inhibits insulin-induced glucose uptake in myoblasts through the activation of the NLRP3 inflammasome. Int J Mol Med 36:839–844. 10.3892/ijmm.2015.2276 [DOI] [PubMed] [Google Scholar]
- 130.Naito Y, Takagi T, Higashimura Y (2014) Heme oxygenase-1 and anti-inflammatory M2 macrophages. Arch Biochem Biophys 564: 83–88. 10.1016/j.abb.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 131.Yu S, Wang X, Lei S, Chen X, Liu Y, Zhou Y, Zhou Y, Wu J, Zhao Y (2015) Sulfiredoxin-1 protects primary cultured astrocytes from ischemia-induced damage. Neurochem Int 19–27. 10.1016/j.neuint.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 132.Riggin CH Jr, Pitha PM (1982) Methylation and a polymorphic restriction site adjacent to human beta-interferon gene. DNA 1: 267–271 [DOI] [PubMed] [Google Scholar]
- 133.Li J, Yang J, Lu YW, Wu S, Wang MR, Zhu JM (2015b) Possible role of staphylococcal enterotoxin B in the pathogenesis of autoimmune diseases. Viral Immunol 28:354–359. 10.1089/vim.2015.0017 [DOI] [PubMed] [Google Scholar]
- 134.Urra FA, Araya-Maturana R (2017) Targeting metastasis with snake toxins: molecular mechanisms. Toxins (Basel) 9 10.3390/toxins9120390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Matsui T, Ingelsson M, Fukumoto H, Ramasamy K, Kowa H, Frosch MP, Irizarry MC, Hyman BT (2007) Expression of APP pathway mRNAs and proteins in Alzheimer’s disease. Brain Res 1161:116–123. 10.1016/j.brainres.2007.05.050 [DOI] [PubMed] [Google Scholar]
- 136.Liu Z, Chen HQ, Huang Y, Qiu YH, Peng YP (2016) Transforming growth factor-β1 acts via TβR-I on microglia to protect against MPP(+)-induced dopaminergic neuronal loss. Brain Behav Immun 51:131–143. 10.1016/j.bbi.2015.08.006 [DOI] [PubMed] [Google Scholar]
- 137.Yanamandra M, Mitra S, Giri A (2015) Development and application of PI3K assays for novel drug discovery. Expert Opin Drug Discov 10:171–186. 10.1517/17460441.2015.997205 [DOI] [PubMed] [Google Scholar]
- 138.Figarska SM, Vonk JM, Boezen HM (2014) NFE2L2 polymorphisms, mortality, and metabolism in the general population. Physiol Genomics 46:411–417. 10.1152/physiolgenomics.00178.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.von Otter M, Bergström P, Quattrone A, De Marco EV, Annesi G, Söderkvist P, Wettinger SB, Drozdzik M et al. (2014) Genetic associations of Nrf2-encoding NFE2L2 variants with Parkinson’s disease—a multicenter study. BMC Med Genet 15: 131 10.1186/s12881-014-0131-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Palma-Gudiel H, Córdova-Palomera A, Leza JC, Fañanás L (2015) Glucocorticoid receptor gene (NR3C1) methylation processes as mediators of early adversity in stress-related disorders causality: a critical review. Neurosci Biobehav Rev 55:520–535. 10.1016/j.neubiorev.2015.05.016 [DOI] [PubMed] [Google Scholar]
- 141.Kulkarni SS, Cantó C (2015) The molecular targets of resveratrol. Biochim Biophys Acta 1852:1114–1123. 10.1016/j.bbadis.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 142.Gupta SC, Patchva S, Aggarwal BB (2013) Therapeutic roles of curcumin: lessons learned from clinical trials. AAPS J 15:195–218. 10.1208/s12248-012-9432-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Deguchi A (2015) Curcumin targets in inflammation and cancer. Endocr Metab Immune Disord Drug Targets 15:88–96. 10.2174/1871530315666150316120 [DOI] [PubMed] [Google Scholar]
- 144.Namura S, Iihara K, Takami S, Nagata I, Kikuchi H, Matsushita K, Moskowitz MA, Bonventre JV et al. (2001) Intravenous administration of MEK inhibitor U0126 affords brain protection against forebrain ischemia and focal cerebral ischemia. Proc Natl Acad Sci U S A 98:11569–11574. 10.1073/pnas.181213498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang X, Wang H, Xu L, Rozanski DJ, Sugawara T, Chan PH, Trzaskos JM, Feuerstein GZ (2003) Significant neuroprotection against ischemic brain injury by inhibition of the MEK1 protein kinase in mice: exploration of potential mechanism associated with apoptosis. J Pharmacol Exp Ther 304:172–178. 10.1124/jpet.102.040246 [DOI] [PubMed] [Google Scholar]
- 146.Freilich RW, Woodbury ME, Ikezu T (2013) Integrated expression profiles of mRNA and miRNA in polarized primary murine microglia. PLoS One 8:e79416 10.1371/journal.pone.0079416 eCollection 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen G, Nie S, Han C, Ma K, Xu Y, Zhang Z, Papa SM, Cao X (2017) Antidyskinetic effects of MEK inhibitor are associated with multiple neurochemical alterations in the striatum of hemiparkinsonian rats. Front Neurosci 11:112 10.3389/finns.2017.00112 eCollection 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, El-Ashry D (2006) Activation of mitogen-activated protein kinase in estrogen receptor alpha-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor alpha-negative human breast tumors. Cancer Res 66:3903–3911. 10.1158/0008-5472.CAN-05-4363 [DOI] [PubMed] [Google Scholar]
- 149.Hoogland IC, Houbolt C, van Westerloo DJ, van Gool WA, van de Beek D (2015) Systemic inflammation and microglial activation: systematic review of animal experiments. J Neuroinflammation 12:114 10.1186/s12974-015-0332-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ahn KO, Lim SW, Li C, Yang HJ, Ghee JY, Kim JY, Kim SH, Kim J et al. (2007) Influence of angiotensin II on expression of toll-like receptor 2 and maturation of dendritic cells in chronic cyclosporine nephropathy Transplantation 83:938–947. 10.1097/01.tp.0000258589.39006.94132 [DOI] [PubMed] [Google Scholar]
- 151.Barakat W, Safwet N, El-Maraghy NN, Zakaria MN (2014) Candesartan and glycyrrhizin ameliorate ischemic brain damage through downregulation of the TLR signaling cascade. Eur J Pharmacol 724:43–50. 10.1016/j.ejphar.2013.12.032 [DOI] [PubMed] [Google Scholar]
- 152.Biancardi VC, Stranahan AM, Krause EG, de Kloet AD, Stern JE (2016) Cross talk between AT1 receptors and toll-like receptor 4 in microglia contributes to angiotensin II-derived ROS production in the hypothalamic paraventricular nucleus. Am J Physiol Heart Circ Physiol 310:H404–H415. 10.1152/ajpheart.00247.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cheng XW, Song H, Sasaki T, Hu L, Inoue A, Bando YK, Shi GP, Kuzuya M et al. (2011) Angiotensin type 1 receptor blocker reduces intimal neovascularization and plaque growth in apolipoprotein E-deficient mice. Hypertension 57:981–989. 10.1161/HYPERTENSIONAHA.110.168385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rinaldi B, Di Filippo C, Capuano A, Donniacuo M, Sodano L, Ferraraccio F, Rossi F, D’Amico M (2012) Adiponectin elevation by telmisartan ameliorates ischaemic myocardium in Zucker diabetic fatty rats with metabolic syndrome. Diabetes Obes Metab 14: 320–328. 10.1111/j.1463-1326.2011.01527.x [DOI] [PubMed] [Google Scholar]
- 155.Dasu MR, Riosvelasco AC, Jialal I (2009) Candesartan inhibits toll-like receptor expression and activity both in vitro and in vivo. Atherosclerosis 202:76–83. 10.1016/j.atherosclerosis.2008.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang J, Jiang H, Yang J, Ding JW, Chen LH, Li S, Zhang XD (2009) Valsartan preconditioning protects against myocardial ischemia-reperfusion injury through TLR4/NF-kappaB signaling pathway. Mol Cell Biochem 330:39–46. 10.1007/s11010-009-0098-1 [DOI] [PubMed] [Google Scholar]
- 157.Wang Y, Li Y, Shen Q, Li X, Lu J, Li X, Yin D, Peng Y (2014b) Valsartan blocked alcohol-induced, toll-like receptor 2 signaling-mediated inflammation in human vascular endothelial cells. Alcohol Clin Exp Res 38:2529–2540. 10.1111/acer.12532 [DOI] [PubMed] [Google Scholar]
- 158.Lv J, Jia R, Yang D, Zhu J, Ding G (2009) Candesartan attenuates angiotensin II-induced mesangial cell apoptosis via TLR4/MyD88 pathway. Biochem Biophys Res Commun 380:81–86. 10.1016/j.bbrc.2009.01.035 [DOI] [PubMed] [Google Scholar]
- 159.Saravanan PB, Shanmuganathan MV, Ramanathan M (2015) Telmisartan attenuated LPS-induced neuroinflammation in human IMR-32 neuronal cell line via SARM in AT1R independent mechanism. Life Sci 130:88–96. 10.1016/j.lfs.2015.03.005 [DOI] [PubMed] [Google Scholar]
- 160.Takahashi Y, Satoh M, Minami Y, Tabuchi T, Itoh T, Nakamura M (2010) Expression of miR-146a/b is associated with the toll-like receptor 4 signal in coronary artery disease: effect of renin-angiotensin system blockade and statins on miRNA-146a/b and toll-like receptor 4 levels. Clin Sci (Lond) 119:395–405. 10.1042/CS20100003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.