Abstract
Different genetic patterns have been demonstrated for narrowly distributed taxa, many of them linking rarity to evolutionary history. Quite a few species in young genera are endemics and have several populations that present low variability, sometimes attributed to geographical isolation or dispersion processes. Assessing the genetic diversity and structure of such species may be important for protecting them and understanding their diversification history. In this study, we used microsatellite markers and plastid sequences to characterize the levels of genetic variation and population structure of two endemic and restricted species that grow in isolated areas on the margin of the distribution of their respective genera. Plastid and nuclear diversities were very low and weakly structured in their populations. Evolutionary scenarios for both species are compatible with open-field expansions during the Pleistocene interglacial periods and genetic variability supports founder effects to explain diversification. At present, both species are suffering from habitat loss and changes in the environment can lead these species towards extinction.
Keywords: Atlantic Forest, highlands, nuclear diversity, open-field species, plastid variability, Pleistocene, rare species, Solanaceae
In our study, we used microsatellite markers and plastid sequences to characterize the levels of genetic variation and population structure of two endemic and restricted species (Petunia mantiqueirensis and Calibrachoa elegans) that grow in isolated areas on the margin of the distribution of their respective genera. Plastid and nuclear diversities were low and weakly structured in their populations. Evolutionary scenarios for both species are compatible with open-field expansions during the Pleistocene interglacial periods and genetic variability supports founder effects to explain diversification. Both species are suffering from habitat loss and changes in the environment can lead these species towards extinction.
Introduction
Many rare or narrowly distributed species present low levels of genetic diversity (Gitzendanner and Soltis 2000). Additionally, there is a positive association between geographical range size and genetic diversity (Ellstrand and Elam 1993; Hamrick and Godt 1996), especially considering the limits of genera distribution. Species that inhabit the border of the distribution of their respective groups usually have smaller and more fragmented populations and, if there is no immigration between theirs and central populations, they can have fixed locally adapted alleles (Bridle and Vines 2006).
Ecological and evolutionary processes define the distribution of species across space and time. The climatic changes that occurred during the Quaternary impacted the distribution and genetic diversity of most species through cycles of range contraction and expansion in the northern and southern hemispheres (Hewitt 2000; Behling 2002; Beheregaray 2008), and, most recently, acquired heightened relevance in relation to current global warming (Lavergne et al. 2010). Climate change alters local conditions and can cause range expansion and contraction affecting the species distribution and this range dynamics may drive the processes of diversification across time and space (Dawson et al. 2011). Geographic range expansion during interglacial periods and the associated founding effects are expected to reduce allelic richness and increase homozygosity (Pfeifer et al. 2009) compared to species at the centre of genus distribution.
Some studies have focused on the evolutionary history of species adapted to open areas interspersed among regions of higher altitude in the southern section of the Atlantic Forest (Behling 2002; Safford 2007). The south-eastern Brazilian highlands, which are located in the subtropical region of South America, consist of a mosaic of grasslands and Atlantic mixed forest. These formations occur between latitudes 18° and 24°S at elevations greater than 1400 m in small isolated areas (Fig. 1A and B). The relationship between grassland and forest during the late Quaternary in this region was documented through pollen records and probably resembles the general pattern of shifts in vegetation range that took place throughout the Pleistocene period (Ledru 1993; Ledru et al. 2005).
Figure 1.
(A) Rectangle area corresponds to the geographical distribution of Petunia mantiqueirensis and Calibrachoa elegans. The grey area corresponds to the distribution of other species from both genera; (B) collection sites; (C) Petunia mantiqueirensis flower; and (D) Calibrachoa elegans flower (Photos J. R. Stehmann, UFMG).
Species of Petunia and Calibrachoa share several evolutionary constraints and biological characteristics, many times occurring sympatrically, side by side. Based on a proposed origin in open fields (Olmstead et al. 2008) at ca. 8.5 Mya (Särkinen et al. 2013), these genera were strongly influenced by the Pleistocene glacial and interglacial cycles (between ca. 2.8 and 1.3 Mya) during their diversification (Lorenz-Lemke et al. 2010; Fregonezi et al. 2013; Särkinen et al. 2013; Barros et al. 2015). The ancestor for both has Andean origin (Reck-Kortmann et al. 2015) that posteriorly dispersed to the north and colonized the highlands (Reck-Kortmann et al. 2014). Both genera include endemic and restricted taxa, frequently associated with specific micro-environments (Stehmann et al. 2009; Fregonezi et al. 2012).
During the last glaciation, the southern and south-eastern Brazilian highlands were predominantly grasslands. Cold and dry climate conditions allowed the grasslands to expand northwards, where small populations of Araucaria angustifolia were restricted to more humid areas along shadowy ravines (Behling et al. 2001; Ledru et al. 2005). As temperature and humidity increased in the early Holocene, the Araucaria forest expanded over the grasslands and only the highest areas in this region during the interglacial periods were refuges for species previously associated with open areas (Behling 2002), with many cold-adapted species still remaining there (Lorenz-Lemke et al. 2010). Strong influence of global climate changes during the Pleistocene on the evolutionary processes in plants has been described for several species from the south-eastern region of Brazil [Hymenaea stigonocarpa (Ramos et al. 2007); Lychnophora ericoides, Collevatti et al. (2009); Epidendrum fulgens, Pinheiro et al. (2011); Tabebuia aurea, Collevatti et al. (2015); and Eugenia uniflora, Turchetto-Zolet et al. (2016)], as well as to species that occur closest to the Atlantic coast, such as Calibrachoa heterophylla (Mäder et al. 2013), Recordia reitzii (Thode et al. 2014), Petunia integrifolia (Ramos-Fregonezi et al. 2015) and Passiflora contracta (Cazé et al. 2016).
Among the species of Petunia and Calibrachoa, P. mantiqueirensis and C. elegans, respectively, have reached the northernmost distribution of their respective genera. Both species are endemic of a very specific habitat at the edge of each genus distribution and are found in small patches with few individuals.
As genetic drift tends to further reduce genetic diversity (Ellegren and Ellegren 2016; Thurman and Barrett 2016), the fixation of alleles because of low effective sizes is a phenomenon that more commonly occurs on the edge of the range of distribution of a species where expansion and contraction events are frequent (Eckert et al. 2008; Excoffier et al. 2009; Angert 2014; Hargreaves and Eckert 2014). As such, our hypothesis was that P. mantiqueirensis and C. elegans present lower genetic variability compared to their congeners, which is related to their rarity and limited occurrence at the edge of their respective genera distribution owing to a pronounced founder effect.
Within this context, the main goal of this study was to assess the genetic diversity of these two rare and endemic species using plastid DNA sequences and nuclear microsatellite markers. We discuss their genetic diversity, comparing these with congeneric species aiming to provide elementary information about the consequences of the expansion of these genera’s distribution and, indirectly, to contribute to the establishment of conservation strategies for these rare and endangered species.
Methods
Plant material
Petunia mantiqueirensis (Fig. 1C) is endemic to high altitude regions in Serra da Mantiqueira, Minas Gerais Brazilian state (elevations > 1300 m), specifically in mixed ombrophilous forest (Araucaria moist forests), which is the northernmost distribution of the Petunia genus. This species is annual and herbaceous, with bee-pollinated flowers displaying a purple and short corolla tube (Stehmann et al. 2009). The area where the species is found is fragmented because of urbanization and intense agricultural activity (Kamino et al. 2012).
Calibrachoa elegans (Fig. 1D) is a micro-endemic, annual and herbaceous species from the Iron Quadrangle (Minas Gerais, south-eastern Brazil) of which only four occurrence sites are known. The species has the northernmost distribution within the genus and occurs in a mountainous landscape (elevation > 1000 m), strongly corrugated, and dominated by grassland, popularly called ferruginous field from ‘nodular canga’ (Fregonezi et al. 2012). The species has unique adaptations to the climate of the region, which presents a rigorous rainfall regime, with 4 months of dry season. It is self-incompatible and bee-pollinated (Stehmann and Semir 2001).
These two species are classified as federally endangered according the Brazilian Red List (CNCFlora, available at http://cncflora.jbrj.gov.br/portal). We sampled all known populations and adult individuals found in the same flowering season [for P. mantiqueirensis, seven sites (Mant1 to Mant7) totalling 38 individuals, and for C. elegans, 81 individuals from four sites (Eleg1 to Eleg4)]. Vouchers were deposited at the BHCB herbarium (Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil—UFMG) and the geographical coordinates for all collection sites were obtained with a global positioning system (GPS) unit [seeSupporting Information—Table S1]. We collected three or four young leaves from each adult individual of both species while seeking to minimize damage to the plants; we dried leaves in silica gel and then pulverized them in liquid nitrogen for DNA extraction.
DNA extraction, amplification, sequencing and genotyping
The genomic DNA was extracted from the powdered leaves with a cetyl trimethyl ammonium bromide (CTAB) protocol (Roy et al. 1992) and the quality and quantity was evaluated by measuring absorbance at 260 and 280 nm on a Nanodrop spectrophotometer (Thermo Scientific Corp., San Jose, CA, USA).
We estimated the genetic diversity based on the plastid sequences (cpDNA) of intergenic spacers, trnH-psbA and trnS-trnG, were amplified and sequenced for 17 individuals of P. mantiqueirensis and 81 individuals of C. elegans using universal primers (Hamilton 1999; Sang et al. 1997, respectively). The PCR reactions were performed in 25-µL final volumes consisting of 1 U Taq polymerase (Invitrogen, Carlsbad, CA, USA), 1× buffer (Invitrogen), 0.2 mM each dNTP, 2 mM MgCl2, 0.2 µM of each primer and 10 ng of genomic DNA. The amplification conditions for trnH-psbA and trnS-trnG were conducted as described previously for Petunia species (Lorenz-Lemke et al. 2006). The PCR products were verified by horizontal electrophoresis in 1 % agarose gel dyed with GelRed™ (Biotium, Inc., Hayward, CA, USA) and purified with polyethylene glycol 20 % (Dunn and Blattner 1987). Sequencing was performed with a Mega BACE 1000 (GE Healthcare Bio Sciences Corp., Piscataway, NY, USA). We also included in our analyses the sequences of 21 individuals of P. mantiqueirensis as previously described (Lorenz-Lemke et al. 2010).
To evaluate current variability in each population and species, individuals of P. mantiqueirensis were genotyped with 12 microsatellite loci markers (SSR) previously developed for Petunia hybrida (Bossolini et al. 2011) adhering to the protocol described for wild Petunia species (Turchetto et al. 2015). For C. elegans, we analysed 10 microsatellite loci developed for C. heterophylla (Silva-Arias et al. 2015) and four markers developed for C. pygmaea (G. Mäder et al., unpubl. data). We amplified SSR regions in a final volume reaction of 10 µL containing ~10 ng of genomic DNA as a template, 200 µM of each dNTP (Invitrogen), 1.7 pmol of fluorescently labelled M13(-21) primer, 3.5 pmol of reverse primer, 0.35 pmol of forward primer with a 5′-M13(-21) tail, 2.0 mM MgCl2 (Invitrogen), 0.5 U of Platinum Taq DNA polymerase (Invitrogen), and 1× Platinum Taq reaction buffer (Invitrogen). The PCR conditions were as follows: an initial denaturation at 94 °C for 3 min; 30–35 cycles of 94 °C for 20 s, 48–58 °C for 45 s and 72 °C for 1 min; and a final extension cycle at 72 °C for 10 min.
The forward primers were FAM-, NED-, HEX-, VIC- and PET-labelled. The amplified product was visualized on a 2.5 % ultra-resolution agarose gel stained with 2 µL 0.001 % GelRed™. The amplified DNA was denatured and size-fractionated employing capillary electrophoresis on an Applied Biosystems (Foster City, CA, USA) with a LIZ (500) molecular size standard (Applied Biosystems). GeneMarker 1.97 software (Softgenetics LLC, State College, PA, USA) was utilized to determine the alleles. Additionally, all alleles were visually verified and scored. Repeat motifs, respective annealing temperatures and fluorescent dyes used in each fragment are presented in Supporting Information—Table S2.
Plastid variability
For each plastid marker, both forward and reverse strands were assembled using ChromasPro 1.7.5 (Technelysium Pty Ltd, Australia) and haplotype sequences were deposited at GenBank (available at https://www.ncbi.nlm.nih.gov/genbank/) under numbers: C. elegansJQ082471.1, JQ082474.1, KM982182.1, KM982262.1; P. mantiqueirensisAY772921.1, DQ792340.1. DNA sequences were aligned using MEGA7 software (Kumar et al. 2016) with the ClustalW algorithm, and manually edited when necessary. Haplotype (h) and nucleotide (π) diversity indexes and the number of variable sites were calculated with Arlequin 3.5.1.2 (Excoffier and Lischer 2010). The two plastid intergenic spacers were concatenated and treated as a single sequence for all analyses. The numbers of variable and informative sites in the manually edited alignment were obtained from Mega7 software. Haplotypes were identified via DNAsp 5.10.01 (Rozas et al. 2003).
Nuclear diversity
For both species, we used Fstat 2.9.3.2 software (Goudet 1995) to evaluate statistics, such as allelic frequencies, the number of alleles per locus (A), gene diversity (GD), allelic richness (AR) and inbreeding coefficient (FIS) for each locus. The frequencies of the null alleles, polymorphic index content (PIC), levels of observed (Ho) and expected (He) heterozygosity, and any significant deviations from the Hardy–Weinberg equilibrium (HWE), in order to evaluate the informativeness of the markers, were estimated using Cervus 3.0.3 software (Marshall et al. 1998; Kalinowski et al. 2007).
We performed locus-by-locus analysis of molecular variance (AMOVA) (Excoffier et al. 1992) and the overall FST to evaluate the partition of the genetic variation among populations. The AMOVA analysis was performed in Arlequin employing 104 permutations. We also estimated pairwise FST to evaluate the level of genetic differentiation between populations of both species.
Genetic relationships among populations of each species were also examined by applying the discriminant analysis of principal components (DAPC; Jombart et al. 2010) on the SSR data set with the Adegenet package (Jombart 2008) without locational priors included in the analysis.
Test of alternative demographic scenarios
An approximate Bayesian computation (ABC) approach (Bertorelle et al. 2010) implemented in the program Diyabc 2.1.0 (Cornuet et al. 2014) was used to test four plausible colonization scenarios for each of these two species based on previous studies (Stehmann et al. 2009; Fregonezi et al. 2012; Reck-Kortmann et al. 2014) employing microsatellite variation. Scenario 1 is of a long-term constant size population (no bottleneck), scenario 2 consisted of a population that is still experiencing a bottleneck, scenario 3 is a population that expanded recently from a bottleneck and scenario 4 is a population that experienced a transitory bottleneck (Fig. 2). The priors for all demographic parameters were uniformly distributed between specified minimum and maximum values [seeSupporting Information—Table S3], which were based on the available information surrounding Petunia and Calibrachoa.
Figure 2.
Demographic scenarios for Petunia mantiqueirensis and Calibrachoa elegans. The four demographic scenarios tested with the DIYABC approach: 1—constant population; 2—bottlenecked population; 3—expanded population; 4—population with a transitory bottleneck. Demographic parameters: Ne—long-term; Ne2, Ne3 and Ne4—current; Na2 and Na4—pre-bottleneck; Na3 and Nb—during bottleneck; t, time of single demographic change; t2 and t1, beginning and end of bottleneck. Prior values for the parameters are in Supporting Information—Table S3. The posterior probability (logistic approach with the 8000 closest simulations) of each scenario is given at the bottom.
The demographic parameters were: scenario 1: Ne (long-term Ne); scenario 2: Ne2 (current Ne), Na2 (pre-bottleneck Ne); scenario 3: Ne3 (current Ne), Na3 (Ne during bottleneck); t (time since demographic change in scenarios 2 and 3); scenario 4: Ne4 (current Ne), Nb (Ne during bottleneck), Na4 (pre-bottleneck Ne), t2 and t1 (time since the beginning and end of the bottleneck, respectively). Demographic prior values are presented in Supporting Information—Table S3. For population size parameters we used broad priors as we do not have much previous information on them. As priors for the times for the bottleneck scenario (t2 and t1, scenario 4), we first used as reference the maximum the influence of the Last Glacial Maximum (LGM, ~21 kya) and as the minimum the first record of Araucaria forest expansion in the region [~8 kya (Ledru et al. 2005), plus or minus 4 kya for uncertainties]. After the first results, we adjusted prior values to the following: extended t and t2 maximum time to 50 kya and reduced time t to a minimum of 10 years ago. Mutational parameter priors were set to evolve under the generalized stepwise mutation model (GSM; Estoup et al. 2002) with mean mutation rate (μ) uniformly ranging from 10−4 to 10−2 and the individual locus mutation rate per generation range from 10−5 to 10−2 (Zhang and Hewitt 2003) with gamma distribution with shape equal to 2. Generation time was set to 1 year. Motif sizes and allele ranges followed the empirical data of each locus [seeSupporting Information—Table S2]. The four single sample summary statistics available were used (mean number of alleles, mean gene diversity, mean allele size and mean M index all across loci). A total of 8 000 000 simulations (2 000 000 for each scenario) were carried out to generate the reference table. The posterior probability of each scenario was assessed employing both logistic regression and direct estimate approaches utilizing 40 000 and 500 best simulations, respectively. For the best scenario, the posterior distribution of the parameters was estimated using logit transformation for the 20 000 best simulations. We also used several options in DIYABC to evaluate the confidence in scenario choices, posterior model checking and determination of bias and precision of the parameter estimation. The details of these analyses are described in Supporting Information—Table S4.
Ecological niche modelling
Ecological niche modelling (ENM) was used to compare niche preferences and potential range shifts that the species can suffer from in response to future climatic oscillations. Localities from the two species were obtained from field observations by the authors, SpeciesLink (http://www.splink.org.br) and the Global Biodiversity Information Facility (http://www.gbif.org). After excluding duplicated, false positive records and spatially autocorrelated points, we used information for 13 localities of P. mantiqueirensis and 11 of C. elegans[seeSupporting Information—Table S5]. We employed Maxent 3.4.1 (Phillips et al. 2006) to run models for each species varying the feature classes and the regularization multiplier (Galante et al. 2018). We used five different feature classes (linear; hinge; linear and quadratic; linear, hinge and quadratic; default) and five values for the regularization multiplier (1–5, with increments of 1) for each species, for a total of 50 models. To evaluate the models, we used ENMTools (Warren et al. 2010; Warren and Seifert 2011) and the best model was chosen according the AICc value. The best model indicated was used to project two future conditions, and three past conditions using 3-fold cross-validation. To model future distributions, we used 2050 (average for 2041–60) and 2070 (average for 2061–80) conditions, with the Community Climate System Model (CCSM) and the Representative Concentration Pathways (RCP 6.0). The RCPs are calculated considering a possible range of radioactive forcing values in the year 2100 relative to pre-industrial values, and RCP 6.0 considers a value of +6.0 W m−2. To model past distributions, we used Mid-Holocene (~6 kya), Last Glacial Maximum (LGM; ~21 kya) and Last Interglacial (LIG; ~120–140 kya) conditions. Explanatory variables included a set of 19 bioclimatic Raster layers at a 30 arc-second resolution (ca. 1 km2 at the Equator) from the WorldClim website, version 1.4 (available at: http://www.worldclim.org/). The Raster package (Hijmans and van Etten 2010) implemented in R software (available at: http://www.R-project.org) was used to calculate Pearson’s correlation between variables. For P. mantiqueirensis, four variables (06—Minimum Temperature of Coldest Month; 09—Mean Temperature of Driest Quarter; 15—Precipitation Seasonality; and 17—Precipitation of Driest Quarter) with pairwise Pearson correlation coefficients below 0.7 and with a high percentage of model contribution (in preliminary runs with all variables) were used to reconstruct the taxon distribution. For C. elegans, we employed seven variables (02—Mean Diurnal Range; 05—Maximum Temperature of Warmest Month; 06—Minimum Temperature of Coldest Month; 07—Temperature Annual Range; 09—Mean Temperature of Driest Quarter; 15—Precipitation Seasonality; and 16—Precipitation of Wettest Quarter).
Results
Plastid diversity
The plastid intergenic spacers, trnH-psbA and trnS-trnG, for 38 individuals of P. mantiqueirensis resulted in a concatenated alignment of 1121 base pairs (bp) long (471 bp corresponding to trnH-psbA and 650 bp to trnS-trnG). The sequences did not present any variable site, producing a single haplotype. The GC content was 28.1 % in trnH-psbA and 30.5 % in trnS-trnG. For C. elegans, the sequences of 81 individuals produced a concatenated alignment with 1191 bp (445 bp for trnH-psbA and 746 bp for trnS-trnG), leading to two different haplotypes. The GC content was 27.2 % in trnH-psbA and 30.7 % in trnS-trnG. Haplotype diversity in C. elegans was h = 0.0299 ± 0.0287, and nucleotide diversity was π = 0.004 ± 0.014 %. The frequency of each haplotype was CEh1 = 0.98 and CEh2 = 0.02; CEh2 was observed only at one collection site (Eleg1).
Nuclear diversity and structure
The 38 individuals of P. mantiqueirensis were genotyped using 12 SSR loci, being 10 polymorphic and two monomorphic (PM110 and PM195) for this species. All loci were in linkage equilibrium (P < 0.001; Bonferroni’s adjusted value for a nominal level of 5 %). The 81 individuals of C. elegans were genotyped based on 14 SSR loci, from which five markers were polymorphic and nine monomorphic (CHE12, CHE46, CHE48, CHE114, CHE119, CPY29, CPY31, CPY58 and CPY144). All analyses were conducted based exclusively on polymorphic markers for each species, although the monomorphic loci are suggestive of low genetic variability and high homogeneity in these species as all of them are polymorphic among other Petunia and Calibrachoa species, respectively.
Petunia mantiqueirensis presented a total of 39 alleles, with an average of 3.9 alleles per locus (2 to 7). The mean PIC value was 0.39 (0.05 to 0.75), and considering all loci and gene diversity, it was, on average, 0.36 (0.06 to 0.80). Allele richness was, on average, 3.33 (2.00 to 7.00). Six loci exhibited a significant deviation from HWE across all samples, three of them had a deficit of heterozygotes and the other three displayed an excess of heterozygotes. Tests for HWE indicated no significant FIS values for all loci and populations and the mean estimated frequency of null alleles was lower than 0.5 % (Table 1). When we analysed each population as a unit, we observed four loci with a significant deviation from HWE in two populations (Mant1 and Mant3), and three loci presented a deficit of heterozygotes and one an excess of heterozygotes. Four populations presented private alleles (Mant1, Mant3, Mant5 and Mant6) [seeSupporting Information—Table S6]. When we consider the species’ global values we observed no significant HWE deviation or FIS values (Table 2).
Table 1.
Genetic diversity indices based on the microsatellite profile in Petunia mantiqueirensis. A, number of alleles per locus; PIC, polymorphic index content; GD, gene diversity; AR, allele richness; Ho, observed heterozygosity; He, expected heterozygosity; FIS, inbreeding coefficient; NUL, frequency of null alleles. *Significant HWE deviation after Bonferroni correction at P = 0.05.
| Locus | A | PIC | GD | AR | H o | H e | F IS | NUL (%) |
|---|---|---|---|---|---|---|---|---|
| PM8 | 4 | 0.388 | 0.429 | 3.912 | 0.400 | 0.429 | 0.068 | 0.023 |
| PM167 | 7 | 0.582 | 0.619 | 6.621 | 0.667 | 0.619 | −0.077 | −0.057 |
| PM101 | 3 | 0.195 | 0.213 | 2.923 | 0.128* | 0.211 | 0.397 | 0.232 |
| PM117 | 3 | 0.578 | 0.662 | 3.000 | 0.625 | 0.661 | 0.055 | 0.020 |
| PM177 | 3 | 0.232 | 0.251 | 2.998 | 0.175* | 0.250 | 0.302 | 0.159 |
| PM21 | 2 | 0.187 | 0.211 | 2.000 | 0.237 | 0.212 | −0.121 | −0.058 |
| PM184 | 5 | 0.579 | 0.650 | 4.969 | 0.775* | 0.651 | −0.193 | −0.115 |
| PM191 | 3 | 0.055 | 0.057 | 2.600 | 0.029* | 0.057 | 0.500 | 0.410 |
| PM173 | 7 | 0.754 | 0.798 | 7.000 | 0.929* | 0.800 | −0.164 | −0.091 |
| PM63 | 2 | 0.354 | 0.462 | 2.000 | 0.714* | 0.466 | −0.545 | −0.217 |
| Mean | 3.9 | 0.390 | 0.363 | 3.335 | 0.390 | 0.363 | −0.075 | 0.031 |
Table 2.
Characterization of the 10 microsatellites for Petunia mantiqueirensis. All populations values considering pooled populations; N, number of alleles; E, number of private alleles; He, expected heterozygosity; Ho, observed heterozygosity; FIS, inbreeding coefficient. *Significant HWE deviation after Bonferroni correction at P = 0.05; – not estimated.
| Mant1 | Mant2 | Mant3 | Mant4 | Mant5 | Mant6 | Mant7 | All populations | ||
|---|---|---|---|---|---|---|---|---|---|
| Petunia mantiqueirensis | N | 27 | 21 | 30 | 14 | 23 | 14 | 14 | 39 |
| E | 3 | – | 3 | – | 1 | 1 | – | – | |
| H e | 0.492 | 0.616 | 0.441 | 0.625 | 0.617 | 0.667 | 0.667 | 0.435 | |
| H o | 0.579 | 0.667 | 0.524 | 0.875 | 0.667 | 1.000 | 1.000 | 0.468 | |
| F IS | −0.033 | −0.130 | −0.199 | −0.750 | −0.109 | −1.000 | −1.000 | −0.075 |
The AMOVA indicated that the percentage of variation within populations (91 %) was higher than among populations (9 %), and the fixation index (FST) over all loci was 0.087 (P < 0.05). Five FST pairwise comparisons had significant genetic differentiation, ranging from 0.07 to 0.25 (P < 0.05). The Mant3 had significant differentiation from four other populations (Mant1, Mant4, Mant5 and Mant6), as well as Mant4 vs. Mant1 (FST = 0.25) [seeSupporting Information—Table S7]. With the DAPC analysis, the individuals from a same collection site did not preferentially form groups and individuals from Mant4 and Mant7 were the most differentiated [seeSupporting Information—Fig. S1A].
Calibrachoa elegans also presented a total of 39 alleles, with an average of 7.8 alleles per locus (4 to 13) and the PIC values for these markers ranged from 0.47 to 0.84 (average 0.63). The gene diversity was, on average, 0.54 (0.45 to 0.73) and allele richness 5.32 (3.06 to 8.42). All loci exhibited a significant departure from HWE across all samples with heterozygote deficits (mean Ho = 0.40 and He = 0.68 across all samples). Moreover, three of them had significant and positive FIS values, whereas one presented a significant and negative FIS value. The estimated frequency of null alleles was lower than 0.5 % in C. elegans (Table 3) as in P. mantiqueirensis (Table 1). When the analysis was performed per population, we observed that a different number of loci exhibited a significant departure from HWE on populations with a positive and significant inbreeding coefficient (FIS) [seeSupporting Information—Table S8]. Eleg2 had the highest average of FIS with one locus presenting significantly positive values. Furthermore, the populations had different numbers of loci with significant deviations from HWE, and all presented a deficit of heterozygotes at least in one locus. All populations also featured private alleles. Note that Eleg2 was the site with the lowest genetic diversity as measured by number of alleles (A) and allele richness (AR) [seeSupporting Information—Table S8]. When we consider the species’ global values (Table 4) we found negative and significant FIS.
Table 3.
Genetic diversity indices based on the microsatellite profile in Calibrachoa elegans. A, number of alleles per locus; PIC, polymorphic index content; GD, gene diversity; AR, allele richness; Ho, observed heterozygosity; He, expected heterozygosity; FIS, inbreeding coefficient; NUL, frequency of null alleles. *Significant HWE deviation after Bonferroni correction at P = 0.05.
| Locus | A | PIC | GD | AR | H o | H e | F IS | NUL (%) |
|---|---|---|---|---|---|---|---|---|
| CHE33 | 13 | 0.841 | 0.729 | 8.422 | 0.473* | 0.863 | 0.351* | 0.287 |
| CHE34 | 6 | 0.471 | 0.498 | 4.163 | 0.390* | 0.506 | 0.305* | 0.105 |
| CHE59 | 4 | 0.478 | 0.463 | 3.061 | 0.421* | 0.565 | 0.081 | 0.140 |
| CHE85 | 10 | 0.682 | 0.452 | 5.981 | 0.481* | 0.724 | −0.095* | 0.197 |
| CHE 126 | 6 | 0.668 | 0.547 | 4.983 | 0.254* | 0.721 | 0.604* | 0.488 |
| Mean | 7.8 | 0.628 | 0.538 | 5.322 | 0.404 | 0.676 | 0.249 | 0.243 |
Table 4.
Characterization of the five microsatellites for Calibrachoa elegans. All populations values considering pooled populations; N, number of alleles; E, number of private alleles; He, expected heterozygosity; Ho, observed heterozygosity; FIS, inbreeding coefficient. *Significant HWE deviation after Bonferroni correction at P = 0.05; – not estimated.
| Eleg1 | Eleg2 | Eleg3 | Eleg4 | All populations | ||
|---|---|---|---|---|---|---|
| Calibrachoa elegans | N | 27 | 10 | 28 | 22 | 39 |
| E | 3 | 2 | 8 | 1 | – | |
| H e | 0.653 | 0.241 | 0.653 | 0.630 | 0.675 | |
| H o | 0.620 | 0.051 | 0.494 | 0.503 | 0.404 | |
| F IS | 0.052 | 0.794 | 0.251 | 0.214 | 0.404* |
The AMOVA analysis revealed that the percentage of variation within populations (72 %) was higher than among populations (28 %) and the fixation index (FST) over all loci was 0.281. FST pairwise comparisons had significant and strong genetic differentiation in just the Eleg2 compared to the other sites (Eleg2 vs. Eleg1 = 0.53; Eleg2 vs. Eleg3 = 0.55 and Eleg2 vs. Eleg4 = 0.49; P < 0.05). With the DAPC, individuals from Eleg2 were completely differentiated from the others, whereas a high superimposition was observed among the other collection sites [seeSupporting Information—Fig. S1B].
Demographic scenarios
The comparison of the four demographic scenarios for P. mantiqueirensis and C. elegans using the ABC approach reflected that the most likely scenario for both species is that in which the population size in the past was much greater than in the present (Fig. 2). Support for this scenario in C. elegans was very high according to the logistic approach (above 0.98), and a little smaller with the direct approach, above 0.90 (95 % CI = ~0.65–1.0), and extremely high for P. mantiqueirensis above 0.99 and 1.0, respectively. Confidence in the choice of these scenarios were high for the C. elegans and very high for P. mantiqueirensis, in which the predictive errors were 0.16 and 0.0, respectively [seeSupporting Information—Table S4]. The posterior model checking for scenario 2 in P. mantiqueirensis presented a good fit while for C. elegans the fit for one of the summary statistics was not good [seeSupporting Information—Table S4]. The posterior density estimates of the demographic parameters for scenario 2 are found in Supporting Information—Table S4 and Table 5. The bias and error in scenario 2 were very low for all parameters in C. elegans while it is higher in t for P. mantiqueirensis[seeSupporting Information—Table S4]. The estimated time since the population reduction presented a wide confidence interval, the model time for P. mantiqueirensis was older than for C. elegans (~19 and ~11 kya, respectively) and it was not younger than ~5 kya for either (Table 5).
Table 5.
Posterior parameter estimates based on ABC scenario 2.
| Parameter | Mode | q050 | q950 | |
|---|---|---|---|---|
| Petunia mantiqueirensis | Ne2 | 534 | 353 | 1330 |
| t | 2000 | 901 | 22 620 | |
| Na2 | 77 200 | 33 200 | 468 000 | |
| Calibrachoa elegans | Ne2 | 4880 | 3180 | 13 000 |
| t | 25 200 | 9030 | 47 400 | |
| Na2 | 477 000 | 36 700 | 483 000 |
Niche preferences and future adequacy
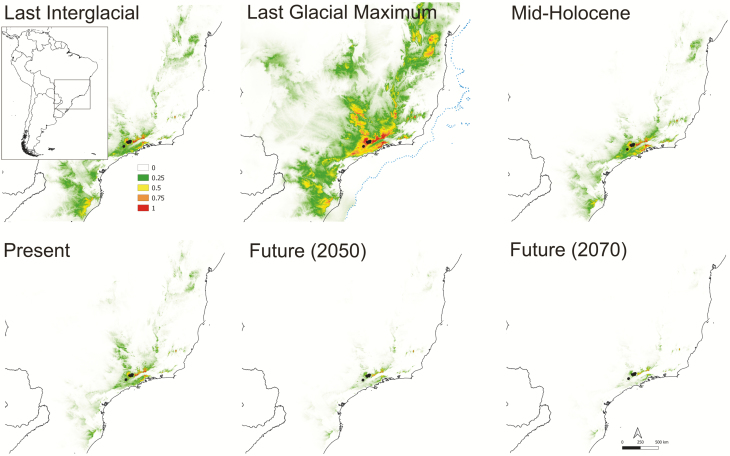
Considering the area under the ‘receiver operating characteristic (ROC) curve’ (AUC) (Peterson et al. 2008; Elith and Leathwick 2009) all tested models had AUC values above 0.9 and could be considered robust model performance (Fielding and Bell 1997). However, when the AICc was analysed the best model was the one using default feature classes and regularization multiplier equal to one for both species [seeSupporting Information—Table S9]. Precipitation seasonality was the most important ecological variable to model P. mantiqueirensis niche adequacy, whereas temperature annual range was the most imperative for C. elegans. In the simulated model for P. mantiqueirensis at present time (Fig. 3), suitable habitats were indicated slightly further to the north than the current distribution, still in the Serra da Mantiqueira (Minas Gerais, Brazil), and even less pronounced to the west in Serra do Mar (Rio de Janeiro, Brazil). The new suitable habitats indicated for C. elegans (Fig. 4) were further to the north and south from the present range still in the canga soils. ENM for future scenarios showed a slight increase in distribution for C. elegans and a slight reduction when P. mantiqueirensis is considered. When projected to past conditions, the largest distribution was during the LGM for both species. For P. mantiqueirensis the distribution during the LIG resembled that on the present but including areas with elevated occurrence probability further south. However, for C. elegans the distribution during this period was the smallest considering all models.
Figure 3.
Ecological niche modelling for Petunia mantiqueirensis obtained with Maxent. Black dots represent species localities in which the models were based. Colour bars indicate suitability scores represented for P. mantiqueirensis. The coast line in the LGM is indicated by a dotted line.
Figure 4.
Ecological niche modelling for Calibrachoa elegans obtained with Maxent. Black dots represent species localities in which the models were based. Colour bars indicate suitability scores represented for C. elegans. The coast line in the LGM is indicated by a dotted line.
Discussion
Here, we evaluated the genetic diversity based on plastid sequences and nuclear microsatellites [see Supporting Information—Table S10] of two narrowly distributed and rare species, P. mantiqueirensis and C. elegans, which represent the northern limit of these genera’s distribution. Each one of these species grows in disjunct habitats from the rest of species in their respective genera, in micro-environments located at extremes of elevation (Stehmann and Semir 2001; Stehmann et al. 2009), and, for C. elegans, a unique and particularly inhospitable soil type (Skirycz et al. 2014).
Petunia and Calibrachoa are two genera with recent diversification histories dating back to the Pleistocene (Lorenz-Lemke et al. 2010; Fregonezi et al. 2013; Särkinen et al. 2013). Both genera are distributed in South America and present widespread and narrowly distributed species, several of them being associated with particular habitats. Phylogeographic and phylogenetic studies of these two genera resulted in the proposal of two main patterns of distribution of genetic diversity and species diversification: (i) historical events, such as climate changes over glacial periods that affected the distribution of species driving allopatric speciation as proposed for Petunia species from the highlands (Lorenz-Lemke et al. 2010), and C. heterophylla (Mäder et al. 2013) and P. integrifolia (Ramos-Fregonezi et al. 2015) from the Atlantic Coastal Plain in south Brazil; and (ii) ecological factors, such as selective pressure of pollinators on morphological traits as indicated by the highly specialized interactions with their pollinators (Fregonezi et al. 2013; Sheehan et al. 2016; Rodrigues et al. 2018) or even climatic conditions (Barros et al. 2015). Both genera originated from an ancestor that inhabited the Pampas region at sea level and, secondarily, migrated to highlands, thereby colonizing the northern part of the genera’s distributions in south-eastern Brazil (Reck-Kortmann et al. 2015) during the Pleistocene glacial cycles as the open-field areas advanced (Behling 2002). In that time, during the cold and dry periods (glacial periods), species from open fields expanded to the north and, when climate turns warmer and more humid (interglacial periods), these populations were fragmented because forest growth and were restricted to the higher elevations (Lorenz-Lemke et al. 2010). Our demographic modelling of colonization for C. elegans and P. mantiqueirensis supports this.
In comparison with other Petunia species, P. mantiqueirensis has lower variability indices based on both plastid and nuclear markers than, for example, the also rare and endemic P. secreta (π = 0.17 % and nine cpDNA haplotypes; Turchetto et al. 2016) or the widespread P. integrifolia subsp. depauperata (π = 0.13 % and 25 haplotypes; Ramos-Fregonezi et al. 2015) and P. axillaris (π = 0.22 % and 35 haplotypes; Turchetto et al. 2014). Based on nuclear diversity, especially considering AR, other endemic species [P. secreta, AR = 5.91; and P. exserta, AR = 4.80 (Turchetto et al. 2016)] feature higher genetic variability than P. mantiqueirensis but are not significantly different from those that are widespread [P. axillaris, AR = 6.30 (Turchetto et al. 2015); P. integrifolia subsp. depauperata, AR = 4.30 (Silva-Arias et al. 2017)], suggesting that the reduced variability in P. mantiqueirensis is not a common characteristic of endemic species in the Petunia genus.
Calibrachoa elegans presented even lower genetic diversity indices compared to other Calibrachoa species as well as P. mantiqueirensis. The four micro-endemic highland species, C. eglandulata, C. sendtneriana, C. serrulata and C. spathulata, which have population sizes and geographical ranges similar to C. elegans, possess greater genetic diversity indices for both plastid and nuclear markers (John et al. 2019), whereas based on plastid diversity, the widespread species C. sellowiana (π = 0.088 %; J. N. Fregonezi et al., unpubl. data) and C. heterophylla (π = 0.41 %; Mäder et al. 2013) also have greater genetic diversity than C. elegans.
The diversification of highland species in Petunia and Calibrachoa has been discussed based on allopatric speciation as the result of isolation and fragmentation from a widespread ancient population during interglacial periods when forest species advanced on the open fields. The consecutive cycles of expansion and contraction of grasslands and forests, which changed the geographical distribution of the species, probably led to population fragmentation and possible local adaptation and population differentiation. More so than the north-east limit for the genera distribution, P. mantiqueirensis and C. elegans are the species that reached the highest elevation. Their restricted distribution probably corresponds to relics of a once widespread population that was fragmented by forest expansion during interglacial periods in the Pleistocene (Behling 2002), which suggests their requirement for a cold habitat and dispersion during cooler periods of the Pleistocene, compatible with previous findings for highlands petunias (Lorenz-Lemke et al. 2010). As commonly expected for rare and endemic plant species (Cole 2003), P. mantiqueirensis and C. elegans display low levels of genetic diversity, which is consistent with long periods as small and isolated populations.
The single haplotype found for P. mantiqueirensis is shared with P. altiplana and P. bonjardinensis (Lorenz-Lemke et al. 2010) and a very short genetic distance was observed between this and remained haplotypes found in other highlands species, reinforcing the recent diversification hypothesis for this group. Although low genetic diversity was found based on nuclear data, no indication of inbreeding was observed, which is consistent with the status of being self-incompatible and bee-pollinated proposed for P. mantiqueirensis (F. F. Araújo, UFMG, pers. comm.).
Petunia mantiqueirensis features a strict relationship with its bee pollinator (Pseudagaspostemon fluminensis) that forages pollen on plants returning at the same plot between 5 and 13 days after the first visit (F. F. Araújo, UFMG, pers. comm.). The maximum foraging distance for this genus of small and solitary bees is a maximum of 300 m (Gathmann and Tscharntke 2002; Zurbuchen et al. 2010). The distance among populations of P. mantiqueirensis ranges from ~0.9 to 14.2 km, and it is unlikely that this small bee promotes pollen flow between different patches, restricting it to an inside-patch flow. Thus, considering the absence of inbreeding and the improbability of gene flow among populations, a widely spread ancestor population that became recently fragmented is the most probable explanation for the observed genetic diversity of this species.
The reduced pollen flow among fragmented, isolated and discrete populations composed of relatively few individuals and the restricted seed dispersal are common characteristics of the majority of Petunia species (Stehmann et al. 2009) that can contribute to a depletion of overall genetic diversity over evolutionary time (Ellstrand and Elam 1993).
Calibrachoa elegans is an example of a rare species with habitat specificity, occurring only in four known populations. The severe environmental conditions (rigorous rainfall regime with 4 months of dry season) combined with the oscillations in the size of the species distribution areas based on the past modelling distribution may be a probable explanation for the lack of variation in C. elegans. Despite C. elegans being described as self-incompatible and bee-pollinated (Stehmann and Semir 2001), a significant deficit of heterozygotes and inbreeding values account for the distance among populations. Two main reasons can explain these results: mates preferentially occur between related individuals and/or self-incompatibility breaks. Self-incompatibility can be transitional in some populations dependent on environmental or biological conditions, like pollinator availability or even pollen sources (Nason and Hamrick 1997; Dobeš et al. 2017), or by fixation of mutations that result in non-self-incompatibility (Nasrallah et al. 2004). In addition, the positioning of reproductive organs in C. elegans is favourable to selfing.
We found stronger genetic differentiation among C. elegans populations than among populations of P. mantiqueirensis. Calibrachoa elegans is pollinated by Hexantheda missionica bees (Stehmann and Semir 2001), a species in which females collect pollen and nectar and effectively pollinate flowers, whereas males only use the flowers as a source of nectar, a place to land between patrol flights in search of females for mating, and as night shelter. This pollinator behaviour combined with the distance among plant patches can lead to low pollen flow and consequently strong differentiation and isolation.
Here, we add genetic information to several other criteria (CNCFlora) to confirm the endangered status for these two species, P. mantiqueirensis and C. elegans: their small and fragmented local populations, habitat specificity, restricted geographical range and low genetic diversity. Our results suggest a strong association between rarity and low genetic diversity related to the evolutionary history of these species. Species with a restricted distribution that are rare and with low diversity can be susceptible to depression because of the inbreeding and loss of natural genetic variability. Genetic drift tends to further diminish genetic diversity (Ellegren and Ellegren 2016; Thurman and Barrett 2016) resulting in allele fixation because of the low effective size. This phenomenon is more common on the edge of species distributions, where expansion and contraction are common (Eckert et al. 2008; Excoffier et al. 2009; Angert 2014; Hargreaves and Eckert 2014). Our findings are in accordance with this.
As in P. secreta (Turchetto et al. 2016), the main risk to P. mantiqueirensis and C. elegans is the fragmented habitat, population size and anthropization of their environment. These two species are adapted to specific habitats and changes in their environment can lead these species to extinction. They are currently threatened and their preservation in situ is necessary, especially because they are suffering from habitat loss. In an extreme situation, an ex situ intervention could be carried out, with the creation of a seed bank to re-introduce the plant to their natural environment. Through this contrasting pattern of rare species, which is associated with evolutionary history, it is important to produce detailed studies evaluating the genetic diversity of these species to create an efficient conservation plan for these two species that represent live relics from a past distribution of their respective genera.
Sources of Funding
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and the Programa de Pós Graduação em Genética e Biologia Molecular da Universidade Federal do Rio Grande do Sul (PPGBM-UFRGS). A.B. was recipient of National Institutes for Science and Technology (INCT) in Ecology, Evolution and Biodiversity Conservation scholarship. G.M. was supported by PNPD-CAPES/PPGBot-UFRGS and C.T. was supported by PNPD-CAPES/PPGBM-UFRGS.
Contributions by the Authors
A.B. and L.B.F. planned, designed and led the project; A.B., G.M., J.N.F. conducted the experiments; A.B, C.T., A.L.A.S., G.M. and S.L.B. ran the analyses; A.B. and L.B.F. wrote most of the text; L.B.F. provided reagents and equipment to develop the experiments. All authors contributed in the preparation of the study and have commented on and approved the final manuscript.
Supporting Information
The following additional information is available in the online version of this article—
Table S1. Petunia mantiqueirensis and Calibrachoa elegans collection points.
Table S2. Characteristics and PCR conditions of the microsatellite loci amplified for Petunia mantiqueirensis and Calibrachoa elegans.
Table S3. Prior values (minimum and maximum with uniform distribution) for the parameters employed for the four demographic scenarios (Fig. 2) with the DIYABC approach. Effective sizes are in number of individuals and times are in number of generations (generation time of 1 year).
Table S4. Additional information of approximate Bayesian computation (ABC) analyses.
Table S5. Geographic coordinates of Petunia mantiqueirensis and Calibrachoa elegans occurrence records used in the ENM.
Table S6. Characterization of 10 microsatellites analysed on Petunia mantiqueirensis collection sites.
Table S7. Pairwise FST values estimated for seven Petunia mantiqueirensis collection sites. (*) Significant values (P < 0.01).
Table S8. Characterization of five microsatellites analysed on Calibrachoa elegans collection sites.
Table S9. AICc scores and area under the receiver operating characteristic curve (AUC) values of the tested models.
Table S10. Data availability: Individual allele per microsatellite loci.
Figure S1. Clustering of individuals according to a discriminant analysis of principal components (DAPC) scatterplot performed based on SSR genotypes for: (A) seven populations of Petunia mantiqueirensis and (B) four populations of Calibrachoa elegans.
Supplementary Material
Acknowledgements
The authors thank P.D. Togni for assistance with sequencing and J.R. Stehmann for help in plant identification and collection.
Conflict of Interest
None declared.
Literature Cited
- Angert LA. 2014. Demography of central and marginal populations of monkey flowers (Mimulus cardinalis and M. lewisii). Ecology 87:2014–2025. [DOI] [PubMed] [Google Scholar]
- Barros MJ, Silva-Arias GA, Fregonezi JN, Turchetto-Zolet AC, Iganci JR, Diniz-Filho JAF, Freitas LB. 2015. Environmental drivers of diversity in subtropical highland grasslands. Perspectives in Plant Ecology, Evolution and Systematics 17:360–368. [Google Scholar]
- Beheregaray LB. 2008. Twenty years of phylogeography: the state of the field and the challenges for the Southern Hemisphere. Molecular Ecology 17:3754–3774. [DOI] [PubMed] [Google Scholar]
- Behling H. 2002. South and southeast Brazilian grasslands during late Quaternary times: a synthesis. Palaeogeography, Palaeoclimatology, Palaeoecology 177:19–27. [Google Scholar]
- Behling H, Bauermann SG, Neves PCP. 2001. Holocene environmental changes in the São Francisco de Paula region, southern Brazil. Journal of South American Earth Sciences 14:631–639. [Google Scholar]
- Bertorelle G, Benazzo A, Mona S. 2010. ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Molecular Ecology 19:2609–2625. [DOI] [PubMed] [Google Scholar]
- Bossolini E, Klahre U, Brandenburg A, Reinhardt D, Kuhlemeier C. 2011. High resolution linkage maps of the model organism Petunia reveal substantial synteny decay with the related genome of tomato. Genome 54:327–340. [DOI] [PubMed] [Google Scholar]
- Bridle JR, Vines TH. 2006. Limits to evolution at range margins: when and why does adaptation fail? Trends in Ecology & Evolution 22:140–147. [DOI] [PubMed] [Google Scholar]
- Cazé ALR, Mäder G, Nunes TS, Queiroz LP, de Oliveira G, Diniz-Filho JAF, Bonatto SL, Freitas LB. 2016. Could refuge theory and rivers acting as barriers explain the genetic variability distribution in the Atlantic forest? Molecular Phylogenetics and Evolution 101:242–251. [DOI] [PubMed] [Google Scholar]
- Cole CT. 2003. Genetic variation in rare and common plants. Annual Review of Ecology, Evolution, and Systematics 34:213–237. [Google Scholar]
- Collevatti RG, Rabelo SG, Vieira RF. 2009. Phylogeography and disjunct distribution in Lychnophora ericoides (Asteraceae), an endangered Cerrado shrub species. Annals of Botany 104:655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collevatti RG, Terribile LC, Rabelo SG, Lima-Ribeiro MS. 2015. Relaxed random walk model coupled with ecological niche modeling unravel the dispersal dynamics of a Neotropical savanna tree species in the deeper Quaternary. Frontiers in Plant Science 6:653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuet JM, Pudlo P, Veyssier J, Dehne-Garcia A, Gautier M, Leblois R, Marin JM, Estoup A. 2014. DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30:1187–1189. [DOI] [PubMed] [Google Scholar]
- Dawson TP, Jackson ST, House JI, Prentice IC, Mace GM. 2011. Beyond predictions: biodiversity conservation in a changing climate. Science 332:53–58. [DOI] [PubMed] [Google Scholar]
- Dobeš C, Konrad H, Geburek T. 2017. Potential population genetic consequences of habitat fragmentation in Central European forest trees and associated understory species - an introductory survey. Diversity 9:9. [Google Scholar]
- Dunn IS, Blattner FR. 1987. Charons 36 to 40: multi enzyme, high capacity, recombination deficient replacement vectors with polylinkers and polystuffers. Nucleic Acids Research 15:2677–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert CG, Samis KE, Lougheed SC. 2008. Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Molecular Ecology 17:1170–1188. [DOI] [PubMed] [Google Scholar]
- Elith J, Leathwick JR. 2009. Species distribution models: ecological explanation and prediction across space and time. Annual Review of Ecology, Evolution and Systematics 40:677–697. [Google Scholar]
- Ellegren H, Ellegren N. 2016. Determinants of genetic diversity. Nature Reviews Genetics 17:422–433. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Elam DR. 1993. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics 24:217–242. [Google Scholar]
- Estoup A, Jarne P, Cornuet JM. 2002. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Molecular Ecology 11:1591–1604. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Foll M, Petit RJ. 2009. Genetic consequences of range expansions. Annual Review of Ecology, Evolution, and Systematics 40:481–501. [Google Scholar]
- Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10:564–567. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding AH, Bell JF. 1997. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environmental Conservation 21:38–49. [Google Scholar]
- Fregonezi JN, Freitas LB, Bonatto SL, Semir J, Stehmann JR. 2012. Infrageneric classification of Calibrachoa (Solanaceae) based on morphological and molecular evidence. Taxon 61:120–130. [Google Scholar]
- Fregonezi JN, Turchetto C, Bonatto SL, Freitas LB. 2013. Biogeographical history and diversification of Petunia and Calibrachoa (Solanaceae) in the Neotropical Pampas grassland. Botanical Journal of the Linnean Society 171:140–153. [Google Scholar]
- Galante PJ, Alade B, Muscarella R, Jansa SA, Goodman SM, Anderson RP. 2018. The challenge of modeling niches and distributions for data‐poor species: a comprehensive approach to model complexity. Ecography 41: 726–736. [Google Scholar]
- Gathmann A, Tscharntke T. 2002. Foraging ranges of solitary bees. Journal of Animal Ecology 71:757–764. [Google Scholar]
- Gitzendanner MA, Soltis PS. 2000. Patterns of genetic variation in rare and widespread plant congeners. American Journal of Botany 87:783–792. [PubMed] [Google Scholar]
- Goudet J. 1995. FSTAT version 1.2: a computer program to calculate F-statistics. Journal of Heredity 86:485–486. [Google Scholar]
- Hamilton MB. 1999. Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Molecular Ecology 8:521–523. [PubMed] [Google Scholar]
- Hamrick JL, Godt MJW. 1996. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society B: Biological Sciences 351:1291–1298. [Google Scholar]
- Hargreaves AL, Eckert CG. 2014. Evolution of dispersal and mating systems along geographic gradients: implications for shifting ranges. Functional Ecology 28:5–21. [Google Scholar]
- Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405:907–913. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, van Etten J. 2010. RASTER: package for reading, writing, and manipulating RASTER (grid) type geographic (spatial) data https://cran.r-project.org/web/packages/raster/index.html.
- John ALW, Mäder G, Fregonezi JN, Freitas LB. 2019. Genetic diversity and population structure of naturally rare Calibrachoa species with small distribution in southern Brazil. Genetics and Molecular Biology 42:108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T. 2008. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics 11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology 16:1099–1106. [DOI] [PubMed] [Google Scholar]
- Kamino LHY, Siqueira MF, Sánchez-Tapia A, Stehmann JR. 2012. Reassessment of the extinction risk of endemic species in the Neotropics: how can modelling tools help us. Natureza & Conservação 10:191–198. [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne S, Mouquet N, Thuiller W, Ronce O. 2010. Biodiversity and climate change: integrating evolutionary and ecological responses of species and communities. Annual Review in Ecology, Evolution and Systematics 41:321–350. [Google Scholar]
- Ledru MP. 1993. Late Quaternary environment and climatic changes in Central Brazil. Quaternary Research 39:90–98. [Google Scholar]
- Ledru MP, Rousseau DD, Cruz FW Jr, Riccomini C, Karmann I, Martin L. 2005. Paleoclimate changes during the last 100,000 yr from a record in the Brazilian Atlantic Rainforest region and interhemispheric comparison. Quaternary Research 64:444–450. [Google Scholar]
- Lorenz-Lemke AP, Mäder G, Muschner VC, Stehmann JR, Bonatto SL, Salzano FM, Freitas LB. 2006. Diversity and natural hybridization in a highly endemic species of Petunia (Solanaceae): a molecular and ecological analysis. Molecular Ecology 15:4487–4497. [DOI] [PubMed] [Google Scholar]
- Lorenz-Lemke AP, Togni PD, Mäder G, Kriedt RA, Stehmann JR, Salzano FM, Bonatto SL, Freitas LB. 2010. Diversification of plant species in a subtropical region of eastern South American highlands: a phylogeographic perspective on native Petunia (Solanaceae). Molecular Ecology 19:5240–5251. [DOI] [PubMed] [Google Scholar]
- Mäder G, Fregonezi JN, Lorenz-Lemke AP, Bonatto SL, Freitas LB. 2013. Geological and climatic changes in quaternary shaped the evolutionary history of Calibrachoa heterophylla, an endemic South-Atlantic species of Petunia. BMC Evolutionary Biology 13:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall TC, Slate J, Kruuk LE, Pemberton JM. 1998. Statistical confidence for likelihood-based paternity inference in natural populations. Molecular Ecology 7:639–655. [DOI] [PubMed] [Google Scholar]
- Nason JD, Hamrick JL. 1997. Reproductive and genetic consequences of forest fragmentation: two case studies of Neotropical canopy trees. Journal of Heredity 88:264–276. [Google Scholar]
- Nasrallah ME, Liu P, Sherman-Broyles S, Boggs NA, Nasrallah JB. 2004. Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proceedings of the National Academy of Sciences of the United States of America 101:16070–16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmstead RG, Bohs L, Migid HA, Santiago-Valentin E, Garcia VF, Collier SM. 2008. A molecular phylogeny of the Solanaceae. Taxon 57:159–1181. [Google Scholar]
- Peterson AT, Papes M, Soberón J. 2008. Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecology Modeling 213:63–72. [Google Scholar]
- Pfeifer M, Schatz B, Picó FX, Passalacqua NG, Fay MF, Carey PD, Jeltsch F. 2009. Phylogeography and genetic structure of the orchid Himantoglossum hircinum (L.) Spreng. across its European central–marginal gradient. Journal of Biogeography 36:2353–2365. [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE. 2006. Maximum entropy modelling of species geographic distributions. Ecology Modeling 190:231–259. [Google Scholar]
- Pinheiro F, de Barros F, Palma‐Silva C, Fay MF, Lexer C, Cozzolino S. 2011. Phylogeography and genetic differentiation along the distributional range of the orchid Epidendrum fulgens: a Neotropical coastal species not restricted to glacial refugia. Journal of Biogeography 10:1923–1935. [Google Scholar]
- Ramos AC, Lemos-Filho JP, Ribeiro RA, Santos FR, Lovato MB. 2007. Phylogeography of the tree Hymenaea stigonocarpa (Fabaceae: Caesalpinioideae) and the influence of Quaternary climate changes in the Brazilian Cerrado. Annals of Botany 100:1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Fregonezi AM, Fregonezi JN, Cybis GB, Fagundes NJ, Bonatto SL, Freitas LB. 2015. Were sea level changes during the Pleistocene in the South Atlantic Coastal Plain a driver of speciation in Petunia (Solanaceae)? BMC Evolutionary Biology 15:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Kortmann M, Silva-Arias GA, Segatto AL, Mäder G, Bonatto SL, de Freitas LB. 2014. Multilocus phylogeny reconstruction: new insights into the evolutionary history of the genus Petunia. Molecular Phylogenetics and Evolution 81:19–28. [DOI] [PubMed] [Google Scholar]
- Reck-Kortmann M, Silva-Arias GA, Stehmann JR, Greppi JA, Freitas LB. 2015. Phylogenetic relationships of Petunia patagonica (Solanaceae) revealed by molecular and biogeographical evidence. Phytotaxa 222:17–32. [Google Scholar]
- Rodrigues DM, Caballero-Villalobos L, Turchetto C, Assis-Jacques R, Kuhlemeier C, Freitas LB. 2018. Do we truly understand pollination syndromes in Petunia as much as we suppose? AoB Plants 10:ply057; doi: 10.1093/aobpla/ply057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Frascaria N, MacKay J, Bousquet J. 1992. Segregating random amplified polymorphic DNAs (RAPDs) in Betula alleghaniensis. Theoretical and Applied Genetics 85:173–180. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497. [DOI] [PubMed] [Google Scholar]
- Safford HD. 2007. Brazilian Páramos IV. Phytogeography of the campos de altitude. Journal of Biogeography 34:1701–1722. [Google Scholar]
- Sang T, Crawford D, Stuessy T. 1997. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). American Journal of Botany 84:1120. [PubMed] [Google Scholar]
- Särkinen T, Bohs L, Olmstead RG, Knapp S. 2013. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evolutionary Biology 13:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan H, Moser M, Klahre U, Esfeld K, Dell’Olivo A, Mandel T, Metzger S, Vandenbussche M, Freitas L, Kuhlemeier C. 2016. MYB-FL controls gain and loss of floral UV absorbance, a key trait affecting pollinator preference and reproductive isolation. Nature Genetics 48:159–166. [DOI] [PubMed] [Google Scholar]
- Silva-Arias GA, Mäder G, Bonatto SL, Freitas LB. 2015. Novel microsatellites for Calibrachoa heterophylla (Solanaceae) endemic to the South Atlantic Coastal Plain of South America. Applications in Plant Sciences 3:1500021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Arias GA, Reck-Kortmann M, Carstens BC, Hasenack H, Bonatto SL, Freitas LB. 2017. From inland to the coast: spatial and environmental signatures on the genetic diversity in the colonization of the South Atlantic Coastal Plain. Perspectives in Plant Ecology Evolution and Systematics 28:47–57. [Google Scholar]
- Skirycz A, Castilho A, Chaparro C, Carvalho N, Tzotzos G, Siqueira JO. 2014. Canga biodiversity, a matter of mining. Frontiers in Plant Science 5:653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehmann JR, Lorenz-Lemke AP, Freitas LB, Semir J. 2009. The genus Petunia. In: Gerats T, Strommer J, eds. Petunia: evolutionary, developmental and physiological genetics. New York: Springer, 1–28. [Google Scholar]
- Stehmann JR, Semir J. 2001. Reproductive biology of Calibrachoa elegans (Miers) Stehmann & Semir (Solanaceae). Brazilian Journal of Botany 24:43–49. [Google Scholar]
- Thode VA, Silva-Arias GA, Turchetto C, Segatto ALA, Mäder G, Bonatto SL, Freitas LB. 2014. Genetic diversity and ecological niche modelling of the restricted Recordia reitzii (Verbenaceae) from southern Brazilian Atlantic forest. Botanical Journal of the Linnean Society 176:332–348. [Google Scholar]
- Thurman TJ, Barrett RD. 2016. The genetic consequences of selection in natural populations. Molecular Ecology 25:1429–1448. [DOI] [PubMed] [Google Scholar]
- Turchetto C, Fagundes NJ, Segatto AL, Kuhlemeier C, Solís Neffa VG, Speranza PR, Bonatto SL, Freitas LB. 2014. Diversification in the South American Pampas: the genetic and morphological variation of the widespread Petunia axillaris complex (Solanaceae). Molecular Ecology 23:374–389. [DOI] [PubMed] [Google Scholar]
- Turchetto C, Segatto ALA, Beduschi J, Bonatto SL, Freitas LB. 2015. Genetic differentiation and hybrid identification using microsatellite markers in closely related wild species. AoB Plants 7:plv084; doi: 10.1093/aobpla/plv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchetto C, Segatto ALA, Mäder G, Rodrigues DM, Bonatto SL, Freitas LB. 2016. High levels of genetic diversity and population structure in an endemic and rare species: implications for conservation. AoB Plants 8:plw002; doi: 10.1093/aobpla/plw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchetto-Zolet AC, Salgueiro F, Turchetto C, Cruz F, Veto NM, Barros MJ, Segatto AL, Freitas LB, Margis R. 2016. Phylogeography and ecological niche modelling in Eugenia uniflora (Myrtaceae) suggest distinct vegetational responses to climate change between the southern and the northern Atlantic Forest. Botanical journal of the Linnean Society 182:670–688. [Google Scholar]
- Warren DL, Glor RE, Turelli M. 2010. ENMTolls: a toolbox for comparative studies of environmental niche models. Ecography 33:607–611. [Google Scholar]
- Warren DL, Seifert SN. 2011. Ecological niche modeling in Maxent: the importance of model complexity and the performance of model selection criteria. Ecological Applications 21:335–342. [DOI] [PubMed] [Google Scholar]
- Zhang DX, Hewitt GM. 2003. Nuclear DNA analyses in genetic studies of populations: practice, problems and prospects. Molecular Ecology 12:563–584. [DOI] [PubMed] [Google Scholar]
- Zurbuchen A, Landert L, Klaiber J. 2010. Maximum foraging ranges in solitary bees: only few individuals have the capability to cover long foraging distances. Biological Conservation 143:669–676. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.