Abstract
ATP-competitive Fibroblast Growth Factor Receptor (FGFR) kinase inhibitors, including BGJ398 and Debio1347, show antitumor activity in patients with intrahepatic cholangiocarcinoma (ICC) harboring activating FGFR2 gene fusions. Unfortunately, acquired resistance develops and is often associated with the emergence of secondary FGFR2 kinase domain mutations. Here, we report that the irreversible pan-FGFR inhibitor, TAS-120, demonstrated efficacy in four patients with FGFR2-fusion-positive ICC who developed resistance to BGJ398 or Debio1347. Examination of serial biopsies, circulating tumor DNA (ctDNA), and patient-derived ICC cells revealed that TAS-120 was active against multiple FGFR2 mutations conferring resistance to BGJ398 or Debio1347. Functional assessment and modeling the clonal outgrowth of individual resistance mutations from polyclonal cell pools mirrored the resistance profiles observed clinically for each inhibitor. Our findings suggest that strategic sequencing of FGFR inhibitors, guided by serial biopsies and ctDNA, may prolong the duration of benefit from FGFR inhibition in patients with FGFR2 fusion-positive ICC.
Keywords: cholangiocarcinoma, FGFR, drug resistance, clinical trials, disease models
INTRODUCTION
Intrahepatic cholangiocarcinoma (ICC) is an aggressive malignancy of the liver bile ducts with poor outcomes and rising incidence (1). Most patients are diagnosed with locally advanced or metastatic disease, precluding potentially curative resection. Standard of care palliative chemotherapy with gemcitabine and cisplatin offers these patients a median survival of less than one year (2). ICCs exhibit an array of genomic alterations of known oncogenic drivers and tumor suppressors, suggesting the potential of targeted therapies in subsets of patients (3–6). Recurrent genomic alterations that activate the FGFR pathway are present in ~20% of ICCs (3, 6–12). The most common alterations are chromosomal fusions consisting of FGFR2 exons 1 to 17, encoding the intact extracellular and kinase domains, fused in-frame to a 3′ partner that possesses a protein dimerization domain. The resulting chimeric FGFR2 proteins are constitutively active and promote proliferation or transformation of several cell types (6, 7, 9). The frequency of FGFR2 fusions in ICC is considerably higher than that reported for any other malignancy (13)(data retrieved from http://www.cbioportal.org). Activating FGFR2 point mutations and amplification or overexpression of FGFR1–3 are also observed in subsets of patients with ICC (8, 14).
Multiple FGFR-selective inhibitors are being tested in clinical trials in patients with ICC with FGFR pathway alterations. These second-generation inhibitors represent an improvement over the early generation of multi-kinase inhibitors with activity against FGFR (e.g. dovitinib and ponatinib), which lack sufficient specificity and potency to effectively treat FGFR-driven tumors. The most clinically advanced FGFR-selective compound in cholangiocarcinoma is the ATP-competitive FGFR1–3 inhibitor, BGJ398 (infigratinib), which demonstrated efficacy in a phase II trial of patients with advanced refractory cholangiocarcinoma harboring FGFR fusions, amplifications, or point mutations (14). The overall response rate (ORR) in this heavily pretreated patient population was 14.8% and the disease control rate (DCR) was 75.4% (18.8% and 83.3%, respectively, for patients with FGFR2 fusions only). A Phase 1 dose-escalation trial using another ATP-competitive FGFR1–3 inhibitor, Debio1347 (CH5183284) (15), has also reported early evidence of antitumor activity in a few tumor types including ICC (16). However, rapid emergence of acquired resistance was frequently observed, with a 5.8-month median progression-free survival in the BGJ398 trial (14). We recently reported genomic characterization of pre- and post-progression cell-free circulating tumor DNA (ctDNA) and tumor biopsies in three patients with FGFR2 fusion-positive ICCs treated with BGJ398; this study revealed the emergence of the FGFR2 V565F gatekeeper mutation at progression in all three patients, two of whom also had additional FGFR2 kinase domain mutations (17). Rapid autopsy in one patient revealed three different FGFR2 kinase domain mutations in spatially distinct metastases, highlighting the additional challenge of inter-lesional heterogeneity in addressing acquired resistance to an ATP-competitive FGFR inhibitor in ICC.
The third-generation, irreversible FGFR inhibitor TAS-120 covalently binds to a highly conserved P-loop cysteine residue in the ATP pocket of FGFR (C492 in the FGFR2-IIIb isoform) (18). TAS-120 exhibits in vitro potency at low nanomolar concentrations and high specificity against wild-type FGFR1–4 as well as against some FGFR2 kinase domain mutations (19). Preliminary results from a phase I basket study of TAS-120 in patients with refractory advanced solid tumors showed an ORR of 25.0% and a DCR of 78.6% in 28 patients with ICC harboring FGFR2 fusions (20), including some patients who had received prior therapy with an ATP-competitive FGFR inhibitor.
Here, we report the results of clinical and translational studies of TAS-120 in the treatment of patients with FGFR2 fusion-positive ICC who progressed on BGJ398 or Debio1347, including patients in whom secondary FGFR2 kinase mutations were detected just prior to TAS-120 initiation. We performed complementary studies investigating FGFR2-mediated signaling mechanisms in ICC models and determined the efficacy of these second- and third-generation FGFR inhibitors against clinically observed FGFR2 kinase domain mutations. Our findings reveal genotype-phenotype correlations for drug sensitivity that inform personalized targeted therapy in FGFR-activated ICC.
RESULTS
TAS-120 provides clinical benefit in patients with ICC with acquired resistance to BGJ398 or Debio1347
Among six patients with advanced FGFR2 fusion-positive ICC who received care at our institution after progression on BGJ398 or Debio1347 in clinical trials, four subsequently enrolled in the phase I trial of TAS-120 () between November 2015 and November 2017. Each of the four patients showed benefit on TAS-120: two of these patients achieved a partial response and two achieved stable disease by RECIST v1.1 criteria (Figure 1A) with a duration of benefit of 5.1 to 17.2 months. We highlight these patients to show proof of concept of an irreversible FGFR inhibitor overcoming acquired resistance to an ATP-competitive FGFR inhibitor in the clinic and to elucidate the potential molecular determinants of response for this observation. The patients’ clinical characteristics and FGFR2 gene alterations are summarized in Table 1A and 1B. No additional cancer-relevant genomic alterations were detected in the pre-treatment biopsies, with the exception of copy number increases of the FGFR1 and MYC loci in the biopsy from patient #3 (see Methods for specific genotyping assays used for the different samples).
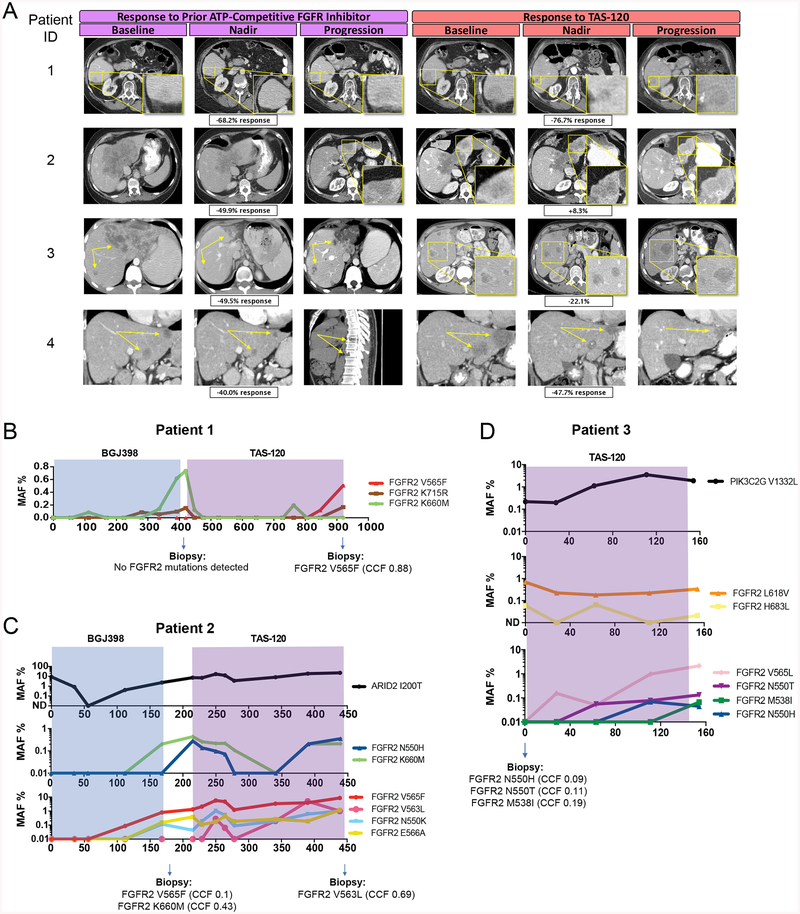
Figure 1. TAS-120 is clinically effective in FGFR2 fusion-positive ICC patients whose tumors acquired resistance to BGJ398 or Debio1347.
A, Radiologic scans of patients 1–4 during the course of FGFR inhibitor therapy.
B-D, ddPCR analysis of serial ctDNA samples from patients 1–3. Time periods of therapy with the specific FGFR inhibitors are indicated by shading. MAF: mutant allele frequency. Mutations identified in tumor biopsies taken at the indicated times are presented at the bottom of each graph. CCF: cancer cell fraction.
Table 1A.
Clinical characteristics and outcomes of patients with FGFR2 fusion-positive cholangiocarcinoma receiving FGFR inhibitors.
| Patient ID | FGFR2 Fusion | 1st FGFR Inhibitor | PFS (Months) | BOR | Intervening Therapies Between 1st and 2nd FGFR inhibitor | Interval Between 1st and 2nd FGFR inhibitor (Months) | 2nd FGFR Inhibitor | PFS (Months) | BOR |
|---|---|---|---|---|---|---|---|---|---|
| 1 | FGFR2-SORBS1 | BGJ398 | 12.6 | −68.2% | None | 1.2 | TAS-120 | 15.8 | −76.7% |
| 2 | FGFR2-ZMYM4 | BGJ398 | 5.6 | −49.9% | None | 1.6 | TAS-120 | 7.2 | +8.3% |
| 3 | FGFR2-INA | Debio1347 | 11.4 | −49.5% | Gemcitabine/Docetaxel, T11 palliative radiation | 3.0 | TAS-120 | 5.1 | −22.1% |
| 4 | FGFR2-NRAP | BGJ398 | 7.1 | −40.0% | T8 palliative radiation, Pembrolizumab, Resection of T8 metastasis, FOLFOX | 7.4 | TAS-120 | 17.2 | −47.7% |
PFS = Progression Free Survival, BOR = Best Overall Survival
Table 1B.
Detection of FGFR2 mutations in ctDNA and tumor biopsies
| Patient ID | FGFR2 Fusion | Post-Progression BGJ398/Debio1347, Prior to TAS-120 | Post-Progression TAS-120 | ||
|---|---|---|---|---|---|
| cfDNA | Tumor Biopsy | cfDNA | Tumor Biopsy | ||
| 1 | FGFR2-SORBS1 | K660M, K715R | None Detected | V565F& | V565F&& |
| 2 | FGFR2-ZMYM4 | V565F, K660M, E566A, N550H, N550K | V565F&&, K660M&& | V565F, K660M, E566A, N550H, N550K, V563L | V563L |
| 3 | FGFR2-INA | H683L&, L618V& | Biopsy #1: H683L Biopsy #2: N550H, N550T, M538I |
V565L, E566A, N550H, L618V, N550T&, M538I& | No Biopsy Obtained |
| 4 | FGFR2-NRAP | None Detected | No Biopsy Obtained | N550K | N550K |
All mutations were detected on CLIA-certified assays as a routine part of clinical care except those designated with an &(detected on ddPCR only) or &&(detected on WES only).
Patient 1 is a 74-year-old female with recurrent FGFR2-SORBS1 fusion-positive ICC metastatic to her liver and lymph nodes. On third-line BGJ398 treatment, she achieved a maximum response of −68% followed by progression of all three liver lesions at approximately 12 months. ctDNA analysis at that time revealed two new FGFR2 kinase domain mutations, K660M and K715R (Figure 1B) (amino acids are numbered according to FGFR2-IIIb splice isoform [NM_001144913.1] since FGFR2 fusions in ICC are expressed in this context (21); the equivalent mutations in the one amino acid shorter IIIc isoform are K659M and K714R). Biopsy of a single liver lesion at the time of progression showed no FGFR2 kinase domain mutations, suggesting that these mutations were subclonal or that other molecular mechanisms drove resistance in this lesion. The patient subsequently received TAS-120, which resulted in a maximum response of −77% and suppression of K660M and K715R below the level of detection in ctDNA. After nearly 16 months on TAS-120, she had progression in all liver lesions. A third FGFR2 mutation, the gatekeeper V565F, emerged in the ctDNA during the final months of TAS-120 treatment and was detected in a post-progression tumor biopsy.
Patient 2 is a 59-year-old female with a FGFR2-ZMYM4 fusion-positive ICC who presented with a dominant 15 cm liver mass and metastases to her liver and lungs. She achieved a maximum response of −50% on second-line BGJ398 treatment. Scans at 6 months showed a mixed response with regression of the dominant mass and progression of satellite liver lesions. ctDNA analysis at that time revealed five mutations in the FGFR2 kinase domain (N550H, N550K, V565F, E566A, and K660M). Two of these mutations were observed in a tumor biopsy of a progressing satellite liver lesion obtained in parallel — V565F and K660M, as previously reported (17)(amino acid numbering is updated here to reflect expression of the FGFR2-IIIb splice isoform [NM_001144913.1]). Upon next line TAS-120 treatment, she achieved stable disease with a best response of +8%. Progression occurred at approximately 7 months, with a mixed response consisting of rebound growth of a previously responsive lung lesion, stability of the dominant mass, and continued progression of the biopsied left lobe liver lesion. While the spatial location of each mutation was unknown, this heterogeneous response to TAS-120 was reflected in ctDNA analysis where levels of some mutations (N550H, K660M) dropped below the level of detection before eventually rebounding at the time of disease progression, and others stabilized (N550K, E566A) or increased (V565F) during therapy (Figure 1C). A sixth FGFR2 mutation (V563L) emerged in ctDNA during TAS-120 therapy and was detected in a biopsy obtained upon disease progression.
Patient 3 is a 28-year-old male with Crohn’s disease and FGFR2-INA fusion-positive ICC who presented with a 5.4 cm liver mass concurrently with liver, lung, peritoneal, and lymph node metastases. He received second-line Debio1347 treatment to a maximum response of −50% followed by disease progression at all sites at nearly 12 months. He then had two post-progression liver biopsies obtained 2.5 months apart on distinct liver lesions with intervening cytotoxic chemotherapy — the first revealed an FGFR2 H683L mutation (CCF=0.23) and the second revealed three FGFR2 mutations (N550H, CCF=0.093; N550T, CCF=0.108; and M538I, CCF=0.19). TAS-120 treatment was initiated immediately after this second biopsy, and ctDNA analysis of plasma collected at this baseline timepoint revealed one of these five mutations (H683L) and one additional mutation (L618V). The patient achieved a maximum response of −22% on TAS-120 treatment and exhibited disease progression at 5.1 months with a mixed response in the liver and growth of lung and bone lesions. ctDNA analysis during treatment showed a modest decline of L618V and H683L levels (Figure 1D). As the tumor progressed, ctDNA analysis revealed the gradual emergence of mutations seen on baseline biopsy (N550H, N550T, M538I) and other previously undetectable mutations (V565L, E566A).
Patient 4 is a 46-year-old male with chronic hepatitis B and recurrent metastatic FGFR2-NRAP fusion-positive ICC involving his liver. Second-line BGJ398 led to a maximum response of −40% but at approximately 7 months, scans showed a mixed response with continued tumor shrinkage in the liver and emergence of osseous metastases. No ctDNA sample or tumor biopsy was available immediately post-progression to assess for mechanisms of resistance. He received palliative spinal radiation, pembrolizumab, T8 metastasectomy, and FOLFOX, with progression after each of these treatments. The patient then initiated TAS-120 with a 7-month interval between FGFR inhibitors. Analysis of ctDNA just prior to receiving TAS-120 did not reveal any detectable molecular alterations, potentially reflecting low levels of shedding of tumor DNA. On TAS-120, this patient achieved a maximum response of −48%, although this benefit could not be correlated with the ability of the drug to overcome specific resistance mechanisms. The patient eventually experienced growth of a single liver lesion at 17.2 months, and at that time, analysis of ctDNA and tumor biopsy demonstrated the emergence of FGFR2 N550K (Table 1B).
These findings extend our prior observations that acquired resistance to FGFR inhibition in ICC is associated with the emergence of multiple, heterogeneous tumor subclones harboring distinct secondary FGFR2 kinase domain mutations. Importantly, in this setting, TAS-120 demonstrated marked clinical benefit, highlighting the critical dependence of these tumors on sustained FGFR signaling and pointing to the importance of these FGFR2 kinase domain mutations as a common mechanism of clinical acquired resistance to FGFR inhibition. Collectively, the assessment of clonal dynamics in ctDNA suggests that TAS-120 has differential activity against individual FGFR2 secondary mutations compared to ATP-competitive FGFR inhibitors. Understanding the spectrum of activity of various FGFR inhibitors against commonly observed acquired FGFR2 mutations may lead to strategies to overcome or delay resistance.
FGFR signaling is critical for MEK/ERK activity and viability in FGFR+ ICC models
In order to study FGFR-driven signaling and examine candidate resistance mutations in a biologically relevant context, we developed a panel of patient-derived biliary tract cancer cell lines and tested these and established biliary tract cancer lines for response to FGFR inhibitors. Treatment of these cell lines with BGJ398 revealed that ICC13–7 and CCLP-1 cells were highly sensitive (IC50 5–15 nM), whereas the other lines tested were resistant (IC50 200–3000 nM) (Figure 2A). Similar profiles were seen in response to the more potent TAS-120 compound, with ICC13–7 and CCLP1 cells showing increased sensitivity (IC50, 0.6–1.5 nM) compared to the rest of the lines (IC50, 300–8000 nM) (Figure 2B). Accordingly, immunoblot analysis of lysates from 11 ICC cell lines and of immortalized bile duct cells (MMNK-1) showed that only ICC13–7 and CCLP1 cells had detectable levels of phosphorylated Fibroblast Growth Factor Receptor Substrate 2 (pFRS2 Y196), consistent with constitutive FGFR signaling (Supplementary Figure S1A). Genomic analysis revealed that ICC13–7 cells harbored an FGFR2-OPTN fusion (Supplemental Figure S1B), whereas all other cell lines lacked FGFR fusions. Moreover, while CCLP-1 cells lacked fusions, intragenic mutations, or copy number gains of FGFR genes, they showed greatly increased expression of wild type FGFR1 (IIIc isoform) as well as the FGF20 ligand compared to the other cell lines analyzed (Supplementary Figure S1C–E). Thus, biliary tract cancer cell lines with activating molecular alterations in the pathway are specifically dependent on FGFR signaling for growth in vitro.
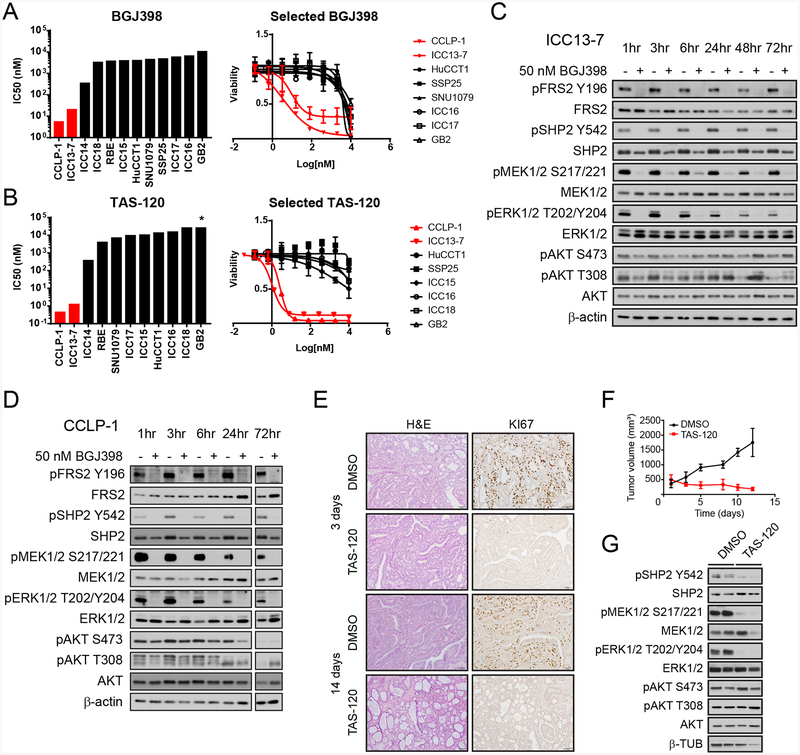
Figure 2. FGFR-activated ICC models show FGFR2-dependent growth and MEK/ERK signaling in vivo and in vitro.
A. Graph of IC50 data and dose response curves for BGJ398 in biliary tract cancer cell lines that show constitutive FGFR activation (red) or lack FGFR activity (black). p < 0.0002 for IC50 difference.
B. Graph of IC50 data and dose response curves for TAS-120 in biliary tract cancer cell lines. p < 0.002 for FGFR-activated versus non-FGFR-activated lines. * denotes IC50 was not reached.
C and D, Immunoblot of signaling effects of 50 nM BGJ398 treatment versus vehicle control in ICC13–7 cells (C) and CCLP-1 cells (D). Cells were treated for the indicated times before harvesting.
E-G. Fragments of an ICC PDX harboring an FGFR2-KIAA1217 fusion were implanted in NSG mice. Mice were randomized for treatment with TAS-120 (25 mg/kg) or vehicle once tumors reached ~500 mm3. E, Histologic images (H&E staining) and measurement of proliferation (Ki67 staining) of tumors isolated at the indicated times. F, Serial measurement of tumor volumes. G, Immunoblot data showing signaling inhibition upon TAS-120 treatment (samples are from 14 days treatment).
FGFR signaling engages a series of downstream effectors in different normal and pathologic contexts (22). We examined the principle pathways controlled by FGFR signaling in the ICC13–7 and CCLP-1 cell lines by BGJ398 treatment and immunoblot analysis using phospho-specific antibodies. BGJ398 treatment (50 nM) led to rapid inhibition of the MEK/ERK pathway as reflected by decreased pFRS2 (Y196), pSHP2 (Y542), pMEK1/2 (S217/221), and pERK1/2 (T202/Y204), whereas minimal effects were observed on the PI3K pathway, as determined by pAKT (T308 and S473) (Figure 2C, D). Dose-response studies showed effective targeting of FGFR2 signaling and downstream inhibition of MEK/ERK at BGJ398 concentrations consistent with the cell viability IC50 data (Supplemental Figure S1F); comparable data were seen for TAS-120 and Debio1347. In many types of cancer, strong feedback mechanisms exist to restore MEK/ERK signaling in response to loss of upstream activators of the pathway (23), and these may limit benefit of certain therapeutics that involve MEK/ERK inhibition. Notably, the inhibition of MEK/ERK signaling was durable in both cell lines, with no evidence of pathway reactivation for up to 3 days for BGJ398 treatment (Figure 2C, D).
To corroborate these results in vivo, we screened a collection of patient-derived xenograft (PDX) models of ICC for FGFR alterations, and identified a model harboring a FGFR2-KIAA1217 fusion (designated MG69) (Supplemental Figure S1G). Treatment of MG69 PDX tumors with TAS-120 (starting when the volume reached ~500 mm3) led to tumor regression and complete proliferative arrest, with prominent effects evident within three days and persisting over a 14-day course (Figure 2E, F). Moreover, FGFR inhibition suppressed MEK/ERK and SHP2 activity, but not PI3K signaling, in MG69 PDX tumors (Figure 2G). Thus, FGFR activated ICC models are highly dependent on FGFR activity to sustain growth and maintain MEK/ERK signaling in vitro and in vivo.
TAS-120 overcomes multiple clinically observed FGFR kinase domain mutations
To gain insight into the clinical landscape of secondary FGFR2 resistance mutations, we subsequently leveraged our FGFR-driven ICC cell line models to study the spectrum of FGFR2 kinase domain mutations emerging upon clinical acquired resistance to BGJ398 (N550K, V565F, E566A, K660M, and K715R) or Debio1347 (M538I, H683L), or to both (N550H, L618V). We engineered these mutations into a retroviral vector expressing the FGFR2-PHGDH fusion, which we observed in an ICC (see Methods). CCLP-1 cells were infected with retroviruses expressing the FGFR2-PHGDH fusion with a wild type or mutant FGFR2 kinase domain or empty vector control. Of the mutations that arose in patients treated with BGJ398, N550K, L618V, and K660M resulted in prominent resistance to the drug in vitro (25- to 39-fold increase in IC50), with the V565F gatekeeper conferring the greatest level of resistance (326-fold) (Figure 3A, top panel; Supplementary Figures S2A and B show immunoblots for expression of the FGFR2 fusions and crystal violet staining of cells at a single drug concentration). The N550H and E566A mutants caused weaker effects (7- to 8-fold) and K715R did not affect BGJ398 sensitivity. The latter variant involves a residue located outside the BGJ398 binding pocket and not implicated in the conformational dynamics of the kinase (17), and thus may not represent a functionally relevant mutation. Finally, BGJ398 remained effective against the M538I and H683L mutations (3- to 4-fold increase in IC50), which were found in the setting of clinical resistance to Debio1347 treatment and have not been observed clinically upon BGJ398 therapy.
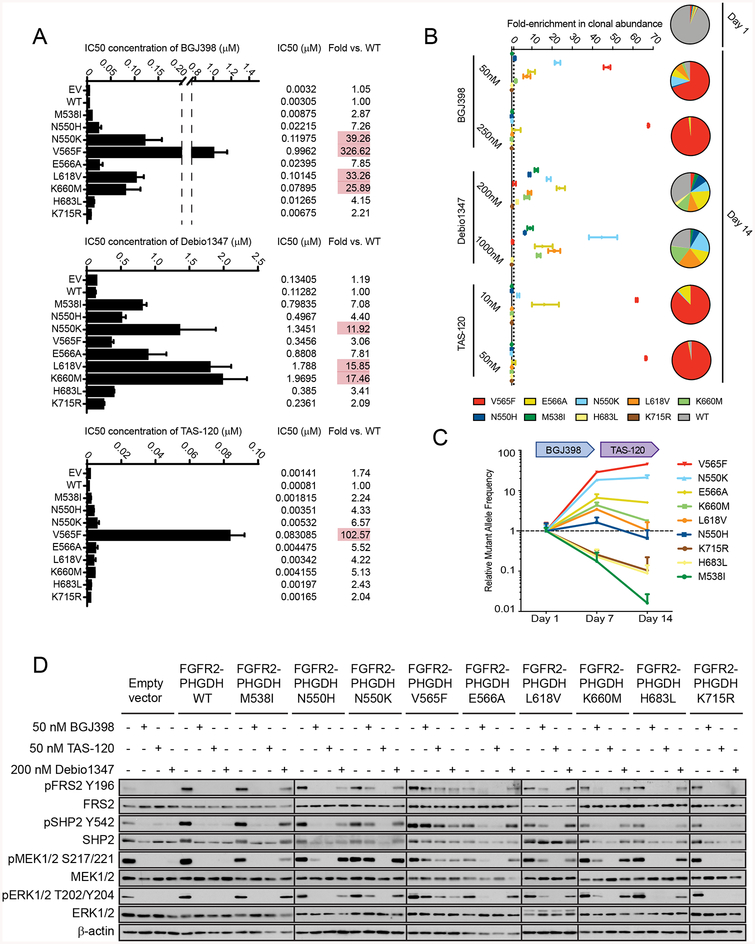
Figure 3. FGFR inhibitors have distinct activity profiles against secondary FGFR2 kinase mutations in ICC cell lines that correlate with clinical data.
A-D, CCLP-1 cells were engineered by retroviral transduction to express the FGFR2-PHGDH fusion with a wild type kinase domain or harboring the indicated mutations, or empty vector. The fusions contain the FGFR2-IIIb splice isoform [NM_001144913.1], and the amino acids are numbered accordingly. A, Graphs of IC50 measurements upon treatment with the indicated FGFR inhibitors. The measured IC50 is also indicated numerically at the right along with the fold-change in IC50 of each cell line relative to cell lines expressing the WT fusion. Red shading highlight mutants conferring a greater than 10-fold increase in IC50. B, Pooled CCLP-1 cell clones of all FGFR2 fusion variants were treated with BGJ398, Debio1347, or TAS-120 at the indicated concentrations over 14 days. The individual clones were monitored using genomic DNA extracted at 14 days, using a ddPCR assay specific to each mutation. Data are mean ± SEM of triplicate determinants of relative change in clonal abundance compared with the start of treatment and are generated from two independent experiments. C, Clonal pools as in (B) were treated sequentially with 50 nM BGJ398 and 10nM TAS-120 to mimic the treatment course of patients. Cells were monitored at 7 and 14 days. Data are expressed in relative mutant allele frequency compared with the start of treatment. Data are mean ± SD of triplicate determinants of relative change in clonal abundance compared with the start of treatment and are generated from two independent experiments. D, Immunoblot of CCLP-1 cells expressing the different FGFR2-PHGDH alleles following treatment with the indicated inhibitor concentrations.
Debio1347 had a distinct profile of sensitivity (Figure 3A, middle panel). The magnitude of resistance provoked by the different mutants was lower than that observed for BGJ398, although this drug is considerably less potent against FGFR signaling overall. The most pronounced resistance to Debio1347 was seen with the N550K, L618V, and K660M mutations (12- to 17-fold increase in IC50), while M538I, N550H, and E566A produced intermediate effects (4- to 8-fold), H683L had a modest effect, and K715R did not significantly affect responsiveness to the drug. Moreover, Debio1347 was relatively effective against the V565F gatekeeper mutation (only 3-fold IC50 increase). Notably, TAS-120 showed only minimal or modest changes in activity against each of the acquired FGFR2 mutations (2- to 7-fold IC50 increase) with the exception of V565F (103-fold) (Figure 3A, bottom panel).
To extend these findings, we modeled clonal outgrowth during acquired resistance using a pooled clone system, in which all nine mutant clones were pooled at an initial abundance of 1% amidst a background of cells expressing the WT FGFR2 fusion (Figure 3B and Supplementary Figure S2C). Clonal pools were exposed to different concentrations of each FGFR inhibitor for 14 days, and the change in relative clonal abundance under the selective pressure of therapy was determined by ddPCR (24). Outgrowth of K715R was not observed under any treatment condition, again suggesting that this mutation is not a functional resistance alteration. Notably, treatment with 50 nM BGJ398 led to outgrowth of the resistance mutations observed in patients 1 and 2 (N550H, N550K, V565F, E566A, K660M) or previously observed (17) in the setting of BGJ398 resistance (e.g. L618V). By contrast. BGJ398 prevented the outgrowth of M538I detected only in Patient 3 who was treated with Debio1347. Conversely, outgrowth of each of these mutations was observed upon treatment with 200 nM Debio1347, with the exception of V565F, consistent with the clinical course of Patient 3. Finally, in the presence of 10 nM TAS-120 only outgrowth of V565F, and to a lesser extent, E566A, and N550K, were observed. Of note, these were the same three mutations that did not decrease in abundance in Patient 2 during TAS-120 therapy (Figure 1C). Interestingly, higher concentrations of BGJ398 or TAS-120 were able to suppress outgrowth of all resistance mutations with the exception of V565F, highlighting the potential importance of drug exposure in suppressing resistant clones.
We next used the pooled clone system to model the effects of sequential FGFR inhibitor therapy, treating clonal pools sequentially with BGJ398 and then TAS-120 to mirror the clinical course of Patients 1, 2, and 4 (Figure 3C). Three of the mutations (K660M, N550H, and L618V) that emerged during BGJ398 treatment decreased in abundance when treatment was switched to TAS-120, consistent with our ctDNA analyses showing that TAS-120 led to decreases in the clonal abundance in K660M (Patient 1 and 2), N550H (in Patient 2), and L618V (in Patient 3). Conversely, V565F continued to increase and E566A and N550K levels stabilized, but failed to decrease upon TAS-120 treatment, similar to the clinical observations in ctDNA from Patient 2. Thus, our model systems accurately mirrored the clonal dynamics of individual resistance mutations observed in ctDNA analysis from patients treated with TAS-120 after progression on BGJ398 or Debio1347.
Signaling studies corroborated the cell viability findings. CCLP-1 cells expressing N550K, V565F, L618V, and K660M retained robust levels of pFRS2, pSHP2, pMEK, and pERK upon treatment with 50 nM BGJ398, whereas signaling by the other mutants was inhibited partially (N550H, E566A) or strongly (H683L) (Figure 3D and Supplemental Figure S2A). Treatment with TAS-120 (50 nM) effectively suppressed signaling by all mutants except V565F. Finally, Debio1347 (200 nM) showed reduced potency against most of the mutants but remained relatively active against the V565F gatekeeper mutation compared to the other two inhibitors. All three inhibitors were effective against K715R. We confirmed our findings for a subset of the FGFR2 mutants in ICC13–7 cells via cell viability assays and immunoblot for signaling proteins (Supplemental Figure S2D–F). Thus, we demonstrate in relevant in vitro ICC models that TAS-120 has activity against multiple secondary FGFR2 resistance mutations, which likely accounts for the benefit of TAS-120 seen in patients who previously progressed on BGJ398 or Debio1347.
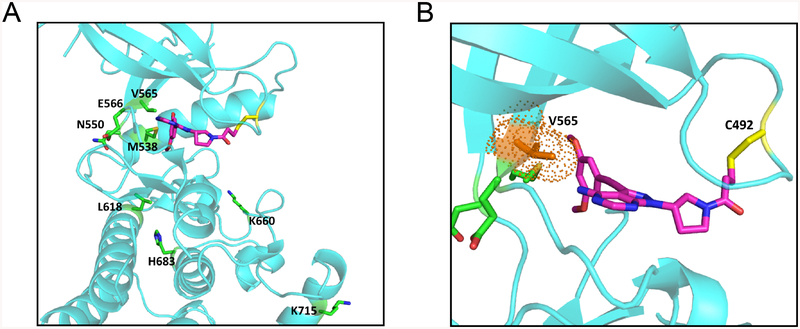
We conducted in silico structural modeling to gain insight into the molecular basis for the drug response profiles. TAS-120 docks into the ATP binding pocket of FGFR2, with its acrylamide group forming a covalent bond with the sulfhydryl group of FGFR2-C492 (Figure 4A and B). As with BGJ398 (17, 25), the dimethoxy phenyl group of TAS-120 is in close contact with the V565 gatekeeper residue. Accordingly, modeling data indicate that TAS-120 and BGJ398 resistance to V565F is due to steric clash preventing access of these drugs into the ATP-binding pocket. TAS-120 remains effective against V565I (19), likely due to less severe hindrance caused by the smaller isoleucine side chain. Debio1347 lacks the bulky dimethoxy phenyl group, and rather possesses a benzimidazole moiety predicted to have stabilizing contacts with V565F, which may account for its relative potency against FGFR2 V565F (15). Notably, TAS-120 retained activity against several mutations that confer BGJ398 and Debio1347 resistance by altering conformational dynamics of FGFR2 rather than directly interacting with mutated residues. In particular, N550H/K and E566A stabilize the active conformation of the kinase by disrupting a network of hydrogen bonds that serve as an autoinhibitory molecular break, K660M forces the A loop of the kinase into an active conformation, and L618V disrupts stabilizing interactions between this residue and an Asp–Phe–Gly (DFG) motif that otherwise favors binding of BGJ398 and Debio1347 (17, 26). Thus, BGJ398 and Debio1347 appear not to act on the active kinase conformation, whereas the covalent binding mode of TAS-120 may permit effective target engagement irrespective of conformation, as observed for the irreversible pan-FGFR inhibitor, FIIN-2 (27). Finally, the specific impairment of Debio1347 activity versus FGFR2 M538I may relate to interactions with the adjacent M539 residue that contribute to the binding of this drug. Overall, the distinct structural features and binding modes of these FGFR inhibitors are in keeping with their specific activity profiles suggested by the clinical data and observed in preclinical models. A recent report defining the binding mode of TAS-120 with FGFR1 based on mass spectrometry and X-ray crystallography analyses is in line with our in silico structural modeling study (18).
Figure 4. Structural modeling of secondary FGFR2 kinase domain mutations with TAS-120.
A, Model showing TAS-120 docked into ATP-binding pocket of wild type FGFR2. Amino acid residues corresponding to mutations conferring resistance to ATP competitive FGFR inhibitors are highlighted. Structural representations were prepared using PyMOL.
B, A close-up view of TAS-120 in ATP-binding pocket of wild type FGFR2. The gatekeeper residue (V565) is in close proximity to dimethoxy phenyl group of TAS-120.
DISCUSSION
In this study, we report that the irreversible FGFR inhibitor, TAS-120, can overcome acquired resistance to the ATP-competitive inhibitors, BGJ398 and Debio1347, and provide clinical benefit in patients with advanced refractory FGFR2 fusion-positive ICC previously treated with these agents. We also find that the spectrum of secondary FGFR2 resistance mutations differs across agents and that structural studies of these agents bound to FGFR provide a molecular basis for these differences. Finally, we demonstrate that preclinical ICC models with activation of the pathway are specifically dependent on FGFR signaling for growth and sustained SHP2/MEK/ERK signaling, and that TAS-120 retains efficacy against FGFR2 kinase domain mutations in this setting. Collectively, these data highlight the FGFR-driven oncogene addiction of a defined subset of ICC and support the clinical utility of TAS-120 in patients with acquired resistance to second generation FGFR inhibitors.
The efficacy seen across several early phase clinical trials of FGFR2 inhibitors in patients with advanced refractory ICC (14, 28–30) represents a breakthrough in a disease with no FDA-approved targeted therapies to date. However, as seen with other tyrosine kinase inhibitors, the rapid emergence of resistance associated with recurrent acquired mutations in the target’s kinase domain has limited the durability of benefit to ATP-competitive inhibitors. TAS-120 was designed to overcome FGFR kinase domain mutations, taking advantage of the improved potency and specificity afforded by its covalent binding mode and distinct orientation in the ATP-binding pocket of FGFRs. This irreversible binding also permanently disables FGFR2 enzymatic activity, thus providing the potential advantage of extended pharmacodynamic duration without the need for maintaining high drug levels. Covalent small molecule kinase inhibitors have demonstrated success in multiple malignancies and have gained FDA approval in EGFR mutant non-small cell lung cancer (afatinib, osimertinib), ERBB2/HER2 mutant breast cancer (neratinib), and BTK mutant chronic lymphocytic leukemia, Waldenstrom’s macroglobulinemia, and mantle cell lymphoma (ibrutinib)(31).
We evaluated the efficacy of TAS-120, BGJ398, and Debio1347 against the spectrum of nine clinically observed secondary FGFR2 kinase domain mutations using ICC cell lines and serial ctDNA analysis. The inhibitors exhibit unique in vitro profiles, and the key findings included: a) the mutations that conferred greatest resistance to BGJ398 were N550K, V565F, L618V, and K660M; b) the mutations that conferred greatest resistance to Debio1347 were N550K, L618V, and K660M, c) Debio1347 largely retained activity against the V565F gatekeeper mutation; and d) TAS-120 remained active against all mutations except V565F, with modest reduction in activity against E566A and N550K. Additional studies will be required to determine the impact of these kinase domain mutations on FGFR2 fusion protein stability and turnover and also on the kinetics of signaling re-activation upon inhibitor withdrawal. Moreover, it will be important to establish the extent to which pre-existing FGFR2 mutations impact time to treatment failure, as observed in EGFR mutant non-small-cell lung cancer (32).
The clonal dynamics observed with serial ctDNA analysis may hold important implications for the clinical management of patients with these resistance alterations. ddPCR analysis of ctDNA showed that the mutation allele frequencies for several FGFR2 mutations decreased upon TAS-120 treatment — K660M in patient 1, N550H and K660M in patient 2, and L618V and H683L in patient 3 — pointing to the activity of TAS-120 against these alleles in the clinic. Similarly, the sustained increase or emergence of V565F upon TAS-120 in patients 1–3 is consistent with the in vitro resistance studies, as was the lack of reduction in levels of E566A and N550K. These data, if validated prospectively in larger clinical cohorts, may provide support for a new paradigm in which particular FGFR resistance mutations, detected in serial ctDNA or tumor biopsies, could inform the choice of subsequent FGFR targeted therapies. The precedent for this is emerging in advanced ALK fusion positive NSCLC where specific ALK kinase domain mutations that arise at the time of crizotinib resistance determine which second-generation inhibitor should be used for next line treatment (33). To guide such strategies in ICC, it will be important to also establish the full spectrum of mechanisms of resistance to TAS-120, including validating the functional impact of V563L, which emerged upon progression on TAS-120 treatment in patient 2. Notably, whereas resistance to other irreversible kinase inhibitors frequently arises due to mutations of cysteine residues that mediate covalent binding (34, 35), no mutations at the covalent binding site of TAS-120 (C492) were identified in any of the four patients studied in the present report.
A key challenge in the administration of pan-FGFR inhibitors remains hyperphosphatemia-related dose holds and dose reductions. One patient in this study had such a dose hold on TAS-120 (see Methods) and two had such dose holds on BGJ398. Hyperphosphatemia is a class effect of FGFR inhibitors arising from on-target pathway blockade of FGF23-FGFR1 signaling in the renal tubule (22, 36). Notably, we found that clonal outgrowth of multiple mutations occurred more readily at lower concentrations of BGJ398 (Fig. 3B), highlighting that reduced drug exposure may play an important role in the emergence of resistance. While further studies are needed to establish the impact of toxicity-related drug modifications on treatment response, the clinical experience highlights the importance of aggressive hyperphosphatemia management and the urgency to develop FGFR2-selective agents.
While ctDNA analysis serves as a useful, non-invasive tool for diagnosing resistance and monitoring response to therapy, our studies also illustrate the importance of a comprehensive approach to studying drug resistance. In patient 3, the three FGFR2 mutations identified in the baseline TAS-120 liver biopsy sample went undetected by both targeted sequencing and ddPCR in the corresponding plasma sample, possibly reflecting low tumor shedding and emphasizing the complementary benefits of tumor biopsy and ctDNA analysis. In patient 1, two FGFR2 kinase domain mutations arose at the time of progression on BGJ398 but only one conferred resistance in functional modeling, underscoring the importance of functionally validating putative resistance mutations discovered on ctDNA or tumor tissue analysis.
In keeping with the clinical data, our preclinical studies demonstrate that ICC models with constitutive activation of FGFR signaling are strongly dependent on the pathway. FGFR inhibitor treatment of FGFR-driven ICC cell lines and a PDX model led to growth inhibition as well as potent and durable inactivation of the SHP2/MEK/ERK pathway. Unlike breast and gastric cancers with high level FGFR2 amplification (37), FGFR signaling in these ICC models was not additionally coupled to the PI3K/AKT pathway. These findings highlight the distinct signaling outputs of oncogenic FGFR signaling in different cancer contexts and point to SHP2/MEK/ERK as a likely major effector of the pathway in ICC. The data are consistent with SHP2/MEK/ERK activation being the principle effector of FGFR in normal physiology (38) and with the frequent presence of concurrent PIK3CA activating mutations with FGFR2 fusions in ICC indicating the potential independence of these pathways (14). While our studies suggested that FGFR2 fusions with different partners had comparable outputs, further studies will be required to fully address the potential differential impact of N-terminus partners on oncoprotein localization, inhibitor sensitivity, and downstream signaling targets, as reported for fusions involving the ROS1 RTK (39).
In summary, we demonstrate that strategically sequencing FGFR inhibitors can prolong the duration of benefit from FGFR inhibition in patients with FGFR2 fusion-positive ICC. Moreover, resistance profiles differ across agents and may evolve under the selective pressure of sequential FGFR inhibitors. However, FGFR2-independent resistance may emerge as an additional issue that can limit the potential of these next-generation inhibitors. As the clinical development of FGFR inhibitors pushes forward, it will be critical to incorporate tumor biopsies at baseline and progression and serial ctDNA analysis into clinical trials. These complementary analyses can facilitate our understanding of resistance and elucidate the biology underlying heterogeneous responses. Overall, this approach of tailoring FGFR-targeted strategies based upon resistance mechanisms detected in serial ctDNA and tumor biopsies may provide a new standard of care for this disease.
METHODS
Patients
Patients provided written informed consent to treatment on the phase II trial of BGJ398 (), phase I trial of Debio1347 (), and phase 1 trial of TAS-120 (). On the phase II trial of BGJ398, the patients received 125mg orally daily on days 1 to 21 of each 28-day cycle. On the phase I trial of Debio1347, the patient received 110 mg orally daily continuously on days 1 to 28 of each 28-day cycle. On the phase I dose escalation or expansion phase of the trial of TAS-120, the patients received 16 to 24 mg orally daily of TAS-120 continuously on days 1 to 21 of each 21-day cycle, and all dose reductions and safety assessments were performed per protocol for all 3 trials. The TAS-120 dosing for each patient was as follows: Patient #1 (16mg); Patient #2 (24mg); Patient #3 (20mg); and Patient #4 (16mg). The timing, reason, and duration of the first dose hold for each patient is as follows: Patient #1 (Cycle 14, Day 1; grade 3 motor neuropathy; 15 days); Patient #2 (Cycle 1, Day 8; hyperphosphatemia; 7 days); Patient #3 (Cycle 3, Day 1; Grade 2 ALT and AST elevation; 7 days); and Patient #4 (Cycle 14, Day 19; palliative XRT; 12 days). The timing, reason, and dosing for the first and subsequent dose reductions for each patient is as follows: Patient #1 (Cycle 15, Day 1, reduced to 12mg PO QD for grade 4 creatine kinase elevation and remained at this dose until progression); Patient #2 (Cycle 3, Day 1, reduced to 16mg PO QD due to hyperphosphatemia and again on Cycle 3, day 15 to 8mg PO QD due to hyperphosphatemia; remained at this dose until progression); Patient #3 (Cycle 3, Day 15, reduced to 16mg PO QD due to grade 2 AST and ALT elevation, remained at this dose until progression); and Patient #4 (no dose reductions).
Computed tomography and/or magnetic resonance imaging scans were performed at baseline and every 6 to 9 weeks to assess for tumor response by RECIST version 1.1 criteria. The clinical and genomic data relating Patient #2’s treatment prior to TAS-120 therapy have been reported previously (17). All biopsies, tumor specimens, and peripheral blood draws for plasma isolation were collected and analyzed in accordance with Institutional Review Board–approved protocols, to which patients provided written informed consent, and all studies were conducted in accordance with the Declaration of Helsinki.
Reporting of FGFR2 mutations
We report FGFR2 kinase domain mutations as the amino acid number of the FGFR2-IIIb splice isoform [NM_001144913.1], which is the primary isoform expressed in FGFR2 fusion-positive ICC (21), Commercial genotyping tests (e.g. Guardant) and our prior report (17) designate mutations using the one amino acid shorter FGFR2-IIIc isoform (NM_000141.4) as the reference sequence.
Targeted Sequencing of Tumor Tissue
DNA derived from the primary tumor and metastases were analyzed using deep-coverage targeted sequencing of key cancer-associated genes. Targeted sequencing was performed in the setting of clinical care via the SNaPshot platform, the FoundationOne platform, or MSK-IMPACT and the methodology has been previously described (40, 41). Clinical genotyping platforms utilized on biopsies after progression on the ATP-competitive inhibitor were as follows: Patients #1 and #2 (FoundationOne), Patients #3 (MSK-IMPACT), and Patient #4 (no biopsy performed). After progression on TAS-120, the assays used on biopsies were as follows: Patient #1 (MGH SNaPshot), Patient #2 (FoundationOne), Patient #3 (no biopsy performed), and Patient #4 (MGH SNaPshot). In Patients #1, #2, and #4, no other cancer-relevant genomic alterations besides the FGFR2 fusion were detected in the treatment-naïve tissue sample taken at initial diagnosis or at diagnosis of advanced disease; MGH SNaPshot was used for Patient #1 and #2 and MSK IMPACT was used for patient #3 and #4.
Solid Fusion Assay
Our internal tumor-profiling assay was performed on RNA extracted from formalin-fixed paraffin-embedded (FFPE) specimens as part of routine clinical care. The SFA is a targeted RNA-sequencing method of Anchored Multiplex PCR to detect FGFR2 fusions, and the methodology has been previously described (42). Mutational profiling was performed at the Clinical Laboratory Improvement Amendments (CLIA)–certified Translational Research Laboratory at the Massachusetts General Hospital Cancer Center.
Droplet Digital PCR
DNA template (8 to 10 μL) was added to 10 μL of ddPCR Supermix for Probes (Bio-Rad) and 2 μL of the custom primer/probe mixture. This reaction mix was added to a DG8 cartridge together with 60 μL of Droplet Generation Oil for Probes (Bio-Rad) and used for droplet generation. Droplets were then transferred to a 96-well plate (Eppendorf) and then thermal cycled with the following conditions: 5 minutes at 95°C, 40 cycles of 94°C for 30 seconds, 55°C for 1 minute followed by 98°C for 10 minutes (Ramp Rate 2°C/sec). Droplets were analyzed with the QX200 Droplet Reader (Bio-Rad) for fluorescent measurement of FAM and HEX probes. Gating was performed based on positive and negative controls, and mutant populations were identified. The ddPCR data were analyzed with QuantaSoft analysis software (Bio-Rad) to obtain Fractional Abundance of the mutant DNA alleles in the wild-type/normal background. The quantification of the target molecule was presented as the number of total copies (mutant plus wild-type) per sample in each reaction. Fractional Abundance is calculated as follows: F.A. % = (Nmut/(Nmut+Nwt))*100), where Nmut is the number of mutant events and Nwt is the number of wildtype events per reaction. ddPCR analysis of normal control plasma DNA (from cell lines) and no DNA template controls were always included. Probe and primer sequences are available upon request.
Targeted Sequencing of Circulating Cell-Free Tumor DNA (ctDNA)
Cell-free DNA was extracted from whole blood, and 5 ng–30 ng of ctDNA was isolated. Sequencing libraries were prepared with custom in-line barcode molecular tagging, and complete sequencing at 15,000× read depth of the critical exons in a targeted panel of 70 genes was performed at a CLIA-certified, College of American Pathologists–accredited laboratory (Guardant Health)(43).
Whole exome sequencing (WES)
WES was performed by the Broad Institute sequencing platform. WES of matched pretreatment and post-progression biopsies and normal blood was performed as previously described (44).
Cell culture
Established cell lines were obtained from the following sources: Riken Bioresource Center (HuCCT1, RBE, SSP-25, HuH-28), Korean Cell Line Bank (SNU1079). We are grateful from the kind gifts of CCLP-1 and CCSW-1 cells from Dr. P.J. Bosma (Academic Medical Center, Amsterdam, the Netherlands), SG231 from Dr. A.J. Demetris (University of Pittsburgh, Pittsburgh, PA), and MMNK-1 from Dr. J. Luyendyk (University of Kansas Medical Center, Kansas City, KS). Cell lines were grown at 37°C under 5% CO2 in their required growth medium (Gibco) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. To generate patient-derived biliary tract cancer cell lines (ICC13–7, ICC14, ICC15, ICC16, ICC17, ICC18, GB2), we utilized resection or autopsy specimens directly or following growth as patient-derived xenografts, as per our Institutional Review Board approved protocol (DFCI, #13–162). Samples were minced with sterile razor blades, digested with trypsin for 30 minutes at 37°C, and then resuspended in RPMI supplemented with 20% fetal bovine serum, 1% L-glutamine (Gibco, #25030–081), 1% MEM Non-Essential Amino Acids Solution (Gibco, #11140–050), 1% Sodium Pyruvate (Gibco, #11360–070), 0.5% penicillin/streptomycin, 10 μg/mL gentamicin (Gibco, #15710–064), and 0.2 Units/mL human recombinant insulin (Gibco, #12585–014) and seeded on plates coated with rat tail collagen (BD Biosciences). Cells were passaged by trypsinization, adapted to RPMI supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin and transferred to uncoated tissue-culture plates prior to functional studies. They were routinely checked to be mycoplasma free. HuCCT1, RBE, SSP-25, HuH-28 and SNU1079 cells were authenticated by short tandem repeat (STR) DNA profiling by the cell line bank from which they were obtained. Authentication by STR DNA profiling through the ATCC was performed for CCLP-1, CCSW-1, SG231, MMNK-1, ICC2, and ICC4 (between December 2015 and March 2016) and ICC13–7, ICC14, ICC15, ICC16, ICC17, ICC18, and GB2 (January and April 2018). All cell lines were used within 20 passages of establishment from patients or receipt from repositories.
Generation of wild type and mutated FGFR2-PHGDH expressing cell lines
A FGFR2-PHGDH fusion construct, containing exons 1–17 of FGFR2-IIIb fused to PHGDH (NM_006623.3) exons 6–12, was amplified from reverse transcribed cDNAs from an ICC patient sample and inserted into the pMSCV vector using the NEBuilder HiFi DNA Assembly (New England Biolabs). All FGFR2 mutations were introduced into the pMSCV vector using the same kit. Targeted Sanger sequencing was done to confirm mutation generated. Retrovirus was generated by transfecting the pMSCV constructs and packaging plasmids into 293T cells. After collection of retrovirus, transfected 293T cells were collected to confirm protein expression from each construct. Viral infections of CCLP-1 and ICC13–7 cells were performed in the presence of polybrene. Infected cells were selected in blasticidin (6–15 ug/ml) for one to two weeks. For both cell lines, he period of time in culture between thawing, infection, selection, recovery, and experimental setup and completion was less than 10 passages.
Cell viability assay
For IC50 measurement using the FGFR inhibitors, cells were dissociated into single cells and seeded into a 384-well tissue culture plate, each well with 200 viable cells and 40μL of growth medium. After 24 hours, compounds were added to each well over a 15-point concentration range, along with DMSO controls, using a Tecan D300e digital drug dispenser. Cells were cultured for 5 days in the presence of compound before assessing viability by adding 15μL of Cell Titer-Glo (Promega) to each well, incubating for 20 minutes at room temperature on a shaker, and measuring luminescence using an Envision plate reader. Each condition was performed in 5 replicates, and each dose point was normalized to DMSO controls to estimate relative viability. At least 2 independent experiments were performed for each compound and cell line. IC50 values were determined by GraphPad Prism using a 4-parameter dose-response model. Crystal violet staining assays were done by seeding cells into 6-well plates one day before addition of drug. Cells were grown in the presence of drug for four days (CCLP-1 cells) or two weeks (ICC13–7 cells), then washed with PBS, fixed with cold methanol, stained with 0.5% crystal violet solution (Sigma-Aldrich), and washed extensively under tap water.
Immunoblot analysis
Cells were treated with drugs in 6-well plates for 5–8 hours or as indicated. Cell protein lysates were prepared in ice-cold RIPA lysis buffer (50mM Tris-HCl, pH 7.4, 150mM NaCl, 2mM EDTA, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, containing Roche protease inhibitors and Calbiochem phosphatase inhibitor cocktail set I and II). Debris was removed by centrifugation in a microfuge at max speed for 10 min at 4 C. Protein concentration in clarified lysate was determined by Pierce BCA protein assay (Thermo Fisher Scientific). Ten micrograms of protein were used to perform analysis by SDS-PAGE, electro-transfer and immunoblotting with specific antibodies. The following antibodies were used: from Cell Signaling Technology (all at 1:1000 dilution): phospho-FRS2 Y196 (3864S), SHP2 (3397S), phospho-MEK1/2 S217/221 (9154S), MEK1/2 (4694S), phospho-ERK1/2 T202/Y204 (9106S), ERK1/2 (4695S), phospho-AKT T308 (13038S), phospho-AKT S473 (4060S), AKT (9272S), FGFR1 (9740S); from Abcam (1:5000 dilution): FRS2 (ab183492), phospho-SHP2 Y542 (ab62322); from Sigma (1:20,000 dilution): b-actin (A5316).
Quantitative RT-PCR
Total RNA from cell lines was extracted using the RNeasy Mini Kit (Qiagen). RNA (1 μg) was reverse transcribed using SuperScript™ II Reverse Transcriptase (Invitrogen) or reagents from the QuantiTect Rev. Transcription Kit (Qiagen), according to the manufacturer’s instructions. Quantitative RT-PCR (qRT-PCR) was performed on a CFX384 Real-Time PCR Detection System (Bio-Rad) with iTaq Universal SYBR Green Supermix, 2– (Bio-Rad). FGFR1-3 and FGF20 were analyzed by qRT-PCR. Data was normalized for expression of the housekeeping gene ribosomal 18S. Primer sequences are provided in Supplementary Table S1.
PDX treatment studies
Mice were housed in pathogen-free animal facilities. All experiments were conducted under protocol 2005N000148 approved by the Subcommittee on Research Animal Care at Massachusetts General Hospital. To develop an FGFR2 fusion human PDX, we obtained tissue from a fresh resection specimen from a patient with an FGFR2-KIAA1217 fusion ICC tumor, per our IRB-approved protocol (DFCI# 13–162). The tissue was rinsed in HBSS and cut into 0.3–0.5 mm3 pieces with sterile razor blades. These tumor pieces were implanted subcutaneously into 6–8-week old female NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, 00557, The Jackson Laboratory). Tumor size was measured with a digital caliper. Upon reaching ~500 mm3, mice were randomized to either vehicle control or 25 mg/kg TAS-120 (in hydroxypropyl methyl cellulose solution) by oral gavage daily for three and fourteen days prior to harvest. Tumor samples were collected for biochemical analysis and histology processing. For histology processing, tissue samples were fixed overnight in 4% buffered formaldehyde, embedded in paraffin, and then sectioned and stained with haematoxylin and eosin by the MGH Pathology Core. Immunohistochemistry (IHC) was performed on paraffin-embedded sections (5 μ m thickness). After deparaffinization and dehydration, slides were incubated for 10 min with 3 % H2O2 at room temperature to block endogenous peroxidase activity. Specimens were brought to the boil in 10 mM sodium citrate buffer (pH 6.0, 5 min, pressure cooker) for antigen retrieval. Slides were blocked for 1 hour in TBS-0.05 % Tween 20 (Fisher Scientific), 1 drop per 1 ml of Protein Block (Dako X0909) and incubated with primary antibody for 1 hour at room temperature. Primary anti-Ki-67 antibody (Abcam, ab15580) was diluted in with PBS-Protein Block (1 drop/ml) at a ratio of 1:400 and incubated with the tissue sections at 4°C. Specimens were reacted for 30 min with the ImmPRESS HRP polymer reagent (Vector Labs) combined with secondary antibodies. Slides were then washed with PBS and stained for peroxidase for 1–2 min with the Betazoid DAB Chromogen reagent, washed with water and counterstained with haematoxylin. Stained slides were photographed with an Olympus DP72 microscope.
Population growth modeling with clonal pool system
Cell pools containing 1% of each mutant cell line and 90% of FGFR2-PHGDH WT cells were seeded at low confluency in 6-well plates. We used 1% as an empirically chosen concentration which allowed growth of cells in the presence of different FGFR inhibitors for 2-weeks without individual mutant clones overtaking the entire population. This percentage was also sufficiently high to enable clone detection by ddPCR. Every experimental condition was performed in triplicate. Two independent experiments were used to generate data for Figures 3B and C. Drug incubation (or DMSO-treated controls) started 24h after cell seeding and drug treatment was refreshed every 3–4 days. After 1, 7 or 14 days in culture, the remaining cells were trypsinized and collected for genomic DNA extraction. Genomic DNA extracted from cells using the DNeasy Blood and Tissue Kit (Qiagen) was subjected to enzymatic fragmentation with either MseI or HaeIII, and amplified using ddPCR Supermix for Probes (Bio-Rad) using FGFR2 assays (PrimePCR ddPCR Mutation Assay, Bio-Rad, and custom-designed). DNA template (20–40ng) was added to 12.5 μL of ddPCR Supermix for Probes (Bio-Rad) and 1.25 μL of the primer/probe mixture. This reaction mix (final volume = 25μL) was added to a DG8 cartridge together with 70 μL of Droplet Generation Oil for Probes (Bio-Rad) and used for droplet generation. Droplets were then transferred to a 96-well plate (Eppendorf) and then thermal cycled with the following conditions: 10 minutes at 95°C, 40 cycles of 94°C for 30 seconds, 55°C for 1 minute followed by 98°C for 10 minutes (ramp rate 2°C/second). Droplets were analyzed with the QX200 Droplet Reader (Bio-Rad) for fluorescent measurement of FAM and HEX probes. Gating was performed based on positive and negative controls, and mutant populations were identified. The ddPCR data were analyzed with QuantaSoft analysis software (Bio-Rad) to obtain fractional abundance of the mutant DNA alleles in the wild-type (WT)/normal background. The quantification of the target molecule was presented as number of total copies (mutant plus WT) per sample in each reaction. Fractional abundance is calculated as follows: F.A. % = (Nmut/(Nmut + Nwt)) × 100), where Nmut is number of mutant events and Nwt is number of WT events per reaction. The number of positive and negative droplets is used to calculate the concentration of the target and reference DNA sequences and their Poisson-based 95% confidence intervals, as previously described (45). Multiple replicates (minimum of three) were performed for each sample. ddPCR analysis of normal control genomic DNA from cell lines and no DNA template (water) controls was performed in parallel with all samples, including multiple replicates as contamination-free controls.
In Silico Structural Modeling
TAS-120 was docked into FGFR2 (PDBID: 1OEC). The loop structure (V488-V496) was modeled so that the S atom of C492 and the terminal carbon of acrylamide in TAS-120 made a covalent bond using the Molecular Operating Environment (MOE) from Chemical Computing Group (https://www.chemcomp.com/).
Supplementary Material
STATEMENT OF SIGNIFICANCE.
ATP-competitive FGFR inhibitors (BGJ398, Debio1347) show efficacy in FGFR2-altered ICC; however, acquired FGFR2 kinase domain mutations cause drug resistance and tumor progression. We demonstrate that the irreversible FGFR inhibitor TAS-120 provides clinical benefit in patients with resistance to BGJ398 or Debio1347 and overcomes several FGFR2 mutations in ICC models.
ACKNOWLEDGEMENTS
This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Cancer Research Program, under Award No. W81XWH-17-1-0491 (N.B., A.X.Z.) and Award No. W81XWH-16-1-0267 (L.Y.L.). Opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense. Additional support was provided by the following: MGH Fund for Medical Discovery Award (L.G., L.S.), Cholangiocarcinoma Foundation Andrea Lynn Scott Memorial Research Fellowship, American Cancer Society Institutional Research Grant, and NIH Loan Repayment Program (L.G.). Cholangiocarcinoma Foundation Christopher J. Wilke Memorial Research Fellowship (L.S). American Cancer Society Postdoctoral Fellowship PF-16-120-01-TBG (L.Y.L.). Hope Funds for Cancer Research Fellowship and Harvard Catalyst KL2/CMeRIT Fellowship (S.R.). TargetCancer Foundation, Evan Schumacher Fund for Rare Cancer Research (S.R, R.N). NIH/NCI 1K08CA194268–01 (S.K.S). The Starr Foundation I11–0040 (R.Y.). Memorial Sloan-Kettering Cancer Center Core Grant P30 CA 008748 (J.J.H, R.Y.). NCI U01 CA176058, U01 CA199253, U01 CA224146 (W.C.H.). HMS Laboratory for Systems Pharmacology Grant (P50GM107618) and the Susan Eid Tumor Heterogeneity Initiative (D.J.). U54CA224068 (R.B.C.). V Foundation for Cancer Research Translational Grant (N.B, C.H.B., L.G.). NCI SPORE P50 CA127003 (N.B., C.H.B, R.B.C). Gallagher Chair in Gastrointestinal Cancer Research and TargetCancer Foundation (N.B.).
Financial support: MGH Fund for Medical Discovery Award (L.G., L.S.), Cholangiocarcinoma Foundation Andrea Lynn Scott Memorial Research Fellowship, American Cancer Society Institutional Research Grant, and NIH Loan Repayment Program (L.G.). Cholangiocarcinoma Foundation Christopher J. Wilke Memorial Research Fellowship (L.S). American Cancer Society Postdoctoral Fellowship PF-16-120-01-TBG and the DOD Peer Reviewed Cancer Research Program Horizon Award W81XWH-16-1-0267 (L.Y.L.). Hope Funds for Cancer Research Fellowship and Harvard Catalyst KL2/CMeRIT Fellowship (S.R.). TargetCancer Foundation, Evan Schumacher Fund for Rare Cancer Research (S.R, R.N). NIH/NCI 1K08CA194268–01 (S.K.S). The Starr Foundation I11–0040 (R.Y.). Memorial Sloan-Kettering Cancer Center Core Grant P30 CA 008748 (J.J.H, R.Y.). NCI U01 CA176058, U01 CA199253, U01 CA224146 (W.C.H.). HMS Laboratory for Systems Pharmacology Grant (P50GM107618) and the Susan Eid Tumor Heterogeneity Initiative (D.J.). U54CA224068 (R.B.C.). V Foundation for Cancer Research (N.B, L.G, C.H.B.). DOD Translational Team Science Award W81XWH-17-1-0491 (N.B., A.X.Z). NCI SPORE P50 CA127003 (N.B., C.H.B, R.B.C). Gallagher Chair in Gastrointestinal Cancer Research and TargetCancer Foundation (N.B.).
Abbreviations list
- ctDNA
circulating tumor DNA
- DCR
disease control rate
- FGFR
Fibroblast Growth Factor Receptor
- FRS2
Fibroblast Growth Factor Receptor Substrate 2
- ICC
intrahepatic cholangiocarcinoma
- ORR
overall response rate
Footnotes
Conflict of interest disclosure statement: L.G. is a consultant/advisory board member for Debiopharm, H3 Biomedicine, Agios Pharmaceuticals, Taiho Pharmaceuticals, and Pieris Pharmaceuticals. T.S., S.O, and H.H. are employees of Tsukuba Research Institute, Taiho Pharmaceutical Co., Ltd. D.T.T. has received consulting fees from Merrimack Pharmaceuticals, Ventana Roche, and EMD Millipore Sigma, which are not related to this work. D.T.T. is a founder and has equity in PanTher Therapeutics, which is not related to this work. D.J. is a consultant/advisory board member for Novartis, EMD Serono, Natera and Eisai. W.C.H. is a consultant for Thermo Fisher, AjuIB, Paraxel, MPM Capital and is a founder and advisor to KSQ Therapeutics. R.K.R is a consultant/advisory board member of Agios Pharmaceuticals, Astra Zeneca, Bristol Myers Squibb, and Target Pharma Solutions (funding to institution) and Advisory Board/IDMC member of Genentech/Roche and Target Pharma Solutions (funding to individual). A.B. is a consultant/advisory board member for Biocartis, Roche and Guardant Health. A.X.Z. reports research funding from Bayer, Bristol-Myers Squibb, Eli Lilly, Merck, and Novartis, and is a consultant/advisory board member for AstraZeneca, Bayer, Bristol-Myers Squibb, Eisai, Eli Lilly, Exelixis, Merck, Novartis, and Sanofi. R.B.C. is a consultant/advisory board member for Amgen, Array Biopharma, Astex Pharmaceuticals, Avidity Biosciences, BMS, Chugai, Fog Pharma, Genentech, LOXO, Merrimack, N-of-one, Novartis, nRichDx, Roche, Roivant, Shire, Spectrum Pharmaceuticals, Symphogen, Taiho, and Warp Drive Bio; holds equity in Avidity Biosciences and nRichDx; and has received research funding from AstraZeneca and Sanofi. N.B. receives research funding from Taiho Pharmaceutical Co. Ltd.
REFERENCES
- 1.Valle JW, Lamarca A, Goyal L, Barriuso J, Zhu AX. New Horizons for Precision Medicine in Biliary Tract Cancers. Cancer Discov. 2017;7(9):943–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362(14):1273–81. [DOI] [PubMed] [Google Scholar]
- 3.Farshidfar F, Zheng S, Gingras MC, Newton Y, Shih J, Robertson AG, et al. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell reports. 2017;19(13):2878–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov. 2017;7(10):1116–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45(12):1470–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, et al. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47(9):1003–10. [DOI] [PubMed] [Google Scholar]
- 7.Arai Y, Totoki Y, Hosoda F, Shirota T, Hama N, Nakamura H, et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology. 2014;59(4):1427–34. [DOI] [PubMed] [Google Scholar]
- 8.Javle M, Bekaii-Saab T, Jain A, Wang Y, Kelley RK, Wang K, et al. Biliary cancer: Utility of next-generation sequencing for clinical management. Cancer. 2016;122(24):3838–47. [DOI] [PubMed] [Google Scholar]
- 9.Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3(6):636–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sia D, Losic B, Moeini A, Cabellos L, Hao K, Revill K, et al. Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nature communications. 2015;6:6087. [DOI] [PubMed] [Google Scholar]
- 11.Borad MJ, Champion MD, Egan JB, Liang WS, Fonseca R, Bryce AH, et al. Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS Genet. 2014;10(2):e1004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham RP, Barr Fritcher EG, Pestova E, Schulz J, Sitailo LA, Vasmatzis G, et al. Fibroblast growth factor receptor 2 translocations in intrahepatic cholangiocarcinoma. Hum Pathol. 2014;45(8):1630–8. [DOI] [PubMed] [Google Scholar]
- 13.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Javle M, Lowery M, Shroff RT, Weiss KH, Springfeld C, Borad MJ, et al. Phase II Study of BGJ398 in Patients With FGFR-Altered Advanced Cholangiocarcinoma. J Clin Oncol. 2018;36(3):276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakanishi Y, Akiyama N, Tsukaguchi T, Fujii T, Sakata K, Sase H, et al. The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor. Molecular cancer therapeutics. 2014;13(11):2547–58. [DOI] [PubMed] [Google Scholar]
- 16.Voss MH, Hierro C, Heist RS, Cleary JM, Meric-Bernstam F, Gandhi L, et al. Debio 1347, an oral FGFR inhibitor: Results from a first-in-human, phase I dose-escalation study in patients with FGFR genomically activated advanced solid tumors. J Clin Oncol. 2017;35:15_suppl; 2500. [Google Scholar]
- 17.Goyal L, Saha SK, Liu LY, Siravegna G, Leshchiner I, Ahronian LG, et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion-Positive Cholangiocarcinoma. Cancer Discov. 2017;7(3):252–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalyukina M, Yosaatmadja Y, Middleditch MJ, Patterson AV, Smaill JB, Squire CJ. TAS-120 Cancer Target Binding: Defining Reactivity and Revealing the First Fibroblast Growth Factor Receptor 1 (FGFR1) Irreversible Structure. ChemMedChem. 2019. [DOI] [PubMed] [Google Scholar]
- 19.Sootome H, Fujioka Y, Miura A, Fujita H, Hirai H, Utsugi T. Abstract A271: TAS-120, an irreversible FGFR inhibitor, was effective in tumors harboring FGFR mutations, refractory or resistant to ATP competitive inhibitors. Molecular cancer therapeutics. 2013;12(11 Supplement):A271. [Google Scholar]
- 20.Meric-Bernstam F, Arkenau H, Tran B, Bahleda R, Kelley R, Hierro C, et al. O-001Efficacy of TAS-120, an irreversible fibroblast growth factor receptor (FGFR) inhibitor, in cholangiocarcinoma patients with FGFR pathway alterations who were previously treated with chemotherapy and other FGFR inhibitors. Annals of Oncology. 2018;29(suppl_5):mdy149–mdy. [Google Scholar]
- 21.Gallo LH, Nelson KN, Meyer AN, Donoghue DJ. Functions of Fibroblast Growth Factor Receptors in cancer defined by novel translocations and mutations. Cytokine Growth Factor Rev. 2015;26(4):425–49. [DOI] [PubMed] [Google Scholar]
- 22.Babina IS, Turner NC. Advances and challenges in targeting FGFR signalling in cancer. Nature reviews Cancer. 2017;17(5):318–32. [DOI] [PubMed] [Google Scholar]
- 23.Sun C, Bernards R. Feedback and redundancy in receptor tyrosine kinase signaling: relevance to cancer therapies. Trends in biochemical sciences. 2014;39(10):465–74. [DOI] [PubMed] [Google Scholar]
- 24.Hazar-Rethinam M, Kleyman M, Han GC, Liu D, Ahronian LG, Shahzade HA, et al. Convergent Therapeutic Strategies to Overcome the Heterogeneity of Acquired Resistance in BRAF(V600E) Colorectal Cancer. Cancer Discov. 2018;8(4):417–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guagnano V, Furet P, Spanka C, Bordas V, Le Douget M, Stamm C, et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamin o]-pyrimidin-4-yl}−1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. Journal of medicinal chemistry. 2011;54(20):7066–83. [DOI] [PubMed] [Google Scholar]
- 26.Byron SA, Chen H, Wortmann A, Loch D, Gartside MG, Dehkhoda F, et al. The N550K/H mutations in FGFR2 confer differential resistance to PD173074, dovitinib, and ponatinib ATP-competitive inhibitors. Neoplasia. 2013;15(8):975–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan L, Wang J, Tanizaki J, Huang Z, Aref AR, Rusan M, et al. Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc Natl Acad Sci U S A. 2014;111(45):E4869–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cleary JM, Voss MH, Meric-Bernstam F, Hierro C, Heist RS, Ishii N. Safety and efficacy of the selective FGFR inhibitor Debio1347 in phase I study patients with FGFR genomically activated advanced biliary tract cancer (BTC). J Clin Oncol 2018;36:suppl_4S; 447. [Google Scholar]
- 29.Mazzaferro V, El-Rayes BF, Droz Dit Busset M, Cotsoglou C, Harris WP, Damjanov N, et al. Derazantinib (ARQ 087) in advanced or inoperable FGFR2 gene fusion-positive intrahepatic cholangiocarcinoma. British journal of cancer. 2019;120(2):165–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollebecque A, Lihou C, Zhen H, Abou-Alfa GK, Borad M, Sahai V, et al. Interim results of FIGHT-202, a phase II, open-label, multicenter study of INCB054828 in patients (pts) with previously treated advanced/metastatic or surgically unresectable cholangiocarcinoma (CCA) with/without fibroblast growth factor (FGF)/FGF receptor (FGFR) genetic alterations. Annals of Oncology. 2018;29:suppl_8; viii258. [Google Scholar]
- 31.Zhao Z, Bourne PE. Progress with covalent small-molecule kinase inhibitors. Drug Discov Today. 2018;23(3):727–35. [DOI] [PubMed] [Google Scholar]
- 32.Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22(3):262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin JJ, Riely GJ, Shaw AT. Targeting ALK: Precision Medicine Takes on Drug Resistance. Cancer Discov. 2017;7(2):137–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woyach JA, Furman RR, Liu TM, Ozer HG, Zapatka M, Ruppert AS, et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370(24):2286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21(6):560–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Degirolamo C, Sabba C, Moschetta A. Therapeutic potential of the endocrine fibroblast growth factors FGF19, FGF21 and FGF23. Nature reviews Drug discovery. 2016;15(1):51–69. [DOI] [PubMed] [Google Scholar]
- 37.Pearson A, Smyth E, Babina IS, Herrera-Abreu MT, Tarazona N, Peckitt C, et al. High-Level Clonal FGFR Amplification and Response to FGFR Inhibition in a Translational Clinical Trial. Cancer Discov. 2016;6(8):838–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brewer JR, Mazot P, Soriano P. Genetic insights into the mechanisms of Fgf signaling. Genes Dev. 2016;30(7):751–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neel DS, Allegakoen DV, Olivas V, Mayekar MK, Hemmati G, Chatterjee N, et al. Differential Subcellular Localization Regulates Oncogenic Signaling by ROS1 Kinase Fusion Proteins. Cancer Res. 2019;79(3):546–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dias-Santagata D, Akhavanfard S, David SS, Vernovsky K, Kuhlmann G, Boisvert SL, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2(5):146–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD. 2015;17(3):251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20(12):1479–84. [DOI] [PubMed] [Google Scholar]
- 43.Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PloS one. 2015;10(10):e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahronian LG, Sennott EM, Van Allen EM, Wagle N, Kwak EL, Faris JE, et al. Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations. Cancer Discov. 2015;5(4):358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21(7):795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.