Abstract
Leukemia inhibitory factor receptor (LIFR) and its ligand LIF play a critical role in cancer progression, metastasis, stem cell maintenance, and therapy resistance. Here, we describe a rationally designed first-in-class inhibitor of LIFR, EC359 that directly interacts with LIFR to effectively block LIF/LIFR interactions. EC359 treatment exhibits anti-proliferative effects, reduces invasiveness and stemness, and promotes apoptosis in TNBC cell lines. The activity of EC359 is dependent on LIF and LIFR expression and treatment with EC359 attenuated the activation of LIF/LIFR driven pathways including STAT3, mTOR, and AKT. Concomitantly, EC359 was also effective in blocking signaling by other LIFR ligands (CTF1, CNTF, and OSM) that interact at LIF/LIFR interface. EC359 significantly reduced tumor progression in TNBC xenografts, patient derived xenografts (PDX) and reduced proliferation in patient derived primary TNBC explants. EC359 exhibits distinct pharmacologic advantages including oral bioavailability, and in vivo stability. Collectively, these data support EC359 as a novel targeted therapeutic that inhibits LIFR oncogenic signaling.
Keywords: LIFR, LIF, LIFR inhibitor, STAT3, breast cancer, TNBC
Introduction
Leukemia inhibitory factor (LIF) is the most pleiotropic member of the interleukin-6 family of cytokines (1). LIF signaling is mediated via the LIF receptor (LIFR) complex, which is comprised of LIFR and glycoprotein 130 (gp130) (2). The LIFR does not have intrinsic tyrosine kinase activity. Both LIFR and gp130 constitutively associate with the JAK-Tyk family of cytoplasmic tyrosine kinases. Consequently, LIF binding to the LIFR complex activates multiple signaling pathways including JAK, STAT, MAPK, AKT, and mTOR (2-4). LIF and LIFR are widely expressed in many solid tumors (1,5-7) and their overexpression is often associated with poor patient prognosis (8,9). Additionally, high circulating LIF levels correlate with tumor recurrence (10).
The LIF/LIFR axis acts on multiple aspects of cancer biology to promote tumor growth, metastasis, and therapy resistance (11). LIF is a key regulator of cancer stem cells (CSCs) (11), plays a role in stem cell maintenance (12,13), regulates self-renewal and pluripotency (12), and is associated with chemoresistance (10,14). LIF functions as a growth factor to promote growth and invasion (15). Recent evidence indicates upregulated LIF-JAK-STAT3 signaling via autocrine and paracrine mechanisms in tumors (10,16,17). However, lack of any small molecule inhibitors (SMIs) that block LIF/LIFR signaling represents a major knowledge gap and critical barrier for advancement of LIF/LIFR targeted cancer therapy.
Among the different sub-types of breast cancer, 60–70% are estrogen receptor (ER) positive (ER+BC), and 15–24% are triple negative breast cancer (TNBC) (18). TNBC is more aggressive, and due to the lack of targeted therapies, represents a disproportional share of the breast cancer mortality (19,20). TNBC exhibit high propensity for metastasis, with some subtypes such as claudin-low that are highly enriched for cancer stem cells, and frequently exhibits therapy resistance (19,20). In breast cancer cells, LIF/LIFR signaling activates multiple signaling pathways including STAT3, AKT, and mTOR pathways and contribute to activation of mTOR downstream targets such as p70S6K and 4EBP1 (4). LIF/LIFR signaling promote tumor progression of both ER+BC and TNBC cells (21-23). Additionally, LIF mRNA levels were elevated in invasive breast carcinomas compared with the normal breast tissues (24). Overexpression of LIF significantly associated with a poorer relapse free survival in breast cancer patients (4).
In this study, we report the development of a novel LIFR inhibitor EC359 that selectively binds LIFR and blocks binding of ligands attenuating LIFR oncogenic signaling. Using molecular modeling, in vitro, and in vivo assays, we demonstrated that EC359 interacts with LIFR and inhibits cell viability of TNBC cells which express both LIF and LIFR. Additionally, EC359 reduced the invasion and stemness of TNBC cells, and promoted apoptosis. In xenograft and patient derived xenograft (PDX) assays, EC359 significantly reduced the tumor progression. This study represents the first report detailing the development of a first-in-class inhibitor of LIF/LIFR.
Methods
Cell lines and reagents
Human breast cancer cells MCF7, T47D, MDA-MB-231, BT-549, SUM-159, HCC1937, MDA-MB-468, HCC1806, and normal mammary epithelial cells (HMEC) were obtained from American Type Culture Collection (ATCC, Manassas, VA), were maintained as per ATCC guidelines and used from early passages (<10 passages after thawing). All model cells utilized were free of mycoplasma contamination and were confirmed by using Mycoplasma PCR Detection Kit purchased from Sigma (St. Louis, MO). Short tandem repeat polymorphism analysis (STR) of the cells was used to confirm the identity using University of Texas Health San Antonio (UTHSA) core facilities. CSCs isolated from TNBC cells were maintained in MammoCult medium along with the supplements according to manufacturer’s instructions (STEMCELL Technologies, Cambridge, MA). The GAPDH, p-ERK½, ERK½, p-p70S6K, p70S6K, p-S6, S6, p-Akt(S473), Akt, p-p38 MAPK, p38 MAPK, p-mTOR(S2448), mTOR, p-STAT3(Y705), STAT3 antibodies were purchased from Cell Signaling Technology (Beverly, MA). LIF and LIFR antibodies were purchased from Santa Cruz Biotechnology (Dallas, Texas). β-actin and all secondary antibodies were purchased from Sigma. ALDEFLUOR assay kit was obtained from StemCell Technologies. The Ki-67 antibody was purchased from Abcam (Cambridge, MA). LIFR Knockout (KO) model cells were generated using Genescript CRISPR gRNA Constructs (Genescript-s64729-LIFR CRISPR guide RNA 1; Genescript-s64731-LIFR CRISPR guide RNA 2) and transfecting them into Cas9 stably expressing BT-549 cells followed by puromycin selection. EC359 and EC330 was synthesized using the detailed synthetic protocol described in the patent WO 2016/154203 A1 (Evestra Inc.). Characterization of EC330 and EC359 produced was described in the Supplementary Methods.
Western blotting and biotin pull down assays
For western blotting, cells were lysed in RIPA buffer (Thermo Fisher Scientific, Waltham, MA) containing protease and phosphatase inhibitors. Total cellular lysates were mixed with 4X SDS sample buffer and run on SDS-PAGE, and transferred onto nitrocellulose membranes and blots were developed using antibodies and the ECL kit (Thermo Fisher Scientific, Waltham, MA). Avidin-biotin pull down was performed as described previously (25). Briefly, BT-549 total cellular lysates and purified LIFR was incubated with Biotin-control or Biotin-EC359 for overnight and incubated with NanoLink™ Streptavidin Magnetic Beads (Solulink) for 1 h at room temperature. The binding of EC359 to LIFR was confirmed by western blotting. Intensity of signaling bands in western blots were quantitated using Image J program.
Cell invasion assays
The effect of EC359 on cell invasion of TNBC cells was determined by using the Corning BioCoat Growth Factor Reduced Matrigel Invasion Chamber assay. MDA-MB-231 and BT-549 cells were treated with vehicle or EC359 (25 nM) for 22 h and invaded cells in all the treatment conditions were determined according to manufacturer protocols.
Extreme limiting dilution assays (ELDA)
Cancer stem cells (CSCs) from MDA-MB-231 and BT-549 cells were sorted using established stem cell marker ALDH using the ALDEFLUOR kit and flow cytometry. CSCs were cultured in MammoCult medium with the supplements as per manufacturer’s instructions. The effect of EC359 on self-renewal of CSCs was determined by ELDA. Briefly, CSCs were seeded in decreasing numbers (100, 50, 20, 10, 5, and 1 cells/well) in 96 well ultra-low attachment plates and treated with vehicle or EC359. After 10 days, the number of wells containing spheres per each plating density was recorded and stem cell frequency between control and treatment groups was calculated using ELDA analysis software (http://bioinf.wehi.edu.au/software/elda/).
Cell viability, clonogenic and apoptosis assays
The effect of EC359 on cell viability of TNBC cells was assessed by using MTT assay as previously described (25). TNBC cells were seeded in 96-well plates (1 × 103 cells/well) and after overnight incubation cells were treated with varying concentrations of EC359 for 5 days. To test the effect of EC359 on the viability of CSCs and non CSCs, Cell Titer-Glo assays were performed (Promega, Madison, WI). Briefly, cells were seeded in 96-well, flat, clear-bottom, opaque-wall microplates and treated with vehicle or EC359 for 3 days. The total ATP content as an estimate of total number of viable cells was measured on an automatic Fluoroskan Luminometer. For clonogenic survival assays, cells were seeded in triplicates in 6 well plates (500 cells/well), after overnight incubation cells were treated with vehicle or EC359 for 5 days and after 2 weeks, colonies that contain ≥ 50 cells were counted and used in the analysis. The effect of EC359 on apoptosis was measured by Annexin V/PI staining and Caspase-3/7 activity assay as described previously (25,26). Briefly, MDA-MB-231 and BT-549 cells were seeded in 96 well plates (2 × 103/well), after overnight incubation cells were treated with vehicle or EC359 (20 nM) for 72 h. After treatment, equal amount of Caspase-3/7 substrate containing solution was added to the media, and luciferase activity was measured using luminometer according to manufacturer protocol (Promega, Madison, WI).
RT-qPCR
Reverse transcription (RT) reactions were performed by using SuperScript III First Strand kit (Invitrogen, Carlsbad), according to manufacturer’s protocol. Real-time PCR was done using SybrGreen on an Illumina Real-Time PCR system. Primer sequences were included in the Supplementary Table S1.
Surface plasmon resonance (SPR) studies
Binding profiles of EC359 to LIF/LIFR were evaluated using SPR. Recombinant human LIF was purchased from R&D Systems, Minneapolis, MN (Cat# 7734-LF-500) and human LIFR-Fc was purchased from Speed Biosystems, Gaithersburg, MI (Cat#YCP1132). Sensor chips were purchased from ForteBio (www.fortbio.com). Detailed SPR protocol was provided in the Supplementary Methods.
Microscale thermophoresis (MST) assays
A serial dilution of the ligand (EC359) was prepared in a way to match the final buffer conditions in the reaction mix (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005 % Tween-20, 10% DMSO). The highest concentration of ligand was 2.00 μM and the lowest 61.0 pM. Five μl of each dilution step was mixed with 5 μl of the fluorescent molecule. The final reaction mixture, which was loaded in capillaries, contained a respective amount of ligand (max. conc: 1.00 μM; min. conc: 30.5 pM) and constant 5 nM fluorescent molecule (protein target LIFR labelled florescent dye- NHS chemistry). Thermophoretic movement of fluorescently labeled protein with EC359 was performed using on a Monolith NT.115 Pico at 25°C, with 7% LED power and 60% Laser power (Nanotemper Technologies, München, Germany).
Molecular modeling studies
The atomic level of interactions of EC359 against human LIFR (hLIFR) were studied by molecular modeling. The existing structural information of LIFR was utilized for the studies. The partial structure of human LIFR (hLIFR) (domains D1-D5) (PDB ID: 3E0G) and structure of human LIF (hLIF) in complex with the partial murine LIFR (mLIFR) (domains D1-D5) has been reported in the Protein Data Bank (PDB ID: 2Q7N) (27,28). As a preliminary step, the sequence and structural similarities of both of these LIFRs were deduced. Further, the three-dimensional structure of hLIF-hLIFR complex was constructed from hLIF-mLIFR by replacing mLIFR. The complex was energy minimized to avoid the residue clashes between the hLIFR and hLIF. From the minimized complex, the hLIFR was again separated and prepared for the docking studies. Since there was no information available on the ligand binding sites, the whole receptor was probed using Sitemap from Schrödinger (Schrodinger, LLC, San Diego, CA) to detect possible binding sites (29). Two steps of molecular docking were performed such as standard precision (SP) and induced fit (IFD) on the identified binding sites. The purpose of SP docking was to detect the binding strength and orientations of ligand at respective binding sites. Based on the docking scores, the sites were ranked. Later an appropriate ligand pose was selected and flexible docking (IFD) was performed by allowing flexibility to the surrounding amino acids (around 6 Å from the center of the ligand). Based on the MM-GBSA (30) score and visual inspection an appropriate pose was selected and subjected to molecular dynamics simulation (MDS) to estimate the residence time of the ligand over a period of 25 ns. The detailed description of methods used in the study was included in the Supplementary Methods and Fig. 2,3.
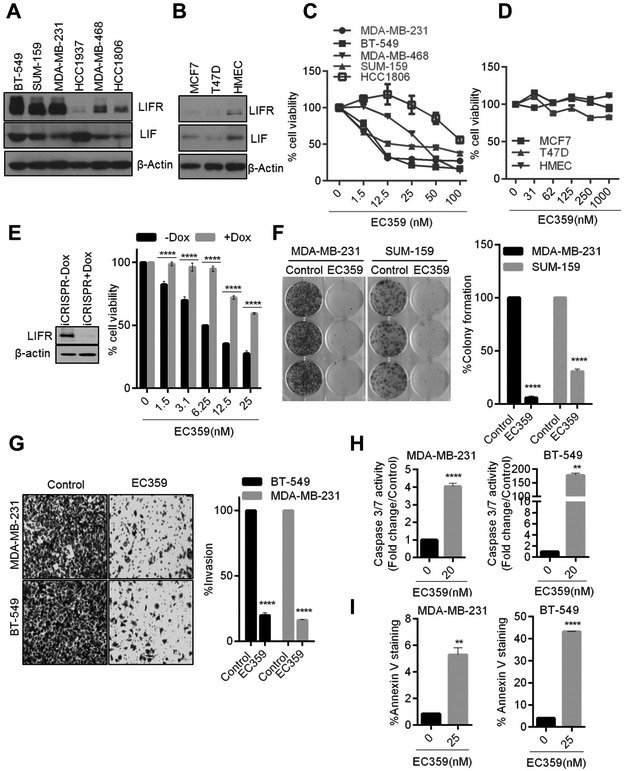
Figure 2.
EC359 decrease cell viability, colony formation, invasion and promote apoptosis of TNBC cells that express LIF and LIFR. The expression of LIF and LIFR in TNBC (A), ER+ BC and normal mammary epithelial cells (B) was determined by western blotting. Effect of increasing doses of EC359 on the cell viability of TNBC (C) ER+BC and normal mammary epithelial cells (D) was determined using the MTT cell viability assay (n=3). E, Effect of inducible CRISPR/Cas9 mediated KO of LIFR on EC359 induced cell viability was determined using MTT assays in BT-549 cells (n=3). F, Effect of EC359 (20 nM) on cell survival was measured using colony formation assays. G, Effect of EC359 (25 nM) on cell invasion of MDA-MB-231 and BT-549 model cells was determined using matrigel invasion chamber assays (n=3). Representative images of invaded cells are shown and the number of invaded cells in five random fields was quantitated. H, Effect of indicated doses of EC359 on caspase 3/7 activity (Caspase-Glo3/7 assay) and I, Annexin V staining in MDA-MB-231 and BT-549 cells (n=3) was determined. ** P<0.01, ****p<0.0001.
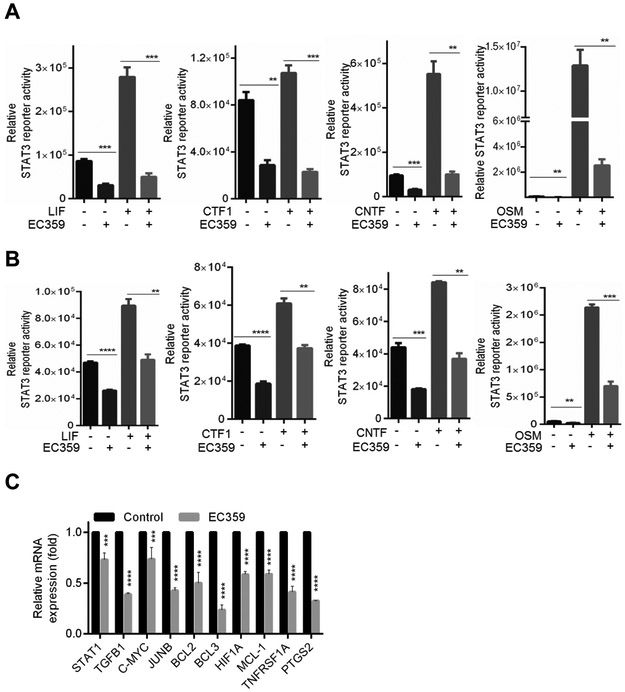
Figure 3.
Effect of EC359 on LIFR mediated transcription in TNBC cells. BT-549 (A) and MDA-MB-231 (B) cells stably expressing STAT3-luc reporter were serum starved for 24 h, pretreated with EC359 (50 nM) for 1 h and then stimulated with indicated concentrations of LIF, CTF1, OSM, CNTF (n=3). Reporter activity was measured after 24 h. C, Effect of EC359 (100 nM) treatment (12 h) on STAT3 targeted genes was measured using RT-qPCR analysis (n=3). ** P<0.01, *** P<0.001, ****p<0.0001
Reporter gene assays
For STAT3-luc assays, MDA-MB-231 and BT-549 cells were stably transduced with STAT3-firefly luciferase reporter lentivirus purchased from Cellomic Technology (Helethrone, MD). STAT3-luc reporter expressing cells were serum starved for 24 h, pretreated with EC359 for 1 h, and then stimulated with LIF or other indicated ligands for 24 h, and reporter activity was measured. Cells were lysed in passive lysis buffer, and the luciferase activity was measured by using the dual-luciferase reporter assay system (Promega, Madison, WI) using luminometer.
In Vivo xenograft studies
All animal experiments were performed after obtaining UTHSA IACUC approval, and all the methods were carried out in accordance with IACUC guidelines. MDA-MB-231 cells (2 ×106) were mixed with equal volume of growth factor reduced matrigel and implanted in the mammary fat pads of 8-week-old female athymic nude mice as described previously (31). After tumor establishment, and achievement of measurable size, mice were randomized into control and treatment groups (n = 8 tumors per group). Control group received vehicle (hydroxy methyl cellulose) and the treatment group received EC359 (5 mg/kg/day) 3 days per week subcutaneously. All mice were monitored daily for adverse toxic effects. Tumor growth was measured with a caliper at 3–4 day intervals, and volume was calculated using a modified ellipsoidal formula: tumor volume = ½(L × W2), where L is the longitudinal diameter and W is the transverse diameter. At the end of the experiment, mice were euthanized, and tumors were excised, and processed for histological and biochemical studies.
Patient derived xenograft (PDX) model
The TNBC tissue was obtained from a deidentified surgical specimen (F0) just after surgery from a patient with invasive ductal carcinoma (pT3 pN2a pM) via UTHSA PDX Core. The tumor tissues were divided in to three parts; the first part was snap frozen and stored in liquid nitrogen, the second part was fixed in 10% buffered formalin and processed for histological characterization, and the third part was placed in ice cold PBS, cut into small pieces (3–5mm3) and engrafted into mammary fat pad of NCI SCID/NCr mice. PDX tumor was confirmed negative for ER, PR, HER2 by the Pathology core. Tumors from early passages were dissected into small pieces and implanted into the flanks of SCID mice. The mice were then randomized when they reached tumor volume of ~150 mm3 into control or treatment groups (n=6 tumors per group). The control group received vehicle (hydroxy methyl cellulose) and the treatment group received EC359 (10 mg/kg/day) 3 days per week subcutaneously. At the end of the treatment, tumors were excised and processed for histological studies, protein and RNA analysis.
Patient-derived explant (PDEx) studies
TNBC tissues were collected from discarded surgical samples from UT Southwestern Medical Center (UTSW) patients for research purposes after obtaining the written informed consent and in accordance with institutional board-approved protocol (STU-032011–187). All the studies were conducted in accordance with the Declaration of Helsinki. Tissues were processed and excised into small pieces and cultured on gelatin sponges for 24 hours in medium containing 10% FBS as described previously (25). Tissues were treated with vehicle or EC359 in culture medium for 72 hours and fixed in 10% buffered formalin at 4°C overnight and subsequently processed into paraffin blocks. Sections were then proceeded for immunohistochemical analysis for Ki-67.
Immunohistochemistry
Immunohistochemical analysis was performed as described previously (25). Briefly, sections were blocked with normal goat serum (Vector Labs, Burlingame, CA) followed by incubation overnight with Ki-67 (1:100) primary antibody and subsequent secondary antibody incubation for 30 minutes at room temperature. Immunoreactivity was visualized by using the DAB substrate and counterstained with hematoxylin (Vector Lab, Inc.). Percent of Ki-67 positive proliferating cells was calculated in five randomly selected microscopic fields.
PK studies and bioavailability studies
A Pharmacokinetic (PK) study of EC359 was conducted in both mice and rats following intravenous (IV) and oral administration of the compound (GVK Bio, Hyderabad, India). IV formulation (5 mg/kg) was prepared as described: A required volume 0.1mL of DMSO stock (20 mg/mL) was taken in an Eppendorf tube then 0.100 mL of DMSO was added and vortexed, then sonicated, followed by addition of 1.800 mL 10% Solutol in PBS, vortexed, and probe sonicated ≈1–2 min. to make a final solution of 1 mg/mL concentration. T½, AUC0-last, AUC0-inf, AUCextra, CL, Vd, MRT0-last, and RSQ were measured using LC-MS/MS. For oral dosing (10 mg/kg) volume 0.200 mL of DMSO stock (20 mg/mL) was taken in an Eppendorf tube then 1.800 mL 10% Solutol in PBS was added, vortexed, and probe sonicated ≈1–2 min. to make a solution of 2 mg/mL concentration. Cmax, Tmax, AUC0-last, AUC0-inf, AUCextra, F%, MRT0-last, and RSQ were measured. GR antagonism assays were performed using SelectScreen™ Biochemical Nuclear Receptor Profiling Service (Invitrogen, Carlsbad,CA).
Statistical analysis
All the statistical analyses were carried out using GraphPad Prism 6 software (GraphPad Software, SanDiego, CA). A Student’s t-test was used to assess statistical differences between control and EC359-treated groups. All the data represented in bar graphs are shown as mean ± SE. A p-value less than 0.05 was considered significant.
Results
Optimization and generation of lead LIFR inhibitor EC359
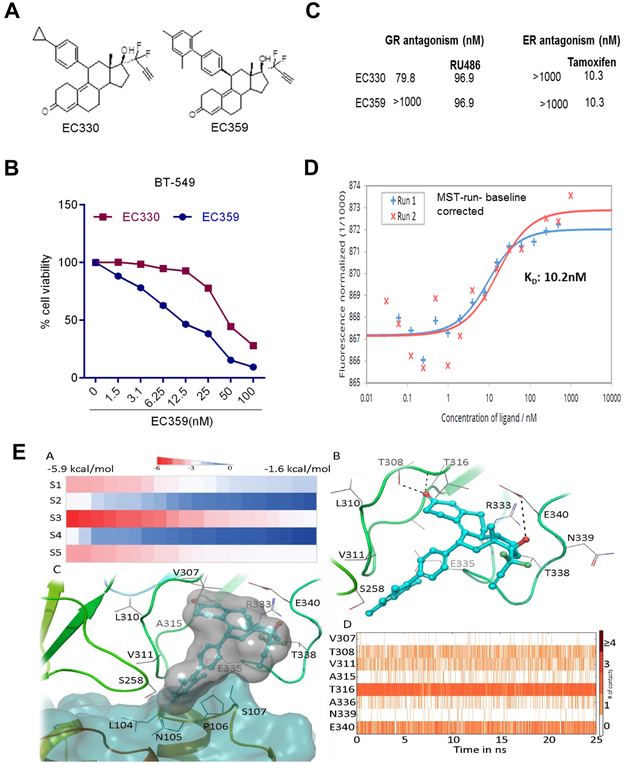
We initially synthesized several compounds to target LIFR signaling using rationalized design based on crystal structure of LIF/LIFR. Within this series of compounds, one compound (EC330) showed higher potency (Fig. 1A, left panel). In cell viability assays (Cell Titer Glo luminescent assay) using cancer cells, EC330 inhibited growth at approximately IC50 ~50 nM (Fig. 1B). The reported X-ray crystallographic studies of LIF suggested a four α-helix bundle topology with a compact core predominantly composed of hydrophobic residues contributed by the four α-helices (32). Initial structure activity relationship studies in our laboratory have shown the following structural features are necessary for the LIF inhibitory action: 1) difluro-acetylenic function at the 17-alpha position and 2) 4’-substituition at the 11-phenyl ring. Since the EC330 has a steroidal backbone, we investigated the binding of EC330 to steroid receptors such as glucocorticoid receptor (GR). EC330 showed some antagonism to GR (79.8 nM), which may elicit unwanted toxicity. Therefore, we pursued medicinal chemistry modifications, which retained its potency on LIFR, while reduced steroidal receptor interactions. Additional SPR studies and subsequent synthetic efforts resulted in the development of EC359 (Fig. 1A, right panel). To examine whether optimization of EC359 retained its activity on par with the initial lead compound EC330, we conducted several studies (Fig. 1B). Receptor binding studies revealed EC359 has more desirable characteristics than EC330 including lack of affinity to GR (Fig. 1C). In cell viability assays, EC359 showed significant inhibitory activity on par with EC330 in BT-549 model cells (Fig. 1B).
Figure 1.
Characterization of EC359. A, schematic representation of structure of EC330 and EC359. B, Dose response curve of EC330 and EC359 determined using MTT assay on BT-549 cells (n=3). C, Ability of EC359 to interact with other steroid receptors were analyzed by in vitro binding assays. D, Binding of EC359 to LIFR was confirmed using MST assays as described in methods. EA: The docking scores of EC359 at different binding sites. The sites were mentioned as S1 to S5. EB: Binding of EC359 (represented in blue ball and stick model) with hLIFR (represented in cartoon and line model). The dotted lines representing the hydrogen bonds. EC: The binding of EC359 in the presence of LIF (represented in blue surface). The binding creates close contacts with residues of LIF. ED: The protein ligand contacts over a 25 ns of MD.
SPR studies confirmed EC359 direct interaction with LIFR
To test whether EC359 directly bind to LIFR complex, binding profiles of EC359 to LIF/LIFR were evaluated using SPR. Two sets of studies were performed: 1) to verify the integrity of recombinant proteins, the interaction between LIFR and LIF was studied; 2) small molecule binding to LIF/LIFR by either immobilizing LIFR or LIF onto a sensor chip was tested. Results from the first set of studies confirmed the integrity of recombinant LIF and LIFR; LIF bound to immobilized LIFR-Fc with a binding constant of 7µM (Supplementary Fig. S1A). In the second set of studies, results showed EC359 binding to LIFR, but not LIF. Further, EC359 bound to LIFR in a dose dependent manner with KD = 81µM (Supplementary Fig. S1B). The results confirmed that EC359 is a specific inhibitor of LIF/LIFR complex.
MST assays revealed high affinity interaction of EC359 with LIFR
SPR studies use immobilized receptor complex ligand induced structural changes may obscure true (in vivo equivalent) affinity of the drug. Hence, we conducted an orthogonal assay, namely MST where the receptor is not immobilized to verify EC359 binding to the receptor complex. MST is a powerful technique to quantify biomolecular interactions. By combining the precision of fluorescence detection with the variability and sensitivity of thermophoresis, MST provides a flexible, robust and fast way to dissect molecular interactions (33,34). MST analysis confirmed direct interaction of EC359 with LIFR with an estimated KD of 10.2 nM (Fig. 1D). To further, demonstrate that EC359 directly interacts with LIFR, we generated biotinylated-EC359. Biotin addition did not affected EC359 biological activity (Supplementary Fig. S1C). Using Biotin-EC359, we examined whether it interacts with LIFR. Purified LIFR protein or BT-549 cellular lysate was incubated with Biotin-EC359 and its ability to interact with LIFR was determined using avidin pull down assay followed by western blotting. Results elucidated that EC359 interact with LIFR. (Supplementary Fig. S1D,E).
Docking studies suggested EC359 can interact at the LIF-LIFR binding interface
The site predictions revealed five potential binding sites on the hLIFR (Supplementary Fig S2-4) in which two sites (2 and 3) are close to the LIF binding interface (Supplementary Fig. S4). The SP docking was performed on all the five sites and docking scores were deduced (Fig. 1EA). The docking scores towards different binding sites range from −5.8 to −1.6 kcal/mol. It was also observed that EC359 has exhibited more promising scores towards site-3 compared to other sites. The binding poses obtained from the docking were superimposed to the hLIF-hLIFR complex to see the potential clashes between the ligand and LIF. As expected, the binding poses at site-3 are making steric clashes with residues of LIF (Supplementary Fig. S5). Since the SP docking is a rigid docking method, the ligand induced conformational changes were also studied using IFD by applying flexibility to the surrounding residues. Using standard protocol, the side chains were optimized and 28 poses were generated. The binding energies of all the 28 poses range from −80 kcal/mol to −33 kcal/mol (Supplementary Fig. S6). It was observed that all the generated ligand poses are potentially making steric clashes with hLIF. One of the top scored poses (binding energy = −77 kcal/mol) was critically analyzed for the detailed atomic level of interactions (Fig. 1EB). In the selected pose, ligand induced conformational changes were observed for the loops close to the LIF binding region (Supplementary Fig. S7). The ligand EC359 was found to sandwich between two loops at the N-terminal of D4 domain by orienting the difluro-acetylenic group to the bulk solvent. The keto group of the EC359 was found to be involved in two hydrogen bonds with the side chain of T308 and the backbone of T316. Similarly, the hydroxyl group of the ligand was also found to mediate a hydrogen bond with the sidechain of E340. Moreover, van der Waals contacts with the surrounding residues were also found to contribute to the ligand binding. It was observed that EC359 binding to hLIFR would prevent hLIF binding due to steric clashes (Fig. 1EC). As a final step, the snapshots obtained from the MD simulation were superimposed with the initial pose and RMSD was calculated for protein and ligand separately. The snapshots were analyzed and found that the structural distortions are affected mainly to the loops (connecting separate domains in the LIFR) and the terminal regions. At the same time the ligand is found to remain bound at the binding site even after 25 ns of MD simulation. The protein ligand contacts over 25 ns of MD simulation are shown in Fig. 1ED and Supplementary Fig. S8.
EC359 has favorable pharmacological features
We then conducted Pharmacokinetics (PK) analysis of EC359 using various established tests (Supplementary Fig. S9). Results from IV dosing studies using 5 mg/kg in rats indicated a mean C0 of 74669.11 ng/ml, T½(h) of 3.86 h, AUC0-last (ng·h/mL) of 15544.36, and AUC0-inf (ng·h/mL) of 15573.91. Results from oral dosing studies using 10 mg/kg in rats indicated a mean Cmax (ng/mL) of 919.50, Tmax (h) of 2.67, AUC0-last (ng·h/mL) of 3792.26, and AUC0-inf (ng·h/mL) of 3876.82. Ames test confirmed that EC359 did not induce an evident (significant) >2 fold increase in the revertant counts at the doses tested (dose related), in the tester strains both with and without metabolic activation according to the evaluation criteria mentioned in OECD guideline no.471. Hence, the compound EC359 is considered non-mutagenic with salmonella typhimurium strains TA98, TA100, TA1535, TA1537 and E.coli combo, both with and without metabolic activation. In hERG cardiotoxicity screening, up to 30 µM concentration of EC359 didn’t show 50% inhibition, hence no liability. Among CYP enzymes EC359 inhibits 2D6, therefore, caution is warranted in concurrent administration of drugs that inhibits 2D6 such as Prozac. Metabolic stability and plasma stability are moderate in human with high plasma protein binding. Good bioavailability (PK) was observed in both intraperitoneal and oral dosing (Supplementary Fig S9). Collectively, the data from these studies indicate EC359 has specific on-target activity (PD) and suggests EC359 as a druggable candidate for further development.
EC359 reduced the cell viability of LIF and LIFR expressing cells
We first examined the expression of LIF and LIFR in cells that represent various subtypes of TNBC (BT-549, SUM-159, MDA-MB-231, HCC1937, MDA-MB-468, and HCC1806), ER+ BC (MCF7, and T47D) as well as normal mammary epithelial cells (HMEC). We found that five of the six TNBC cells expressed high levels of LIF and LIFR when compared to ER+BC cells and normal cells (Fig. 2A,B). Next, we examined the efficacy of EC359 on cell viability of TNBC and ER+BC cells. Treatment with EC359 resulted in a significant dose-dependent reduction in the cell viability of TNBC cells (IC50 10–50 nM) and their inhibition is well correlated with LIF and LIFR expression levels (Fig. 2C). Interestingly ER+BC cells which express low levels of LIF and LIFR exhibited low sensitivity to EC359 treatment (IC50 >1000 nM) when compared with TNBC cells (Fig. 2D). To further confirm the target specificity of EC359, we generated doxycycline inducible LIFR-KO cells using Cas9 stably expressing TNBC cells. Results indicated a reduction of LIFR expression in BT-549 models contributed to the resistance of the EC359 mediated decrease in cell viability (Fig. 2E). Collectively, this data suggests that EC359 activity depends on presence of functional LIF/LIFR signaling axis in cells.
EC359 reduced invasion, and induced apoptosis of TNBC cells
We next examined the efficacy of the EC359 on the survival of TNBC cells. In clonogenic survival assays, EC359 significantly reduced the colony formation ability of MDA-MB-231 and SUM-159 cells (Fig. 2F). Given the important role of the LIF axis in the invasiveness of cancer cells, we examined the effect of EC359 in reducing the invasion of TNBC cells. Matrigel invasion assays demonstrated that EC359 significantly reduced the invasion potential of MDA-MB-231 and BT-549 cells (Fig. 2G). Furthermore, we examined whether EC359 induced apoptosis in TNBC cells using caspase 3/7 activity assay and Annexin V staining assay. EC359 treatment significantly increased caspase 3/7 activity (Fig. 2H) and Annexin V positive cells (Fig.2I) in both MDA-MB-231 and BT-549 cells. Collectively, these results suggest that EC359 exhibits significant inhibitory activity on invasion and promotes apoptosis of TNBC cells.
EC359 inhibited LIFR mediated transcriptional changes
LIF/LIFR activates multiple signaling pathways including JAK/STAT3, MAPK, AKT, and mTOR; all of which are implicated in TNBC progression. To confirm the inhibitory effect of EC359 on LIF/LIFR mediated STAT3 activation, BT-549 cells that stably express STAT3-Luc reporter were pretreated with vehicle or EC359 followed by stimulation with LIF. As expected, LIF treatment significantly increased the STAT3 reporter activity and this activation was inhibited by EC359 treatment (Fig. 3A). Since our modeling studies predicted EC359 interaction with the ligand binding interface of LIFR, we examined whether EC359 also blocks signaling by other LIFR ligands such as Oncostatin M (OSM), Ciliary Neurotrophic Factor (CNTF), and Cardiotrophin 1 (CTF1). Results showed that EC359 blocked the OSM, CNTF and CTF1 mediated STAT3 activity in BT-549 cells (Fig. 3A). We also confirmed that EC359 has the ability to block LIF, OSM, CNTF and CTF1 mediated STAT3 activation using MDA-MB-231 cells stably expressing STAT3-Luc reporter (Fig. 3B). In RT-qPCR assays using BT-549 cells, EC359 treatment significantly reduced the expression of several known STAT3 target genes (Fig. 3C).
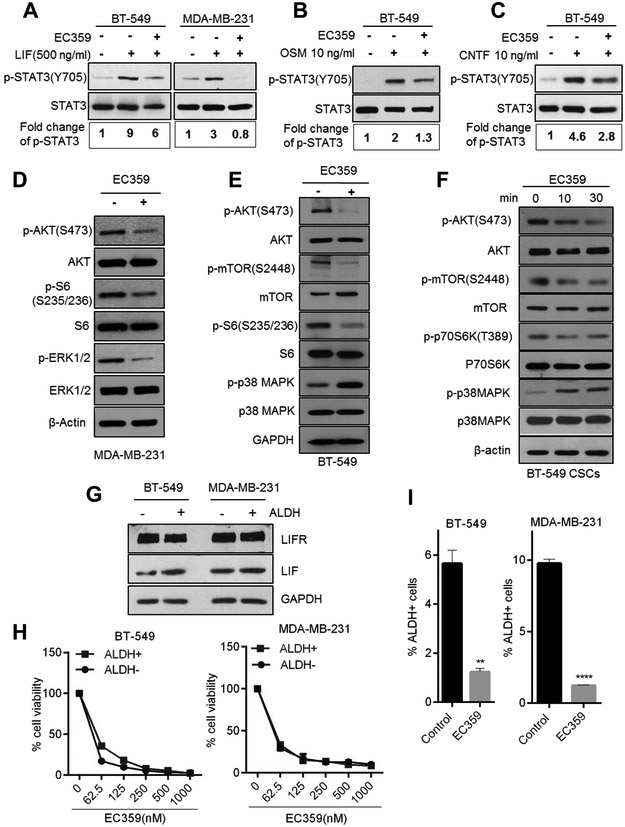
EC359 reduced LIFR mediated activation of downstream signaling pathways
To further confirm the effect of EC359 on LIF/LIFR downstream signaling pathways, MDA-MB-231 and BT-549 cells were pretreated with vehicle or EC359 and subsequently stimulated with LIF. STAT3 activation was examined using western blotting. EC359 treatment substantially reduced the LIF activation of STAT3 in both BT-549 and MDA-MB-231 cells (Fig. 4A). EC359 also reduced the STAT3 activation by OSM and CNTF (Fig. 4B,C). Additionally, EC359 treatment substantially decreased the phosphorylation of AKT, mTOR, S6 and ERK½ in MDA-MB-231 and BT-549 cells (Fig. 4D,E). EC359 treatment also increased the phosphorylation of proapoptotic p38MAPK in BT-549 cells (Fig. 4E). We confirmed whether alteration in downstream signaling seen upon EC359 treatment such as STAT3 occurs in the cell line that has a Dox -inducible deletion of the LIFR. Results showed that KO of LIFR significantly reduced the STAT3 activation. Further, stimulation of LIFR KO cells with LIF did not activated STAT3 in this model. However, EC359 is able to block LIF mediated STAT3 activation in LIFR expressing control cells. These results confirm that the downstream effects seen in EC359 are due to its effects on LIFR and that STAT3 is a downstream effector of LIFR in TNBC cells (Supplementary Fig. S10A). These results suggest that EC359 acts as a LIFR inhibitor and attenuates LIF and other LIFR ligand mediated signaling in TNBC cells.
Figure 4.
EC359 inhibits LIFR downstream signaling and reduce stemness of TNBC cells. A, MDA-MB-231 and BT-549 cells were serum starved for 24 h, pretreated with EC359 (100 nM) for 1 h and then stimulated with LIF (10 min) and the status of STAT3 phosphorylation was measured using western blotting. STAT3 phosphorylation was quantitated using image J program, normalized to total STAT3 and shown as fold induction over control cells. B, BT-549 cells were serum starved for 24 h, pretreated with EC359 (100 nM) for 1 h and then stimulated with OSM (10 ng), and C, CNTF (10ng) for 10 min, and the status of STAT3 phosphorylation was measured using western blotting. D, E, F, MDA-MB-231, BT-549, and CSCs were treated with EC359 (100 nM) and status of LIFR downstream signaling was measured using western blotting. G, ALDH + and ALDH− cells were isolated by FACS and the expression levels of LIFR and LIF were measured by western blotting. H, Effect of EC359 on the viability of ALDH+ (CSCs) and ALDH− (non CSCs) cells was determined using cell titer glo assay (n=3). I, BT-549 and MDA-MB-231 cells were treated with EC359 (100 nM) and the status of ALDH+ cells was determined by FACS analysis **P<0.01, ****P<0.0001.
EC359 reduced the cell viability and self-renewal of TNBC stem cells
The LIF/LIFR axis plays a vital role in stemness (6,12). To test the effect of EC359 on stemness, CSCs were isolated from MDA-MB-231 and BT-549 using ALDH+ flow cytometry sorting. EC359 treatment substantially decreased the phosphorylation of AKT, mTOR, p70S6K, and increased phosphorylation of proapoptotic p38MAPK in CSCs (Fig. 4F). Western blot analysis showed that ALDH+ (CSCs) and ALDH− (differentiated) cells have similar levels of LIFR (Fig. 4G). Further, in cell viability assays, EC359 similarly inhibited both ALDH+ and ALDH− cells (Fig. 4H). To further study the effect of EC359 on the self-renewal ability of CSCs, extreme limiting dilution assays (ELDA) were performed. Results showed that EC359 significantly reduced the self renewal of CSCs compared to control (Supplementary Fig. S10B). Further, pretreatment of TNBC cells with EC359 significantly reduced the abundance of ALDH+ cells (Fig. 4I).
EC359 reduced TNBC xenograft tumor growth in vivo
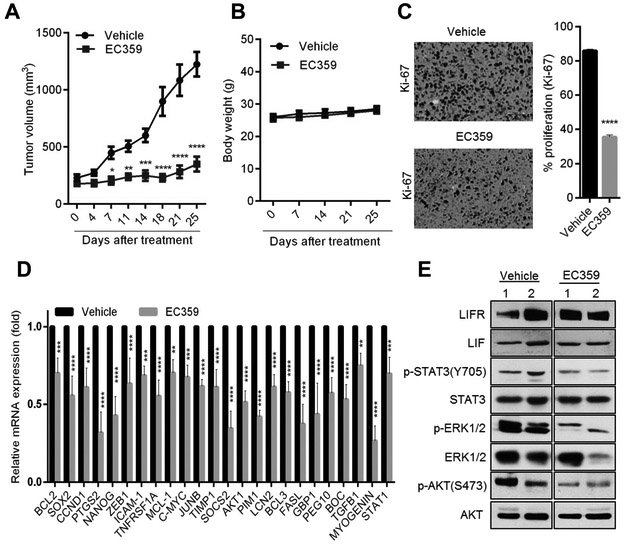
To test the efficacy of EC359 on in vivo tumor progression, we established MDA-MB-231 xenograft tumors in the mammary fat pad of nude mice. Mice were randomized to vehicle (hydroxy methyl cellulose) and EC359 (5 mg/kg/day via subcutaneous injection) 3 days/week. EC359 treatment significantly reduced the tumor progression compared to vehicle (Fig. 5A). The body weights of mice in the vehicle and EC359 treated groups remained unchanged (Fig. 5B) confirming the low toxicity of EC359. Moreover, EC359 treated tumors exhibited fewer proliferating cells (Ki-67 positive cells) compared to vehicle treated tumors (Fig. 5C). Additionally, RT-qPCR analysis confirmed significant decrease in the activation of STAT3 target genes in EC359 treated tumors compared to vehicle (Fig. 5D). Western blot analysis confirmed that xenograft tumors express LIFR and LIF (Fig. 5E). Further, EC359 treatment substantially reduced the phosphorylation of STAT3, ERK½ and Akt in tumors compared to vehicle treated tumors (Fig. 5E). Collectively, these results suggest that EC359 has potent anti-tumor activity on TNBC in preclinical models.
Figure 5.
EC359 inhibits the growth of TNBC xenograft tumors. A, MDA-MB-231 xenografts (n=8) were treated with vehicle or EC359 (5mg/kg/s.c./3 days/week). Tumor volumes are shown in the graph. B, Body weights of vehicle and EC359 treated mice are shown. C, Ki-67 expression as a marker of proliferation was analyzed by IHC and quantitated. D, Status of STAT3 target genes were measured by using RT-qPCR analysis (n=3). E, LIFR downstream signaling was measured using western blotting (data using two different xenograft tumors is shown). * P<0.05, ** P<0.01, *** P<0.001, ****p<0.0001.
EC359 has activity against primary patient derived TNBC explants and reduced in vivo tumor progression in PDX model.
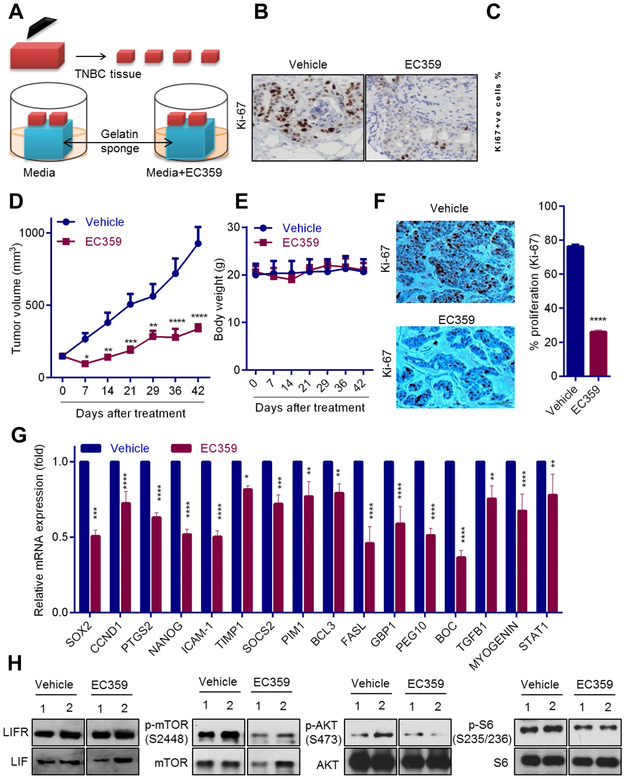
We tested the utility of EC359 using an ex vivo culture model of primary breast tumors, which allowed for the evaluation of drugs on human tumors while maintaining their native tissue architecture (Fig. 6A). Briefly, surgically extirpated de-identified TNBC tissues were cut into small pieces and placed on gelatin sponge soaked in the culture medium and grown for a short term in the presence of vehicle or EC359 (Fig. 6A). Treatment of TNBC explants with EC359 substantially decreased their proliferation (Ki-67 positivity) compared to vehicle treated tumors (Fig. 6B,C). Next, we tested the effect of EC359 on PDX tumor growth in vivo. EC359 treatment significantly reduced the tumor progression compared to the vehicle treated control group (Fig. 6D) and did not affected body weight (Fig. 6E). EC359 treated PDX tumors exhibited fewer proliferating cells compared to vehicle treated tumors (Fig. 6F). RT-qPCR analysis confirmed a significant decrease in the activation of STAT3 target genes in EC359 treated mice (Fig. 6G). Western blot analysis confirmed that PDX tumors express LIFR and LIF (Fig. 6H). Furthermore, EC359 treatment substantially reduced the phosphorylation of mTOR, S6 and AKT in tumors compared to vehicle treated tumors (Fig. 6H). These results suggest that EC359 has therapeutic activity on primary patient derived TNBC explants and PDX tumors.
Figure 6.
EC359 decreases the growth of patient-derived explants (PDEx) ex vivo and PDX tumors in vivo. A, Schematic representation of ex vivo culture model. B, TNBC explants were treated with EC359 for 72 h and the proliferation was determined using Ki-67 immunostaining. Representative Ki-67 staining from one tumor treated with vehicle or EC359 is shown. C, The Ki67 expression in TNBC explants (n=3) is quantitated. D, TNBC PDX tumors (n=6) were treated with vehicle or EC359 (10mg/kg/s.c./3 days/week). Tumor volumes are shown in the graph. E, Body weights of vehicle and EC359 treated mice are shown. F, Ki-67 expression as a marker of proliferation was analyzed by IHC and quantitated. G, STAT3 target genes were measured by using RT-qPCR analysis (n=3). H, Status of LIFR downstream signaling was measured using western blotting (data using two different PDX tumors is shown). * P<0.05, ** P<0.01, *** P<0.001, ****p<0.0001.
Discussion
Leukemia inhibitory factor (LIF) is the most pleiotropic member of the interleukin-6 family of cytokines (4) that signals via the LIFR (5). Recent evidence suggested tumors exhibit upregulated LIF/LIFR signaling via autocrine and paracrine mechanisms (1,5-7). However, lack of specific inhibitors targeting LIF/LIFR axis represents a critical barrier in the field. In this study, we rationally designed a small organic molecule, EC359 that emulates the LIF/LIFR binding site and functions as a first-in-class LIFR inhibitor from a library of compounds. Using multiple TNBC cells, we demonstrated that EC359 decreases cell viability, invasion, and promotes apoptosis. Mechanistic studies using Western blot, reporter gene assays, and RT-qPCR confirmed significant reduction of activation of LIF/LIFR mediated pathways. Utilizing, xenograft, patient-derived xenograft (PDX), and patient-derived explant (PDEx) models, we demonstrated the in vivo efficacy of EC359.
The molecular modeling and SPR suggests the putative binding site is at the interface of LIF and LIFR. EC359 may display longer resident time (i.e. slower koff) in the LIF/LIFR complex as suggested by the molecular model. Reasonable (about 30min) residence time (1/koff) suggests it may display biological activity in vitro and in vivo. However, potency of EC359 may depend on the concentration of LIF. During the SPR assay, we noted that EC359 was incompletely dissolved in the running buffer. Thus, due to poor solubility of the inhibitor in running buffer (inhibitor precipitates at concentrations 25–50 μM in 5% DMSO) derived kinetic constants must be considered approximate; spiking at 2µM (Green color) suggest that the compound may be aggregated; micro-aggregation affect not only the transport property of the ligands, but also the off-rate. Thus, the weaker binding affinity derived from SPR studies are likely due to poor solubility of the compound, which may be attributed as limitation of SPR technique. Nonetheless, results from SPR show that EC359 is specific to LIFR.
Recently, the MST technique has been widely used for characterizing protein-ligand interactions. MST offers a unique advantage over conventional isothermal titration calorimetry (ITC); unlike SPR, in MST the target is not immobilized, and ligand-binding is independent of size or physical properties of ligands. MST analysis indicated higher binding affinity (KD) between LIF and LIFR (1.36 nM) than LIFR and EC359 (10.2 nM). Also the longer residence time / slower koff in SPR demonstrates this pertinent biological effect. These values were consistent with high nM potency of EC359 in vitro and in vivo. The discrepancy in the binding affinity measured between SPR and MST assays may be due to either difference in steady-state binding (MST) vs kinetic binding (SPR), or drug-induced structural changes upon binding; structural changes at the binding site, or both. Despite the differeces in SPR and MST techniques, our results show that EC359 directly binds and disrupts the LIFR signaling complex.
The LIF/LIFR axis exhibits differential effects, which depended on the cell type include stimulating or inhibiting cell proliferation, differentiation and survival (4,11,24). LIFR is also reported to function as a metastasis suppressor through the Hippo-YAP pathway (35) and confer a dormancy phenotype in breast cancer cells disseminating to bone (36). It should be noted that presence of LIF is important for LIFR activation as we have determined in our SPR analysis. Hence studies using low LIF expressing cell lines such as MCF-7 or T47D may not have an overly active LIF/LIFR signaling. However, LIFR signaling is complex as multiple ligands activate LIFR including LIF, CNTF, OSM and CTF1. Despite the ability of LIF to activate JAK1/STAT3, PI3K/AKT and MAPK pathways in these cell line, differences in signaling outcome may in part arise from differential levels of activation of these three pathways, multiple ligands to LIFR and differences in tumor micro environment (TME) (1,37).
Earlier studies revealed that LIF, CTF1, and OSM share an overlapping binding site located in the Ig-like domain of LIFR and different behaviors of LIF, CTF1, and OSM can be related to the different affinity of their site for LIFR (38). Our modelling studies predicted that EC359 will interact at the LIF-LIFR binding interface and block interaction of LIF to LIFR. In agreement with published studies, our reporter assays and western blot analyses showed that EC359 has the ability to block the signaling mediated by other cytokines (CTF1, CNTF, and OSM) that interact LIFR at LIF/LIFR interface. Blockage of LIFR by EC359 can leverage additional benefit of interfering the LIFR-JAK-STAT pathway by all known four LIFR ligands. We speculate that the unique ability of EC359 to bind the common ligand binding site blocks multiple ligand’s interactions with LIFR, offers an advantage over other biologics or small molecules that can only target either of these ligands alone. This may also account the apparent differences in the activity seen by EC359 in TNBC and ER+BC tumors as TNBC expresses higher levels LIFR ligands compared to ER+BC. In SPR studies, we found that the presence of ligand (LIF) further enhanced LIFR interaction with EC359 compared to LIFR alone. Since the ER+BC cells lack or possess low levels of LIF and LIFR, the increased fold difference in activity (sensitivity) of EC359 towards TNBC cells may reflect presence of increased ligand/receptor levels in TNBC. Further, EC359 is unable to block OSM, CTF1, and CNTF, interactions with their natural receptors (OSMR/gp130, LST/gp130, CNTFR/gp130 respectively); therefore, EC359 is less likely to affect the physiological signaling of CTF1, CNTF, and OSM. As a consequence, the issue of toxicity is less likely occur. Accordingly, in xenograft studies we did not observed toxicity over the course of EC359 treatment. However, future studies using formal toxicity protocols are needed to address the toxicity concerns and is beyond the current scope of this work.
Breast cancer cells often exhibit autocrine stimulation of LIF-LIFR axis. Some subtypes of TNBC such as claudin-low are highly enriched for CSC markers (39,40). The LIF promoter is hyper-methylated in normal breast epithelial cells, but extensively demethylated during breast cancer progression (5). TNBC cells have higher expression of LIF and LIFR compared to ER+BC cells and overexpression of LIF is significantly associated with a poorer relapse free survival in breast cancer patients (4). Together, these emerging findings strongly suggest that LIF signaling in TNBC may be clinically actionable and that disruption of the LIF signaling cascade has potential to block progression of subtypes of TNBC that exhibit a LIF/LIFR autocrine loop.
LIF activates multiple signaling pathways via LIFR including STAT3, MAPK, AKT, and mTOR (3,4) all are implicated in cancer progression. Tumors exhibit upregulated LIF-JAK-STAT3 signaling via autocrine and paracrine mechanisms (1,5-7). LIF signaling also plays a role in crosstalk between tumor cells and fibroblasts, and mediates the pro-invasive activation of stromal fibroblasts (9). LIF/LIFR signaling is implicated in modulation of multiple immune cell types present in tumor micro environment (TME) including T-eff, T-reg, macrophages (41), and myeloid cells which results in immune suppression (42). In our studies using TNBC model cells, we found that EC359 substantially reduced the activation of STAT3, MAPK, AKT, and mTOR; and significantly delayed tumor progression in vivo. However, our mechanistic studies are limited to EC359 effects on epithelial cells; future studies are needed to clearly examine the EC359 on TME.
LIF and LIFR are over-expressed in multiple solid tumors (5,7,43). While LIF can act on a wide range of cell types, LIF knockout mice have revealed that many of these actions are not apparent during ordinary development (1), indicating a potential therapeutic window for LIF/LIFR axis inhibitors in addition to less toxicity in normal adult tissues. Considering the importance of the LIF/LIFR axis in cancer, humanized Anti LIF antibody (MSC-1) that blocks LIF signaling is being tested in a phase I clinical trial mode to determine its safety and tolerability (ClinicalTrials.gov,). Given the wide deregulation of the LIF/LIFR axis in multiple tumors, the small molecule LIFR inhibitor EC359 may have utility in treating other solid tumors including glioblastoma, ovarian cancer, colon cancer, and pancreatic cancer all of which exhibit dysregulated LIF/LIFR signaling. Our studies only examined the utility of EC359 using TNBC models. Future studies are needed to further evaluate the effects of EC359 in other cancer models and to examine any potential beneficial effects of EC359 on TME.
In summary, our data demonstrated that EC359 is a highly potent and specific LIFR inhibitor. EC359 blocked LIF/LIFR physical and functional interaction, signaling and reduced cell viability of LIF/LIFR expressing TNBC cells both in vitro and in vivo. EC359 represents an exciting new mechanism to modulate LIF/LIFR oncogenic functions. Since EC359 is a small, stable molecule, it is amenable for translation to clinical trials for patients with TNBC as either monotherapy or in combination with current standard of care.
Supplementary Material
Acknowledgments
General: We thank Perry Jessica (Ob/Gyn UT Health San Antonio) for proofreading of the manuscript. MST studies were performed by 2bind GmbH, Germany. K.V. Dileep, thanks Japan Society for the Promotion of Science for a postdoctoral fellowship.
Funding: This study was supported by the DOD BCRP grant W81XWH-18–1-0016 (R.K. Vadlamudi; K.J. Nickisch); and DOD BCRP grant W81XWH-16–0294 (R.R.Tekmal); NCI Cancer Center Support Grant P30CA054174–17; Max and Minnie Tomerlin Voelcker Fund (G.R.Sareddy) and NIH grant 1R01CA179120–01 (R.K.Vadlamudi; M. Rao).
Footnotes
Data and materials availability: All data supporting the conclusions is included in the paper and/or in the Supplementary Materials.
Competing interests: Authors including Bindu Santhamma, Alejandra Chávez-Riveros, Vijaya Manthati, Hareesh B. Nair, and Klaus J. Nickisch are either current or former employees of Evestra. All other authors declare no potential conflicts of interest.
References
- 1.Nicola NA, Babon JJ. Leukemia inhibitory factor (LIF). Cytokine Growth Factor Rev 2015;26(5):533–44 doi 10.1016/j.cytogfr.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stahl N, Boulton TG, Farruggella T, Ip NY, Davis S, Witthuhn BA, et al. Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL-6 beta receptor components. Science 1994;263(5143):92–5. [DOI] [PubMed] [Google Scholar]
- 3.Liu SC, Tsang NM, Chiang WC, Chang KP, Hsueh C, Liang Y, et al. Leukemia inhibitory factor promotes nasopharyngeal carcinoma progression and radioresistance. J Clin Invest 2013;123(12):5269–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Yang Q, Yu H, Wu L, Zhao Y, Zhang C, et al. LIF promotes tumorigenesis and metastasis of breast cancer through the AKT-mTOR pathway. Oncotarget 2014;5(3):788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin JE, Park SH, Jang YK. Epigenetic up-regulation of leukemia inhibitory factor (LIF) gene during the progression to breast cancer. Mol Cells 2011;31(2):181–9 doi 10.1007/s10059-011-0020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penuelas S, Anido J, Prieto-Sanchez RM, Folch G, Barba I, Cuartas I, et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell 2009;15(4):315–27 doi 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Wu L, Yu H, Zhao Y, Zhang C, Wang J, Yue X, et al. HIF-2alpha mediates hypoxia-induced LIF expression in human colorectal cancer cells. Oncotarget 2015;6(6):4406–17 doi 10.18632/oncotarget.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H, Cheng Y, Martinka M, McElwee K. High LIFr expression stimulates melanoma cell migration and is associated with unfavorable prognosis in melanoma. Oncotarget 2015;6(28):25484–98 doi 10.18632/oncotarget.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albrengues J, Bourget I, Pons C, Butet V, Hofman P, Tartare-Deckert S, et al. LIF mediates proinvasive activation of stromal fibroblasts in cancer. Cell Rep 2014;7(5):1664–78. [DOI] [PubMed] [Google Scholar]
- 10.Morton SD, Cadamuro M, Brivio S, Vismara M, Stecca T, Massani M, et al. Leukemia inhibitory factor protects cholangiocarcinoma cells from drug-induced apoptosis via a PI3K/AKT-dependent Mcl-1 activation. Oncotarget 2015;6(28):26052–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kellokumpu-Lehtinen P, Talpaz M, Harris D, Van Q, Kurzrock R, Estrov Z. Leukemia-inhibitory factor stimulates breast, kidney and prostate cancer cell proliferation by paracrine and autocrine pathways. Int J Cancer 1996;66(4):515–9 doi . [DOI] [PubMed] [Google Scholar]
- 12.Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 2005;132(5):885–96 doi 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 13.Kuphal S, Wallner S, Bosserhoff AK. Impact of LIF (leukemia inhibitory factor) expression in malignant melanoma. Exp Mol Pathol 2013;95(2):156–65. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Yu H, Hu W. LIF is a new p53 negative regulator. J Nat Sci 2015;1(7):e131. [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B, Lu Y, Li J, Liu Y, Liu J, Wang W. Leukemia inhibitory factor promotes tumor growth and metastasis in human osteosarcoma via activating STAT3. APMIS 2015;123(10):837–46 doi 10.1111/apm.12427. [DOI] [PubMed] [Google Scholar]
- 16.Kamohara H, Ogawa M, Ishiko T, Sakamoto K, Baba H. Leukemia inhibitory factor functions as a growth factor in pancreas carcinoma cells: Involvement of regulation of LIF and its receptor expression. Int J Oncol 2007;30(4):977–83. [PubMed] [Google Scholar]
- 17.Shin JE, Park SH, Jang YK. Epigenetic up-regulation of leukemia inhibitory factor (LIF) gene during the progression to breast cancer. Mol Cells 2011;31(2):181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med 2010;363(20):1938–48. [DOI] [PubMed] [Google Scholar]
- 19.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol 2010;7(12):683–92. [DOI] [PubMed] [Google Scholar]
- 20.Comprehensive molecular portraits of human breast tumours. Nature 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Zhang H, Zhao Y, Wang C, Cheng Z, Tang L, et al. A mandatory role of nuclear PAK4-LIFR axis in breast-to-bone metastasis of ERalpha-positive breast cancer cells. Oncogene 2019;38(6):808–21 doi 10.1038/s41388-018-0456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quaglino A, Schere-Levy C, Romorini L, Meiss RP, Kordon EC. Mouse mammary tumors display Stat3 activation dependent on leukemia inhibitory factor signaling. Breast Cancer Res 2007;9(5):R69 doi 10.1186/bcr1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhingra K, Sahin A, Emami K, Hortobagyi GN, Estrov Z. Expression of leukemia inhibitory factor and its receptor in breast cancer: a potential autocrine and paracrine growth regulatory mechanism. Breast Cancer Res Treat 1998;48(2):165–74. [DOI] [PubMed] [Google Scholar]
- 24.Yue X, Zhao Y, Zhang C, Li J, Liu Z, Liu J, et al. Leukemia inhibitory factor promotes EMT through STAT3-dependent miR-21 induction. Oncotarget 2016;7(4):3777–90 doi 10.18632/oncotarget.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raj GV, Sareddy GR, Ma S, Lee TK, Viswanadhapalli S, Li R, et al. Estrogen receptor coregulator binding modulators (ERXs) effectively target estrogen receptor positive human breast cancers. Elife 2017;6 doi 10.7554/eLife.26857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sareddy GR, Viswanadhapalli S, Surapaneni P, Suzuki T, Brenner A, Vadlamudi RK. Novel KDM1A inhibitors induce differentiation and apoptosis of glioma stem cells via unfolded protein response pathway. Oncogene 2017;36(17):2423–34 doi 10.1038/onc.2016.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skiniotis G, Lupardus PJ, Martick M, Walz T, Garcia KC. Structural organization of a full-length gp130/LIF-R cytokine receptor transmembrane complex. Mol Cell 2008;31(5):737–48 doi 10.1016/j.molcel.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huyton T, Zhang JG, Luo CS, Lou MZ, Hilton DJ, Nicola NA, et al. An unusual cytokine:Ig-domain interaction revealed in the crystal structure of leukemia inhibitory factor (LIF) in complex with the LIF receptor. Proc Natl Acad Sci U S A 2007;104(31):12737–42 doi 10.1073/pnas.0705577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halgren T New method for fast and accurate binding-site identification and analysis. Chem Biol Drug Des 2007;69(2):146–8 doi 10.1111/j.1747-0285.2007.00483.x. [DOI] [PubMed] [Google Scholar]
- 30.Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov 2015;10(5):449–61 doi 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy SS, Gonugunta VK, Bandyopadhyay A, Rao MK, Goodall GJ, Sun LZ, et al. Significance of PELP1/HDAC2/miR-200 regulatory network in EMT and metastasis of breast cancer. Oncogene 2014;33(28):3707–16 doi 10.1038/onc.2013.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson RC, Grey LM, Staunton D, Vankelecom H, Vernallis AB, Moreau JF, et al. The crystal structure and biological function of leukemia inhibitory factor: implications for receptor binding. Cell 1994;77(7):1101–16. [DOI] [PubMed] [Google Scholar]
- 33.Seidel SA, Dijkman PM, Lea WA, van den Bogaart G, Jerabek-Willemsen M, Lazic A, et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 2013;59(3):301–15 doi 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol 2011;9(4):342–53 doi 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen D, Sun Y, Wei Y, Zhang P, Rezaeian AH, Teruya-Feldstein J, et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med 2012;18(10):1511–7 doi 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson RW, Finger EC, Olcina MM, Vilalta M, Aguilera T, Miao Y, et al. Induction of LIFR confers a dormancy phenotype in breast cancer cells disseminated to the bone marrow. Nat Cell Biol 2016;18(10):1078–89 doi 10.1038/ncb3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auernhammer CJ, Melmed S. Leukemia-inhibitory factor-neuroimmune modulator of endocrine function. Endocr Rev 2000;21(3):313–45 doi 10.1210/edrv.21.3.0400. [DOI] [PubMed] [Google Scholar]
- 38.Plun-Favreau H, Perret D, Diveu C, Froger J, Chevalier S, Lelievre E, et al. Leukemia inhibitory factor (LIF), cardiotrophin-1, and oncostatin M share structural binding determinants in the immunoglobulin-like domain of LIF receptor. J Biol Chem 2003;278(29):27169–79 doi 10.1074/jbc.M303168200. [DOI] [PubMed] [Google Scholar]
- 39.D’Amato NC, Ostrander JH, Bowie ML, Sistrunk C, Borowsky A, Cardiff RD, et al. Evidence for phenotypic plasticity in aggressive triple-negative breast cancer: human biology is recapitulated by a novel model system. PLoS One 2012;7(9):e45684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist 2010;15 Suppl 5:39–48. doi: 10.1634/theoncologist.2010-S5-39.:39–48. [DOI] [PubMed] [Google Scholar]
- 41.Duluc D, Delneste Y, Tan F, Moles MP, Grimaud L, Lenoir J, et al. Tumor-associated leukemia inhibitory factor and IL-6 skew monocyte differentiation into tumor-associated macrophage-like cells. Blood 2007;110(13):4319–30 doi 10.1182/blood-2007-02-072587. [DOI] [PubMed] [Google Scholar]
- 42.Zhao X, Ye F, Chen L, Lu W, Xie X. Human epithelial ovarian carcinoma cell-derived cytokines cooperatively induce activated CD4+CD25-CD45RA+ naive T cells to express forkhead box protein 3 and exhibit suppressive ability in vitro. Cancer Sci 2009;100(11):2143–51 doi 10.1111/j.1349-7006.2009.01286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bressy C, Lac S, Nigri J, Leca J, Roques J, Lavaut MN, et al. LIF Drives Neural Remodeling in Pancreatic Cancer and Offers a New Candidate Biomarker. Cancer Res 2018;78(4):909–21 doi 10.1158/0008-5472.CAN-15-2790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.