Abstract
Multiple studies have associated the transcription factor IRF1 with tumor suppressive activities. Here we report an opposite tumor cell–intrinsic function of IRF1 in promoting tumor growth. IRF1-deficient tumor cells showed reduced tumor growth in MC38 and CT26 colon carcinoma and B16 melanoma mouse models. This reduction in tumor growth was dependent on host CD8+ T cells. Detailed profiling of tumor-infiltrating leukocytes did not show changes in the various T cell and myeloid cell populations. However, CD8+ T cells that had infiltrated IRF1-deficieint tumors in vivo exhibited enhanced cytotoxicity. IRF1-deficient tumor cells lost the ability to upregulate PD-L1 expression in vitro and in vivo and were more susceptible to T cell-mediated killing. Induced expression of PD-L1 in IRF1-deficient tumor cells restored tumor growth. These results indicate differential activity of IRF1 in tumor escape.
INTRODUCTION
The Interferon regulatory factors (IRF) are transcription factors involved in cellular stress responses. Depending on the cellular context, specific members of the IRF family are responsible for the induction of Interferons (IFN), lymphocyte development and oncogenic signaling (1–3). Due to the role in inducing type I IFN, which mediates immunosurveillance of tumors, a number of IRFs, such as IRF1, IRF3, IRF7 have been ascribed as anti-tumorigenic factors, whereas both pro and anti-tumor functions have been reported for the other IRFs (4). In fact, in an in vivo genetic screen using the lung metastasis model of mouse B16-F10 melanoma, IRF1 knockout mice were found to have the highest metastasis score; IRF7 knockout mice also had a higher metastatic score than wild-type (WT) (5). IRF1 is lost or reduced in expression in a number of human leukemias (6–8). This and other cellular studies (9,10) have suggested an anti-tumorigenic role of IRF1. However, a tumor cell–intrinsic role of IRF1 in solid tumors to affect tumor progression is not clear.
Despite the success of immune checkpoint blockade (ICB) therapy in different cancers, resistance and relapses are common (11,12). ICB is based on the finding that a majority of intratumoral T cells are ineffective in their effector function due to inhibitory signaling through T-cell receptors such as CTLA4 and PD-1. Therefore, blocking of these inhibitory signaling using neutralizing antibody should reinvigorate the cytotoxic function of the effector T cells to clear the tumor. However, one mechanism of resistance, especially for the ICB therapy targeting the PD-1 axis, is the upregulation of PD-L1, a ligand for the T-cell inhibitory receptor PD-1. PD-L1 is expressed on tumor cells and tumor-associated macrophages, where its transcription is induced by multiple signals including cytokines such as IFNγ, IFNα/β, TNFα, and other various TLR and oncogenic signals (13). Transcriptional regulation of steady-state PD-L1 mRNA expression is controlled through 3’-UTR mediated RNA-decay (14,15). A number of studies have identified correlation between genetic changes in the IFNγ signaling and the ICB therapy resistance (16,17). However, mechanisms for primary and acquired resistance to PD-1/PD-L1 inhibition are varied and can be both multifactorial and overlapping (18).
IRF1 is an early target gene downstream of IFNγ signaling and modulates IFNγ-mediated gene induction (19). IRF1 also regulates constitutive and inducible expression of PD-L1 by IFNγ (20–23). This led us to hypothesize that IRF1 might play a different role in tumor cells than in immune cells in determining the outcome of tumor progression. Here, using syngeneic mouse implantable tumor models, we show a tumor cell–intrinsic pro-tumorigenic role of IRF1. IRF1-deficiency in the tumor cell results in reduced tumor progression. We found that IRF1 is necessary for PD-L1 upregulation in tumor cells and tumor progression in vivo. Although IRF1-deficiency did not result in changes in the tumor infiltrating lymphocyte profiles, the cytotoxicity of the tumor infiltrating CD8+ T cells was increased in tumors arising from IRF1-deficient cells compared to those from the parental cells (here onwards designated as WT cells). These results indicate a tumor enhancing effect of IRF1 in the tumor cell that is distinct from its role in the host immune cell.
MATERIALS AND METHODS
Cells and reagents
CT26 and B16-F10 cells were purchased from ATCC. Murine colon adenocarcinoma cell line MC38 was a generous gift from Dr. Dario Vignali. All cell lines were used within 10 generations. Cell lines were not authenticated, but checked monthly for mycoplasma contamination by DAPI staining and commercial PCR (Roche Diagnostics). MC38 and CT26 cells were maintained in DMEM (Corning, Corning, NY)) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA) and Penicillin/Streptomycin (Lonza, Basel, Switzerland). B16-F10 cells were maintained in RPMI (Corning) supplemented with 10% FBS and Penicillin/Streptomycin.
Generating IRF1-deficient (IRF1-KO) and PD-L1 expressing cell lines
CRISPR-Cas9–mediated genome editing was carried out as described (24–26), with the use of murine IRF1 target sequence GAAGCACGCTGCTAAGCACGG. Briefly, plasmid encoding gRNA, Cas9-mCherry was transfected into MC38, CT26 and B16-F10 parental cell lines with lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA). After 48 h of transient transfection, mCherry+ single cells were sorted into 96-well cell culture plates. Single cell clones were expanded and transferred into 12-well cell culture plate. Cells were treated with 100 ng/mL mouse IFNγ (Biolegend, San Diego, CA) for 6 h before harvesting. We examined the expression of IRF1 via western blot.
Mouse PD-L1 coding region (PD-L1) was PCR amplified from pUNO mouse CD274 (Addgene Plasmid# 107012) using primers 5’- CACCATGAGGATATTTGCTGGCATTATAT TCAC - 3’) and 5’ - GAGTTTGGTGACTACATCTTAAGATCTATCATGTCGTC - 3, TOPO cloned into pENTR D-TOPO and transferred into pInducer20 vector using Gateway Cloning (Thermo Fisher Scientific). B16-F10 IRF1-KO cells were transduced with lentivirus prepared from pInducer20-PD-L1 and selected by G418 (800 μg/mL) to make a stable cell line. Doxycycline (DOX) induced expression of PD-L1 in B16-F10 IRF1-KO cells was confirmed by western blot.
Mice and tumor models
All mice experiments were conducted in accordance with, and with the approval of, University of Pittsburgh Institutional Animal Care and Use Committee (IACUC). All mice were housed in specific pathogen-free conditions and used at 6–8 weeks old. C57BL/6J and BALB/c mice were obtained from The Jackson Laboratory. C57BL/6J mice were subcutaneously inoculated with 106 MC38 cells or intradermally inoculated with 5 × 105 B16-F10 cells. BALB/cJ mice were subcutaneously inoculated with 106 CT26 cells. Each dose of tumor cells was suspended in 100 μL of PBS (Lonza). For CD8+ T-cell depletion studies, mice were intraperitoneal injected with anti-mouse CD8α (Clone YTS 169.4, 250 μg/mouse) (Bio X Cell, West Lebanon, NH) or rat IgG2b isotype control (Clone LTF-2, 250 μg/mouse) (Bio X Cell) 2 days before tumor cell inoculation and then twice a week. For re-challenge studies, after 1 month of complete tumor regression with MC38 IRF1-KO cells, mice were re-inoculated with 106 MC38 cells. In all the tumor studies, tumor volumes were measured and calculated twice a week with the formula V = 0.52*width2*length.
Detection of tumor specific memory T cell
To detect tumor-specific memory T cells, spleens and brachial draining lymph nodes (dLNs) were collected after 1 month of complete tumor regression with CT26 IRF1-KO cells. Naïve mice and CT26 IRF1-KO cell injected tumor bearing mice (day 45 of post-injection) were used as negative and positive controls respectively. Spleens and dLNs were mechanically processed between frosted glass slides to generate single cell suspensions in RPMI 1640 with 10% FBS. Cells were resuspended in flow cytometry staining (FCS) buffer (2% FBS in 1X PBS), blocked with Clear Back FC receptor blocking reagent (MBL Intl., Woburn, MA) for 5 minutes at room temperature and stained with PE-conjugated AH1 MHC Class I tetramer (H-2Ld MuLV gp70 SPSYVYHQF, MBL Intl.) for 30 minutes at 4°C. Antibodies to cell-surface markers [anti-CD8 (clone KT15, MBL Intl., anti-CD44 (clone IM7, Biolegend), anti-CD62L (clone MEL-14, Biolegend), anti-CD45 (clone 30-F11, Biolegend)] were added to a final concertation of 1:500 and cells were stained for 30 minutes at 4°C. Cells were washed twice with FCS buffer, stained with eBioscience Fixable Viability Dye eFluor780 at 1:4000 dilution in 1x PBS for 10 min at 4°C, washed twice with FCS buffer, and fixed in eBioscience Fixation/Permeabilization reagent for 15 minutes at room temperature in the dark. Fixed cells were washed once with 1x eBioscience Permeabilization Buffer prior to a final wash with FSC buffer and storage in FCS buffer at 4°C protected from light. A BD LSRFortessa cytometer and FACSDiva software (BD Biosciences, San Jose, CA) were used to collect uncompensated data. Compensation and analyses were performed using FlowJo (FlowJo, Ashland, OR) v10 software. We designated naïve T cells = CD45+ CD44− CD62L+, central memory T cells (TCM) = CD45+ CD44+ CD62L+, and effector memory T cells (TEM) = CD45+ CD44+ CD62L−.
Tumor infiltrating lymphocyte analysis
Tumor and lymph nodes were collected on post-injection days 12 and 14 when the tumor sizes between B16-F10 WT and IRF1-KO groups did not show significant difference. Collected tumors were injected with 0.5 – 1 mL/tumor digestion mixture [2 mg/mL collagenase IV (Thermo Fisher Scientific), 200 μg/mL Hyaluronidase V (Sigma-Aldrich, St. Louis, MO), and 4 U/mL DNase I (Sigma-Aldrich)] and incubated at 37°C for 20 min. Tumors were dissociated between two frosted glass slides and filtered through 40 μM cell strainer to make single-cell suspensions. Lymph nodes were mechanically smashed through a 40 μM cell strainer to get single-cell suspensions.
Cells from tumors and lymph nodes were resuspended in FCS buffer and stained with the following fluorescence-conjugated Abs: anti-CD8 (clone 53–6.7, Biolegend), anti-Tim3 (clone RMT3–23, Biolegend), anti-CD3 (clone 145–2C11, eBioscience), anti-LAG3 (clone C9B7W, Biolegend), anti-PD-1 (clone 29F.1A12, Biolegend), anti-CD45 (clone 30-F11, Biolegend), anti-PD-L1 (clone 10F.9G2, Biolegend), anti-CD4 (clone RM4–5, eBioscience), anti-CD25 (clone PC61.5, eBioscience), anti-Foxp3 (clone FJK-16s, eBioscience), anti-Ly6c (clone HK1.4, eBioscience), anti-Ly6G (Gr-1) (clone RB6–8C5, eBioscience), anti-CD11b (clone M1/70, eBioscience), anti-CD335 (NKp46) (clone29A1.4, Biolegend), anti-CD19 (clone 6D5, Biolegend), , anti-TNFα (clone MP6-XT22, Biolegend), anti-IFNγ (clone XMG1.2, Biolegend). Cells stained for Foxp3 and/or intracellular cytokines (TNFα and IFNγ) were fixed and permeabilized with the Foxp3/Transcription Factor Staining Buffer Set) (eBioscience). Flow cytometry data was acquired and analyzed as above. We designated, exhausted T cells = CD3+, CD8+, PD-1high, Tim3+; T regulatory cells (Treg) = CD3+, CD4+, CD25+, Foxp3+; granulocytic myeloid derived suppressor cells (GrMDSC) = CD45+, CD11b+, Gr-1+; monocytic MDSC = CD45+, CD11b+, Ly6c+; NK cells = CD3−, C19+, CD335+.
For intracellular cytokine staining, isolated tumor cells and lymph node cells were cultured in 96-well plates with R10 media containing 100 ng/mL PMA and 500 ng/mL Ionomycin (Fisher Scientific, Hampton, NH) overnight. The next day, cells were incubated with BD GolgiPlug Protein Transport Inhibitor (1:1000) for 4 h. After incubation, cells were harvested for intracellular cytokine staining as described above.
IFNγ induced PD-L1 and MHC I expression in vitro
B16-F10 WT and IRF1-KO cells were treated with 100 ng/mL of IFNγ for different time points and then trypsinized for harvesting. Harvested cells were subjected to regular flow staining with anti-PD-L1 and anti-MHC I (H-2Kb) (clone AF6–88.5, Biolegend).
RNA expression analysis
Total RNA was isolated from mouse IFNγ treated B16-F10 WT and IRF1-KO cells at different time points using Trizol (Thermo Fisher Scientific). cDNA was synthesized using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Quantitative real-time PCR using Bio-Rad PrimePCR probes and SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) was conducted in a CFX96 Real Time System (Bio-Rad) following the manufacturer’s instructions. All real-time PCR amplification was normalized to GAPDH. The PrimePCR Probe Assay IDs for mouse CD274 (PD-L1), Gbp2, Icam1 and Stat1 are qMmuCEP0052618, qMmuCEP0056450, qMmuCEP0056911, qMmuCEP0054514.
Immunoblotting
B16-F10 WT and IRF1-KO cells were lysed after mouse IFNγ treatment using cell lysis buffer (150 mM NaCl, 1.5 mM MgCl2, 2 mM EGTA, 2 mM DTT, 10 mM NaF, 12.5 mM β-Glycerophosphate, 1 mM Na3VO4, 1 mM PMSF, 1% Triton-X100, 0.1% Sodium Deoxycholate, and protease inhibitors). The cleared cell lysates were subjected to SDS-PAGE and transferred to nylon membrane. The blots were incubated with anti-mouse PD-L1 (Biolegend) and IRF1 (D5E4) XP Rabbit mAb (Cell Signaling Technology, Danvers, MA) followed by appropriate HRP-conjugated secondary antibody and visualized by ECL detection (GE Healthcare, Chicago, IL).
In vitro cytotoxicity assay
Pmel T cells were harvested from the spleen of B6.Cg-Thy1a/Cy Tg(TcraTcrb)8Rest/J mice and stimulated with gp100 peptide. After the stimulation, Pmel CD8+ T cells were cultured in R10 media with 50 U/mL of mouse IL-2 (PeproTech Inc., Rocky Hill, NJ) for expansion for 7 days. B16-F10 WT and IRF1-KO cells were plated into 96-well plates at 10,000 cells per well and incubated for 5 h. Pmel CD8+ T cells were prepared in R10 media without IL-2 and added into the 96-well plate at T cell : tumor cell ratio of 10:1, 5:1, 2:1, 1:1 and 1:2. Cells were incubated O/N and harvested for cell viability examination using Zombie Aqua Fixable Viability Kit (Biolegend) and anti-CD8 (clone 53–6.7, eBioscience).
Statistics
Results shown are pooled samples from at least twice repeated in vivo infection studies. For each data point, mean and SEM were plotted. Statistical significance was calculated either by Student’s T-test or two-way ANOVA with Sidak’s multiple comparison test as appropriate and represented as * P < 0.03 and *** P < 0.001.
RESULTS
Loss of IRF1 in tumor cells causes tumor regression in mice.
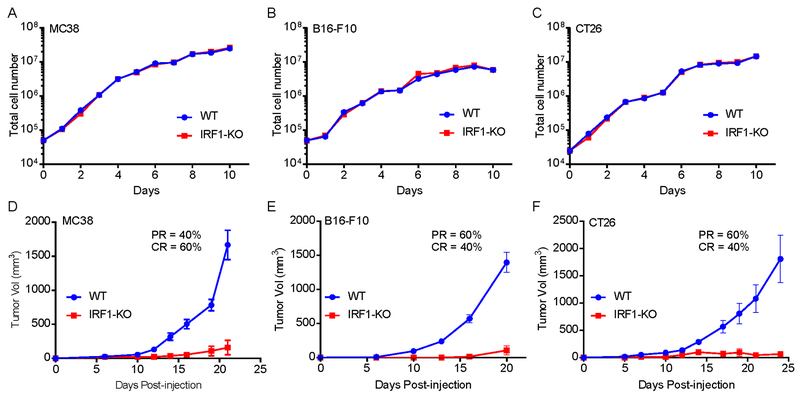
To investigate the tumor intrinsic role of IRF1 during tumor progression, we generated several IRF1-deficient (IRF1-KO) syngeneic murine tumor cell lines (MC38, B16-F10 and CT26) via CRISPR/Cas9-mediated genome editing (Supplementary Fig. S1A–C) and compared their growth rates with WT cells both in vitro and in vivo. As shown in Fig. 1A–C, there was no difference in growth rates between IRF1-KO and WT cells when cultured in vitro. However, when these cells were injected in syngeneic mice (MC38 and B16-F10 in C57BL/6J and CT26 in BALB/c mice respectively) the IRF1-KO cells showed significantly reduced tumor growth than WT cells (Fig. 1D–F). Additionally, in some mice injected with IRF1-KO cells, tumors disappeared from the injection site around the third week of post-injection. For example, in MC38, B16-F10 and CT26 IRF1-KO cell-injected mice, 2/5, 3/5 and 3/5 mice had complete regression of tumors respectively (Supplementary Fig. S1D–F). Taken together these results indicate that in tumor cells IRF1 may positively contribute in immune escape of the tumor cells.
Fig. 1: Knockout of IRF1 in tumor cells causes a loss of in vivo tumorigenicity.
(A), (B) and (C) Cell growth rates of MC38, B16-F10 and CT26 WT and IRF1-KO in vitro. Cells were seeded and cultured in 10 wells of cell culture plates, and cell numbers were counted daily (n=3). For each data point mean and SEM were plotted.
(D), (E) and (F) Tumor growth rates of MC38 (n=5), B16-F10 (n=5) and CT26 (n=5) WT and IRF1-KO cells in C57BL/6 and/or BALBc mice. Mice were subcutaneously or intradermally injected with 106 (MC38 and CT26) or 5 ×105 (B16-F10) cells, followed by tumor growth measurements. Representative results from at least twice repeated experiments are shown.
CD8+ T cells are necessary for the loss of tumorigenicity of IRF1-KO cell.
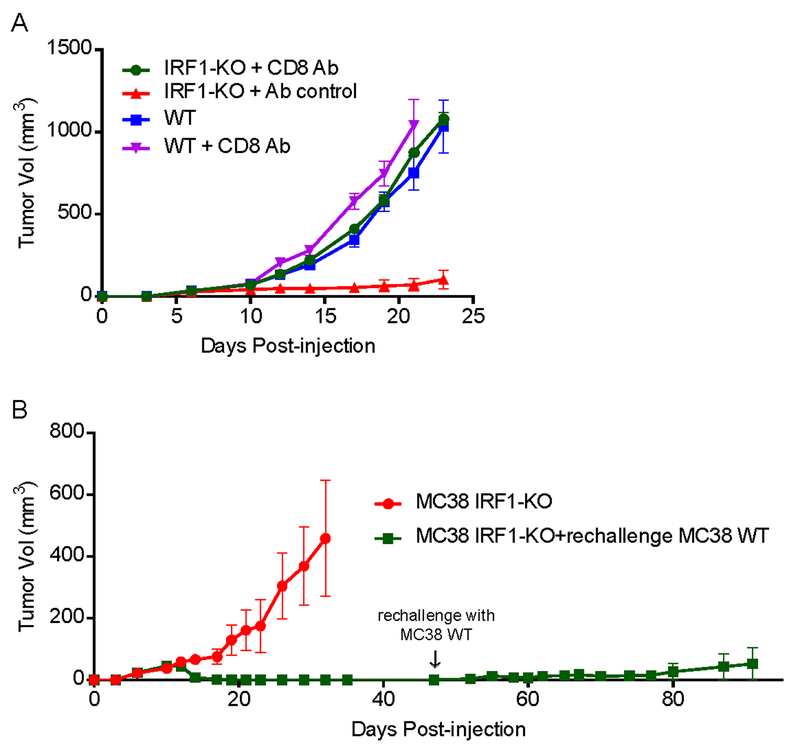
Having observed the regression of IRF1-KO tumor in mice models, we sought to investigate the contribution of host T cells to the loss of tumorigenicity by IRF1-KO tumor cells. We depleted CD8+ T cells in mice inoculated with MC38 IRF1-KO cells by intraperitoneal injection of anti-mouse CD8α and compared the IRF1-KO tumor growth with MC38 WT tumor. We found that the loss of CD8+ T cells in mice allowed the IRF1-KO tumor to grow at the same rate as WT tumor, whereas the IRF-1 KO cell inoculated mice injected with IgG2b isotype control had the same slow tumor growth compared to the other two groups of mice (Fig. 2A). In addition, depletion of CD8+ T cells in MC38 WT tumor cell injected mice, the tumor growth was faster than in normal mice (Fig. 2A). The depletion efficiency of CD8+ T cells was tested and confirmed in spleens (Supplementary Fig. S2A). Similar results were obtained when we repeated this experiment with B16-F10 WT and IRF1-KO cells (Supplementary Fig. S2B and C). These results indicate that CD8+ T cells play a role in the regression of the tumors arising from IRF1-KO cells injection.
Fig. 2: Host CD8+ T cells are required for the loss of tumorigenicity in IRF1-deficient tumor cells.
(A) Tumor growth of MC38 IRF1-KO cells in CD8+ T-cell depletion C57BL/6 mice. 106 of MC38 WT or IRF1-KO cells were subcutaneously injected into C57BL/6 mice (n=8). IRF1-KO cell injection groups of mice were intraperitoneally injected with 250 μg of anti-CD8 or rat IgG2b isotype control 2-day before tumor cell injection, then followed by antibody or isotype control injections twice a week. A group of WT MC38 cell injected mice were intraperitoneally injected with anti-CD8 as control. Representative results from at least twice repeated experiments are shown.
(B) MC38 IRF1-KO cells induced WT tumor regression. Following complete regression of MC38 IRF-1 KO tumors (n=8), mice were re-challenged with MC38 WT tumor cells on day 47 of post-injection. 7 out of 8 mice showed complete tumor regression. Representative results from at least twice repeated experiments are shown.
We also investigated whether the mice developed immunologic memory following the regression of IRF1-KO tumor cells. After 30 days of complete IRF1-KO MC38 tumor regression, we re-challenged those mice with IRF1-sufficient MC38 tumor cells and recorded the tumor growth. Twelve mice were injected with MC38 IRF1-KO cells. 4 out of 12 mice developed palpable tumors on back (red line) whereas the remaining 8 mice had complete tumor regression after post-injection day 17 (green line) (Fig. 2B). After re-challenge of WT MC38 tumor cells, 5 out of 8 mice had never developed any tumor, 2 out of 8 started developing a tumor that completely regressed later, and only one mouse developed a tumor with slow progression (Supplementary Fig. S2B).
To determine whether the tumor rejection in those re-challenged mice was related to tumor specific T cells, we examined the frequencies of tumor specific memory T cells using AH1 MHC class I tetramer (H-2Ld MuLV gp70 SPSYVYHQF), which could bind to anti-CT26 specific TCR in mice. Due to the lack of tetramer that could recognize anti-MC38 or anti-B16-F10 specific TCR, we used the tetramer which could detect anti-CT26 specific TCR. After 30 days of complete IRF1-KO CT26 tumor regression, spleens and lymph nodes were collected from those mice. The frequencies of CD8+ tetramer+ cells in either TCM or TEM cells in spleen and lymph nodes were assessed by flow cytometry. Naïve unchallenged mice were used as naïve controls. Approximately 8% of TEM cells were tetramer+ in the spleen of IRF1-KO CT26 cell challenged mice (Supplementary Fig. S2D), whereas less than 1% of tetramer+ TEM cells were found in lymph nodes (dLN) from those mice (Supplementary Fig. S2E). In addition, there were no tetramer+ TCM cells in both spleens and lymph nodes from those mice. The data suggested that IRF1-KO tumor cells caused the development of tumor specific memory T cells. And those T cells would contribute to the rejection of WT tumor cells in the re-challenge experiment. Taken together these observations showed that CD8+ T cells were responsible for the IRF-1 KO tumor regression and they could develop memory responses.
Absence of IRF1 in tumor does not change profiles of tumor infiltrating lymphocytes.
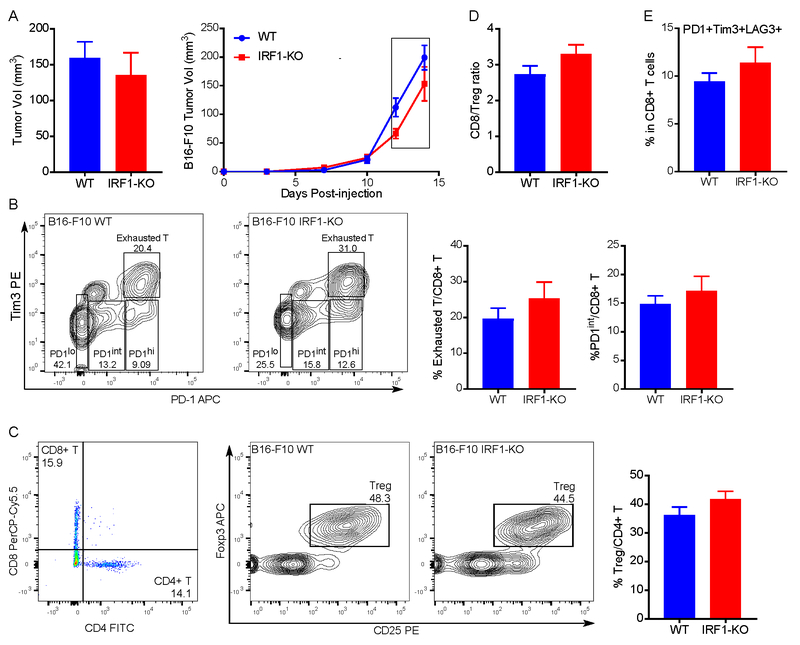
Having found that CD8+ T cells are necessary in IRF1-KO tumor regression, we examined the profile of the tumor infiltrating lymphocytes (TIL) in our model system. Following injection of the B16-F10 WT and IRF1-KO cells into C57BL/6J mice we profiled the TIL via flow cytometry. As we have shown that approximately 50% of the mice injected with IRF1-KO cells had tumor regression (Supplementary Fig. S1) and some mice never developed any measurable size of tumor after the injection, we collected the IRF1-KO tumor as early as possible before the tumor regression. In addition, we compared the TIL between WT and IRF1-KO tumors of similar sizes, days 12 – 14 post-injection (Fig. 3A).
Fig. 3: Loss of IRF1 in tumor does not influence the frequencies of tumor infiltrating lymphocytes populations.
5 × 105 of B16-F10 WT or IRF1-KO cells were intradermally injected into C57BL/6 mice (n=13, 12). Tumors were collected around size 100–200 mm3 on days 12–14 post-injection (A). (B) Representative flow cytogram (left) and tabulated (right) percentage of exhausted T cells and PD-1 intermediate expression CD8+ T cells (activated T cells) in B16-F10 WT and IRF1-KO tumor groups. (C) Representative flow cytogram (left) and tabulated (right) percentage of regulatory T cells (Tregs) in B16-F10 WT and IRF1-KO tumor groups. The ratio of CD8+ T cells : Tregs (D) and the frequencies of PD1+, Tim3+ LAG3+ in CD8+ T cells (E) in the two groups. For each column mean and SEM were plotted. Statistical significance was calculated by unpaired student t test.
Exhaustion is a state of T cells with decreased effector cytokine production, reduced cytolytic activity and overexpressed inhibitory receptors (27). Exhausted T cells are usually associated with poor tumor outcome (12). Thus, we examined the percentage of exhausted T cells defined as CD3+, CD8+, PD-1hi and Tim3+ and activated CD8+ T cells as CD3+, CD8+, PD-1int and Tim3ˉ. As shown in Fig. 3B, there was no difference in the percentage of exhausted T cells and activated CD8+ T cells between WT and IRF1-KO tumors. As regulatory T cells (Tregs) are associated with tumor progression and poor clinical outcome (28), we next compared the percentages of Tregs between these two groups. The percentage of Tregs in CD4+ T cells did not show any difference (Fig. 3C). In addition, the ratio of CD8+ T cell to Treg in IRF1-KO tumors was the same as WT tumors (Fig. 3D). Because LAG3 is also a marker to define exhausted T cells, we also assessed the frequencies of PD1+, Tim3+ and LAG3+ cells in the CD8+ T-cell population. There was no difference between the two groups (Fig. 3E). Combined with previous CD8+ T-cell depletion experiment data, it seemed that CD8+ T cells were indispensable during tumor regression. But the regression was not due to changes in percentages of exhausted T cell, activated T cells, or Tregs. This led us to investigate changes in the other lymphocyte populations, such as myeloid derived suppressor cells (MDSCs). However, we did not observe any statistically significant difference in the percentages of granulocytic MDSC and monocytic MDSC in WT and IRF1-KO tumors (Supplementary Fig. S3A and B). Additionally, both groups had the same amount of NK cells (Supplementary Fig. S3C). In summary, the results indicated that IRF1 deficiency in tumor did not quantitatively affect the TIL profile.
IRF1 deficiency reduces expression of PD-L1 on tumor cells in vitro and in vivo.
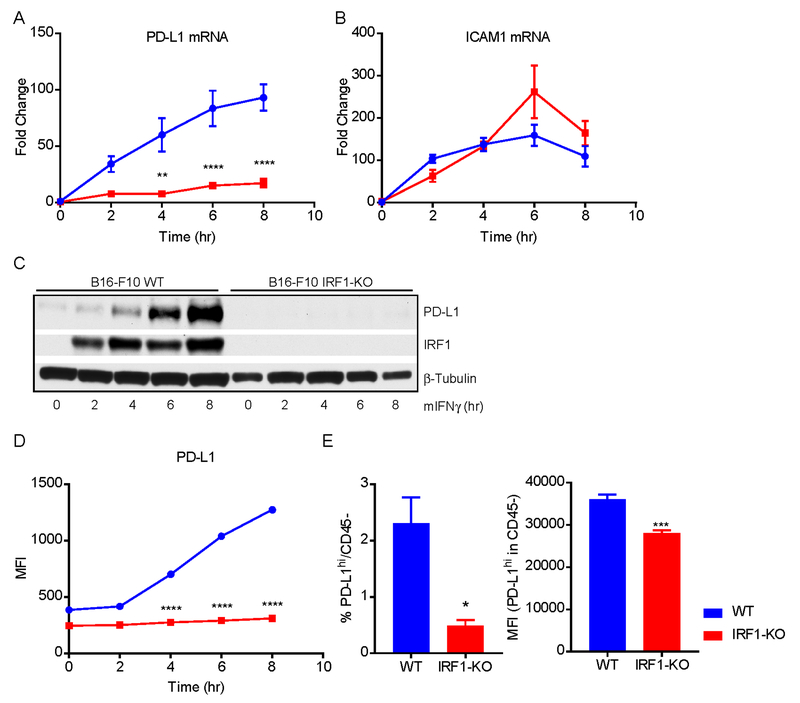
Since tumor intrinsic IRF1 did not change the percentage of TILs, we sought to investigate whether the loss of IRF1 changed the properties of tumor cells. First, we compared a common set of IFNγ-induced mRNA expression between B16-F10 WT and IRF1-KO cells in vitro. IFNγ treatment induced the mRNA expression of PD-L1, ICAM1, STAT1 and GBP2 in B16-F10 WT in a time dependent manner, such that expression peaked between 6–8 h (Fig.4A and B, Supplementary Fig. S4A and B). However, a number of genes, e.g. PD-L1, STAT1 and GBP2 showed significantly reduced induction in B16-F10 IRF1-KO cells (Fig.4A, Supplementary Fig. S4A and B). Loss of IRF1 did not affect the induction of ICAM1 mRNA (Fig. 4B), Next, we confirmed the loss of IFNγ-induced PD-L1 mRNA by measuring protein. We found almost a complete loss of total (Fig. 4C) as well as cell surface expressed PD-L1 protein in B16-F10 IRF1-KO cells after IFNγ treatment (Fig. 4D). These results indicated that IRF1 is necessary for IFNγ-induced expression of PD-L1 in tumor cells whereas IRF1 does not affect ICAM1 induction by IFNγ.
Fig. 4: PD-L1 expression in vitro and in vivo is specifically affected by IRF1 loss.
B16-F10 WT and IRF-1 KO cells were treated with mouse IFNγ for 0, 2, 4, 6 and 8 hrs, and collected for the detection of expression of PD-L1. (A-B) The mRNA expression of PD-L1 and ICAM1 were detected using TaqMan real-time PCR. (C) Total protein expression of PD-L1 was examined by immunoblotting. (D) The cell surface expression of PD-L1 was assessed by flow cytometry. (E) 5 × 105 of B16-F10 WT or IRF1-KO cells were intradermally injected into C57BL/6 mice (n=5). Tumor were collected on day 12 of post-injection. The percentage of and geometric mean (MFI) of PD-L1high in CD45− cells (tumor cells) were tested by flow cytometry. In (A) and (B) each data point represents mean and SEM from 3 independent replicates. Representative results from twice repeated experiment are shown in (C - F).
PD-L1 is an inhibitory ligand that binds to the inhibitory receptor PD-1 on T cells and inhibits T-cell function (27). Accordingly, reduced PD-L1 expression on tumor cells may result in the reduction of inhibitory effects on the tumor-infiltrating T cells. Therefore, we examined PD-L1 expression in tumor cells in vivo. We defined CD45ˉ cells as tumor cells and determined the percentage of CD45ˉ PD-L1+ cells, as well as the mean fluorescence intensity (MFI) of PD-L1 signal. As shown in Fig. 4E, WT tumors had a significantly higher percentage of PD-L1 expressing cells than IRF1-KO tumors, whereas the MFI of PD-L1 signal was reduced in the IRF1-KO tumor cells compared to the WT cells. Taken together these results indicate that the loss of IRF1 expression in tumor cells affects PD-L1 expression on the tumor cells leading to tumor regression.
Enhanced killing of IRF1-deficient tumor cells by T cells.
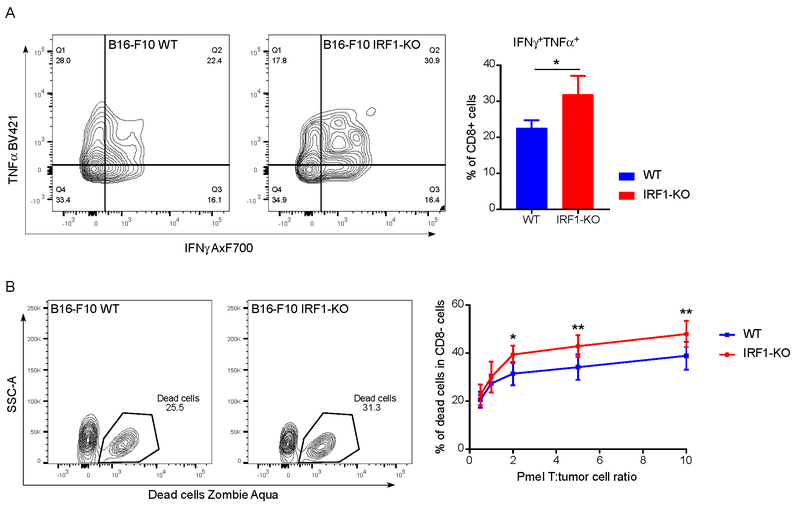
Having found that the loss of IRF1 expression in tumor cells decreased their PD-L1 expression, we examined whether this would alter the effectiveness of tumor infiltrating T cells. After collecting and preparing the single cell suspension of tumors, we stimulated the mixture of tumor cells and immune cells with PMA and ionomycin overnight, then measured intracellular cytokines following GolgiPlug (protein transport inhibitor which contains brefeldin A) treatment for 4 hrs. The intracellular cytokine (IFNγ, and TNFα) expression was assessed using flow cytometry. We found that there were significantly more IFNγ, TNFα dual-expressing CD8+ cells in IRF1-KO tumors than in WT tumors (Fig. 5A).
Fig. 5: IRF1 deficient tumor cells affect infiltrating T cell functions ex vivo and are more vulnerable to CD8+ T-cell cytotoxicity in vitro.
(A) Intracellular cytokine production of infiltrating CD8+ T cells in B16-F10 WT and IRF1-KO tumors (n=9, 8). Single cell suspension was prepared after tumor collection. The mixture of tumor cells and infiltrated lymphocytes was stimulated overnight with PMA and ionomycin, then cytokine secretion was blocked with GolgiPlug™ for 4 hrs. Representative flow-cytogram for intracellular cytokine expression is shown. For each type of cytokine expression, cumulative mean and SEM were plotted from twice repeated experiments, statistical significance was calculated by unpaired student t test, and presented as * P < 0.05. (B) The percentage of dead tumor cells at different Pmel T : tumor cell ratios. 10,000 tumor cells in each 96-well plate were cocultured with different amount of Pmel CD8+ T cells overnight. Cells were harvested and examined for viability using Zombie Aqua Fixable Viability Kit. The experiment was repeated 3 times. For each data point mean and SEM from twice repeated experiments were plotted, statistical significance calculated by two-way ANOVA with Sidak’s multiple comparison test, and presented as * P < 0.05, ** P < 0.01.
We next examined whether IRF1-deficient tumor cells were more sensitive to T-cell killing. We utilized activated Pmel T cells specific for gp100 antigen expressed on B16-F10 tumor cells (29). Pmel T cells were mixed with B16-F10 WT and IRF1-KO cells at different ratios followed by the measurement of cell killing. The results showed that there were significantly more cell killing in B16-F10 IRF1-KO cells than WT cells (Fig. 5B). Since IRF1-KO tumor cells had less PD-L1 expression than WT tumor cells, they showed less inhibitory effects on the Pmel T cells and were more vulnerable to T-cell cytotoxic activity compared with WT tumor cells. In summary, the results indicated that loss of IRF1 in tumor cells impaired the PD-L1-mediated inhibitory effects on tumor infiltrated T cells to increase the vulnerability of the tumor cells to the T cell-mediated cytotoxicity.
Expression of PD-L1 in IRF1-KO cells restores tumor progression in mice.
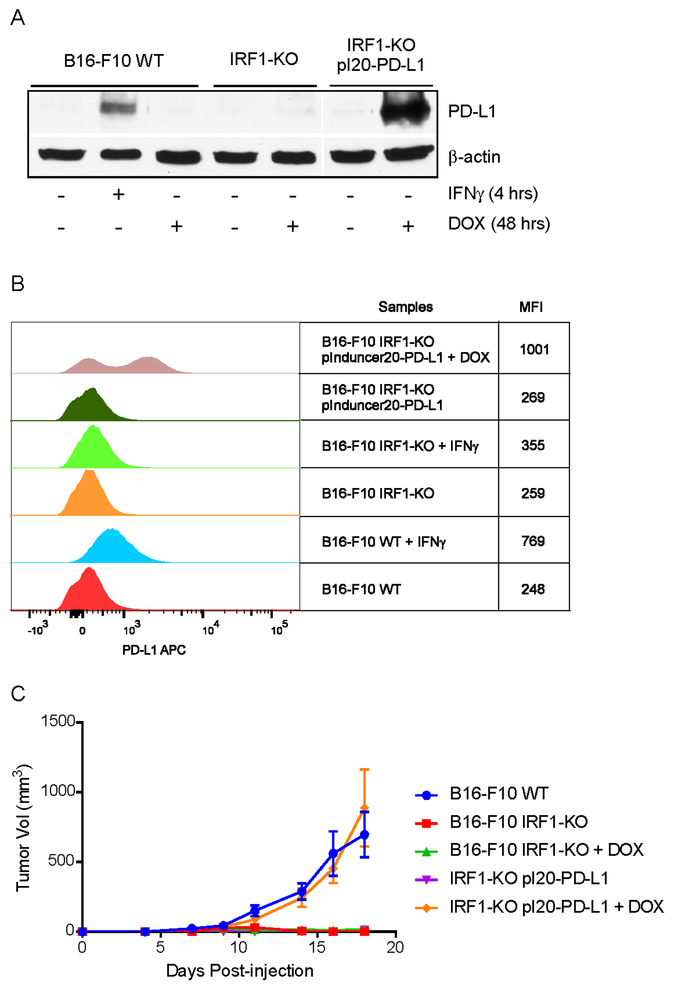
To confirm the role of PD-L1 in tumor progression, we made a Tetracycline-inducible PD-L1 expressing cell line in B16-F10 IRF1-KO (called B16-F10 IRF1-KO pInducer20-PD-L1) and monitored the in vivo tumor growth of this tumor cell line. To avoid 3’ UTR-mediated post-transcriptional repression (14) we used we used the coding region of PD-L1. Total protein expression of PD-L1 and cell surface expression of PD-L1 were confirmed using western blot and flow cytometry (Fig. 6AandB). Mice were fed with doxycycline (DOX) containing food (200 mg/kg) from 3 days before tumor cells injection. Mice injected with B16-F10 IRF1-KO pInducer20-PD-L1 cell and maintained on DOX food had similar tumor progression as B16-F10 WT cell–injected mice. Without the DOX food, the tumor growth rate of B16-F10 IRF1-KO pInducer20-PD-L1 tumor cells was the same as B16-F10 IRF1-KO cells (Fig. 6C). The results confirmed that the reduction of PD-L1 expression is the cause of tumor regression in the IRF1-KO tumor cells.
Fig. 6: DOX-induced PD-L1 expression rescues tumorigenicity of IRF1-KO cells in mice.
B16-F10 WT cells were treated with mouse IFNγ for 4 hrs for positive control. B16-F10 WT, IRF1-KO and IRF1-KO pInducer20-PD-L1 cells were treated with DOX for 48 hrs and collected for the detection of PD-L1expression. (A) Total protein expression of PD-L1 was examined by immunoblotting. (B) The cell surface expression of PD-L1 was assessed by flow cytometry. Representative histogram from twice repeated experiments are shown. (C) 5 × 105 of B16-F10 WT, IRF1-KO or IRF1-KO pInducer20-PD-L1 cells were intradermally injected into C57BL/6 mice (n=5). One group of either B16-F10 IRF1-KO or IRF1-KO pInducer20-PD-L1 cell injected mice were fed with DOX containing food (200 mg/kg) from 3 days before tumor cell injection, then continued being fed with DOX containing food. Tumor growth measurements were taken with all groups of mice.
DISCUSSION
Based on studies using IRF1-deficient mice as well as genomic loss observed in human leukemia, IRF1 has been described as a tumor suppressor gene (4). In contrast to this notion, here we describe tumor cell–intrinsic tumor promoting activity of IRF1. Loss of IRF1 in the tumor cells resulted in loss of in vivo tumor growth without affecting in vitro cell growth in multiple syngeneic tumor models. Mechanistically, we found that the absence of IRF1 led to a loss of PD-L1 transcription, resulting in increased killing of tumor cells by CD8+ T cells. The reduced PD-L1 expression on the tumor cells did not change the TIL profile of the IRF1-KO cell injected tumors compared with the WT. However, the cytotoxic activity of the TILs from IRF1-KO tumors were higher than WT. This indicated that the cell intrinsic loss of PD-L1 can result in loss of tumorigenicity, as reported (30–32). However, similar to previous reports, we also observed occasional escape of IRF1-KO cell injected tumors, which may be attributed to the PD-L1 expression on the myeloid cells resulting in the inhibition of T cell-mediated cytotoxicity.
Multiple signaling pathways regulate PD-L1 expression both through transcription and mRNA stability (33). IFNγ and the transcription factor IRF1 are established as the primary drivers for PD-L1 expression (20–22). The present study establishes that IRF1 was crucial for tumor cell–intrinsic expression of PD-L1 in vivo. We investigated the induction of PD-L1 in tumor cell lines after treatment with several cytokines that have been reported to induce PD-L1. However, only IFNγ significantly induced PD-L1in the B16-F10 cell line. In this case, loss of IRF1 resulted in almost complete loss of PD-L1 induction.
IRF1 is an inducible but ubiquitously expressed transcription factor. In many cell types IRF1 transcription is induced strongest by IFNγ and less so with IFNβ (34). Additionally, IRF1 transcription has been reported downstream of NF-κB activation in a number of cell types (35). Besides transcription, the steady state amount of IRF1 protein is also regulated through post-translational modification and degradation (19). A number of IFNγ-inducible genes require IRF1 in addition to STAT1 for sustained induction (36). Our results indicate PD-L1 to be one such gene in vivo in the context of tumor cells. Conversely, IRF2 is a negative regulator of IRF1-mediated transcriptional activation (9) and has been reported to inhibit PD-L1 expression and tumorigenesis (37). Although a role of IRF1 in mediating the PD-L1 regulation by IRF2 was not demonstrated in this report (30), this observation again points towards a role of IRF1 in PD-L1 induction and tumorigenicity.
The primary and acquired resistance to PD-1/PD-L1-targeted therapy can arise due to multiple factors. Among these, poor tumor immunogenicity, T-cell exclusion and tumor cell–intrinsic resistance to IFNγ result in the nonresponder phenotype. IFN resistance can evolve through acquisition of loss of function mutations in the JAK/STAT pathway (16–18). In both cases, loss of IFNγ-mediated tumor cell growth inhibition is the primary driver of the resistance (16,17). However, our results imply a negative role for IFNγ signaling and induction of IRF1-mediated PD-L1 upregulation in the tumor cell resulting in reduced tumor cell killing. In fact, similar to our results, IFNGR loss was found to decrease tumor growth in mice (38). This again points to the complex interaction that exists in the tumor microenvironment. Nonetheless, it may still be possible to modulate the IRF1/IRF2 axis in the tumor cell for targeted tumor therapy.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by AI118896 (SNS), CA178766 (SNS) and CA204173 (CJB) from National Institutes of Health. This project used a number of UPCI core facilities that are supported by award P30CA047904 and R50CA211241.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interests.
References
- 1.Lohoff M, Mak TW. Roles of interferon-regulatory factors in T-helper-cell differentiation. Nat Rev Immunol. 2005;5:125–35. [DOI] [PubMed] [Google Scholar]
- 2.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF Family Transcription Factors in Immunity and Oncogenesis. Annu Rev Immunol. 2008;26:535–84. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka N, Taniguchi T. The interferon regulatory factors and oncogenesis. Semin Cancer Biol. 2000;10:73–81. [DOI] [PubMed] [Google Scholar]
- 4.Yanai H, Negishi H, Taniguchi T. The IRF family of transcription factors. Oncoimmunology. 2012;1:1376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Der Weyden L, Arends MJ, Campbell AD, Bald T, Wardle-Jones H, Griggs N, et al. Genome-wide in vivo screen identifies novel host regulators of metastatic colonization. Nature. 2017;541:233–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willman CL, Sever CE, Pallavicini MG, Harada H, Tanaka N, Slovak ML, et al. Deletion of IRF-1, mapping to chromosome 5q31.1, in human leukemia and preleukemic myelodysplasia. Science (80- ). 1993;259:968–71. [DOI] [PubMed] [Google Scholar]
- 7.Tzoanopoulos D, Speletas M, Arvanitidis K, Veiopoulou C, Kyriaki S, Thyphronitis G, et al. Low expression of interferon regulatory factor-1 and identification of novel exons skipping in patients with chronic myeloid leukaemia. Br J Haematol. 2002;119:46–53. [DOI] [PubMed] [Google Scholar]
- 8.Harada H, Kondo T, Ogawa S, Tamura T, Kitagawa M, Tanaka N, et al. Accelerated exon skipping of IRF-1 mRNA in human myelodysplasia/leukemia; a possible mechanism of tumor suppressor inactivation. Oncogene. 1994;9:3313–20. [PubMed] [Google Scholar]
- 9.Harada H, Kitagawa M, Tanaka N, Yamamoto H, Harada K, Ishihara M, et al. Anti-oncogenic and oncogenic potentials of interferon regulatory factors-1 and −2. Science. 1993;259:971–4. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka N, Ishihara M, Lamphier MS, Nozawa H, Matsuyama T, Mak TW, et al. Cooperation of the tumour suppressors IRF-1 and p53 in response to DNA damage. Nature. 1996;382:816–8. [DOI] [PubMed] [Google Scholar]
- 11.Sharma P, Allison J. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardoll D The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun C, Mezzadra R, Schumacher TN. Regulation and Function of the PD-L1 Checkpoint. Immunity. 2018;48:434–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature. 2016;534:402–6. [DOI] [PubMed] [Google Scholar]
- 15.Moore C, Molina-Arcas M, East P, Zecchin D, Downward J, Snijders AP, et al. Oncogenic RAS Signaling Promotes Tumor Immunoresistance by Stabilizing PD-L1 mRNA. Immunity. 2017;47:1083–1099.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, Shi LZZLZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-γ Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy Cell. 2016;167:397–404.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016;375:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nowicki TS, Hu-Lieskovan S, Ribas A. Mechanisms of Resistance to PD-1 and PD-L1 Blockade. Cancer J. 2018;24:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kröger A, Köster M, Schroeder K, Hauser H, Mueller PP. Review: Activities of IRF-1. J Interf Cytokine Res. 2002;22:5–14. [DOI] [PubMed] [Google Scholar]
- 20.Ebine K, Kumar K, Pham TN, Shields MA, Collier KA, Shang M, et al. Interplay between interferon regulatory factor 1 and BRD4 in the regulation of PD-L1 in pancreatic stellate cells. Sci Rep. 2018;8:13225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hugo W, Parisi G, Sun L, Comin-Anduix B, Lo RS, Shin DS, et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017;19:1189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi I-H, Park Y-M, Oh S, Yao S, Lee S-J, Lee S-W, et al. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-γ-induced upregulation of B7-H1 (CD274). FEBS Lett. 2006;580:755–62. [DOI] [PubMed] [Google Scholar]
- 23.Lai Q, Wang H, Li A, Xu Y, Tang L, Chen Q, et al. Decitibine improve the efficiency of anti-PD-1 therapy via activating the response to IFN/PD-L1 signal of lung cancer cells. Oncogene. 2018;37:2302–12. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt T, Schmid-Burgk JL, Ebert TS, Gaidt MM, Hornung V. Designer Nuclease-Mediated Generation of Knockout THP1 Cells. Methods Mol Biol. 2016. page 261–72. [DOI] [PubMed] [Google Scholar]
- 25.Cuevas RA, Ghosh A, Wallerath C, Hornung V, Coyne CB, Sarkar SN. MOV10 Provides Antiviral Activity against RNA Viruses by Enhancing RIG-I–MAVS-Independent IFN Induction. J Immunol. 2016;196:3877–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, Zhang Y, Ghosh A, Cuevas RA, Forero A, Dhar J, et al. Antiviral Activity of Human OASL Protein Is Mediated by Enhancing Signaling of the RIG-I RNA Sensor. Immunity. 2014/06/17 2014;40:936–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Facciabene A, Santoro S, Coukos G. Know thy enemy: Why are tumor-infiltrating regulatory T cells so deleterious? Oncoimmunology. 2012;1:575–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Overwijk WW, Theoret MR, Finkelstein SE, Surman DR, de Jong LA, Vyth-Dreese FA, et al. Tumor Regression and Autoimmunity after Reversal of a Functionally Tolerant State of Self-reactive CD8 + T Cells. J Exp Med. 2003;198:569–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juneja VR, McGuire KA, Manguso RT, LaFleur MW, Collins N, Haining WN, et al. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med. 2017;214:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau J, Cheung J, Navarro A, Lianoglou S, Haley B, Totpal K, et al. Tumour and host cell PD-L1 is required to mediate suppression of anti-tumour immunity in mice. Nat Commun. 2017;8:14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noguchi T, Ward JP, Gubin MM, Arthur CD, Lee SH, Hundal J, et al. Temporally Distinct PD-L1 Expression by Tumor and Host Cells Contributes to Immune Escape. Cancer Immunol Res. 2017;5:106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun C, Mezzadra R, Schumacher TN. Regulation and Function of the PD-L1 Checkpoint. Immunity. 2018. page 434–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95:15623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson CM, Hale PT, Carlin JM. NF-κB activation contributes to indoleamine dioxygenase transcriptional synergy induced by IFN-γ and tumor necrosis factor-α. Cytokine. 2006; [DOI] [PubMed] [Google Scholar]
- 36.Chatterjee-Kishore M, Wright KL, Ting JPY, Stark GR. How Stat1 mediates constitutive gene expression: A complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorand RD, Nthale J, Myers JT, Barkauskas DS, Avril S, Chirieleison SM, et al. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science (80- ). 2016;353:399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell. 2016;167:1540–1554.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.