Abstract
Background:
Hormone receptor-positive/HER2−negative (HR+/HER2−) breast cancer is associated with low levels of stromal tumor-infiltrating lymphocytes (sTIL) and PD-L1, and demonstrates poor responses to checkpoint inhibitor therapy. Evaluating the effect of standard chemotherapy on the immune microenvironment may suggest new opportunities for immunotherapy-based approaches to treating HR+/HER2− breast tumors.
Methods:
HR+/HER2− breast tumors were analyzed before and after neoadjuvant chemotherapy. sTIL were assessed histologically; CD8+ cells, CD68+ cells, and PD-L1 staining were assessed immunohistochemically; whole transcriptome sequencing and panel RNA expression analysis (NanoString) were performed.
Results:
96 patients were analyzed from two cohorts (N=55 Dana-Farber cohort; N=41 MD Anderson cohort). sTIL, CD8, and PD-L1 on tumor cells were higher in tumors with basal PAM50 intrinsic subtype. Higher levels of tissue-based lymphocyte (sTIL, CD8, PD-L1) and macrophage (CD68) markers as well as gene expression markers of lymphocyte or macrophage phenotypes (NanoString or CIBERSORT) correlated with favorable response to neoadjuvant chemotherapy, but not with improved distant metastasis-free survival in these cohorts or a large gene expression dataset (N=302). In paired pre-/post-chemotherapy samples, sTIL and CD8+ cells were significantly decreased post-treatment while expression analyses (NanoString) demonstrated significant increase of multiple myeloid signatures. Single gene expression implicated increased expression of immunosuppressive (M2-like) macrophage-specific genes post-chemotherapy.
Conclusions:
The immune microenvironment of HR+/HER2−tumors differs according to tumor biology. This cohort of paired pre-/post-chemotherapy samples suggests a critical role for immunosuppressive macrophage expansion in residual disease. The role of macrophages in chemo-resistance should be explored and further evaluation of macrophage-targeting therapy is warranted.
Keywords: Breast cancer, chemotherapy, tumor microenvironment, biomarkers, intrinsic subtype
Introduction
Hormone receptor-positive/HER2−negative (HR+/HER2−) tumors make up over two-thirds of all breast cancer(1). Examination of the breast cancer tumor microenvironment (TME) suggests that HR+ breast tumors may be more immunologically “cold” than their triple-negative and HER2−positive counterparts. These observations are primarily based on assessment of tumor-infiltrating lymphocytes (TIL) and expression of the T cell checkpoint protein programmed death-ligand 1 (PD-L1; or its receptor, PD-1) in treatment-naïve tumors. TIL infiltration is lower in ER-positive as compared to ER-negative tumors(2), and there is an inverse correlation between estrogen receptor (ER) positivity and PD-1/PD-L1 expression(3–5). Chemotherapy can elicit anti-tumor immune responses through a variety of mechanisms, including triggering antigen release from dying tumor cells, stimulation of immune effector cells, and inhibition of immune regulatory cells(6). The immunomodulatory effects of chemotherapy on HR+/HER2− breast tumors specifically have not been widely explored.
Immune checkpoint inhibitor therapy has revolutionized the treatment landscape and improved prognosis for many malignancies. To date, however, responses to immune checkpoint inhibitor monotherapy have been marginal in HR+ breast cancer. In patients with refractory HR+/HER2− metastatic breast cancer, the anti-PD-L1 antibody avelumab led to an overall response rate of 2.8% (among patients unselected for PD-L1 status), and the anti-PD-1 antibody pembrolizumab led to an overall response rate of 12% (among patients with PD-L1-positive tumors)(7,8). Given the prevalence of HR+/HER2− breast cancer, identifying even a small subset of immunologically “hot” HR+/HER2− tumors could be clinically significant and allow identification of patients likely to benefit from immunotherapy. While most analysis of the TME in breast cancer to date has focused on T lymphocytes and T cell checkpoints, evidence from both preclinical models and clinical specimens suggests that macrophages and their associated regulatory pathways play a particularly important role in pathogenesis and treatment response of HR+ breast tumors(9–11). A broader assessment of the TME in HR+/HER2− breast tumors is needed.
The goal of this study was to examine both tissue-based and RNA-based immune biomarkers in patients with HR+/HER2− non-metastatic breast cancer treated with chemotherapy. By incorporating and extending analyses beyond investigation of lymphocyte-mediated immune responses, with a particular focus on macrophages, we sought to identify candidate biomarkers of response to therapy, to gain insights into immune-related biological processes that may be harnessed for therapeutic potential, and to analyze the heterogeneity that exists in the HR+ TME. Two cohorts of patients treated with standard neoadjuvant chemotherapy (plus the anti-vascular endothelial growth factor (VEGF) antibody bevacizumab in one cohort), were used to examine how the TME changes following treatment. Results from a variety of methods used to assess the TME (including histology, protein expression, and gene expression), were integrated to capture multiple aspects of tumor-immune interactions and explore the overlap and complementation of distinct analytical tools.
Materials and Methods
Patient Cohorts
The first cohort, from Dana-Farber Cancer Institute (DFCI), consisted of patients enrolled on a prospective clinical trial (DFCI #07–130); see REMARK diagram (Figure S1). Patients enrolled on trial with HR+/HER2− breast cancer had node-positive or high-risk node-negative tumors without distant metastases. Details of trial design, eligibility, treatment regimen, and clinical results have been reported previously(12). Briefly, patients received a single dose of bevacizumab followed by dose-dense adriamycin/cyclophosphamide (ddAC) then dose-dense paclitaxel/bevacizumab, all delivered in the neoadjuvant setting. Patients included in this post-hoc analysis had HR+ (ER ≥1% or progesterone receptor (PR) ≥1%) and HER2− breast cancer, received at least one dose of on-trial therapy, and had at least one tissue specimen available for evaluation. Pathologic complete response (pCR) and residual cancer burden (RCB)(13) at surgery were determined by central pathology review. Long-term disease-free survival (DFS) and overall survival (OS) outcomes were determined by retrospective chart review. DFS events were defined as local/regional invasive breast cancer recurrence, contralateral invasive breast cancer, distant recurrence, or death from any cause. The second cohort, from the University of Texas MD Anderson Cancer Center, consisted of patients with HR+ (ER ≥1% or progesterone receptor (PR) ≥1%) and HER2− breast cancer who received neoadjuvant chemotherapy with adriamycin, cyclophosphamide, and paclitaxel (T-AC) without bevacizumab, and had tumor tissue available for analysis from both pre-treatment and post-treatment (surgical) timepoints. All analyses were approved by the institutional review boards of DFCI or MD Anderson Cancer Center. Research was conducted in accordance with the US Common Rule, and written informed consent was obtained from each subject.
Tissue-based biomarker assessment
Stromal TILs were scored on an hematoxylin and esosin (H&E) stained slide by a breast pathologist according to international consensus guidelines(14). Immunohistochemical (IHC) staining of formalin-fixed paraffin-embedded (FFPE) slides was performed using monoclonal antibodies against CD8 (DAKO M7103), CD68 (DAKO M0876), and PD-L1 (clone 9A11, Cell Signaling). Scoring for PD-L1 was measured by percentage of cells stained in tumor and stromal tissue compartments. For image analysis, three to five representative pictures were taken for each case at 200× magnification. The images were analyzed using Inform2.3 Image Analysis Software (Mantra Software/PerkinElmer). Positive staining was assessed quantitatively, and thresholds used to bin data were selected based on individual biomarker distribution and existing standards.
NanoString immune gene expression panel analysis
RNA was isolated from areas of invasive tumor on FFPE slides. Two hundred ng of RNA per sample was loaded and run on the HuV1_CancerImmu_v1_1 Nanostring for analysis of the NanoString PanCancer Immune Profiling Panel (PCI; DFCI cohort) or the PanCancer IO 360 Profiling Panel (IO360; MD Anderson cohort). Raw gene counts for both cohorts were log2 transformed and normalized to the geometric mean of 10 housekeeper genes common between panels. For gene signatures, 20 NanoString signatures previously found to be highly correlated between PCI and IO360 panels were analyzed. A modified version of the Tumor Inflammation Signature (TIS; using 16 of 18 genes in the published signature) was used for samples in the PCI/DFCI cohort. Additional signatures were computed after normalization to 20 housekeeping genes and removal of samples with low housekeeper expression. Signature scores range from approximately 0 to 10, with an average value of 5 in pancancer tumor samples from The Cancer Genome Atlas(15).
DFCI cohort whole transcriptome sequencing and analysis
Gene expression profiles for the clinical trial samples were generated by mRNA sequencing using the Illumina TruSeq Kit to create libraries for paired-end sequencing on an Illumina HiSeq 2000. Data was aligned and genes quantified as previously described(16), as upper quartile normalized RSEM abundance estimates that were log2 transformed. Gene expression samples and patients were also derived from a large previously published meta-analysis of patients with HR+/HER2− breast cancer treated with neoadjuvant chemotherapy(17). The Parker et al. predictor was used for subtype categorizations using a nearest centroid procedure which returned calls as a five-level classifier (Basal-like, Luminal A, Luminal B, HER2−enriched, and Normal-like)(18). Proportion of infiltrating immune cell subsets was calculated using the CIBERSORT algorithm(19).
Statistical methods
Association between biomarker assessments, gene expression profiles and patient characteristics are made under a general linear model with Student t-tests, ANOVA, and Pearson correlation coefficients (r) as indicated in the text. Changes in quantitative biomarkers from pre- to post-neoadjuvant treatment were made using Wilcoxon signed rank test. Kaplan-Meier plots were generated using ‘packHV’ package(20). Differences in the survival function were calculated by log rank test. A nominal p < 0.05 was used to declare statistical significance. For each gene expression analysis, the Benjamini-Hochberg step-up procedure was used to control the false discovery rate (FDR)(21). Statistical analyses were performed using STATA version 14.2 and R version 3.4.1.
Results
Patient characteristics
96 patients with HR+/HER2− breast tumors were analyzed from two separate cohorts, one from DFCI (N=55) and one from MD Anderson (N=41). Characteristics of patients from both cohorts are summarized in Table 1. All patients in both cohorts had tumors positive for ER and/or PR expression, and the large majority had either grade 2 (54–55%) or 3 (29–40%) tumors. At the time of surgery following pre-operative therapy, most patients (82% in DFCI cohort; 93% in the MD Anderson cohort) had RCB(13) class of II or III, and only 4 patients from the DFCI cohort had pCR (RCB score 0). Of note, since only patients with pre- and post-chemotherapy tumor tissue available were selected for the MD Anderson cohort, there were no patients with pCR in this cohort.
Table 1.
Patient/tumor characteristics and surgical outcomes (DFCI and MD Anderson cohorts)
| DFCI cohort (N=55) | MD Anderson cohort (N=41) | |||
|---|---|---|---|---|
| Category | Patients: # (%) | Category | Patients: # (%) | |
| Age (median (IQR)) | 47 yrs (39–51 yrs) | 57 yrs (46–62 yrs) | ||
| HR status | ER+/PR+ | 45 (82%) | ER+/PR+ | 32 (78%) |
| ER+/PR- | 9 (16%) | ER+/PR- | 7 (17%) | |
| ER-/PR+ | 1 (2%) | ER-/PR+ | 0 | |
| Unknown | 0 | Unknown | 1 (2%) | |
| Stage at diagnosis | 1 | 2 (4%) | 1 | 1 (2%) |
| 2 | 41 (75%) | 2 | 24 (59%) | |
| 3 | 12 (22%) | 3 | 14 (34%) | |
| 4 | 0 (0%) | 4 | 1 (2%) | |
| Unknown | 0 (0%) | Unknown | 1 (2%) | |
| Grade | 1 | 3 (6%) | 1 | 2 (5%) |
| 2 | 30 (55%) | 2 | 22 (54%) | |
| 3 | 22 (40%) | 3 | 12 (29%) | |
| Unknown | 0 | Unknown | 5 (12%) | |
| Intrinsic subtype (assessed in N=38 pts from DFCI cohort) | Luminal A | 24 (63%) | ||
| Luminal B | 7 (18%) | |||
| Basal | 6 (16%) | NA | NA | |
| Normal | 1 (3%) | |||
| Not assessed | 17 | |||
| RCB* | 0 | 4 (7%) | 0 | 0 |
| 1 | 5 (9%) | 1 | 3 (7%) | |
| 2 | 22 (40%) | 2 | 22 (54%) | |
| 3 | 23 (42%) | 3 | 16 (39%) | |
| Unable to assess | 1 (2%) | Unable to assess | 0 | |
Characteristics of patients included in two separate cohorts. Stage at diagnosis indicates clinical anatomic stage according to American Joint Committee on Cancer Staging, 8th edition.
Patients with RCB 0 were excluded from MD Anderson cohort.
Abbreviations: DFCI, Dana-Farber Cancer Institute; IQR, interquartile range; NA, not available/not applicable; RCB, Residual Cancer Burden.
Immune biomarkers and tumor intrinsic subtype
Stromal TIL, PD-L1 protein expression on tumor and stromal cells, CD8, and CD68 protein expression were measured in treatment-naïve breast biopsy specimens from all patients in the DFCI cohort; distribution of each biomarker is shown in Table S1. The majority of patients (82%) had <10% sTIL infiltration; no patients had lymphocyte-predominant breast cancer (≥60% TIL)(14). The large majority (78%) of patients were negative (defined as <1%) for tPD-L1, and an even greater percentage (88%) had negative PD-L1 expression on stromal cells (sPD-L1), though remaining patients did demonstrate expression of tumor or stromal PD-L1 at a level (≥1%) shown to be clinically significant in breast cancer(22).
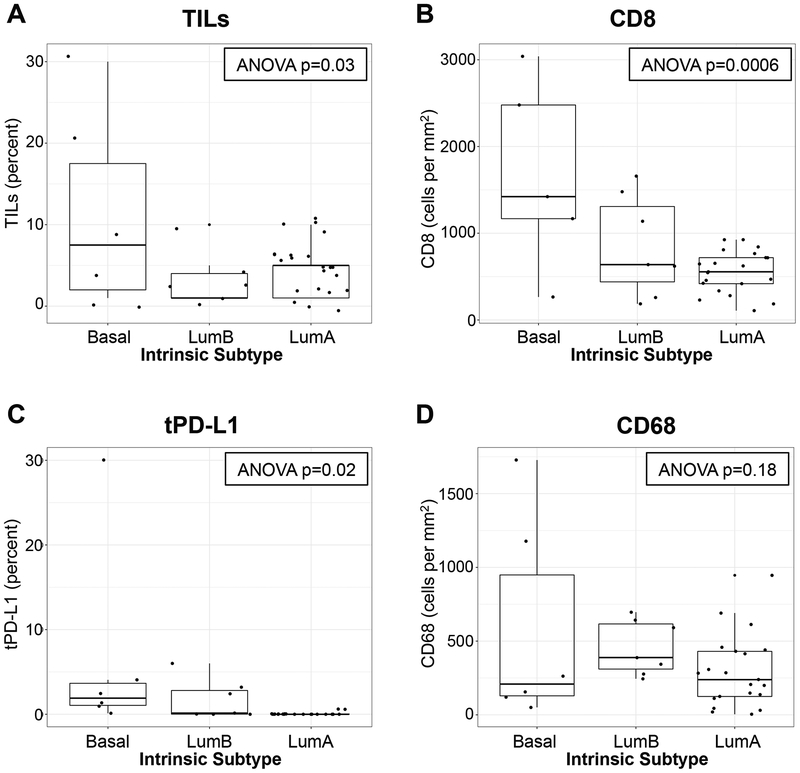
Available primary tumors (N=38 patients) from the DFCI cohort underwent RNA sequencing and tumor intrinsic subtype was classified by PAM50 gene signature (Table 1). The majority (63%) were classified as luminal A. Of note, all 6 tumors determined to be basal subtype by PAM50 had ER expression ≤15% (≤10% in 5 of 6 tumors), and PR expression ≤10%. There was significant variation between luminal A, luminal B, and basal tumors in terms of sTIL, CD8, and tPD-L1 (ANOVA p=0.03, p=0.0006, p=0.02, respectively) but not CD68 (ANOVA p=0.18; Figure 1). For all biomarkers except CD68, median levels were highest in basal tumors. In addition, the highest values observed for each biomarker were clustered in the basal subtype. However, luminal A and luminal B tumors also displayed a range of values (including some higher values) for all biomarkers, with the exception of tPD-L1 in luminal A tumors, which was universally low (only 2 patients had expression ≥1%; maximum 1.2%). The modified TIS (a gene expression profile denoting interferon-gamma-related signaling that has been shown to predict clinical response to immune checkpoint blockade(15)) also differed significantly across tumor intrinsic subtypes (ANOVA p=0.0016; Figure S2). Basal tumors had the highest median TIS (9.68). However, luminal A tumors spanned a range of TIS values, with 2/14 samples scoring ≥9, in the same range as basal tumors.
Figure 1. Tissue-based immune biomarkers and tumor intrinsic subtype.
In the DFCI cohort, stromal tumor-infiltrating lymphocytes (TILs; A) were evaluated histologically, and CD8+ cells (B), tumor cell PD-L1 expression (tPD-L1; C), and CD68+ cells (D) were stained immunohistochemically on full slide sections from treatment-naïve breast tumors. The distribution of each biomarker was assessed according to tumor intrinsic subtype (Luminal A, Luminal B, or Basal-like) based on the PAM50 gene signature. Each boxplot represents the 25th to 75th percentile with the median indicated as the central line and whiskers indicating 1.5 × interquartile range.
Immune biomarkers, response to neoadjuvant chemotherapy, and distant metastasis-free survival
In both unadjusted models and models adjusted for tumor size and grade, all tissue-based immune-related biomarkers assessed in the DFCI cohort (sTIL, CD8, CD68, tPD-L1, sPD-L1) were significantly associated with more favorable RCB score (Table S1). Of the 4 patients with pCR in the DFCI cohort, all had either the highest or among the highest values in the cohort for multiple tissue-based immune biomarkers; they also had basal or luminal B intrinsic subtype (Table S2). All 3 patients with sPD-L1 expression ≥5% at baseline experienced a pCR. Correlations between biomarkers and pCR status were not assessed given the low number of patients with pCR. There was no significant association between RCB and pre-to-post-treatment change in sTIL, CD8, CD68, tPD-L1, or sPD-L1.
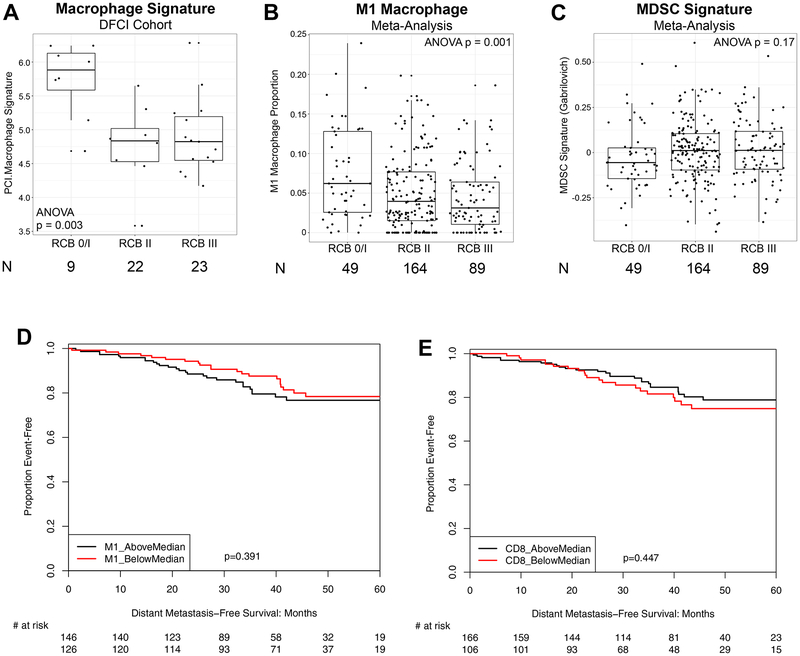
Since CD68 can stain some non-macrophage cell types(23), and since macrophages can take on a variety of different biological roles which CD68 staining does not distinguish, we used RNA expression data to investigate the relationship between macrophages and response to neoadjuvant chemotherapy. Higher expression of the Nanostring PanCancer Immune Panel macrophage signature (PCI.Macrophage) was significantly correlated with more favorable response to neoadjuvant chemotherapy (as assessed by RCB score) (Figure 2A). The PCI.Macrophage signature was positively correlated with the proportion of macrophages identified by the M1 macrophage signature (pro-inflammatory/anti-tumor) as measured by expression-based inferred immune subset proportion, determined via CIBERSORT(19) (Pearson’s r=0.57). Using gene expression data from a large cohort of patients with HR+/HER2− breast tumors who received neoadjuvant chemotherapy, collected for a previously published meta-analysis (17), we similarly found that macrophages identified by the CIBERSORT M1 signature correlated with more favorable RCB response to chemotherapy (N=302 patients with RCB status; Figure 2B). Taken together, these data suggest that anti-tumor M1-like macrophages behave like lymphocyte-related immune markers (sTIL, CD8, and PD-L1) with respect to predicting response to neoadjuvant chemotherapy. By contrast, a signature representing immune suppressive myeloid-derived suppressor cells (MDSCs)(24) showed a non-statistically significant visual trend in the opposite direction, with lower MDSC signature values corresponding to more favorable RCB response to chemotherapy (ANOVA p=0.17; N=302 pts with RCB status and adequate signature gene representation; Figure 2C).
Figure 2. Gene-expression based biomarker correlation with chemotherapy response at surgery and long-term breast cancer outcomes.
Macrophage signature in the DFCI cohort was calculated using NanoString PanCancer Immune (PCI) Panel, and correlation with RCB status was assessed (A). M1-like macrophage inferred proportion by CIBERSORT and MDSC signature score(43) was calculated from microarray data from our prior meta-analysis of HR+/HER2− tumors treated with neoadjuvant chemotherapy(17), and correlation with RCB status was assessed (B-C). For A-C, due to a paucity of complete responses, pCR (RCB 0) and RCB-I were grouped together. Each boxplot represents the 25th to 75th percentile with the median indicated as the central line and whiskers indicating 1.5 × interquartile range. Association of M1-like macrophage or CD8 cell proportion by CIBERSORT with distant metastasis-free survival was assessed (D-E).
Given the positive correlation between both lymphocyte-related and macrophage-related immune markers and favorable response to neoadjuvant chemotherapy, we assessed the correlation between immune markers and long-term breast cancer outcomes. In the DFCI cohort, there were 14 DFS events (12 distant recurrences, 1 loco-regional recurrence, 1 non-breast cancer death) and median DFS was 9.7 years (median OS not reached; median follow-up 7.9 years). There were no DFS events among the patients with pCR. There was no significant correlation of DFS with any tissue-based biomarker measured before or after treatment, in both adjusted and unadjusted models, with Cox regression hazard ratios near 1 in all cases (data not shown). Since this analysis was underpowered due to the small sample size of the cohort, we calculated the correlation between M1-like or M2-like macrophages, MDSCs, or CD8 lymphocytes (by CIBERSORT) and distant metastasis-free survival (DMFS) in patients from the large meta-analysis cohort (N=272 with outcome data; median follow-up 3.0 years). There was no significant correlation between macrophages identified by either the M1 or M2 CIBERSORT signatures, MDSC gene signature, or CIBERSORT CD8 lymphocyte proportion (stratified by above/below median) with DMFS (Figure 2D–2E; data not shown for M2 macrophages and MDSC signature).
Changes in the immune microenvironment after neoadjuvant chemotherapy
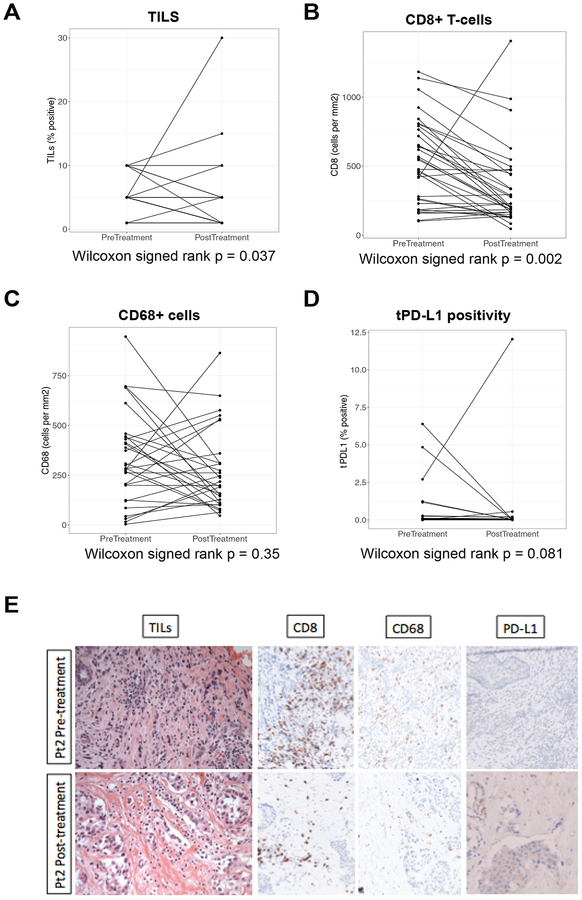
In the DFCI cohort, tissue-based immune biomarkers were scored both before and after neoadjuvant treatment with ddAC-T plus bevacizumab. sTIL and CD8+ T cells both decreased significantly post-treatment in paired analyses (Wilcoxon signed rank p=0.037 and p=0.002, respectively); there was a trend toward a decrease in tPD-L1 expression post-treatment (Wilcoxon signed rank p=0.081), and there was no significant change in the number of CD68+ cells (Figure 3A–3D). Histologic images from a representative patient with pre-to-post-treatment decrease in sTIL, CD8, and CD68 are shown (Figure 3E).
Figure 3. The tissue-based immune microenvironment before and after chemotherapy (DFCI cohort).
Tissue-based immune biomarkers were scored on paired slides from pre- versus post-preoperative therapy with dose-dense adriamycin/cyclophosphamide plus bevacizumab (A-D). Histology images (400×) from a patient with pre-to-post-treatment decrease in stromal tumor-infiltrating lymphocytes (sTIL), CD8, and CD68 (10% to 5%, 842 cells/mm2 to 206 cells/mm2, and 414 cells/mm2 to 145 cells/mm2 pre- to post-treatment, respectively) are shown. PD-L1 expression on tumor (tPD-L1) and stromal cells was 0% both pre- and post-treatment for this patient (E).
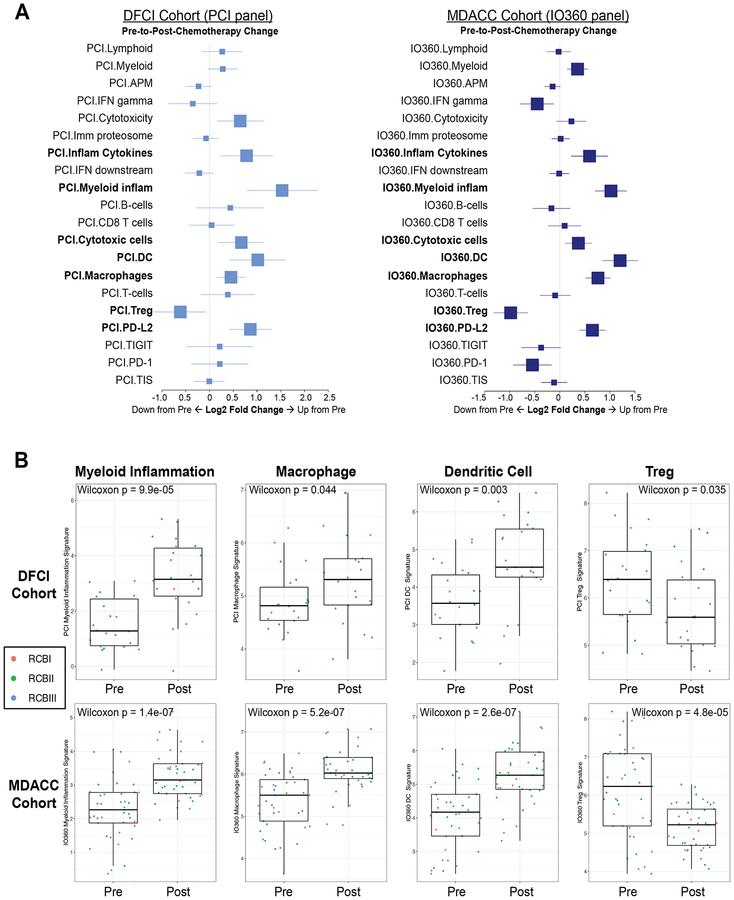
For a more complete assessment of changes in the TME following chemotherapy, we used NanoString gene expression analysis to compare pre- and post-chemotherapy immunologic features in the separate patient cohorts (DFCI N=22 patients with paired samples, treated with ddAC-T plus bevacizumab; and MD Anderson N=41 patients with paired samples, treated with T-AC without bevacizumab). Changes in NanoString gene expression signatures pre-to-post-chemotherapy in paired samples were examined (Figure 4A). Upregulation of lymphoid, T cell, or CD8 T cell signatures was not observed, consistent with tissue-based biomarker data, though a decrease in T regulatory cells was seen. There were significant increases in macrophages, dendritic cells, and a myeloid inflammation signature (which captures functional myeloid markers) post-chemotherapy in both the DFCI and MD Anderson cohorts (Figure 4B).
Figure 4. NanoString gene expression signatures before and after chemotherapy.
Immune gene expression signatures by PanCancer Immune Profiling Panel (PCI; DFCI cohort) or the PanCancer IO 360 Profiling Panel (IO360; MD Anderson cohort) were assessed pre- and post-neoadjuvant chemotherapy. Signatures that changed significantly in both cohorts are indicated in bold (A). Individual box-and-whisker plots highlight signatures of particular interest in both cohorts, with associated statistical test shown for significant change and points colored according to RCB status of each individual patient (B). Each boxplot represents the 25th to 75th percentile with the median indicated as the central line and whiskers indicating 1.5 × interquartile range. Abbreviations: MDACC, MD Anderson Cancer Center.
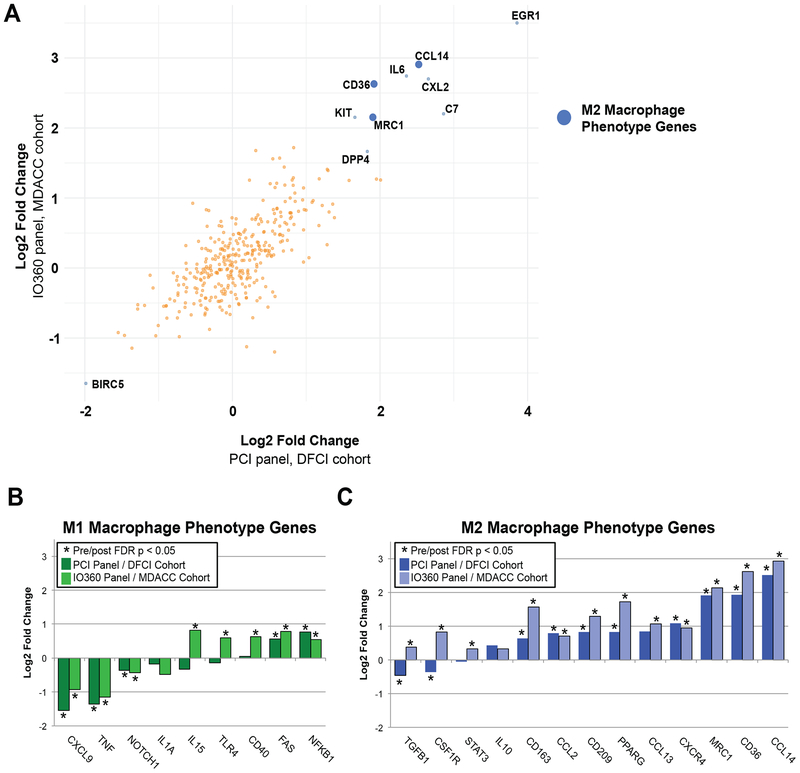
Expression level changes in individual genes pre-to-post-chemotherapy were also examined. Figure 5A shows genes whose expression changed significantly post-chemotherapy (by false discovery rate (FDR) p-value <0.05) in both the DFCI and MD Anderson cohorts. The most significantly upregulated gene in post-treatment samples was EGR1 (early growth response 1), a master regulator transcription factor with highly diverse biological roles(25). The most significantly downregulated gene in both cohorts was BIRC5, which is a component of the PAM50 proliferation signature and highly correlated to tissue Ki-67 staining(26) and mammary epithelial proliferation in 3D culture(17); reduced proliferation in residual tumor cells is a known effect of chemotherapy(26). Other upregulated genes in both cohorts included MRC1 and CCL14, both genes that are important for and specific to macrophage functions. Gene expression changes were highly concordant between the two cohorts; results for each cohort individually are shown in Figure S3. Because signature analysis suggested a significant increase in macrophages post-chemotherapy, but macrophages are a heterogeneous group of immune cells with many different biological roles, we used individual gene expression data to infer changes in M1-like (pro-inflammatory/anti-tumor) versus M2-like (immunosuppressive/pro-tumor) macrophages (Figure 5B–C). We hand curated a list of genes associated with M1-like versus M2-like macrophage phenotypes based on comprehensive literature review (Table S3). Only 2/9 genes associated with M1-like macrophage phenotype increased significantly post-chemotherapy in both DFCI and MD Anderson cohorts, while in contrast, 8/13 genes associated with a M2-like macrophage phenotype increased significantly post-chemotherapy in both cohorts. Overall, M2-like macrophage-associated genes were upregulated more frequently and displayed higher fold-change post-chemotherapy than M1-like-associated genes.
Figure 5. NanoString individual gene expression before and after chemotherapy.
Individual gene expression by PanCancer Immune Profiling Panel (PCI; DFCI cohort) or the PanCancer IO 360 Profiling Panel (IO360; MD Anderson cohort) were assessed pre- and post-neoadjuvant chemotherapy. Genes were ranked by fold-change (averaged across both cohorts) and the top 10 genes by absolute value of average fold-change, with false discovery rate p-value <0.05 in both cohorts, are labeled. Blue dots indicate genes that relate to M2-like macrophage phenotype; there were no genes in the top 10 that related to M1-like macrophage phenotype (A). Individual genes associated with M1-like versus M2-like phenotype were hand curated after comprehensive literature search (Table S3) and plotted according to log2 fold change (B). Abbreviations: MDACC, MD Anderson Cancer Center.
Discussion
In this study, we used histology, protein expression, and gene expression analyses to examine the TME of HR+/HER2− breast cancer patients treated with neoadjuvant chemotherapy. Immunohistochemical profiles of HR+/HER2− tumors at baseline demonstrate significant variation by intrinsic subtype. In detailed pre-/post-neoadjuvant chemotherapy analyses, we demonstrated reduction in sTIL and CD8+ T cells with concurrent upregulation of macrophage-related gene expression—specifically the M2-like, pro-tumor macrophage phenotype.
sTIL, CD8, tPD-L1, and immune gene expression signatures varied significantly between HR+ tumors with different intrinsic subtypes, with the highest levels clustering within the basal-like subtype (where ER expression was low, and PR expression was often negative). This suggests that with respect to the immune microenvironment and thus, potentially, responsiveness to immunotherapy, tumors with low (≤10%) ER and PR expression may be better grouped with triple-negative breast cancer than with more strongly ER/PR-positive tumors. This assertion has potential implications for future clinical trial design, but given small numbers reported here, requires further exploration. Differences in the TME between luminal A and luminal B tumors were not substantial in our dataset (with the exception of tPD-L1 expression which was universally low in luminal A tumors). Interestingly, by both immunohistochemical- and expression-based metrics, a subset of luminal A tumors appeared to have elevated immune activity, including high TIS scores (a marker of responsiveness to immune checkpoint blockade)(15). This implies that luminal A tumors should be investigated in greater depth to understand the biological underpinnings of the full spectrum of TMEs. Despite infrequent HR+ tumors with elevated immune activity, most tumors reflect a relatively immune suppressed microenvironment. The role of immunosuppressive cells in the induction or maintenance of this immune suppressed state warrants further investigation.
Our results also add to the literature regarding the utility of tissue- and RNA-based immune biomarkers for predicting short-term response to chemotherapy versus long-term outcome from breast cancer. Importantly, higher levels of all tissue biomarkers (sTIL, CD8, CD68, tPD-L1, and sPD-L1) and a macrophage gene expression signature assessed in pre-treatment specimens in the DFCI cohort had a significant positive correlation with favorable pathologic response to preoperative therapy, yet none of the tissue biomarkers measured either pre- or post-neoadjuvant therapy had a significant correlation with DFS. Similar analysis of gene expression-based immune biomarkers from a significantly larger cohort of patients with HR+/HER2− breast cancer treated with neoadjuvant chemotherapy demonstrated the same pattern: higher anti-tumor immune activity at baseline was associated with favorable pathologic response to chemotherapy, but had no effect on distant metastasis-free survival. By contrast, expression of an immune-suppressive MDSC signature showed a non-statistically significant correlation with less favorable response to preoperative chemotherapy, still with no relationship to distant metastasis-free survival. These findings corroborate and build on prior work in HR+/HER2− breast cancer. More sTIL have been shown to predict pCR to neoadjuvant chemotherapy in all three breast cancer subtypes(27–29), but in HR+/HER2− disease, this has not translated into improved long-term outcomes(29).
The value of immune biomarkers as predictive in the short-term but not prognostic in the long-term in HR+/HER2− breast cancer, in contrast to HER2+ and triple-negative tumors, is intriguing. The contribution of estrogen-driven oncogenesis to tumor growth may ultimately be the primary determinant of long-term outcomes in HR+/HER2− breast cancer. One possibility is that the HR+/HER2− tumors with higher immune biomarkers at baseline may be less estrogen-driven, explaining both their better response to neoadjuvant chemotherapy and less favorable long-term outcomes(30). Also, higher expression of an inflammatory gene signature as well as more TIL in the TME have been shown to correlate with poorer antiproliferative response to aromatase inhibitors in HR+ breast cancer(30). The interplay between the TME of HR+/HER2− tumors, estrogen-driven tumor growth, and hormonal therapy responsiveness is an important area for further investigation.
Our analysis of tissue-based immune biomarkers in HR+/HER2− breast cancer patients treated with neoadjuvant ddAC-T plus bevacizumab (DFCI cohort) showed that sTIL and CD8+ cells decrease significantly in post-treatment residual disease relative to pre-treatment specimens, with a trend toward decreased post-treatment tPD-L1 expression. Gene expression analysis of immune signatures following AC-T with and without bevacizumab (DFCI and MD Anderson cohorts) demonstrated no significant change in lymphoid, total T cell, or CD8 T cell signatures following chemotherapy, with a significant decrease in T regulatory cells. Of note, some differences between tissue-based and expression-based analyses (for example, CD8 cells decreased by immunohistochemistry, but did not change significantly by NanoString) may be related to the slightly different areas of tissue assessed by each method. While NanoString analysis was performed on RNA extracted from circled areas of tumor, analysis of immunostaining was performed only on representative captured images. Importantly, changes in NanoString signatures post-treatment were highly concordant between DFCI and MD Anderson cohorts, suggesting that bevacizumab (a component of therapy in DFCI cohort only) was not a major contributor to the observed changes.
Given evidence for the immunogenic effects of cytotoxic chemotherapy(6,31), the decrease in sTIL and CD8 staining post-chemotherapy was somewhat surprising. However, a similar trend toward decreased TILs following neoadjuvant anthracycline-containing chemotherapy has previously been shown in triple-negative breast cancer(32), and in HER2+ breast cancer patients treated with neoadjuvant anthracycline-containing chemotherapy with or without trastuzumab(33). It is difficult to fully rule out direct hematologic toxicity from chemotherapy partially contributing to decreased post-treatment lymphocytes, and it is likely that analysis at a single timepoint following completion of neoadjuvant chemotherapy is not the optimal way to assess chemotherapy’s effect on the TME. Serial on-treatment biopsies over the course of chemotherapy may provide additional information, but in practice are difficult to obtain. We posit that cancer cells evolve to become more immunologically silent over the course of cancer therapy. This is supported by prior evidence showing that metastatic tumors contain lower levels of anti-tumor immune components including TIL, CD8 cells, and PD-L1 compared to primary tumors(34), and that response rates to single agent checkpoint inhibitors are higher in earlier as opposed to later lines of therapy for metastatic breast cancer(35). The mechanisms of post-treatment immune editing are unclear and may vary depending on the type of therapy administered, individual breast tumor biology, and patient-specific immune factors. Examining biomarkers of antigen presentation (for example, HLA-ABC, HLA-DR, and dendritic cell markers), which have been shown to account for some immunologic differences between triple-negative and luminal-type breast cancers(36), may help to elucidate biology associated with the post-treatment immune-suppressed TME.
NanoString gene expression analysis of both cohorts showed a significant increase post-chemotherapy in signatures of the innate immune system and myeloid cell types, including functional markers of myeloid inflammation, dendritic cells, and macrophages. While there was no significant change in CD68 staining pre- to post-chemotherapy in the DFCI cohort, CD68 has been shown to be expressed by other cells besides macrophages(23) and furthermore does not distinguish between different macrophage subtypes. An increase in myeloid cells following neoadjuvant chemotherapy in breast cancer, including increased macrophages, has been shown previously in a small cohort(23), and treatment of breast cancer cells with chemotherapy has been shown to increase CSF1, a macrophage recruitment factor(37). Macrophages are a broad group of immune cells with highly diverse biological roles that are not easily divided in a purely binary fashion, but can function in both pro-tumor and anti-tumor capacities. We found that increased expression of genes associated with M2-like (pro-tumor) macrophages was predominant post-chemotherapy, with particularly large increases in CCL14, CD36, and MRC1, whereas M1-like (anti-tumor) macrophage-associated genes changed heterogeneously but were more likely to decrease post-chemotherapy. This suggests increased presence and/or activity of M2-like macrophages in the chemotherapy-resistant residual tumor cells, which in turn implies a possible biological role for M2-like macrophages in chemo-resistance in HR+/HER2− breast tumors. Several approaches targeting pro-tumor macrophages and/or immune suppressive myeloid precursors have been associated with improved response to immunotherapy and/or chemotherapy and are in pre-clinical or clinical development(38–41). Our data, while preliminary, would support combined regimens of chemotherapy plus M2 macrophage-targeting agents in HR+/HER2− breast cancer.
There are several limitations of our study. First, it is notable that our sample consists exclusively of HR+/HER2− breast cancers that were treated with neoadjuvant chemotherapy. A large proportion of HR+/HER2− early stage breast cancer patients are treated with hormonal therapy alone and are not represented in our study. Second, in our investigation of tissue-based immune biomarker changes that occurred following neoadjuvant chemotherapy plus bevacizumab in the DFCI cohort, it is difficult to know what effects are attributable to chemotherapy, bevacizumab, or both. VEGF, the target of bevacizumab, is a pro-angiogenic cytokine that has been shown to mediate an immunosuppressive effect on the TME through a range of mechanisms(42). However, side-by-side analysis of NanoString gene expression data from the DFCI cohort (who received bevacizumab) and the MD Anderson cohort (who did not) demonstrates very high concordance between the two cohorts in expression level changes for both individual genes and gene signatures, which is particularly notable given that all gene expression assays were performed separately for the two cohorts.
In summary, we have demonstrated that the TME of HR+/HER2− breast tumors differs according to tumor biology. Furthermore, our data point to an important role for macrophages and, potentially, other myeloid cell types such as dendritic cells and immune-suppressive myeloid precursors, in response and resistance to chemotherapy for this very common breast cancer subtype. Additional studies of tumor-immune interactions and the immunologic effects of standard cancer therapies are warranted in a dedicated HR+/HER2− breast cancer patient population. Our data support investigating the combination of cytotoxic chemotherapy with M2 macrophage-targeting strategies in HR+/HER2− breast cancer.
Supplementary Material
Statement of Translational Relevance:
Hormone receptor-positive/HER2−negative (HR+/HER2−) breast cancer is associated with low levels of stromal tumor-infiltrating lymphocytes (sTIL) and PD-L1 expression, and has shown only marginal responses to single agent checkpoint blockade immunotherapy. Further work is needed to better understand the tumor microenvironment in HR+/HER2− breast cancer to inform immunotherapy regimens that will bring clinical benefit in this common breast cancer subtype. In this study, we analyzed the tumor microenvironment before and after neoadjuvant combination chemotherapy in 96 patients with HR+/HER2− breast cancer. After chemotherapy, there was a significant decrease in sTILs and CD8 T cells, but a significant increase in gene signatures associated with myeloid cell types such as macrophages and dendritic cells. Single gene expression implicated increased expression of pro-tumor (M2-like) macrophage-specific genes post-chemotherapy. Our results suggest that regimens combining chemotherapy with macrophage-targeting agents should be investigated in HR+/HER2− breast cancer.
Financial support:
This work was supported by National Institutes of Health Division of Loan Repayment (AGW and DGS); Susan G. Komen Leadership Grant (EPW, CMP); Terri Brodeur Breast Cancer Foundation Grant (DGS); Breast Cancer Research Foundation Grant (CMP), Pink Ribbons Project (EAM), and Nancy Owens Memorial Foundation (EAM). In addition, we acknowledge the National Institutes of Health (NIH) grant Dana-Farber/Harvard Cancer Center Breast Cancer SPORE 5P50CA168504.
Conflict of interest disclosure statement: SMT receives institutional research funding from Genentech, Novartis, Pfizer, Eli Lilly, Merck, Nektar, AstraZeneca, Bristol Meyers Squibb, Exelixis, Eisai and has served on advisory boards for Genentech, Novartis, Pfizer, Eli Lilly, Merck, Nektar, Eisai, Nanostring, Immunomedics, Bristol Meyers Squibb, and AstraZeneca. CMP is an equity stock holder, consultant, and Board of Director Member of BioClassifier LLC. CMP is also listed an inventor on patent applications on the Breast PAM50 Subtyping assay. EAM serves on advisory boards for Merck, SELLAS Lifesciences, AstraZeneca/MedImmune, TapImmune, and Peregrine Pharmaceuticals, and received institutional research funding from Genentech, Astra Zeneca/MedImmune, and SELLAS Lifesciences. RW receives grant research funding from Astra Zeneca and Acerta. WTB receives research funding from Pfizer. HAB, AS, and PD are employed by NanoString. IK serves as a consultant for Daiichi Sankyo and has consulting/contracted research relationships with Genentech/Roche. EPW has received honorarium from Genentech, Leap, InfiniteMD, and Lilly. DD has consulted for Axela, Genomic Health Inc, and Novartis, and serves on the academic advisory board for Oncology Analytics, Inc. JLG is a consultant for GSK and receives sponsored research support from GSK and Eli Lilly.
References
- 1.Howlader N, Altekruse SF, Li CI, Chen VW, Clarke CA, Ries LA, et al. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. Journal of the National Cancer Institute 2014;106(5) doi 10.1093/jnci/dju055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Issa-Nummer Y, Darb-Esfahani S, Loibl S, Kunz G, Nekljudova V, Schrader I, et al. Prospective validation of immunological infiltrate for prediction of response to neoadjuvant chemotherapy in HER2−negative breast cancer--a substudy of the neoadjuvant GeparQuinto trial. PloS one 2013;8(12):e79775 doi 10.1371/journal.pone.0079775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muenst S, Schaerli AR, Gao F, Daster S, Trella E, Droeser RA, et al. Expression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancer. Breast cancer research and treatment 2014;146(1):15–24 doi 10.1007/s10549-014-2988-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vidula N, Yau C, Goga A, Rugo HS. Programmed cell death 1 (PD-1) receptor and prograamed death ligand 1 (PD-L1) expression in primary breast cancer (BC); correlations with clinical characteristics and patient outcomes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2015;33:suppl; abstr 1090. [Google Scholar]
- 5.Ghebeh H, Mohammed S, Al-Omair A, Qattan A, Lehe C, Al-Qudaihi G, et al. The B7-H1 (PD-L1) T lymphocyte-inhibitory molecule is expressed in breast cancer patients with infiltrating ductal carcinoma: correlation with important high-risk prognostic factors. Neoplasia 2006;8(3):190–8 doi 10.1593/neo.05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bracci L, Schiavoni G, Sistigu A, Belardelli F. Immune-based mechanisms of cytotoxic chemotherapy: implications for the design of novel and rationale-based combined treatments against cancer. Cell death and differentiation 2014;21(1):15–25 doi 10.1038/cdd.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rugo HS, Delord JP, Im SA, Ott PA, Piha-Paul SA, Bedard PL, et al. Safety and Antitumor Activity of Pembrolizumab in Patients with Estrogen Receptor-Positive/Human Epidermal Growth Factor Receptor 2-Negative Advanced Breast Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2018;24(12):2804–11 doi 10.1158/1078-0432.Ccr-17-3452. [DOI] [PubMed] [Google Scholar]
- 8.Dirix LY, Takacs I, Jerusalem G, Nikolinakos P, Arkenau HT, Forero-Torres A, et al. Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1b JAVELIN Solid Tumor study. Breast cancer research and treatment 2018;167(3):671–86 doi 10.1007/s10549-017-4537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svensson S, Abrahamsson A, Rodriguez GV, Olsson AK, Jensen L, Cao Y, et al. CCL2 and CCL5 Are Novel Therapeutic Targets for Estrogen-Dependent Breast Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2015;21(16):3794–805 doi 10.1158/1078-0432.CCR-15-0204. [DOI] [PubMed] [Google Scholar]
- 10.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer cell 2015;27(4):462–72 doi 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelekanou V, Villarroel-Espindola F, Schalper KA, Pusztai L, Rimm DL. CD68, CD163, and matrix metalloproteinase 9 (MMP-9) co-localization in breast tumor microenvironment predicts survival differently in ER-positive and -negative cancers. Breast cancer research : BCR 2018;20(1):154 doi 10.1186/s13058-018-1076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tolaney SM, Boucher Y, Duda DG, Martin JD, Seano G, Ancukiewicz M, et al. Role of vascular density and normalization in response to neoadjuvant bevacizumab and chemotherapy in breast cancer patients. Proceedings of the National Academy of Sciences of the United States of America 2015;112(46):14325–30 doi 10.1073/pnas.1518808112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2007;25(28):4414–22 doi 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 14.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 2015;26(2):259–71 doi 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. The Journal of clinical investigation 2017;127(8):2930–40 doi 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Network TCGA. Comprehensive molecular portraits of human breast tumours. Nature 2012;490(7418):61–70 doi 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stover DG, Coloff JL, Barry WT, Brugge JS, Winer EP, Selfors LM. The Role of Proliferation in Determining Response to Neoadjuvant Chemotherapy in Breast Cancer: A Gene Expression-Based Meta-Analysis. Clinical cancer research : an official journal of the American Association for Cancer Research 2016;22(24):6039–50 doi 10.1158/1078-0432.Ccr-16-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. Journal of Clinical Oncology 2009;27(8):1160–7 doi Doi 10.1200/Jco.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nature methods 2015;12(5):453–7 doi 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varet H. 2016. packHV: A few Useful Functions for Statisticians. <https://CRAN.R-project.org/package=packHV>.
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 1995;57(1):289–300. [Google Scholar]
- 22.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med 2018;379(22):2108–21 doi 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 23.Ruffell B, Au A, Rugo HS, Esserman LJ, Hwang ES, Coussens LM. Leukocyte composition of human breast cancer. Proceedings of the National Academy of Sciences of the United States of America 2012;109(8):2796–801 doi 10.1073/pnas.1104303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emens LA, Esteva F, Beresford M, Saura C, De Laurentiis M, Kim S-B, et al. Results from KATE2, a randomized phase 2 study of atezolizumab (atezo)+trastuzumab emtansine (T-DM1) vs placebo (pbo)+T-DM1 in previously treated HER2+ advanced breast cancer (BC). San Antonio Breast Cancer Symposium 2018. [Google Scholar]
- 25.Zhao Y, Xia Q, Liu Y, Bai W, Yao Y, Ding J, et al. TCF7L2 and EGR1 synergistic activation of transcription of LCN2 via an ERK1/2-dependent pathway in esophageal squamous cell carcinoma cells. Cellular signalling 2018;55:8–16 doi 10.1016/j.cellsig.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2010;16(21):5222–32 doi 10.1158/1078-0432.Ccr-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denkert C, von Minckwitz G, Brase JC, Sinn BV, Gade S, Kronenwett R, et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J Clin Oncol 2015;33(9):983–91 doi 10.1200/JCO.2014.58.1967. [DOI] [PubMed] [Google Scholar]
- 28.Denkert C, Loibl S, Noske A, Roller M, Muller BM, Komor M, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol 2010;28(1):105–13 doi 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 29.Denkert C, von Minckwitz G, Darb-Esfahani S, Lederer B, Heppner BI, Weber KE, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. The Lancet Oncology 2018;19(1):40–50 doi 10.1016/s1470-2045(17)30904-x. [DOI] [PubMed] [Google Scholar]
- 30.Dunbier AK, Ghazoui Z, Anderson H, Salter J, Nerurkar A, Osin P, et al. Molecular profiling of aromatase inhibitor-treated postmenopausal breast tumors identifies immune-related correlates of resistance. Clinical cancer research : an official journal of the American Association for Cancer Research 2013;19(10):2775–86 doi 10.1158/1078-0432.CCR-12-1000. [DOI] [PubMed] [Google Scholar]
- 31.Zitvogel L, Apetoh L, Ghiringhelli F, Andre F, Tesniere A, Kroemer G. The anticancer immune response: indispensable for therapeutic success? The Journal of clinical investigation 2008;118(6):1991–2001 doi 10.1172/JCI35180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loi S, Dushyanthen S, Beavis PA, Salgado R, Denkert C, Savas P, et al. RAS/MAPK Activation Is Associated with Reduced Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancer: Therapeutic Cooperation Between MEK and PD-1/PD-L1 Immune Checkpoint Inhibitors. Clinical cancer research : an official journal of the American Association for Cancer Research 2016;22(6):1499–509 doi 10.1158/1078-0432.CCR-15-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamy AS, Pierga JY, Sabaila A, Laas E, Bonsang-Kitzis H, Laurent C, et al. Stromal lymphocyte infiltration after neoadjuvant chemotherapy is associated with aggressive residual disease and lower disease-free survival in HER2−positive breast cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 2017;28(9):2233–40 doi 10.1093/annonc/mdx309. [DOI] [PubMed] [Google Scholar]
- 34.Szekely B, Bossuyt V, Li X, Wali VB, Patwardhan GA, Frederick C, et al. Immunological differences between primary and metastatic breast cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 2018;29(11):2232–9 doi 10.1093/annonc/mdy399. [DOI] [PubMed] [Google Scholar]
- 35.Emens LA, Cruz C, Eder JP, Braiteh F, Chung C, Tolaney SM, et al. Long-term Clinical Outcomes and Biomarker Analyses of Atezolizumab Therapy for Patients With Metastatic Triple-Negative Breast Cancer: A Phase 1 Study. JAMA oncology 2018. doi 10.1001/jamaoncol.2018.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michea P, Noel F, Zakine E, Czerwinska U, Sirven P, Abouzid O, et al. Adjustment of dendritic cells to the breast-cancer microenvironment is subset specific. Nature immunology 2018;19(8):885–97 doi 10.1038/s41590-018-0145-8. [DOI] [PubMed] [Google Scholar]
- 37.DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer discovery 2011;1(1):54–67 doi 10.1158/2159-8274.Cd-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guerriero JL. Macrophages: The Road Less Traveled, Changing Anticancer Therapy. Trends in molecular medicine 2018;24(5):472–89 doi 10.1016/j.molmed.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christmas BJ, Rafie CI, Hopkins AC, Scott BA, Ma HS, Cruz KA, et al. Entinostat Converts Immune-Resistant Breast and Pancreatic Cancers into Checkpoint-Responsive Tumors by Reprogramming Tumor-Infiltrating MDSCs. Cancer Immunol Res 2018;6(12):1561–77 doi 10.1158/2326-6066.CIR-18-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim K, Skora AD, Li Z, Liu Q, Tam AJ, Blosser RL, et al. Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proceedings of the National Academy of Sciences of the United States of America 2014;111(32):11774–9 doi 10.1073/pnas.1410626111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orillion A, Hashimoto A, Damayanti N, Shen L, Adelaiye-Ogala R, Arisa S, et al. Entinostat Neutralizes Myeloid-Derived Suppressor Cells and Enhances the Antitumor Effect of PD-1 Inhibition in Murine Models of Lung and Renal Cell Carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research 2017;23(17):5187–201 doi 10.1158/1078-0432.CCR-17-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukumura D, Kloepper J, Amoozgar Z, Duda DG, Jain RK. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nature reviews Clinical oncology 2018. doi 10.1038/nrclinonc.2018.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Condamine T, Dominguez GA, Youn JI, Kossenkov AV, Mony S, Alicea-Torres K, et al. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Science immunology 2016;1(2) doi 10.1126/sciimmunol.aaf8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.