Abstract
We have previously shown that expression levels of 48 long non-coding RNAs (lncRNAs) can generate a prognostic lncRNA score that independently associates with outcome of older patients with cytogenetically normal acute myeloid leukemia (CN-AML). However, the techniques used to identify and measure prognostic lncRNAs (i.e., RNA sequencing and microarrays) are not tailored for clinical testing. Herein we report on an assay (based on the nCounter platform) that is designed to produce targeted measurements of prognostic lncRNAs in a clinically applicable manner. We analyzed a new cohort of 76 older CN-AML patients and found that the nCounter assay yielded reproducible measurements and that the lncRNA score retained its prognostic value; patients with high lncRNA scores had lower complete remission (CR) rates (P=0.009; 58% vs. 87%), shorter disease-free (P=0.05; 3-year rates: 0% vs. 21%), overall (OS, P=0.02; 3-year rates: 10% vs. 29%) and event-free survival (EFS; P=0.002, 3-year rates: 0% vs. 18%) than patients with low lncRNA scores. In multivariable analyses, the lncRNA score independently associated with CR rates (P=0.02), OS (P=0.02) and EFS (P=0.02). To gain biological insights, we examined our initial cohort of 71 older CN-AML patients, previously analyzed with RNA sequencing. Genes involved in immune response and B-cell receptor signaling were enriched in patients with high lncRNA scores. We conclude that clinically applicable lncRNA profiling is feasible and potentially useful for risk stratification of older CN-AML patients. Furthermore, we identify potentially targetable molecular pathways that are active in the high-risk patients with high lncRNA scores.
Keywords: Acute myeloid leukemia, prognosis, long non-coding RNAs, transcriptome profiling
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease with regard to the underlying molecular abnormalities and its clinical course (1, 2). The outcome of patients with AML is generally poor (2), especially in the case of patients who are 60 years of age or older. Only 10% of older AML patients who are fit to receive induction chemotherapy will remain alive and leukemia-free five years after their diagnosis (2). Thus, it is important to identify molecular markers that could distinguish between the patients who will respond to standard treatment from those who will not and who could benefit from experimental therapeutic approaches. Currently, chromosomal alterations, which are detected in approximately 55–60% of all AML cases, are used in the clinic to guide treatment decisions (2–5). In patients who lack microscopically detectable chromosomal abnormalities and thus have cytogenetically normal AML (CN-AML), recurrent gene mutations that associate with clinical outcome have been identified and are currently used to risk-stratify the treatment of CN-AML patients (6–8).
Long non-coding RNAs (lncRNAs) comprise a novel class of non-coding RNA molecules, which are equal to or longer than 200 nucleotides (9). LncRNAs have been shown to regulate many key cellular functions (10–12) and have been implicated in cancer pathogenesis (13–15). Our group has previously shown that expression levels of lncRNAs have prognostic significance in older patients with CN-AML (16). Specifically, we have demonstrated that a weighted summary expression score of 48 lncRNAs (called lncRNA score) provides independent prognostic information in this patient population.
Although two independent techniques of transcriptome interrogation (i.e., a microarray platform and total RNA sequencing) were used to generate and validate the prognostic lncRNA score in the aforementioned study (16), these techniques are not suitable for patient testing in the clinic. Consequently, there is a need for development of a fast, reproducible and clinically applicable assay that would enable the translation of lncRNA profiling from the bench to the bedside. To address this need, we designed an assay allowing targeted measurements of prognostic lncRNAs using the nCounter analysis system (NanoString Technologies, Inc.). The nCounter platform has been developed to provide RNA measurements in a single reaction without amplification and is compatible with real-life clinical testing. This technology is currently used as the basis of an FDA-approved assay that measures the expression of RNA molecules for risk stratification of breast cancer patients (17, 18). Herein, we analyzed a cohort of 76 older patients with CN-AML, and report on the prognostic value of the lncRNA score, as measured by the nCounter lncRNA assay. In addition, to identify potentially targetable molecular pathways in the subset of patients with high lncRNA scores, we performed transcriptome analyses in our initial cohort of older CN-AML patients who had been previously analyzed with total RNA sequencing (RNA seq) (16).
Materials and Methods
Patients and treatment
In this study, we analyzed pretreatment bone marrow (BM) or blood of older patients (aged ≥60 years) with de novo CN-AML, who received intensive cytarabine/anthracycline-based therapy on Cancer and Leukemia Group B (CALGB)/Alliance for Clinical Trials in Oncology (Alliance) frontline clinical trials. We studied only patients who were alive 30 days after initiation of induction chemotherapy. Per protocol, no patient received allogeneic stem cell transplantation in first complete remission (CR). Details regarding treatment protocols are provided in the Supplementary Data. There were no patients who were selected for the study but not included in the final analyses because of poor RNA quality or failure of the profiling experiments. All patients provided written informed consent for the analyses of their samples and data. All study protocols were in accordance with the Declaration of Helsinki and approved by institutional review boards at each center.
Cytogenetic and molecular analyses
Cytogenetic analyses were performed in CALGB/Alliance-approved institutional laboratories and the results were confirmed by central karyotype review (19). The diagnosis of normal karyotype was based on at least 20 metaphase cells analyzed in BM specimens subjected to short-term (24- or 48-hour) unstimulated cultures.
Targeted amplicon sequencing using the Miseq platform (Illumina) was used to analyze DNA samples for the presence of gene mutations that have been reported to associate with clinical outcome of CN-AML patients [i.e., mutations in the ASXL1, DNMT3A (R882 and non-R882), IDH1, IDH2 (R140 and R172), NPM1, RUNX1, TET2 and WT1 genes, and FLT3-tyrosine kinase domain (FLT3-TKD) mutations], as described previously (16, 20). A variant allele frequency of ≥10% was used as the cut-off to distinguish between mutated versus wild-type alleles of these genes. The presence of mutations in the CEBPA gene and FLT3-internal tandem duplications (FLT3-ITD) were evaluated using Sanger sequencing (21) and fragment analysis (22), respectively as described previously. As recent publications (2, 23, 24) indicate that only biallelic CEBPA mutations confer prognostic significance, we considered patients with this genotype as mutated.
Transcriptome analyses
The nCounter analysis system from NanoString allows direct profiling of individual RNA molecules in a single reaction without amplification. The custom assay we designed includes 46 of the 48 lncRNAs that are used to generate the prognostic lncRNA score, as well as selected mRNAs and microRNAs (miRs) whose expression were previously reported as associated with clinical outcome of CN-AML patients. These include BAALC (25), ERG (26), MN1 (27), miR-155 (28), miR-3151 (29) and miR-181a (30), and the seven genes that comprise the integrated genetic-epigenetic score (31) [CD34, MIR155HG, RHOC, SCRN1, F2RL1, FAM92A1 and VWA8]. Internal controls for normalization (e.g., GAPDH, ABL) were also included per NanoString guidelines. These probes were designed and synthesized by NanoString Technologies and the experiments were performed at The Ohio State University using the nCounter Diagnostic analysis system. Total RNA extracted with Trizol reagent was used as input material for the assay. For the details concerning calculation of the prognostic lncRNA score with the nCounter assay measurements see the Supplementary Data. The nCounter profiling experiments have been submitted to the GEO repository under the accession number GSE130923.
The expression status (i.e., high or low expresser) of each of the aforementioned prognostic mRNA and miR transcripts (e.g., BAALC, miR-155) was determined for each patient using the median expression value measured by the nCounter assay as the cut-off.
Statistical analyses
Clinical endpoint definitions are given in the Supplementary Data. Baseline demographic, clinical, and molecular features were compared between patients with high and those with low lncRNA scores using the Wilcoxon rank sum and Fisher’s exact tests for continuous and categorical variables, respectively (32). The estimated probabilities of disease-free (DFS), overall (OS) and event-free survival (EFS) were calculated using the Kaplan–Meier method, and the log-rank test evaluated differences between survival distributions (33). Cox proportional hazards models were used to calculate hazard ratios for DFS, OS and EFS (34). Multivariable Cox proportional hazards models were constructed using a forward selection procedure. All statistical analyses were performed by the Alliance Statistics and Data Center. The researchers who conducted the laboratory profiling experiments were blinded to patient outcome results.
Results
Design of nCounter assay and reproducibility of the measurements
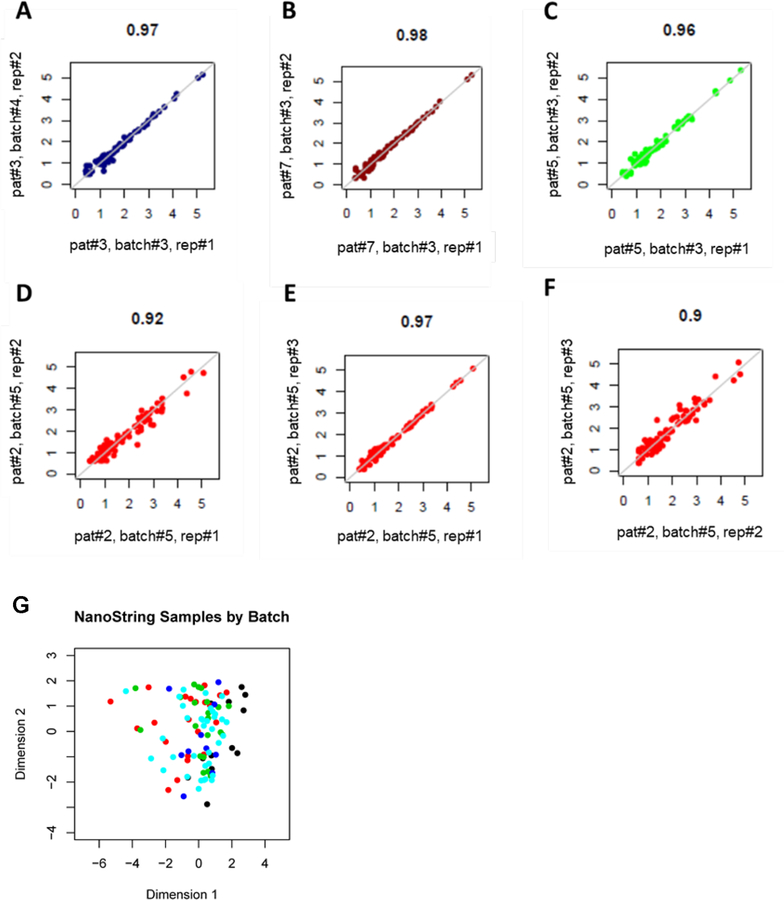
nCounter probes capable of interrogating the lncRNA expression levels could be generated for 46 of the 48 prognostic lncRNAs described previously (16) (Supplementary Table S1). The sequences of the designed probes are provided in the Supplementary Data section (Supplementary Table S2). The assay also included mRNA and miRNA transcripts that have been previously reported to be prognostic in AML and reference genes for quality control of the analyzed material (Supplementary Table S3). Samples of 76 older patients with de novo CN-AML were measured with the nCounter platform in five independent experiments. To evaluate the reproducibility of lncRNA measurements across time and measured batches, we measured 16 of the 76 samples repeatedly. Eleven samples were measured twice in separate batches, three samples were measured 3 times in the same batch, and 2 samples measured four times in the same batch, yielding a total of 39 replicate runs. Analyses of samples that were measured repeatedly showed satisfactory reproducibility within and across measured batches in all but one case. The median of the Pearson’s r squared values for the correlation of the nCounter measurements for each pair of replicate runs was 0.94 (range: 0.44–0.98, Supplementary Figures S1 and S2). The samples of three patients that were measured twice in the same or separate batches (Fig. 1A–C) and of one patient, which were measured three times in the same batch (Fig. 1D–F) are depicted in Figure 1 as examples.
Figure 1.
Scatterplots depicting the correlation of nCounter results in repeated measurement of samples. A-C, Measurements of three samples that were analyzed twice in separate batches, D-F, Measurements of one sample that was analyzed three times in one batch. The Pearson’s r squared value is annotated on the top of each plot. G, Multidimensional scaling plots showing relationships between the total measurements of the 76 samples analyzed with the nCounter assay. Physical distance between samples indicates similarity: the shorter the distance, the higher the similarity of the measurements. Each dot represents the combined measurements of an individual sample. Measurements are colored by batch: black indicates batch 1; red, batch 2; green, batch 3; deep blue, batch 4 and light blue, batch 5.
To study whether the separation of the measurements in different batches impacted on the nCounter results, we generated multidimensional scaling plots. These plots display the pairwise Euclidean distance between the samples in two dimensions, and the physical distance between samples represents sample similarity (i.e., the shorter the distance between two annotated measurements, the higher the similarity between them). We observed that when all analyzed samples were depicted simultaneously, there was no segregation of the nCounter measurements by batches (Fig. 1G).
Association of lncRNA score with pretreatment characteristics of patients
To examine whether the lncRNA score retained its prognostic value when determined by nCounter assay measurements, we performed outcome analysis in our new cohort of 76 older patients with CN-AML. Specifically, we used the median value of the lncRNA score, as measured with the nCounter assay, to divide our dataset into patients with high and patients with low lncRNA scores. With regard to pretreatment characteristics, patients with high lncRNA scores were older (P=0.04), and harbored mutated NPM1 (P=0.003) less frequently and mutated RUNX1 (P=0.03) and FLT3-ITD (P=0.005) more frequently than patients with low lncRNA scores. Patients with high lncRNA scores were also more frequently classified in the Intermediate or Adverse Risk Groups of the ELN Classification (2) (P<0.001), and were high expressers of ERG (P<0.001), BAALC (P=0.003) and miR-155 (P=0.04), and low expressers of MN1 (P=0.04) more often than patients with low lncRNA scores (Table 1).
Table 1.
Comparison of clinical and molecular characteristics by low and high long non-coding RNA (lncRNA) score in the cohort of 76 older patients (aged ≥60 years) with cytogenetically normal acute myeloid leukemia, who were analyzed with the nCounter assay.
| Characteristic | Low lncRNA Score (n=38) | High lncRNA Score (n=38) | P |
|---|---|---|---|
| Age, years | 0.04 | ||
| Median | 66 | 71 | |
| Range | 60–81 | 60–82 | |
| Sex, n (%) of females | 21 (55) | 19 (50) | 0.82 |
| Race, n (%) | 1.00 | ||
| White | 35 (95) | 34 (92) | |
| Non-white | 2 (5) | 3 (8) | |
| Hemoglobin, g/dL | 0.15 | ||
| Median | 9 | 9.4 | |
| Range | 6.5–11.9 | 6.8–11.3 | |
| Platelet count, x109/L | 0.67 | ||
| Median | 70 | 70 | |
| Range | 18–507 | 5–592 | |
| WBC count, x109/L | 0.54 | ||
| Median | 29.7 | 28.5 | |
| Range | 1.4–343.6 | 1.0–173.1 | |
| Blood blasts, % | 0.18 | ||
| Median | 36 | 58 | |
| Range | 0–90 | 0–95 | |
| Bone marrow blasts, % | 0.56 | ||
| Median | 68 | 64 | |
| Range | 6–99 | 0–95 | |
| Extramedullary involvement, n (%) | 7 (19) | 4 (13) | 0.53 |
| NPM1, n (%) | 0.003 | ||
| Mutated | 28 (80) | 15 (44) | |
| Wild type | 7 (20) | 19 (56) | |
| FLT3-ITD, n (%) | 0.005 | ||
| Mutated | 8 (26) | 20 (63) | |
| FLT3-ITD allelic raio ≥0.50 | 4 | 7 | |
| FLT3-ITD allelic ratio <0.50 | 4 | 13 | |
| Wild type | 23 (74) | 12 (38) | |
| CEBPA, n (%) | 1.00 | ||
| Biallelic mutations | 1 (4) | 0 (0) | |
| Wild type or monoallelic mutations | 27 (96) | 25 (100) | |
| FLT3-TKD, n (%) | 1.00 | ||
| Present | 2 (6) | 2 (6) | |
| Absent | 32 (94) | 31 (94) | |
| WT1, n (%) | 0.71 | ||
| Mutated | 3 (9) | 4 (12) | |
| Wild type | 31 (91) | 29 (88) | |
| TET2, n (%) | 0.59 | ||
| Mutated | 11 (32) | 8 (24) | |
| Wild type | 23 (68) | 25 (76) | |
| IDH1, n (%) | 0.75 | ||
| Mutated | 5 (15) | 6 (18) | |
| Wild type | 29 (85) | 27 (82) | |
| IDH2, n (%) | 0.51 | ||
| Mutated | 4 (12) | 6 (18) | |
| Wild type | 30 (88) | 27 (82) | |
| ASXL1, n (%) | 0.43 | ||
| Mutated | 2 (6) | 4 (12) | |
| Wild type | 32 (94) | 29 (88) | |
| DNMT3A, n (%) | 0.12 | ||
| Mutated | 15 (44) | 8 (24) | |
| Wild type | 19 (56) | 25 (76) | |
| RUNX1, n (%) | 0.03 | ||
| Mutated | 1 (3) | 7 (21) | |
| Wild type | 33 (97) | 26 (79) | |
| ELN Risk Category,a n (%) | <0.001 | ||
| Favorable | 22 (79) | 6 (23) | |
| Intermediate | 5 (18) | 9 (35) | |
| Adverse | 1 (4) | 11 (42) | |
| ERG expression group,b n (%) | <0.001 | ||
| High | 11 (29) | 27 (71) | |
| Low | 27 (71) | 11 (29) | |
| BAALC expression group,b n (%) | 0.003 | ||
| High | 12 (32) | 26 (68) | |
| Low | 26 (68) | 12 (32) | |
| MN1 expression group,b n (%) | 0.04 | ||
| High | 24 (63) | 14 (37) | |
| Low | 14 (37) | 24 (63) | |
| miR-181a expression group,b n (%) | 0.82 | ||
| High | 20 (53) | 18 (47) | |
| Low | 18 (47) | 20 (53) | |
| miR-3151,b n (%) | 0.82 | ||
| High | 20 (53) | 18 (47) | |
| Low | 18 (47) | 20 (53) | |
| miR-155 expression group,b n (%) | 0.04 | ||
| High | 14 (37) | 24 (63) | |
| Low | 24 (63) | 14 (37) | |
Abbreviations: n, number; WBC, white blood cell; ELN, European LeukemiaNet; FLT3-ITD, internal tandem duplication of the FLT3 gene; FLT3-TKD, tyrosine kinase domain mutation in the FLT3 gene.
Among patients with cytogenetically normal acute myeloid leukemia (CN-AML), the ELN Favorable Risk Category comprises patients with biallelic mutations in CEBPA and patients with mutated NPM1 without FLT3-ITD or with FLT3-ITDlow. The ELN Intermediate Risk Category includes patients with wild-type CEBPA and either wild-type NPM1 without FLT3-ITD, wild-type NPM1 and FLT3-ITDlow or mutated NPM1 and FLT3-ITDhigh. The ELN Adverse Risk Category comprises patients with wild-type CEBPA and wild-type NPM1 with FLT3-ITDhigh, patients with mutated TP53, and patients with mutated RUNX1 and/or mutated ASXL1 (if these mutations do not co-occur with Favorable-risk AML subtype). FLT3-ITDlow is defined by a FLT3-ITD/FLT3 wild-type allelic ratio of less than 0.5 and FLT3-ITDhigh is defined as by a FLT3-ITD/FLT3 wild-type allelic ratio of equal to or more than 0.5.
The median expression value was used as the cut point.
Association of lncRNA score with clinical outcome
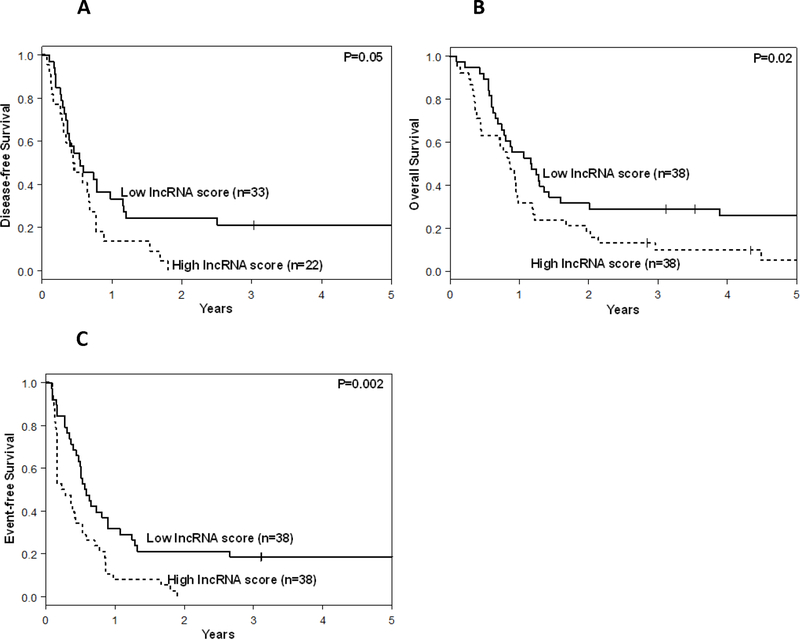
Survival analyses in the new cohort of older CN-AML patients showed that patients with high lncRNA scores were less likely to achieve a CR than those with low lncRNA scores (58% vs. 87%, P=0.009). High lncRNA score status associated with shorter DFS (P=0.05; Fig. 2A). None of the patients with high lncRNA scores were alive and leukemia free three years after diagnosis in contrast to 21% of the patients with low lncRNA scores who were. Patients with high lncRNA scores also had shorter OS (P=0.02, 3-year rates: 10% vs. 29%; Fig. 2B). In addition, high lncRNA scores associated with shorter EFS (P=0.02; Fig. 2C, Table 2). Three years after diagnosis 18% of the patients with low lncRNA scores were alive and had not experienced an event in comparison to none of the patients with high lncRNA scores.
Figure 2.
Outcomes of older patients (aged ≥60 years) with cytogenetically normal acute myeloid leukemia with low and high long non-coding RNA (lncRNA) scores. A, Disease-free survival, B, overall survival and C, event-free survival. The lncRNA score of each individual patient was computed as a weighted score, based on the nCounter assay measurements of 46 prognostic lncRNAs.
Table 2.
Outcome of older patients (aged ≥ 60 years) with cytogenetically normal acute myeloid leukemia by low and high long non-coding RNA (lncRNA) score status
| End point | Low lncRNA score (n=38) | High lncRNA score (n=38) | P |
|---|---|---|---|
| Complete remission | 0.009 | ||
| n, (%) | 33 (87) | 22 (58) | |
| Disease-free survival | 0.05 | ||
| Median, years | 0.5 | 0.5 | |
| Disease-free at 3 years, % (95% CI) | 21 (9–36) | 0 | |
| Disease-free at 5 years,% (95% CI) | 21 (9–36) | 0 | |
| Overall survival | 0.02 | ||
| Median, years | 1.2 | 0.9 | |
| Alive at 3 years, % (95% CI) | 29 (16–44) | 10 (3–22) | |
| Alive at 5 years, % (95% CI) | 26 (13–40) | 5 (0–18) | |
| Event-free survival | 0.002 | ||
| Median, years | 0.6 | 0.3 | |
| Event-free at 3 years, % (95% CI) | 18 (8–32) | 0 | |
| Event-free at 5 years,% (95% CI) | 18 (8–32) | 0 | |
Abbreviations: n, number; CI, confidence interval.
Multivariable analyses
To assess whether the prognostic lncRNA score provides independent prognostic information in the context of other established prognostic markers, we constructed multivariable proportional hazards models. High lncRNA scores independently associated with a lower CR rate (P=0.02), after adjusting for BAALC expression status (P=0.02). High lncRNA score status also independently associated with shorter OS (P=0.02), after adjusting for white blood cell (WBC) counts (P=0.005) and the sex of patients (P=0.02). Finally, high lncRNA score was an independent marker of shorter EFS (P=0.02); patients with high lncRNA scores had approximately a two-fold increase in their risk of experiencing an event than those with low lncRNA score, after adjusting for WBC counts (P=0.002) and BAALC expression status (P=0.02, Table 3). The lncRNA score status did not remain significantly associated with DFS duration in multivariable analysis.
Table 3.
Multivariable analyses for outcome in the dataset of older (aged ≥ 60 years) cytogenetically normal acute myeloid leukemia patients
| Variables in final modelsa | Complete Remission | Overall survival | Event-free survival | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | |
| lncRNA score, high versus low | 0.18 (0.04–0.75) | 0.02 | 1.84 (1.09, 3.10) | 0.02 | 1.88 (1.08, 3.27) | 0.02 |
| BAALC, high versus low | 0.17 (0.04, 0.74) | 0.02 | - | - | 1.88 (1.10, 3.21) | 0.02 |
| White blood cell counts, continuous | - | - | 1.34 (1.09, 1.64) | 0.005 | 1.38 (1.13, 1.68) | 0.002 |
| Sex, male versus female | - | - | 1.90 (1.14, 3.17) | 0.01 | - | - |
Abbreviations: OR, odds ratio; HR, hazard ratio; CI, confidence interval; lncRNA, long non-coding RNA.
The lncRNA score status did not remain significantly associated with disease-free survival in multivariable analysis.
NOTE: Odds ratios less than one indicate lower chances for achieving complete remission. Hazard ratios greater than one indicate higher risk for failure to achieve complete remission or death (overall survival) or for failure to achieve complete remission, relapse or death (event-free survival) for the first category listed. Variables considered for model inclusion were: lncRNA score status (high vs. low), age (as a continuous variable, in 10-year increments), sex (male vs. female), race (white vs. non-white), white blood cell count [as a continuous variable, in 50-unit increments], hemoglobin (as a continuous variable, in 1-unit increments), platelet count (as a continuous variable, in 50-unit increments), extramedullary involvement (present vs. absent), ASXL1 mutations (mutated vs. wild-type), CEBPA mutations (biallelic mutations vs. monoallelic mutations or wild-type), DNMT3A mutations (mutated vs. wild-type), FLT3-ITD (present vs. absent), FLT3-TKD (present vs. absent), IDH1 mutations (mutated vs. wild-type), IDH2 mutations (mutated vs. wild-type), NPM1 mutations (mutated vs. wild-type), RUNX1 mutations (mutated vs. wild-type), TET2 mutations (mutated vs. wild-type), WT1 mutations (mutated vs. wild-type), BAALC expression levels (high vs. low), ERG expression levels (high vs. low), MN1 expression levels (high vs. low), miR-181a expression levels (high vs. low), miR-3151 (expressed vs. not expressed), and miR-155 expression levels (high vs. low). For BAALC, ERG, MN1, miR-181a and miR-155 the median expression value, as calculated by the nCounter assay, was used as the cut point to divide patients into high and low expressers.
Biological insights regarding the molecular pathways that associate with the lncRNA score
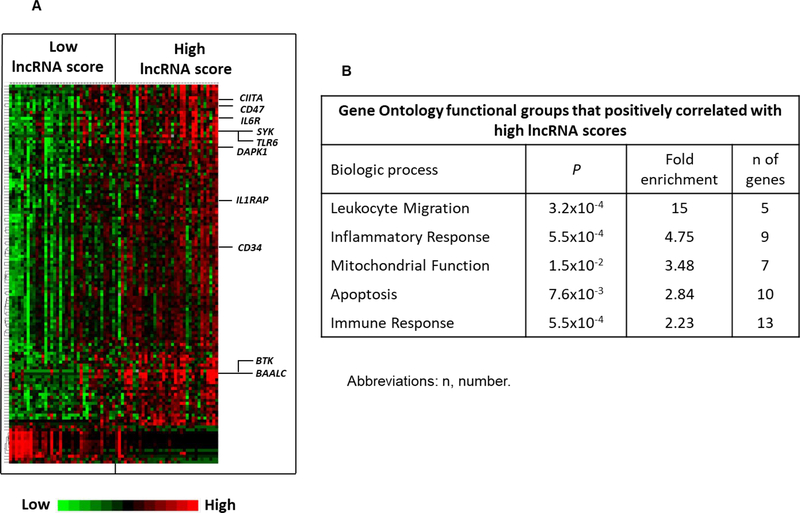
Using the nCounter assay, we could demonstrate that lncRNA score profiling could distinguish between the patients who were more likely to respond to standard chemotherapy from those who were not. However, older patients often have comorbidities that preclude intensification of chemotherapy and allogeneic stem cell transplantation as therapeutic options. It is therefore important to identify targetable molecular pathways, in particular in those patients that are predicted not to benefit from standard therapy. To this end, we performed correlation analysis in our initial cohort of 71 older CN-AML patients, who were analyzed with total RNA sequencing (16). This sequencing technique provides comprehensive information of both coding and non-coding fractions of the transcriptome. We applied stringent criteria [P value of <0.001 and false discovery rate (FDR) of <0.05] and identified 115 transcripts that were upregulated and three transcripts which were downregulated in patients with high lncRNA scores (Fig. 3A and Supplementary Table S4)
Figure 3.
Messenger RNA (mRNA) transcripts which associate with the prognostic long non-coding RNA (lncRNA) score in older patients (aged ≥60 years) with cytogenetically normal acute myeloid leukemia (CN-AML). A, Heat map of the gene-expression signature associated with the lncRNA score. Rows represent protein-coding genes and columns represent patients. Patients are grouped by lncRNA score: low on the left and high on the right. The lncRNA score of each individual patient was computed as a weighted score of 46 prognostic lncRNAs. Expression values of the mRNA transcripts are represented by color: green, expression less than median value; red, expression greater than median value. B, Gene Ontology functional groups that positively correlate with high lncRNA scores in older patients with CN-AML. Gene Ontology functional groups are ranked according to fold enrichment.
Among transcripts overexpressed in patients with high lncRNA scores, we identified genes that are key regulators of the immune system, such as CD74 and CIITA that are implicated in peptide processing and presentation in antigen presenting cells; BTK and SYK implicated in B-cell receptor signaling; and IL1RAP, IL6R and TLR6, which are important cytokine receptors. We also found aberrant overexpression of CD34 in patients with high lncRNA scores, a surface marker of leukemic stem cells whose expression has been related to chemotherapy resistance and poor outcome in AML (35). In addition, genes implicated in leukemogenesis such as DAPK1 (36) and IDH1 (37) were also upregulated in patients with high lncRNA scores. Finally, in keeping with the adverse prognostic impact of high lncRNA score, we found mRNAs, which are established markers of poor outcome in CN-AML, such as BAALC (25) to be enriched in the subset of patients with high lncRNA scores.
To further characterize and classify the genes and molecular pathways that are active in patients with high lncRNA scores we performed Gene Ontology analysis (38). Gene Ontology analysis revealed enrichment for genes involved in leukocyte migration, inflammatory response, mitochondrial function, apoptosis and immune response in patients with high lncRNA scores (Fig. 3B).
Discussion
LncRNAs are gaining increasing recognition as key regulators of important cellular functions including imprinting, cell cycle regulation and apoptosis (39–43). Over the past years, it has become evident that lncRNAs are functionally associated with malignant diseases (43–45) and that they affect clinical outcome of cancer patients. In CN-AML, we reported a prognostic score, which is based on the expression levels of 48 lncRNAs and provides independent prognostic information in older CN-AML patients (16). Importantly, we found that the prognostic lncRNA score showed no association with recurrent prognostic gene mutations that are currently used for risk stratification of CN-AML patients, such as biallelic CEBPA mutations, NPM1 mutations or FLT3-ITD. For this reason, we hypothesized that the lncRNA score could further refine risk stratification of older CN-AML patients. However, the techniques that were used to identify and measure the prognostic lncRNA molecules are not clinically applicable. To acquire fast and reproducible transcriptome measurements, which would facilitate translation of lncRNA profiling to the clinic, we designed a prognostic lncRNA-measuring assay using the nCounter technology. The nCounter platform has been specifically developed to serve as the basis of clinically applicable tests, and is currently used in FDA-approved transcriptome profiling assays (17, 18).
To test the efficacy and reproducibility of our nCounter lncRNA assay, we analyzed a new cohort of 76 older CN-AML patients, who were treated on frontline CALGB/Alliance studies. We performed a total of five experiments using standard RNA extraction techniques and methods. We sought to evaluate the performance of the assay in real-life conditions and, therefore, did not discard any samples on the basis of RNA yield or quality. We performed multiple measurements of individual samples, so as to evaluate the robustness and reproducibility of our assay. We found a satisfactory correlation of the repeated measurements when these were conducted within the same run of the assay or in independent experiments in all but one case.
To examine whether the nCounter-based lncRNA score retained its prognostic value, we performed outcome analyses in our new cohort of older CN-AML patients. We found that the lncRNA score was significantly associated with outcome and that patients with high lncRNA scores were less likely to achieve a CR and had shorter DFS, OS and EFS than patients with low lncRNA scores. We also detected associations of the lncRNA score status with prognostic mutations, such as those in the RUNX1 gene and FLT3-ITD. Despite these associations, in multivariable analyses, the nCounter assay lncRNA score was shown to be an independent prognosticator for achievement of CR, as well as OS and EFS duration after adjusting for other co-variates.
Our current study was conducted in a cohort of older CN-AML patients of relatively small size and was designed to evaluate the feasibility and utility of lncRNA profiling by the use of a nCounter assay in the clinical setting. LncRNA score-based risk assessment in the current study was dependent on profiling of a group of patients to establish the median lncRNA score value for this group that was then used to distinguish low- from high-risk patients. Nevertheless, because the nCounter platform allows individualized transcriptomic measurements it could be potentially used for risk-assignment of individual patients in the future. To achieve this goal, a larger number of older CN-AML patients should be analyzed to establish the optimal lncRNA score value that should be used as a widely accepted cut-off between patients with a low and those with a high lncRNA score in the clinic.
While it is important to identify those older CN-AML patients that will respond to conventional therapeutic modalities, those who will not represent a therapeutic challenge. Confounding comorbidities often preclude the use of such options as intensification of chemotherapy or allogeneic stem cell transplantation that have been proven to be efficacious in younger adult AML patients. To gain biological insights and identify potentially targetable pathways active in patients with high lncRNA scores, we examined which mRNA transcripts correlate with unfavorable lncRNA profiles and performed gene ontology analyses in our initial cohort of 71 older CN-AML patients, previously analyzed with RNA seq (16). We found genes involved in the regulation of the immune response and B-cell receptor signaling, such as BTK and SYK, to be overexpressed in patients with high lncRNA scores. High expression levels of immune response-related genes are reminiscent of the mRNA expression signature associated with RUNX1 mutations in CN-AML patients (46). The relatively small number of patients with RUNX1 mutations in our initial cohort (n=8) renders it unlikely that these mutations are the sole drivers of the detected lncRNA score-related gene expression signature. It could be hypothesized instead that high expression of the prognostically unfavorable lncRNAs has a similar impact on the transcriptome to RUNX1 mutations.
In recent years, targeting BTK with inhibitory molecules has proven to be a successful therapeutic approach for certain lymphoid malignancies (47, 48) and BTK inhibitors are currently included among the standard-of-care therapeutic agents for these diseases. Use of BTK inhibitors has also yielded encouraging preclinical results in AML (49). The high expression of BTK in patients with high lncRNA score that we detected could provide the rationale for exploring the efficacy of BTK-targeting agents in these patients. Thus, lncRNA profiling could be potentially used not only to risk-stratify treatment of older CN-AML patients but also to guide novel therapeutic approaches in patients who are at high risk of treatment failure.
In summary, we demonstrate the technical feasibility of using the nCounter assay for prognostic lncRNA profiling in a clinically applicable manner. We have also validated the prognostic value of lncRNA expression in older CN-AML patients, in our new cohort of patients analyzed using a different profiling method than the ones used previously. We believe that the value of the nCounter assay for improving risk stratification of AML patients warrants evaluation in future prospective clinical trials.
Supplementary Material
Acknowledgements
We would like to thank: Donna Bucci and Wacharaphon Vongchucherd of The Alliance NCTN Biorepository and Biospecimen Resource for sample processing and storage services, and Lisa J. Sterling and Christine Finks of The Ohio State University, Comprehensive Cancer Center, Columbus, OH for data management.
Support: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821, U10CA180882 and U24CA196171 (all above mentioned grants were awarded to the Alliance for Clinical Trials in Oncology), P50CA140158 (J.C. Byrd), U10CA180850, U10CA180861 (C.D. Bloomfield), U10CA180866, U10CA180867 and R35CA197734 (J.C. Byrd). This work was also supported in part by the Leukemia Clinical Research Foundation. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med 2015;373:1136–52. [DOI] [PubMed] [Google Scholar]
- 2.Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017;129:424–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood 2002;100:4325–36. [DOI] [PubMed] [Google Scholar]
- 4.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010;116:354–65. [DOI] [PubMed] [Google Scholar]
- 5.Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev 2004;18:115–36. [DOI] [PubMed] [Google Scholar]
- 6.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 2012;366:1079–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metzeler KH, Herold T, Rothenberg-Thurley M, Amler S, Sauerland MC, Görlich D, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood 2016;128:686–98. [DOI] [PubMed] [Google Scholar]
- 9.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629–41. [DOI] [PubMed] [Google Scholar]
- 10.Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008;322:750–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature 1991;351:153–5. [DOI] [PubMed] [Google Scholar]
- 12.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011;43:621–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer 2011;10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell 2014;158:593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garzon R, Volinia S, Papaioannou D, Nicolet D, Kohlschmidt J, Yan PS, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A 2014;111:18679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu MC, Pitcher BN, Mardis ER, Davies SR, Friedman PN, Snider JE, et al. PAM50 gene signatures and breast cancer prognosis with adjuvant anthracycline- and taxane-based chemotherapy: correlative analysis of C9741 (Alliance). NPJ Breast Cancer 2016;2:15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mrózek K, Carroll AJ, Maharry K, Rao KW, Patil SR, Pettenati MJ, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: the Cancer and Leukemia Group B experience. Int J Oncol 2008;33:239–44. [PMC free article] [PubMed] [Google Scholar]
- 20.Kroll KW, Eisfeld AK, Lozanski G, Bloomfield CD, Byrd JC, Blachly JS. MuCor: mutation aggregation and correlation. Bioinformatics 2016;32:1557–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcucci G, Maharry K, Radmacher MD, Mrózek K, Vukosavljevic T, Paschka P, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B study. J Clin Oncol 2008;26:5078–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitman SP, Archer KJ, Feng L, Baldus C, Becknell B, Carlson BD, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a Cancer and Leukemia Group B study. Cancer Res 2001;61:7233–9. [PubMed] [Google Scholar]
- 23.Wouters BJ, Löwenberg B, Erpelinck-Verschueren CAJ, van Putten WLJ, Valk PJM, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 2009;113:3088–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dufour A, Schneider F, Metzeler KH, Hoster E, Schneider S, Zellmeier E, et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J Clin Oncol 2010;28:570–7. [DOI] [PubMed] [Google Scholar]
- 25.Baldus CD, Tanner SM, Ruppert AS, Whitman SP, Archer KJ, Marcucci G, et al. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: a Cancer and Leukemia Group B study. Blood 2003;102:1613–8. [DOI] [PubMed] [Google Scholar]
- 26.Marcucci G, Baldus CD, Ruppert AS, Radmacher MD, Mrózek K, Whitman SP, et al. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: a Cancer and Leukemia Group B study. J Clin Oncol 2005;23:9234–42. [DOI] [PubMed] [Google Scholar]
- 27.Langer C, Marcucci G, Holland KB, Radmacher MD, Maharry K, Paschka P, et al. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol 2009;27:3198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcucci G, Maharry KS, Metzeler KH, Volinia S, Wu YZ, Mrózek K, Nicolet D, et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol 2013;31:2086–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisfeld A-K, Marcucci G, Maharry K, Schwind S, Radmacher MD, Nicolet D, et al. miR-3151 interplays with its host gene BAALC and independently affects outcome of patients with cytogenetically normal acute myeloid leukemia. Blood 2012;120:249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwind S, Maharry K, Radmacher MD, Mrózek K, Holland KB, Margeson D, et al. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol 2010;28:5257–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcucci G, Yan P, Maharry K, Frankhouser D, Nicolet D, Metzeler KH, et al. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J Clin Oncol 2014;32:548–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conover WJ Practical Nonparametric Statistics John Wiley & Sons; 1971; page 406. [Google Scholar]
- 33.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc 1958;53:457–81. [Google Scholar]
- 34.Vittinghoff E, Glidden DV, Shiboski SC, McCulloch CE. Regression methods in biostatistics: linear, logistic, survival and repeated measures models New York: Springer; 2005. [Google Scholar]
- 35.Geller RB, Zahurak M, Hurwitz CA, Burke PJ, Karp JE, Piantadosi S, et al. Prognostic importance of immunophenotyping in adults with acute myelocytic leukaemia: the significance of the stem-cell glycoprotein CD34 (My10). Br J Haematol 1990;76:340–7. [DOI] [PubMed] [Google Scholar]
- 36.Larramendy ML, Niini T, Elonen E, Nagy B, Ollila J, Vihinen M, et al. Overexpression of translocation-associated fusion genes of FGFRI, MYC, NPMI, and DEK, but absence of the translocations in acute myeloid leukemia. A microarray analysis. Haematologica 2002;87:569–77. [PubMed] [Google Scholar]
- 37.Marcucci G, Maharry K, Wu Y-Z, Radmacher MD, Mrózek K, Margeson D, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol 2010;28:2348–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 39.Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008;322:750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature 1991;351:153–55. [DOI] [PubMed] [Google Scholar]
- 41.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011;43:621–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011;147:1537–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 2015;28:529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015;47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwarzer A, Emmrich S, Schmidt F, Beck D, Ng M, Reimer C, et al. The non-coding RNA landscape of human hematopoiesis and leukemia. Nat Commun 2017;8:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mendler JH, Maharry K, Radmacher MD, Mrózek K, Becker H, Metzeler KH, et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and microRNA expression signatures. J Clin Oncol 2012;30:3109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Byrd JC, O’Brien S, James DF. Ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med 2013;369:1278–9. [DOI] [PubMed] [Google Scholar]
- 48.Wang ML, Rule S, Martin P, Goy A, Auer R, Kahl BS, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med 2013;369:507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rushworth SA, Murray MY, Zaitseva L, Bowles KM, MacEwan DJ. Identification of Bruton’s tyrosine kinase as a therapeutic target in acute myeloid leukemia. Blood 2014;123:1229–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.