Abstract
Development of animal models to investigate the complex ecosystem of malignant gliomas using the Cre/loxP recombination system has significantly contributed to our understanding of the molecular underpinnings of this deadly disease. In these model systems, once the tumor is induced by activation of Cre-recombinase in a tissue specific manner, further genetic manipulations to explore the progression of tumorigenesis are limited. To expand the application of mouse models for gliomas, we developed GFAP-FLPo mice that express FLPo recombinase specifically in GFAP-positive cells. Lentivirus-based in vivo delivery of cancer genes conditioned by FLP/FRT-mediated recombination initiated gliomas in GFAP-FLPo mice. Using the Cre-mediated multi-fluorescent protein expressing system, we demonstrated that the GFAP-FLPo mouse model enables the analysis of various stages of gliomagenesis. Collectively, we present a new mouse model that will expand our ability to dissect developmental processes of gliomagenesis, and to provide new avenues for therapeutic approaches.
Keywords: Glioma, glioblastoma, GEMM, animal model, FLP recombinase, GFAP-FLPo, Brainbow
Introduction
Gliomas form the most common and a heterogeneous group of primary brain tumors. The most malignant form of glioma, glioblastoma, is one of the most lethal cancer types with a median survival of less than 15 months (1,2). Gliomas display enormous intra-tumoral heterogeneity which imposes severe limitations for development of therapeutic approaches (3-5). Better understandings of cellular and molecular components embedded in heterogeneous tumors are essential to improve personalized treatment. Therefore, a pressing need for neuro-oncology is the ability to accurately and effectively interrogate cellular and molecular candidates in the complex tumor tissues under controlled circumstances.
Genetically engineered mouse models (GEMMs) serve as front-line model systems to transform our understanding of tumor development, and to provide preclinical platforms for evaluating the efficacy of anti-tumor drugs (6,7). In these models, a sufficient, but usually minimal number of cancer-related genes is delivered to cell(s)-of-origin of tumors to initiate tumorigenesis in mice. Consequently, tumors arise de novo and coevolve with native microenvironment in the organ, thereby resulting in the formation of tumors unlike the canonical transplantation models with established tumor cells which usually exhibit rapid formation of tumors regardless of their microenvironment (8,9). In fact, GEMMs for gliomas recapitulate key aspects of the human disease, including genetic, temporal and histopathological features (10-13), which shed light on the importance of cell-of-origin, progression, heterogeneity, and the tumor stroma that cannot be easily studied in human cancers (14-16).
From the technical perspective, site-specific recombination techniques such as Cre/loxP have been powerful methods centrally used for the studies of gene function, modeling diseases and the labeling and manipulation of specific cell populations in vivo (17). Traditionally GEMMs for cancer research have employed Cre-mediated recombination, which limits further genetic manipulation in these model systems. Although alternative recombination systems such as FLP, PhiC31, Dre and Vika have been identified, and engineered to efficiently process catalytic reactions in the mammalian cells, GEMMs with these alternatives have at present limited availability (18-21).
Here, we describe the development of new transgenic animals, GFAP-FLPo mice, for modeling gliomas using lentiviral vectors. This GFAP-FLPo mouse strain expresses FLPo recombinase exclusively in GFAP-positive cells in the brain, which allows us to restrict the initial sources of cell-of-origin of gliomas with the delivery of a combination of HrasG12V and shRNA against Trp53. Furthermore, stage- and tumor-specific activation of Cre recombinase combined with multi-fluorescent protein expression system visualizes morphologically heterogeneous tumor populations in gliomas initiated even with strong oncogenic events. This model will provide more flexible choice of strategies to study basic principles of the complex glioma ecosystem.
Materials and Methods
Mice.
All mice were maintained under pathogen-free conditions at the Salk Institute, and all procedures performed in this study were approved by the Institutional Animal Care and Use Committee. DNA microinjection method was employed at the Salk Transgenic Core (TGC) to achieve random integration of GFAP-FLPo transgene into the mouse genome in order to generate GFAP-FLPo transgenic mice. For the construction of GFAP-FLPo vector, GFAP-cre vector (Addgene # 24704, (22)) was used as an initial source of 15-kb GFAP promoter cassette and Cre sequence was replaced by FLPo-poly(A). This Gfap promoter cassette contains all components for mouse Gfap gene expression including regulatory elements, exons, introns and 3’ UTR, while the expression of endogenous Gfap gene expression is disrupted by a point mutation at start codon sites (ATG to TTG) (Fig. 1A). FLPo-bGHpoly(A) fragment was constructed on pLV-GFP vector, and a NotI/SalI-digested fragment containing FLPo-bGHpoly(A) was then ligated into the NotI/SalI-digested GFAP-cre vector (into the first exon of Gfap gene). The construction was verified by restriction enzyme digestion and sanger-sequencing method. GFAP-FLPo-bGHpoly(A) vector was then linearized with SfiI digestion and purified for microinjection. Obtained GFAP-FLPo mice were backcrossed into a C57BL/6J (Jackson laboratory) background for at least 8 generations. GFAP-FLPo transgenic line #62 will be available from The Jackson Laboratory as JAX Stock No. 033116. FRT-reporter RC::FLTG (23), FRT-STOP-FRT-KrasG12D (24) and Rosa26-CAG-Brainbow2.1/ Confetti (25,26) mice were obtained from the Jackson Laboratory (Stock No. 026932, 008653 and 013731, respectively). Tamoxifen (T5648, Sigma) was dissolved in sunflower seed oil (S5007, Sigma) and injected intraperitoneally into mice for 3 days at a concentration of 100mg/kg body weight.
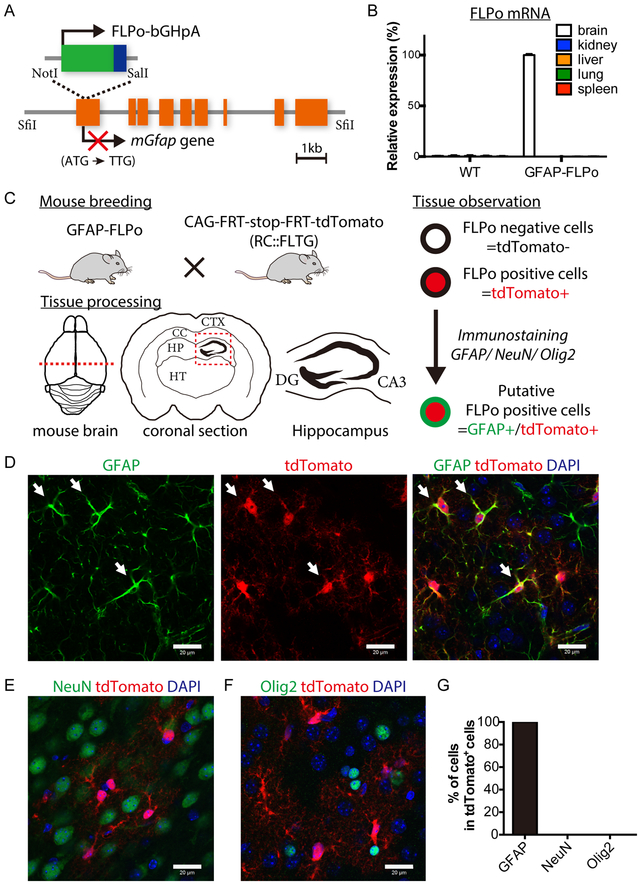
Fig. 1. The development of GFAP-FLPo transgenic animals.
(A) Schematic illustration of GFAP-FLPo construct to generate GFAP-FLPo transgenic animals. FLPo-bGHpA sequence was integrated into first exon of mouse Gfap gene cassette using two restriction enzymes, NotI and SalI. Endogenous expression of GFAP is prevented by a point mutation at the start codon (ATG to TTG). SfiI site to remove the vector backbone.
(B) FLPo mRNA levels in WT and GFAP-FLPo animal (#62) tissues. Data are means with SEM.
(C) GFAP-FLPo mice crossed with CAG-FRT-stop-FRT-tdTomato (RC::FLTG) animals to trace the lineage of FLPo positive cells in the various brain regions of GFAP-FLPo animals. CTX: cortex. CC: corpus callosum. HP: hippocampus. HT: thalamus. DG: dentate gyrus. CA3: Cornu Ammonis 3.
(D) Immunostaining analysis of GFAP (green) and tdTomato (red) expression in the cortex of 3-week-old GFAP-FLPo; CAG-FRT-stop-FRT-tdTomato animal. Arrows mark GFAP-positive/ tdTomato positive cells. Scale bars, 20 μm.
(E) Immunostaining analysis of NeuN (green) and tdTomato (red) expression in the cortex of 3-week-old GFAP-FLPo; CAG-FRT-stop-FRT-tdTomato animal. Scale bars, 20 μm.
(F) Immunostaining analysis of Olig2 (green) and tdTomato (red) expression in the cortex of 3-week-old GFAP-FLPo; CAG-FRT-stop-FRT-tdTomato animal. Scale bars, 20 μm.
(G) Quantified analysis of GFAP, NeuN and Olig2-positive cells in the cortex of 3-week-old GFAP-FLPo; CAG-FRT-stop-FRT-tdTomato animal. 3 animals were subjected to the analysis.
Genotyping PCR analysis.
PCR primers to amplify the conjugation part of GFAP-FLPo transgene were Gfap Fw: CGGAGACGCATCACCTCTG, FLPo Rv: GATCAGCTTCTTCAGGCTG. The expected size, 570bp of amplicon was determined by agarose gel electrophoresis.
Immunostaining.
Thick (100-200um) brain sections were prepared with vibratome and stored in tissue freezing media (25% Glycerol, 30% Ethylene glycol, 1.38g/L NaH2PO4, 5.48g/L Na2HPO4). The primary antibodies used in this study as follows; anti-RFP/ tdTomato (MBS448092; MyBioSource), anti-GFAP (Z0334; DAKO), anti-NeuN (MAB377, MilliporeSigma), anti-Olig2 (AB9610, MilliporeSigma), anti-Sox2 (14-9811-80; eBioscience). The secondary antibodies were as follows (all from Invitrogen): Alexa Fluor 568 anti-goat IgG, Alexa Fluor 647 anti-rat IgG, Alexa Fluor 647 anti-rabbit IgG, Alexa Fluor 647 anti-mouse IgG. The nucleus was stained with DAPI.
Imaging.
Brainbow2.1 tissues with 200-1mm thickness were prepared with vibratome and then cleared with tissue clearing method, ScaleS (27). Briefly, processed brain tissues were transferred into either ScaleSQ(0) or Triton X-100-containing ScaleSQ(5) and incubated at 37 °C for 4 hours, and then transferred into S4(0) to be incubated at room temperature for overnight. The images were acquired with laser scanning microscopy, LSM 810 or 880 with Airyscan (Zeiss) equipped with the laser lines (405, 458, 488, 514, 561, 594 and 633nm). Separation of brainbow2.1 fluorophores was obtained by using a 458nm laser line for mCFP (458-498nm barrier filter), a 488-nm laser line for nGFP (491-518nm barrier filter), a 514-nm laser line for EYFP (518-562nm barrier filter), a 561-nm laser line for RFP (588-635nm barrier filter). The obtained images were processed with Imaris (Bitplane) or Zen (Zeiss) software.
Lentivirus preparation.
VSV-pseudotyped 3rd generation lentiviruses were produced by Polyethylenimine-MAX (Polyscience) or Lipofectamine 2000 (Thermo Fisher Scientific)-based transfection of 293T cells with a transfer plasmid, packaging plasmids (GAG/POL, RSV-Rev) and an envelope plasmid (VSV-G). 2μM sodium butyrate was added into medium on the occasion of transfection and medium change to increase the viral production (28). Supernatant was collected 48 and 72 h post-transfection, and lentiviruses were concentrated by the ultracentrifugation. Biological titer of lentiviruses was evaluated with 293T-based on fluorescence expression. 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM: Corning, 10-013-CV) supplemented with 10% heat-inactivated fetal bovine serum (FBS). FBS was evaluated and selected based on the assays measuring 293T proliferation.
Stereotaxic injection of lentivirus.
Lentiviruses (1μl, 4×10^8-10^9 IFU/mL, 0.1uL/30sec-1min) were stereotaxically injected into GFAP-FLPo mice at the age of 6-16 weeks old under anesthesia. The following coordinates were used (in mm posterior, lateral and dorsal to the bregma): HP (2.0, 1.5, 2.3) and CTX (1, 1, 0.5).
Statistical analyses.
Statistical analyses were performed using GraphPad Prism software. The Mann-Whitney U-, Student’s t-, and Fisher’s exact tests were used for statistical evaluations. Log-rank tests were used to evaluate statistical differences in Kaplan-Meier analyses. Data are represented as mean ± SEM or ± SD; P-values < 0.05 were considered to be significant.
Results
Establishment of GFAP-FLPo mice.
Glial fibrillary acidic protein (GFAP), a class-III intermediate filament, is a cell-specific marker expressed in astrocytes and neural stem cells that have experimentally been shown to be one of the cell-of-origin of gliomas in the adult mouse brain (10,29). Detailed in vivo tracing experiments have shown that 15kb of entire mouse Gfap gene cassette restricts transgene expression to GFAP-expressing cells in the CNS more effectively than 2kb of conventional mouse/ human GFAP-5’ flanking region containing cis-acting regulatory elements (30,31). To direct the expression of an optimized FLP recombinase (FLPo), specifically to GFAP-expressing cells, we employed mouse Gfap gene cassette with a disrupted start codon of endogenous Gfap gene (22,32). FLPo recombinase was integrated into first exon of GFAP cassette (Fig. 1A). After the confirmation of FLPo recombination activity in vitro, GFAP-FLPo founder lines were established by pronuclear injection of linearized GFAP-FLPo construct, and GFAP-FLPo transgene carried by 4 GFAP-FLPo founder lines (#32, #41, #62 and #71) was confirmed by genotyping PCR analysis to detect the junction region of FLPo cDNA and GFAP gene cassette (Fig. S1A). To confirm in vivo expression of FLPo recombinase, we next analyzed FLPo mRNA expression levels in the brain, kidney, liver, lung and spleen from 4 GFAP-FLPo lines and WT animals. While no FLPo mRNA was detected in any tissues from #71 mice, FLPo expression was detected predominantly in the brains from #32, #41 and #62 founder strains. We noticed that, relatively higher expression of FLPo in the non-brain tissues such as the spleen from #32 and #41 mice unlike endogenous Gfap gene expression (Fig. 1B, and Fig. S1B-D). These data suggest that insertional site of transgene construct may affect level and tissue specificity of transgene expression and we, therefore, employed #62 mice for further analysis.
Specific expression of FLPo in GFAP-positive cell lineage.
To investigate FLP-mediated recombination and it’s cell-type specificity in the mouse brain, we next crossed GFAP-FLPo mice with reporter mice that express tdTomato after FLP-mediated deletion of a FRT-flanked transcriptional stop cassette (RC::FLTG) (23) (Fig. 1C). We first examined whether tdTomato-expressing cells are GFAP-positive in the various brain regions such as cortex (CTX), subgranular zone (SGZ) of the hippocampal dentate gyrus (DG), corpus callosum (CC) and thalamus (TH) (Fig. 1C). GFAP protein is not expressed in the cell body such as the nucleus whereas the expression of tdTomato can be observed both in the cell body and the dendrites. We, therefore, examined fluorescently labelled cells in thick brain sections (100-200μm) using confocal microscopy (Fig. 1D-F). Immunostaining experiments revealed that tdTomato-expressing cells were all GFAP-positive in the brains of 3week-old animals (Fig. 1D and G, S1E and F, Fig. 2A and Table 1). Additionally, tdTomato-positive cells in the cortex are all negative for neuronal marker NeuN (Fig. 1E and G) and oligodendrocyte lineage marker olig2 (Fig. 1F and G). We also performed the quantitative analysis of FLPo-mediated recombination in GFAP-positive cells in a region-specific manner (Table 1). These data suggest that FLPo expression is tightly controlled and induced in specific GFAP-positive populations such as astrocytes in GFAP-FLPo animals.
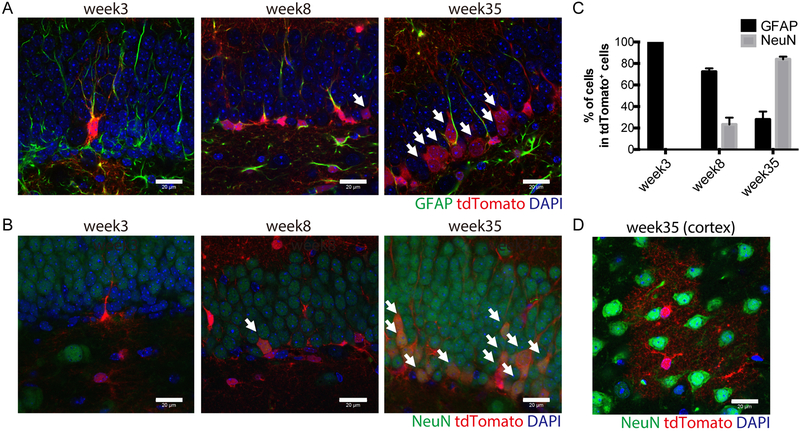
Fig. 2. FLPo-mediated recombination in GFAP-positive neural stem cells.
(A) Immunostaining analysis of GFAP (green) and tdTomato (red) expression in the subgranular zone (SGZ) of the hippocampal dendate gyrus (DG) of 3, 8 and 35-week-old GFAP-FLPo; CAG-FRT-stop-FRT-tdTomato animal. Scale bars, 20 μm. Arrows mark GFAP-negative/ tdTomato positive cells.
(B) Immunostaining analysis of NeuN (green) and tdTomato (red) expression in the SGZ of the hippocampal DG of 3, 8 and 35-week-old GFAP-FLPo; CAG-FRT-stop-FRT-tdTomato animal. Scale bars, 20 μm. Arrows mark NeuN-positive/ tdTomato positive cells.
(C) Quantified analysis of GFAP-positive and NeuN-positive cells in the SGZ of the hippocampal DG of 3, 8 and 35-week-old GFAP-FLPo; CAG-FRT-stop-FRT-tdTomato animal. Data are means with SEM. 3 animals per condition were subjected to the analysis.
(D) Immunostaining analysis of NeuN (green) and tdTomato (red) expression in the cortex of 35-week-old GFAP-FLPo; CAG-FRT-stop-FRT-tdTomato animal. Scale bars, 20 μm.
Table 1.
Quantitative analysis of FLPo-mediated recombination.
| 3 week-old animals analysis under confocal microscopy | |||||
|---|---|---|---|---|---|
| GFAP/tdTomato | tdTomato/GFAP | olig2/tdTomato | NeuN/tdTomato | Sox2/tdTomato | |
| CTX | 100% (71) | 20.2% (350) | 0% (128) | 0% (56) | 100% (16) |
| SGZ (radial glia like) | 100% (36) | 7.3% (491) | 100% (53) | ||
| HP (non-SGZ) | 100% (385) | 69.6% (553) | |||
| CC | 100% (80) | 51.6% (155) | |||
| TH | 100% (25) | 12.4% (201) | |||
GFAP/tdTomato: % of GFAP positive/ tdTomato positive cells in tdTomato positive cells in the brain area shown in a left column. tdTomato/ GFAP: % of GFAP positive/ tdTomato positive cells in GFAP positive cells in the brain area shown in a left column. olig2/tdTomato: % of olig2 positive/ tdTomato positive cells in tdTomato positive cells in the brain area shown in a left column. NeuN/tdTomato: % of NeuN positive/ tdTomato positive cells in tdTomato positive cells in the brain area shown in a left column. Sox2/tdTomato: % of Sox2 positive/ tdTomato positive cells in tdTomato positive cells in the brain area shown in a left column. Number of cells analyzed is shown in brackets. 3 animals were subjected to the analysis.
FLPo-mediated recombination in GFAP-positive NSCs.
GFAP-positive neural stem cells (NSCs) reside in the SGZ of the hippocampal DG, to continuously generate neurons in the adult mouse forebrain (33). We observed that 6.0% of GFAP-positive cells with radial glia cell like morphology was labelled with tdTomato in the SGZ of 3week-old GFAP-FLPo reporter animals, while the FLPo-mediated recombination event is strongly biased toward GFAP-positive astrocytes in the HP (Fig. S2A, Table 1; non-SGZ: 66.2%, SGZ-radial glia like: 6.0% in the HP). Radial glia like tdTomato positive cells in the SGZ are positive for a stem cell marker Sox2, although astrocytes are also positive for Sox2 in the mouse brain (Fig. S2B and Table 1). To investigate whether FLPo expressing cells in the SGZ display NSC-like properties to produce non-GFAP-positive neuronal lineage progeny, we performed long term lineage tracing experiments (Fig. 1C). While tdTomato positive cells were all positive for GFAP and negative for NeuN in the SGZ of 3week-old animals, GFAP negative/ tdTomato positive cells were increasingly observed in the 8week-old and 35week-old animals (Fig. 2A-C). Notably the tdTomato positive cells gave rise to NeuN positive cells during 8weeks and 35weeks in the SGZ, but not in the cortex (Fig. 2B-D). Thus, these data suggest that limited FLPo expression is induced in NSCs in the SGZ of GFAP-FLPo animals.
Glioma formed by FLP inducible lentiviral system.
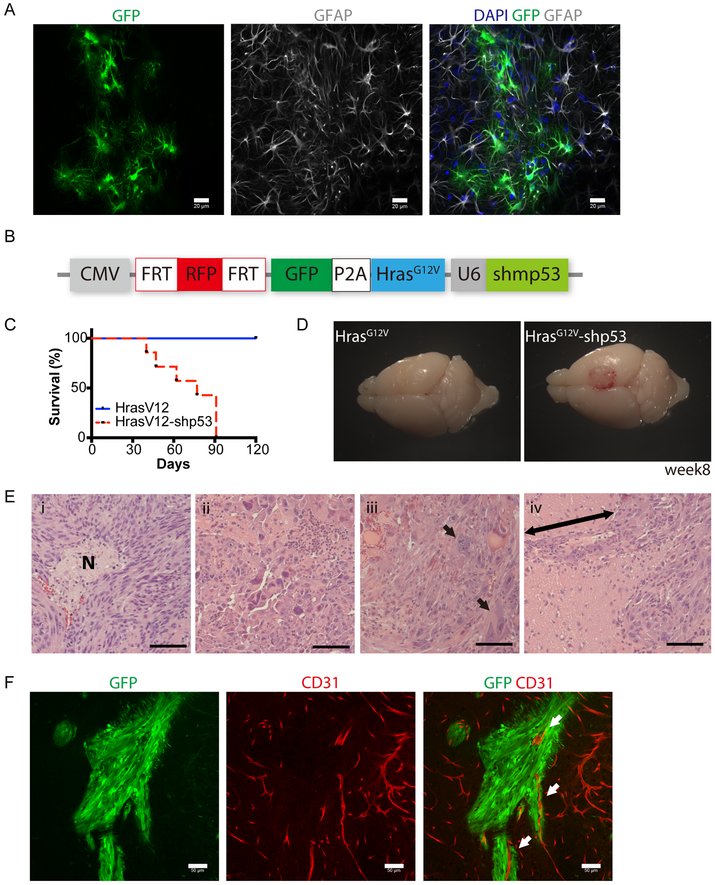
We have previously established a novel mouse model of malignant gliomas using lentiviral vectors (10,11). To evaluate whether lentiviral vectors deliver genetic components specifically into GFAP positive cells in a FLP dependent manner, we injected CAG-FRT-RFP-FRT-GFP into GFAP-FLPo animals. 1 week after a lentiviral injection into the cortex of GFAP-FLPo animals, GFP positive cells were all positive for GFAP and exhibited a sign of hypertrophy that is a key feature of reactive astrocyte seen after acute and focal brain injury (Fig. 3A) (34). To establish FLP inducible glioma models, we generated a new construct backbone, Frt-pTomo, that harbors FLP-mediated transgene expression platform, 2A peptide based multi-gene expression system, and short hairpin RNA (shRNA)-mediated gene depletion (Fig. 3B). As we have successfully modeled gliomas by employing a combination of HrasG12V and p53 suppression in GFAP-Cre animals (11), we generated cytomegalovirus immediate-early promoter (CMV)-FRT-RFP-FRT-GFP-2A-HrasG12V (Flag-tagged)-U6-shp53 and co-tranfected it with either Cre or FLPo recombinase in 293T cells (Fig. S3A). FLP-mediated recombination was confirmed by the presence of GFP and Flag-tagged HrasG12V only after the transduction of FLPo. To induce gliomas with FLP/ FRT system, we next directly transduced GFAP positive cells by injecting Frt-pTomo lentiviral vectors into the CTX or HP of GFAP-FLPo mouse brains. All transduced animals developed tumors when injected either in the CTX or HP (Fig. 3C and D and Fig. S3B and C). No such tumors were found in animals injected with HrasG12V alone (Fig. 3C and D), and in GFAP-FLPo; FRT-stop-FRT-KrasG12D mice during 6-month observation (Fig. S3D). Although GFAP-FLPo model required higher virus titer (4×105- IFU/mouse) than GFAP-cre model (1×105- IFU/mouse), the survival time frame of animals with gliomas was similar to that with tumor generated in GFAP-cre mice (Fig. 3C and Fig. S3E). Histological analysis of established tumors provided highly malignant features seen in high-grade gliomas such as necrosis, cellular pleomorphisms, multinuclear giant cells and highly infiltrative properties along blood vessels, (Fig. 3E and F). We had previously shown psudopalisading necrosis in mice transduced with pTomo lentiviral vectors (10).
Fig. 3. Cell-type specific delivery of oncogenes to model gliomas in GFAP-FLPo.
(A) Immunostaining analysis of GFAP (gray) and GFP (green) expression in the cortex of GFAP-FLPo animals stereotaxically injected with lentiviral vectors harboring CAG-FRT-STOP-FRT-GFP into the cortex. Scale bars, 20 μm. More than 5 animals were subjected to the analysis. Similar results were obtained in two independent experiments.
(B) A diagram of the FLP/ FRT-based lentiviral vector to induce tumorigenesis in GFAP-FLPo animals. CMV: cytomegalovirus immediate-early promoter. P2A: porcine teschovirus-1 derived 2A self-cleaving peptide sequence. U6: RNA polymerase III-based promoter from mouse U6 small nuclear RNA. shp53: shRNA against mouse Trp53.
(C) Survival analysis of GFAP-FLPo mice after stereotaxic injection with either CMV-FRT-RFP-FRT-GFP-P2A-HrasG12V-U6-shp53 or CMV-FRT-RFP-FRT-GFP-P2A-HrasG12V. Similar results were obtained in at least three independent experiments.
(D) Representative images of the brains from GFAP-FLPo animals injected with either CMV-FRT-RFP-FRT-GFP-P2A-HrasG12V-U6-shp53 or CMV-FRT-RFP-FRT-GFP-P2A-HrasG12V into the cortex.
(E) H&E staining analysis of tumors established in the brain of GFAP-FLP mice after stereotaxic injection with CMV-FRT-RFP-FRT-GFP-P2A-HrasG12V-U6-shp53. (i) Necrotic area with the spindle-shaped cells, (ii) pleomorphism, (iii) multinucleated gigantic cells, (iv) perivascular invasion. Arrow marks perivascular infiltration of cells. Scale bars, 50 μm. Similar histological results were observed in sections from more than six animals.
(F) Immunostaining analysis of CD31 (red) and GFP (green) expression in the brain from GFAP-FLPo mice after stereotaxic injection CMV-FRT-RFP-FRT-GFP-P2A-HrasG12V-U6-shp53. Arrows mark CD31-positive blood vessel surrounded by GFP-positive glioma cells. Scale bars, 20 μm.
Gliomas combined with Cre-inducible multi-fluorescent protein expression.
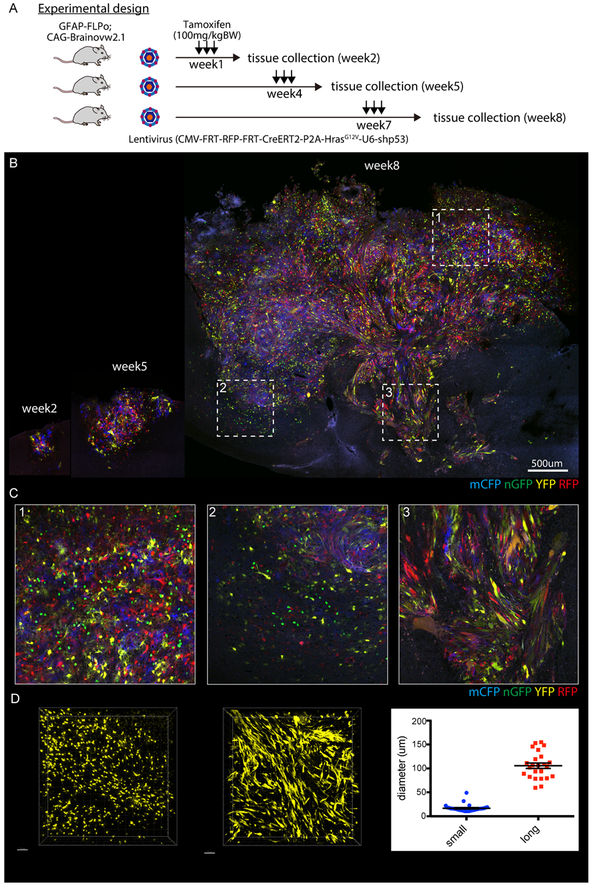
To demonstrate the feasibility of dual recombinase system using Cre and FLPo, we employed CAG-Brainbow2.1 cassette that stochastically expresses either one of four fluorescent proteins after the temporal induction of Cre recombinase activity (Fig. 4A) (25,26). Tumor-specific induction of multi-fluorescent proteins distinguishes adjacent glioma cells with different fluorescent proteins and therefore visualizes morphologically heterogeneous features of tumor cells at in situ 3D tissue context (35). We developed a Frt-pTomo vector which possesses a fusion of a mutated estrogen receptor and Cre (namely CreERT2) in order to induce Cre activity specifically in tumor cells after the treatment of tamoxifen. To test the accessibility of active form of tamoxifen (4-Hydroxytamoxifen) in FLP-induced glioma tissue, CreERT2-mediated recombination, and leakiness of the CreERT2 system, we first infected GFAP-FLPo; CAG-Brainbow2.1 animals with Frt-pTomo vectors expressing CreERT2, HrasG12V and shp53 to induce gliomas. Although there was no evidence of fluorescent proteins expression in the normal brain of GFAP-FLPo; CAG-Brainbow2.1 animal, we did detect small amounts of leakiness of recombination activity in the pathological area even in the absence of tamoxifen (Fig. S4A). High-dose tamoxifen treatment 4 weeks after virus infection induced nearly equal amount of 4 fluorescent protein expression (Fig. S4A). At 1, 4, and 7 weeks post-injection into the CTX or HP, we treated animals with tamoxifen to induce expression of fluorescent proteins and collected tissues with one-week incubation time (Fig. 4A-C, and Fig. S4B and C). With this technique, we observed at least 2 morphologically distinct cell populations, small cells (16.70μm ± SEM 1.373, N=30) that dominated tissues at early time points and elongated cells (105.8μm ± SEM 5.659, N=24) that frequently showed infiltrative phenotype along blood vessels (Fig 4B-D). These cells are consistently observed even with the injection of viruses into the HP (Fig. S4B and C). Although gliomas are initiated synchronously by lentiviruses with defined oncogenic insults most likely into limited number of astrocytes, the established tumor possesses heterogeneous phenotypes.
Fig. 4. Rapid application of Cre-mediated multi-color imaging in FLPo-inducible gliomas.
(A) Experimental design of multi-color imaging of glioma induced in GFAP-FLPo animals. GFAP-FLPo; CAG-Brainobw2.1 animals were stereotaxically received lentiviral vectors harboring CMV-FRT-RFP-FRT-CreERT2-P2A-HrasG12V-U6-shp53. After the injection, animals were treated with tamoxifen at a concentration of 100mg/kgBW for 3days to induce CreERT2-mediated fluorescent protein induction.
(B, C) Gliomagenesis in the cortex of GFAP-FLPo animals, fluorescently labelled by Branibow2.1 cassette containing membrane localized CFP (mCFP), nuclear localized GFP (nGFP), YFP and RFP at indicated weeks. Dashed box highlighted areas with tumor having different characteristics shown in (C). 1. An area dominated by a small cell population, 2. Border area between normal brain and glioma, small cells infiltrate into normal brain part, 3. Border area between normal brain and glioma, elongated cells infiltrated along blood vessels. Scale bar, 500 μm. More than 4 animals were subjected to the analysis. Similar results were observed in at least two independent experiments.
(D) Representative image at week8 and quantification of cellular diameter of glioma cells with distinctive cellular morphology. Scale bars, 70 μm. Data are means with SEM.
Discussion
The development of model systems that facilitates our ability to investigate gliomas at the molecular and cellular level, is an essential process in order to combat such an incurable disease. Among cancer models such as cell-lines, organoids, model organisms and patient-derived xenografts, GEMMs are specifically equipped with the capacity to study progression and drug response under the influence of native microenvironment (8,9,36). In the present study, we describe the development of a novel glioma model using FLP/FRT recombination system. This glioma model should enable additional manipulation of mouse genome mediated by Cre in any cell type at any time in vivo, and therefore, accelerate the precise understanding of cellular and molecular mechanisms in the developmental processes of gliomas. In fact, our observation reveals that tamoxifen treatment sufficiently induces CreERT2-mediated recombination in glioma cells at any stages of gliomagenesis. Thus, various genetic strategies using Cre recombinase, 1) temporal and sequential induction of oncogenic events rather than delivering those to cell-of-origin at the same time, 2) temporal depletion of genes in established tumors to perform a proof-of-concept study for therapeutic feasibility, 3) specific depletion of genes in stroma such as immune cells and endothelial cells that are abundant in the tumor tissues, and 4) lineage tracing of tumor cell/ stroma subpopulations during gliomagenesis and drug treatment, could be employed with this model.
Genetic, epigenetic and microenvironmental factors collectively determine the properties of glioma cells, and any alterations or changes in those regulatory components theoretically provide the opportunity to develop heterogeneous phenotypes in glioma tissues (37-39). In fact, recent longitudinal and multi-regional analyses of clinical biopsies support the presence of multi-directional transitioning of glioblastoma subtypes and phenotypic association of glioblastoma subtypes with their surrounding microenvironment (40,41). Furthermore various experimental evidences provide mechanistic insights of highly plastic nature of glioma cells partly regulated by developmental programs, microRNA and inflammation (42-45). Genetics and chemical based manipulation of factors that are highly associated with heterogeneous phenotypes using model systems would be further required for the precise understandings of tumor heterogeneity and progression.
Although a combination of mutations in Ras and Tp53 is not common for human gliomas, delivering such oncogenic insults to GFAP-positive cell-of-origins constantly develops mouse gliomas which exhibit many features of human malignant gliomas as described previously (11,46). One of advantages of lentivirus-based animal model is rapid and flexible choice of strategies to manipulate cancer genes to initiate gliomas. The development of FLP/FRT-mediated mouse models initiated with different sets of the relevant driver genes will allow for the detailed analysis of tumor-specific molecular and cellular mechanisms, which should provide key insights into intertumoral heterogeneity. Furthermore, even with strong oncogenic drivers for tumor initiation, established tumors exhibit histologically and morphologically heterogeneous phenotypes. These evidences imply that tumor-intrinsic and/or extrinsic changes may be additionally required for glioma cells to shape the trajectory towards malignant gliomas and also to develop heterogeneous phenotypes. Further investigations using transcriptomic analysis to capture gene expression dynamics of single-cells from different stages, and in situ analysis to characterize tumor-microenvironment interaction would provide the mechanistic insights of cause of both of inter- and intra-tumoral heterogeneity underlying therapeutic failure in human gliomas.
Recently, CRISPR (clustered regularly interspaced short palindromic repeats)/ Cas9 system has been applied for modeling of human cancers in mouse (7,47). As the generation of GEMMs and breeding animals are time-consuming processes, use of CRISPR/ CAS9 system to directly mutate cancer genes in vivo provides more flexible and rapid strategies to interrogate candidate genes. However, the variety of insertions and deletions (indels) might be created in the initial pool of cell-of-origins, which potentially can result in the development of established tumors with experimentally generated heterogeneity. Careful recognition of advantage and disadvantage of model systems is required for further application, and lineage tracing strategies to tag the cell(s)-of-origin at single-cell level might be useful to interpret the data (48).
In conclusion, our mouse model expands the utility of GEMMs in neuro-oncology, and will enable a more rapid and complete understandings of the molecular and cellular regulators of all aspects of gliomagenesis.
Supplementary Material
Significance.
This study presents a new glioma mouse model derived using lentiviral vectors and two recombination systems that will expand the ability to dissect developmental processes of gliomagenesis.
Acknowledgements
We thank Dinorah Friedmann-Morvinski, Yasushi Soda, Eugene Ke, Gerald Pao, Junko Ogawa, Xiaoyan Zhu, Nina Tonnu and Mark Schmitt for helpful discussions and supports. This work was supported in part by a Grant-in-Aid for JSPS Fellows from the Japan Society for the Promotion of Science, a grant from the NIH-NCI (5R01CA195613), Cancer Center Support (5P30CA014195), the H. N. and Frances C. Berger Foundation, and Leona M. and Harry B. Helmsley Charitable Trust grant 2017-PG-MED001. This work was also supported by the Transgenic Core Facility of the Salk Institute with funding from NIH-NCI CCSG: 5P30CA014195, and by the Waitt Advanced Biophotonics Core Facility of the Salk Institute with funding from NIH-NCI CCSG: 5P30CA014195 and the Waitt Foundation.
Financial support: Grant-in-Aid for JSPS Fellows from the Japan Society for the Promotion of Science, NIH-NCI (5R01CA195613), NIH-NCI Cancer Center Support Grant (5P30 CA014195), the H. N. and Frances C. Berger Foundation, Leona M. and Harry B. Helmsley Charitable Trust grant 2017-PG-MED001, Waitt Foundation.
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
REFERENCES
- 1.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008-2012. Neuro Oncol. 2015;17:iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Reifenberger G, Deimling von A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathologica. 2016;131:803–20. [DOI] [PubMed] [Google Scholar]
- 3.Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG, et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science. 2017;355:6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sottoriva A, Spiteri I, Piccirillo SGM, Touloumis A, Collins VP, Marioni JC, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110:4009–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenting K, Verhaak R, Laan ter M, Wesseling P, Leenders W. Glioma: experimental models and reality. Acta Neuropathologica. 2017;133:263–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winters IP, Murray CW, Winslow MM. Towards quantitative and multiplexed in vivo functional cancer genomics. Nat Rev Genet. 2018;45:D777. [DOI] [PubMed] [Google Scholar]
- 8.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog Signaling Enhances Delivery of Chemotherapy in a Mouse Model of Pancreatic Cancer. Science. 2009;324:1457–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seehawer M, Heinzmann F, D’Artista L, Harbig J, Roux P-F, Hoenicke L, et al. Necroptosis microenvironment directs lineage commitment in liver cancer. Nature. 2018;562:69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marumoto T, Tashiro A, Friedmann-Morvinski D, Scadeng M, Soda Y, Gage FH, et al. Development of a novel mouse glioma model using lentiviral vectors. Nature Medicine. 2009;15:110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alcantara Llaguno S, Chen J, Kwon C-H, Jackson EL, Li Y, Burns DK, et al. Malignant Astrocytomas Originate from Neural Stem/Progenitor Cells in a Somatic Tumor Suppressor Mouse Model. Cancer Cell. 2009;15:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philip B, Yu DX, Silvis MR, Shin CH, Robinson JP, Robinson GL, et al. Mutant IDH1 Promotes Glioma Formation In Vivo. Cell Reports. 2018;23:1553–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soda Y, Marumoto T, Friedmann-Morvinski D, Soda M, Liu F, Michiue H, et al. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proceedings of the National Academy of Sciences. 2011;108:4274–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozawa T, Riester M, Cheng Y-K, Huse JT, Squatrito M, Helmy K, et al. Most Human Non-GCIMP Glioblastoma Subtypes Evolve from a Common Proneural-like Precursor Glioma. Cancer Cell. 2014;26:288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griveau A, Seano G, Shelton SJ, Kupp R, Jahangiri A, Obernier K, et al. A Glial Signature and Wnt7 Signaling Regulate Glioma-Vascular Interactions and Tumor Microenvironment. Cancer Cell. 2018;33:874–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:654–8. [DOI] [PubMed] [Google Scholar]
- 18.Schönhuber N, Seidler B, Schuck K, Veltkamp C, Schachtler C, Zukowska M, et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nature Medicine. 2014;20:1340–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raymond CS, Soriano P. High-Efficiency FLP and ΦC31 Site-Specific Recombination in Mammalian Cells Marinus M, editor. PLoS ONE. 2007;2:e162–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imayoshi I, Hirano K, Sakamoto M, Miyoshi G, Imura T, Kitano S, et al. A multifunctional teal-fluorescent Rosa26 reporter mouse line for Cre- and Flp-mediated recombination. Neuroscience Research. 2012;73:85–91. [DOI] [PubMed] [Google Scholar]
- 21.Karimova M, Baker O, Camgoz A, Naumann R, Buchholz F, Anastassiadis K. A single reporter mouse line for Vika, Flp, Dre, and Cre-recombination. Sci Rep. 2018;8:504–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia ADR, Doan NB, Imura T, Bush TG, Sofroniew MV. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat Neurosci. 2004;7:1233–41. [DOI] [PubMed] [Google Scholar]
- 23.Plummer NW, Evsyukova IY, Robertson SD, de Marchena J, Tucker CJ, Jensen P. Expanding the power of recombinase-based labeling to uncover cellular diversity. Development. 2015;142:4385–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young NP, Crowley D, Jacks T. Uncoupling Cancer Mutations Reveals Critical Timing of p53 Loss in Sarcomagenesis. Cancer Research. 2011;71:4040–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. [DOI] [PubMed] [Google Scholar]
- 26.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, et al. Intestinal Crypt Homeostasis Results from Neutral Competition between Symmetrically Dividing Lgr5 Stem Cells. Cell. 2010;143:134–44. [DOI] [PubMed] [Google Scholar]
- 27.Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, et al. ScaleS: an optical clearing palette for biological imaging. Nat Neurosci. 2015;18:1518–29. [DOI] [PubMed] [Google Scholar]
- 28.Ellis BL, Potts PR, Porteus MH. Creating Higher Titer Lentivirus with Caffeine. Human Gene Therapy. 2011;22:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hol EM, Pekny M. Glial fibrillary acidic protein (GFAP) and the astrocyte intermediate filament system in diseases of the central nervous system. Current Opinion in Cell Biology. 2015;32:121–30. [DOI] [PubMed] [Google Scholar]
- 30.Johnson WB, Ruppe MD, Rockenstein EM, Price J, Sarthy VP, Verderber LC, et al. Indicator expression directed by regulatory sequences of the glial fibrillary acidic protein (GFAP) gene: in vivo comparison of distinct GFAP-lacZ transgenes. Glia. 1995;13:174–84. [DOI] [PubMed] [Google Scholar]
- 31.Brenner M, Kisseberth WC, Su Y, Besnard F, Messing A. GFAP promoter directs astrocyte-specific expression in transgenic mice. Journal of Neuroscience. 1994;14:1030–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bush TG, Savidge TC, Freeman TC, Cox HJ, Campbell EA, Mucke L, et al. Fulminant jejuno-ileitis following ablation of enteric glia in adult transgenic mice. Cell. 1998;93:189–201. [DOI] [PubMed] [Google Scholar]
- 33.Gonçalves JT, Schafer ST, Gage FH. Adult Neurogenesis in the Hippocampus: From Stem Cells to Behavior. Cell. 2016;167:897–914. [DOI] [PubMed] [Google Scholar]
- 34.Burda JE, Sofroniew MV. Reactive gliosis and the multicellular response to CNS damage and disease. Neuron. 2014;81:229–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanai H, Tanaka T, Ueno H. Multicolor lineage tracing methods and intestinal tumors. J Gastroenterol. 2013;48:423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogawa J, Pao GM, Shokhirev MN, Verma IM. Glioblastoma Model Using Human Cerebral Organoids. Cell Reports. 2018;23:1220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller TE, Liau BB, Wallace LC, Morton AR, Xie Q, Dixit D, et al. Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature. 2017;547:355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nature Medicine. 2013;19:1264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell. 2017;32:42–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin X, Kim LJY, Wu Q, Wallace LC, Prager BC, Sanvoranart T, et al. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nature Medicine. 2017;23:1352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suvà ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rooj AK, Ricklefs F, Mineo M, Nakano I, Chiocca EA, Bronisz A, et al. MicroRNA-Mediated Dynamic Bidirectional Shift between the Subclasses of Glioblastoma Stem-like Cells. CellReports. 2017;19:2026–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–75. [DOI] [PubMed] [Google Scholar]
- 45.Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Breunig JJ, Levy R, Antonuk CD, Molina J, Dutra-Clarke M, Park H, et al. Ets Factors Regulate Neural Stem Cell Depletion and Gliogenesis in Ras Pathway Glioma. Cell Reports. 2015;12:258–71. [DOI] [PubMed] [Google Scholar]
- 47.Sánchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer. 2015;15:387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanay A, Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541:331–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.