Abstract
Insulinoma-associated-1 (IA-1 or INSM1) encodes a zinc-finger transcription factor, which was isolated from a human insulinoma subtraction library, with specific expression patterns, predominantly in developing neuroendocrine (NE) tissues and tumors. INSM1 is key in early pancreatic endocrine, sympatho-adrenal lineage, and pan-neurogenic precursor development. Insm1 gene ablation results in impairment of pancreatic β-cells, catecholamine biosynthesis, and basal progenitor development during mammalian neocortex maturation. Recently, INSM1 has emerged as a superior, sensitive, and specific biomarker for NE tumors. INSM1 regulates downstream target genes and exhibits extra nuclear activities associated with multiple signaling pathways, including Sonic Hedgehog, PI3K/AKT, MEK/ERK1/2, ADK, p53, Wnt, histone acetylation, LSD1, cyclin D1, Ascl1, and N-myc. Novel strategies targeting INSM1-associated signaling pathways facilitate the suppression of NE tumor growth. In addition, INSM1 promoter- driven reporter assay and/or suicide gene therapy are promising effective therapeutic approaches for targeted specific NE tumor therapy. In the present review, the current knowledge of the biological role of INSM1 as an NE tumor biomarker is summarized, and novel strategies targeting multiple signaling pathways in the context of INSM1 expression in NE tumors are further explored.
Implications:
NE transcription factor (INSM1) may serve as an NE biomarker for the development of novel cancer therapeutics against NE tumors.
Keywords: neuroendocrine tumor, biomarker, INSM1, N-myc, Shh
Introduction
Insulinoma-associated-1 (IA-1 or INSM1) is a zinc-finger transcription factor that was identified in a human insulinoma subtraction library, from which it was isolated (1). Insm1 is an intronless gene encoding a 510-amino acid protein that contains 5 zinc-finger DNA-binding motifs and several potential functional domians (2,3). This novel complementary DNA (cDNA) was isolated from a rare human insulinoma due to its unique tissue specificity. However, the restricted specific expression pattern and the biological function of INSM1 were at the time largely unknown, except for its markedly elevated expression levels in human insulinoma tissues (Fig. 1). The first INSM1-specific expression pattern in cells or tissues other than insulinoma was demonstrated in a rat pheochromocytoma cell line (PC-12). Subsequently, normal fetal human brain tissues and primary neuroendocrine (NE) tumor samples were observed to be positive for INSM1 expression by Northern blot analyses, including pituitary tumors, carcinoids, small cell lung carcinoma (SCLC), medullary thyroid carcinoma, neuroblastoma (NB), retinoblastoma, and medulloblastoma (4). An early study on 64 lung cancer cell lines and lung tumor tissues used Northern blot analyses to define IA-1 (later renamed INSM1) as a novel marker for NE differentiation in human lung cancer (5). Importantly, additional studies from other laboratories later confirmed that INSM1 is a specific NE tumor marker that is critical for NE lung differentiation (6-8).
Figure 1. INSM expression in human insulinoma tissues (1).
Human insulinoma tissues were stained with an anti-INSM1 antibody (1:500; A8; sc-271408). INSM1 exhibits a high expression level in insulinoma tumors (magnification, x40). INSM1, insulinoma-associated-1.
Neuroendocrinology was first described as the recognition of hormones secreted from the pituitary gland, which is closely controlled by the brain, particularly the hypothalamus (9). Endocrine glands contain small islands of secretory cells of epithelial origin that secret hormones. Although the endocrine system consists of numerous different glands distributed throughout the body, the total mass of endocrine cells in each gland with functional hormone production is relatively small. These endocrine cells secrete different hormones that target tissues in response to growth and development for maintaining the body’s metabolism. Dysregulation of hormone secretion and abnormal NE cell growth could be due to NE cell transformation. During endocrine organ development, these endocrine/NE cells exhibit a tissue-specific differentiation pathway, which determines various endocrine organs with their specific hormone-producing endocrine cells. Abnormal differentiation or dysregulation of these endocrine/NE cells results in the generation of benign or malignant NE tumors that could have a profound impact on the body’s metabolism. Since INSM1 serves such a critical role in NE cell differentiation, it is conceivable that INSM1 could emerge as a novel NE-specific tumor marker and a critical regulator of NE differentiation (4,10-12). The present review discusses the functional role of INSM1 as a biomarker for NE tumors. INSM1 overexpression in NE tumors could have negative effects on tumor cell growth. Based on our understanding of INSM1 as an NE tumor-specific biomarker and its associated signaling pathways, INSM1 could be a prominent target for designing novel cancer therapeutic strategies.
INSM1 is a unique zinc-finger transcription factor
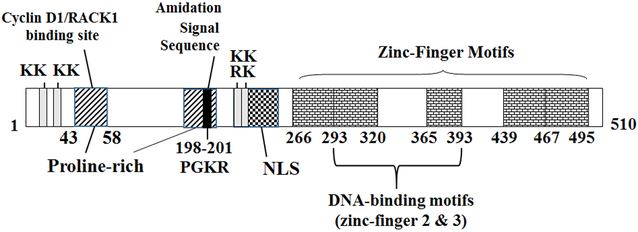
IA-1 was later renamed INSM1 in the GenBank DNA database. The isolated cDNA encodes an intronless gene mapped to chromosome 20p11.2 (2). Its 1530 base-pair open reading frame translates into a protein of 510 amino acids with a predicted molecular weight of 52,923 Da (Fig. 2). The N-terminal portion (~250 amino acids) of INSM1 contains a Snail/Gfi-1 (SNAG) motif, 2 proline-rich regions (20–30% proline content), a putative nuclear localization signaling (NLS) sequence (positions 221–246), 4 dibasic amino acid sites (positions 8–9, 11–12, 221–222 and 227–228) and a potential amidation signal sequence (PGKR). The SNAG motif functions as a transcriptional repressor domain involved in cell cycle arrest, recruitment of histone-modifying factors, and interaction with LSD1 for SCLC cell proliferation (13-16). The presence of dibasic amino acids are characteristics of prohormone convertase processing sites for peptide hormone such as insulin, glucagon, somatostation, and pancreatic polypeptide, while the amidation signal sequence facilitates the addition of an α-amide group in several bioactive NE peptides (17,18). However, no evidence exists thus far that the IA-1 transcription factor can be processed into an NE peptide hormone. The proline-rich region located at the IA-1 N-terminus has been observed in numerous mammalian transcription factors and is essential for protein-protein interactions (19,20). Studies confirmed that the N-terminal proline-rich region of INSM1 (43–58 amino acids) is capable of binding to cyclin D1 and receptor of activated C kinase 1 (RACK1) during cellular signaling (12,21,22). Another unique feature of the encoding sequence of IA-1 is its 5 Cys2-His2-type zinc-finger motifs, which suggest a functional role for IA-1 as a DNA-binding transcription factor. The DNA-binding target sequence, TG/TC/TC/TT/AGGGGG/TCG/A recognized by IA-1 was determined using a selected and amplified random oligonucleotide binding assay (3). Several downstream target genes were subsequently identified with this consensus DNA-binding sequence in their promoter region, including neurogenic differentiation factor 1 (NeuroD1), insulin, RE1 silencing transcription factor (REST), ripply transcription repressor 3 (Ripply3), neurogenin 3 (Ngn3), adherens junction belt-specific protein (Plekha7), and Insm1 itself (autoregulation) (21,23-25). INSM1 encodes a transcription repressor, which is highly conserved among different species. Evolutionary Insm1 homologues in chimpanzee, mouse, rat, Xenopus laevis, zebrafish, Drosophila melanogaster and Caenorhabditis elegans exhibit 99.4, 90.6, 76.7, 55.7, 54.8, 22.6, and 18.2% identity, respectively (4,26). A human homolog named INSM2 was identified with a broader tissue expression pattern and similar functional role in pancreatic islets (27,28). The second zinc-finger motif is the most highly conserved with 96% identity among different species. The second highly conserved zinc-finger motif is particularly important, since only human INSM1 zinc-fingers 2 and 3 are sufficient for target gene binding (3). INSM1 expression is restricted to normal embryonic NE tissues, including the developing forebrain, hindbrain, olfactory epithelium, retina, cerebellum, pancreas, thymus, thyroid, adrenal gland, and endocrine cells of the gastrointestinal tract (26,29-31).
Figure 2. INSM1 structure.
INSM1 complementary DNA encodes a 510-amino acid protein divided into an N-terminal (1-250 amino acids) and a C-terminal (251-510 amino acids) domain. The N-terminal domain contains 2 proline-rich regions (shade line) with 43-58 amino acids such as the cyclin D1/RACK1 binding site (12,21,22). Several dibasic amino acids (KK and KR, grey area), an amidation signal sequence (PGKR, black) and a nuclear localization sequence (diamond box) are located at the N-terminal domain. The C-terminal domain contains 5 equally spaced zinc-finger motifs (brick boxes). Zinc-finger 2 and 3 motifs are required for specific DNA binding (3). INSM1, insulinoma-associated-1.
Biological functions of INSM1 in NE differentiation
Since the identification of INSM1 as a transcription factor, biochemical analyses of its upstream and downstream target genes revealed that INSM1 binds to its own promoter and auto-regulates INSM1 gene expression as a feedback mechanism during embryonic NE cell differentiation (3). Additionally, the Ngn3, Ripply3, NeuroD1, and insulin target genes were identified to be associated with pancreatic endocrine cell differentiation (21,23,24,32). INSM1 transcriptional repressor activity involved the recruitment of cyclin D1 and histone deaceylatase-3 (HDAC-3) that led to the modification of the acetylation state of histone H¾. INSM1 serves an important role in pancreatic cell differentiation. Two of the critical endocrine islet transcription factors, Ngn3 and NeuroD1, activate the E-box elements present in the INSM1 promoter, whereas INSM1 down regulates Ngn3 and NeuroD1 expression during pancreatic development (21,29,31,33). In combination the INSM1 auto-regulatory mechanism and the Ngn3/NeuroD1/INSM1 feedback regulation strongly suggest that INSM1 is critical for the modulation of the transient expression of INSM1 during early pancreatic endocrine cell differentiation (33-36). In vivo analysis of the developing pancreas supports that Insm1 is the immediate downstream target of ngn3 and is upstream of neuroD1, pair-box 4 (pax4), aristaless-related homeobox (arx), homeodomain transcription factor 6.1 (nkx6.1), and pair-box 6 (pax6) gene expression in endocrine pancreas differentiation (29). The Insm1 mutant mouse is embryonic lethal (37). In Insm1 mutants, endocrine pancreatic β-cell development was severely impaired. Additional evidence supports that Insm1 is a transcription factor that acts in a pan-endocrine manner, which is essential for the differentiation of endocrine cells into pancreas, intestine, adrenal gland, and anterior pituitary gland. A recent study suggested that haplo-insufficiency of insm1 impaired postnatal baseline β-cell mass (38). The SNAG domain at the N-terminus of INSM1 recruits histone-modifying factors, lysine (K)-specific demethylase 1A (Kdm1a), histone deacetylase ½ (Hdac1/2) and REST co-repressor 1–3 (Rcor1–3) and other proteins implicated in transcriptional regulation high mobility group 20a/b (Hmg20a/b) and genetic suppressor element 1 (Gse1) in the anterior pituitary gland (15).
Based on the embryonic lethality and deficient catecholamine synthesis observed in Insm1 mutant mice, Insm1 expression appears to be crucial for sympatho-adrenal (SA) lineage differentiation (39). SA lineage cells are derived from the neural crest, which lead to sympathetic neurons and adrenal chromaffin cells. Cell differentiation and catecholamine synthesis involve multiple transcription factors such as achaete-scute complex homolog-like 1 (Mash1), paired-like homeobox 2a/b (Phox2a/b), Hand2, and GATA binding protein 2/3 (Gata2/3), which are critical for mouse fetal development and survival (40,41). Castro et al reported (42) that Insm1 could be a direct target of Mash1 regulation since Insm1 is not correctly initiated from the SA lineage in Phox2b and Mash1-mutant mice (39). Therefore, Insm1 serves a key role in modulating SA lineage development and catecholamine synthesis.
In the developing brain, Insm1 expression serves a critical role in areas where neurogenesis occurs. Insm1 mutant mice failed to develop properly in the granule cell layer of the cerebellum, the dentate gyrus of the postnatal hippocampus, the ventricular zone and the subventricular zone of the neocortex (43). The REST co-repressors and the repressor INSM1 regulate the developing brain proliferation-differentiation balance (44). Insm1 down-regulates adherens junction belt-specific protein Plekha7 through developing neocortex delamination (25). High levels of INSM1 expression were also detected in neuronal progenitors and developing sensory neurons (30,31). A recent study demonstrated that the homeotic cell transformation of outer hair cells without INSM1 into inner hair cells reveals a mechanism by which these neighboring mechanosensory cells begin to differ (45). Consistently, IA-1 constitutes a novel molecule downstream of xenopus homolog of achaete-scute (Xash1) that is involved in the formation of Xenopus noradrenergic primary neuron populations (46).
INSM1/IA-1 is a specific NE tumor marker
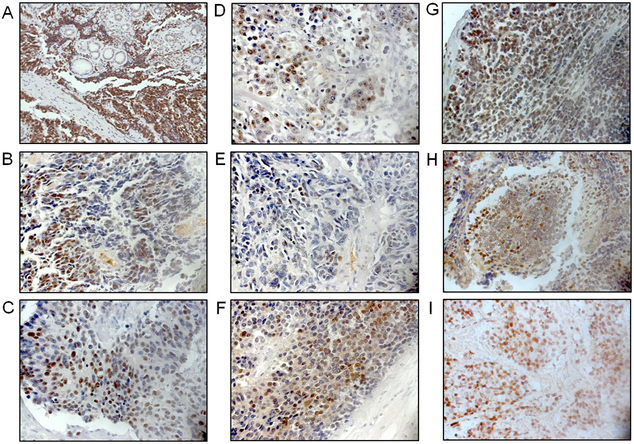
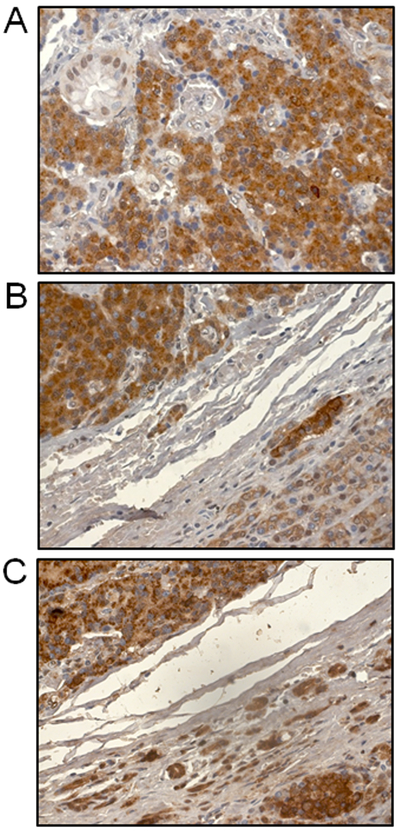
Regarding the structure and biological function of INSM1, this protein is a specific NE tumor marker. INSM1-associated transformation processes are mainly involved in NE tumorigenesis. Elevated IA-1 messenger RNA (mRNA) levels were first detected in a panel of 64 human lung cancer cell lines and multiple human lung tumors (5). Northern blot analysis revealed 97% (30 of 31) positivity in SCLC cell lines. By contrast, IA-1 mRNA was detected in only 13% (4 of 30) of non-SCLC cell lines, including 9 with an NE phenotype (4 of 9 NE positive non-SCLC cells) and 3 carcinoids. Recently, a monoclonal antibody that facilitates the diagnosis of NE tumors, namely clone A8 against INSM1 (sc-271408; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) has become available commercially. Immunohistochemical (IHC) analyses of INSM1 in NE and neuro-epithelial neoplasms (NEN) revealed that INSM1 was detectable by IHC in 88.3% of 129 NEN specimens (47), with only 1 false positive detected in 27 neoplasms without a neuro-epithelial or NE component. Further IHC analysis for INSM1 was conducted with a tissue array containing 35 SCLC samples at different clinical stages and 5 normal lung tissues. All the SCLC tissues were strongly positive for INSM1 expression (48). Additional studies demonstrated that INSM1 is a superior biomarker for diagnosing NE tumors in lung, thoracic cavity, carcinoid tumorlets of the lung, NE of the uterine cervix, pancreatic NE tumors, NE carcinomas of the head and neck, Merkel cell carcinoma, and primary central nervous system neoplasms (49-58). Fig. 3 shows examples of INSM1 IHC staining pattern using multiple NE tumor tissue arrays (US Biomax, Inc., Derwood, MD, USA). INSM1 protein staining displays a clear nuclear pattern in SCLC, esophagus NE, lung carcinoid, rectum carcinoid, colon NE, cardiac NE, retinoblastoma, medulloblastoma, and NB derived from a TH-N-myc transgenic mouse (N-myc activated NB tumor model) (59).
Figure 3. INSM1 immunohistochemical staining of NE tumors.
NE tumor tissue arrays were obtained from US Biomax Inc. (Derwood, MD, USA). Anti-INSM1 antibody staining (1:500) was performed with a MACH 3 biotin-free polymer detection kit (48). Representative NE tumor slides (magnification, x40): (A) SCLC; (B) esophagus NE; (C) lung carcinoid; (D) rectum carcinoid; (E) colon NE; (F) cardiac NE; (G) retinoblastoma; (H) medulloblastoma; and (I) neuroblastoma derived from a TH-N-myc transgenic mouse model (59). INSM1, insulinoma-associated-1; NE, neuroendocrine.
Extra nuclear activities of INSM1 in NE tumors
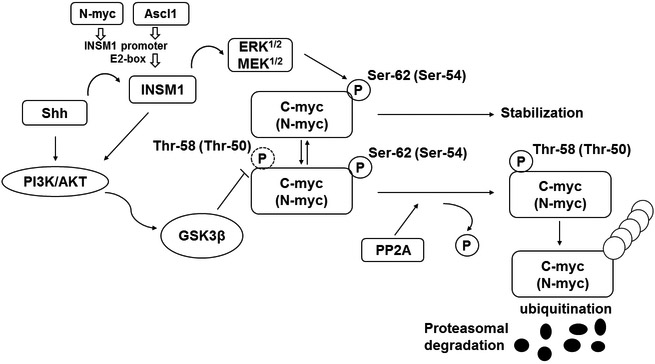
Since INSM1 only is expressed in normal fetal tissues and NE tumors, it was further investigated whether high INSM1 expression levels contribute to NE tumor cell growth. In addition to the transcriptional regulation of target genes by INSM1, the extra nuclear activity of INSM1 in NE tumor cells was further explored. It was postulated that insulin and insulin receptor (InR) signaling could serve a key role in fueling tumors, thus connecting obesity with diabetes and cancer (60). Previous studies using an insulin-lowering drug known as metformin demonstrated that it was associated with the modulation of InR signaling and led to a significant decrease in cancer incidence (61). A study demonstrated that INSM1 induces pancreatic cell trans-differentiation by enhancing the InR-mediated phosphatidylinositol-4,5-bisphosphate 3-kinase/RAC-alpha serine/threonine-protein kinase (PI3K/AKT) signaling pathway (22). INSM1 serves an important role, since it is closely associated with pancreatic endocrine cell differentiation (24,29,37). The biochemical studies revealed that INSM1 binds to cyclin D1 and interrupts cyclin D1 and CDK4 binding, which induces non-NE cell cycle arrest (12). In NB, Sonic hedgehog (Shh) signaling positively correlates with N-myc and INSM1 expression, which contributes to NB cell viability. N-myc binds and activates the E2-box of the INSM1 promoter, whereas INSM1 increases N-myc levels by activating the PI3K/AKT/glucogen synthase kinase 3 beta (GSK3β) signaling pathways and enhances N-myc stability (62). The positive feedback loop of N-myc and INSM1 stimulates NB cell proliferation. Additionally, INSM1 expression enhances cellular invasiveness and oncogenesis in NB cells. A recent study revealed that INSM1 signals through the same sonic hedgehog (Shh) signaling pathway, with N-myc/Ascl1 activation of INSM1 via extracellular signal-regulated protein kinase/mitogen activated protein kinase kinase½ (ERK/MEK1/2) phosphorylation signaling occurring in NE lung cancer (63). Shh facilitates N-myc stability through PI3K/AKT and INSM1 activation in NE lung cancer cells. Both MEK/ERK1/2 and PI3K/AKT phosphorylation signaling are required for Ser-54 (C-myc, Ser-62) phosphorylation and blockage of GSK3β-mediated phosphorylation of N-myc (Thr-50, C-myc/Thr-58), which stabilizes N-myc protein and prevents it from entering the proteasomal degradation pathway (Fig. 4). The negative effects of Shh inhibitor and/or knockdown of INSM1 in NE lung cancer cells were further investigated. Although common treatment of human cancers using an inhibitor of Shh signaling pathway is marginally effective, the combination of a Smoothened inhibitor (cyclopamine or vismodegib) and a Gli-mediated inhibitor (GANT-61) greatly enhanced the inhibition of SCLC in the context of INSM1 expression. Therefore, targeting multiple signaling pathways associated with the activation of the INSM1/N-myc transcription factors could be a prominent approach to delay or interrupt NE tumor growth.
Figure 4. Shh activates the PI3K/AKT and INSM1 signaling pathways to stabilize and prevent N-myc from entering the proteasomal degradation pathway.
N-myc and Ascl1 bind to the E2-box of the INSM1 promoter for activation of INSM1 protein expression (63). Shh activates PI3K/AKT and N-myc via INSM1-mediated activation of MEK/ERK1/2, which phosphorylates N-myc (Ser-54) for stabilization. PI3K/AKT phosphorylates GSK3β and blocks N-myc (Thr-50) phosphorylation to prevent further PP2A-mediated removal of pSer54 for entering the proteasomal degradation pathway. N-myc and INSM1 form a positive feedback loop to promote neuroendocrine tumor cell growth and transformation.
Experimental cancer therapeutics targeting INSM1-associated signaling pathways
INSM1 can be considered as an oncofetal differentiation factor according to its exclusive expression pattern in the fetal stage of NE precursors and tumors, while being silenced in adult normal tissues. Upon transformation, the INSM1 gene is re-expressed strictly in NE tumors, which resembles its fetal NE tissue expression pattern. The re-expression of the INSM1 gene in NB tumors contributes to the aggressive phenotype of a certain subtype of NBs with amplified N-myc oncoprotein expression (62). INSM1 and N-myc counter-regulation promotes NB tumor growth in vivo. Therefore, it is logical to use an INSM1 promoter-driven luciferase screening-platform to investigate the role of INSM1 in the suppression of NE tumors. A novel adenosine kinase (ADK) inhibitor, 5-iodotubercidin (5’-IT), was identified to exert potent inhibition of NB tumor cell growth in parallel with INSM1 suppression (64). The inhibition effect of 5’-IT mediated through the adenosine receptor-3 signaling pathway that suppresses the cyclic AMP (cAMP), Wnt, and pERK1/2 signaling pathways, which are involved in apoptosis. INSM1 expression and ADK signaling could contribute to cell growth and tumorigenicity in human NB tumors. Since elevated INSM1 expression in NB and NE lung cancer is closely associated with Shh signaling and N-myc stability, using inhibitors targeting both the Shh and ADK signaling pathways could be a novel approach to downregulate INSM1 and suppress NE cancer growth. The development of a preclinical test for Shh and ADK signaling inhibitors would be useful. The blockage of upstream or downstream molecules in the Shh signaling pathway downregulates INSM1 expression and restricts SCLC proliferation (63). Since both Shh signaling and increased expression of INSM1/N-myc serve critical roles in the development and maintenance of SCLC (65,66), these molecules could be potential direct therapeutic targets for cancer therapy. A recent study also demonstrated that the lysine-specific demethylase 1A (LSD1) inhibitor T-3775440 inhibits SCLC cell proliferation. T-3775440 disrupts LSD1 interactions with the SNAG domain containing INSM1 and growth factor independent 1B (GFI1B) proteins (16).
Modified INSM1 promoter regulates suicide or reporter gene expression in NE tumors
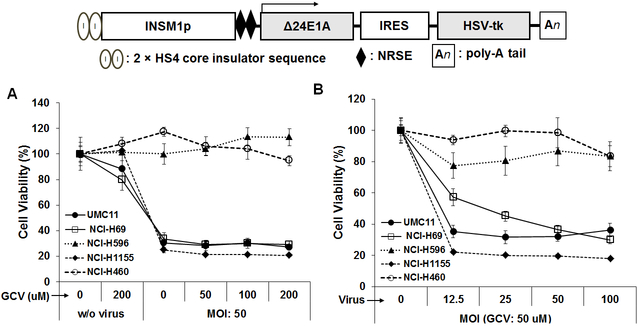
The specific expression of INSM1 in NE tumors indicates that INSM1 could be a prominent target for NE tumor therapy. Instead of directly targeting the INSM1 transcription factor or INSM1-associated signaling pathways, it is also logical to target INSM1-positive tumor cells using INSM1 promoter-specific suicide gene therapy. The expression pattern of INSM1 is tightly controlled via specific INSM1 promoter expression, which is restricted to embryonic NE precursors and tumors (31,67). INSM1 promoter-linked suicide gene therapy led to specific death of NE tumor cells in vivo and in vitro (68-70). In order to maintain INSM1 tissue specificity and to avoid the over powering effects of adenoviral regulatory element, the chicken β-globin HS4 insulator and 2 tandem copies of the neuronal restrictive silencer element (2xNRSE) were successfully constructed, which prevented interference from strong regulatory elements present in the adenoviral genome (71,72). An unexpected benefit of the modification of the Ad-INSM1-promoter construct was 10-fold increase in promoter activity in intratumoral injected xenograft tumors. A conditionally replicating adenovirus (CRAds) was further constructed, which contained the 1.7-kb human INSM1 promoter (which drives the expression of ∆24E1A and HSV-tk) separated by an internal ribosomal entry site (IRES) element. Conditional expression of ∆24E1A in NE tumor cells enhanced safe CRAds replication (oncolytic), which increased the cell killing efficiency (70). In a previous study, the INSM1 promoter was modified with an upstream chicken β-globin HS4 insulator element and 2xNRSE in order to maintain the promoter specificity and increase the sensitivity of the adenoviral vector. The results of cell viability MTS [3-(4,5-dimethyl-2yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetraazolium] assay of CRAd (HS4-INSM1–2xNRSE-∆24E1A-IRES-HSV-tk)-mediated NE lung tumor cell death are shown in Fig. 5. The INSM1 promoter regulated replicating HSV-tk armed oncolytic adenovirus exhibited effective cell killing capacity and sensitivity, similarly to the effects observed when the INSM1 promoter expression pattern is restricted to NE tumors. INSM1-negative adeno-squamous cells (NCI-H596) and large cell lung carcinoma cells (NCI-H460) survived due to lack of INSM1 promoter activity, in contrast to INSM1-positive cells (UMC-11, NCI-H1155 and NCI-H69), which were destroyed effectively. Based the fact that the INSM1 promoter regulates specific gene expression in NE tumors, an NE-specific reporter assay using a modified INSM1 promoter linked to a Gaussia reporter gene was developed for the diagnostic evaluation of NE tumor growth in vivo (73). Therefore, in addition to the therapeutic approach described above, it is possible to monitor NE tumor growth during therapy using an INSM1 promoter-driven Gaussia reporter adenoviral vector.
Figure 5. Modified INSM1 promoter-driven oncolytic adenovirus (CRAd-HS4-INSM1-2xNRSE-∆24E1A-IRES-HSV-tk) suppresses neuroendocrine tumor cell growth (70).
(A) INSM1 non-expressing cell lines, including lung adeno-squamous carcinoma (NCI-H596) and large cell lung carcinoma (NCI-H460), and INSM1-expressing cell lines such as SCLC (NCI-H69), carcinoid (UMC-11) and large cell lung carcinoma (NCI-H1155) were evaluated without or with virus (MOI = 50:1) at different GCV concentrations (μM). The oncolytic virus exhibits basal-killing efficiency due to virus replication in the absence of GCV, in contrast to efficient killing when GCV concentration increased. (B) Responsive killing with a constant GCV (50 μM) and incremental MOI was observed in INSM- expressing cells (UMC-11, NCI-H69 and NCI-H1155), while no effect was observed in INSM1 non-expressing cells (NCI-H596 and NCI-H460).
Conclusions
INSM1 was identified in a human insulinoma subtraction library >25 years ago (1). INSM1 represents a common zinc-finger transcription factor with a unique tissue specific expression pattern. The encoded protein recognizes a specific DNA target sequence and downregulates the expression of multiple target genes associated with NE cell differentiation. Functional analyses revealed that INSM1 serves a critical role in pancreatic endocrine, SA lineage, and pan-neurogenic precursor development, which is consistent with the notion that INSM1 functions as an NE differentiation factor (37,39,43). Another important feature of INSM1 is its high expression level in NE tumors. INSM1 has emerged as a sensitive and specific biomarker for NE tumors, particularly SCLC (6-8). However, it is not clear whether INSM1 solely is a biomarker or whether its presence in NE tumors also contributes to NE tumorigenicity. A previous study on the molecular mechanisms of INSM1 in NE tumors revealed that INSM1 exerts extra nuclear activities that potentiate the tumorigenicity by activating Shh, PI3K/AKT, MEK/ERK1/2, ADK, p53, cyclin D1, β-catenin, LSD1, and N-myc. Although it is generally perceived that direct targeting of transcription factors is relatively difficult when trying to achieve effective tumor suppression, recent studies have shown that targeting INSM1-associated signaling pathways in the context of INSM expression is effective (63,64). Furthermore, fusing the specific INSM1 promoter to guide suicide or reporter genes targeting NE tumors could be a novel approach for INSM1 promoter mediated direct diagnosis or inhibition of NE tumors. The present review reports a unique NE transcription factor (INSM1) that may serve as an NE biomarker for the development of novel cancer therapeutics against NE tumors.
Acknowledgements:
The authors acknowledge the supports received from the Departments of Pediatrics and Genetics, Louisiana State University Health Sciences Center and Children’s Hospital (New Orleans, LA, USA).
Funding:
The research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under the award no. CA218764 (to MSL). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was supported in part by the LSU Research Enhancement Program and LSU Leveraging Innovation for Technology Transfer (LIFT2), grant no. HSCNO-2017-LIFT-008 (to MSL).
Footnotes
Competing interests:
The authors have no conflict of interest.
References
- 1.Goto Y, DeSilva MG, Toscani A, Prabhakar BS, Notkins AL, and Lan MS (1992) A novel human insulinoma-associated cDNA, IA-1, encodes a protein with zinc-finger DNA-binding motifs. J.Biol.Chem. 267, 15252–15257 [PubMed] [Google Scholar]
- 2.Lan MS, Li Q, Lu J, Modi WS, and Notkins AL (1994) Genomic organization, 5’-upstream sequence, and chromosomal localization of an insulinoma-associated intronless gene, IA-1. J.Biol.Chem. 269, 14170–14174 [PubMed] [Google Scholar]
- 3.Breslin MB, Zhu M, Notkins AL, and Lan MS (2002) Neuroendocrine differentiation factor, IA-1, is a transcriptional repressor and contains a specific DNA-binding domain: identification of consensus IA-1 binding sequence. Nucleic Acids Res. 30, 1038–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lan MS, and Breslin MB (2009) Structure, expression, and biological function of INSM1 transcription factor in neuroendocrine differentiation. FASEB J. 23, 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan MS, Russell EK, Lu J, Johnson BE, and Notkins AL (1993) IA-1, a new marker for neuroendocrine differentiation in human lung cancer cell lines. Cancer Res. 53, 4169–4171 [PubMed] [Google Scholar]
- 6.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, and Meyerson M (2001) Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A 98, 13790–13795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedersen N, Mortensen S, Sorensen SB, Pedersen MW, Rieneck K, and Bovin LF (2003) Transcriptional gene expression profiling of small cell lung cancer cells. Cancer Res. 63, 1943–1953 [PubMed] [Google Scholar]
- 8.Taniwaki M, Daigo Y, Ishikawa N, Takano A, Yasui W, Inai K, Kohno N, and Nakamura Y (2006) Gene expression profiles of small-cell lung cancers: molecular signatures of lung cancer. Int.J.Oncol. 29, 567–575 [PubMed] [Google Scholar]
- 9.Harris GW (1965) Development and present status of neuroendocrinology. Dtsch.Med.Wochenschr. 90, 61–65 [DOI] [PubMed] [Google Scholar]
- 10.Fujino K, Motooka Y, Hassan WA, Ali Abdalla MO, Sato Y, Kudoh S, Hasegawa K, Niimori-Kita K, Kobayashi H, Kubota I, Wakimoto J, Suzuki M, and Ito T (2015) Insulinoma-Associated Protein 1 Is a Crucial Regulator of Neuroendocrine Differentiation in Lung Cancer. Am.J.Pathol. 185, 3164–3177 [DOI] [PubMed] [Google Scholar]
- 11.Jia S, Wildner H, and Birchmeier C (2015) Insm1 controls the differentiation of pulmonary neuroendocrine cells by repressing Hes1. Dev Biol 408, 90–98 [DOI] [PubMed] [Google Scholar]
- 12.Zhang T, Liu WD, Saunee NA, Breslin MB, and Lan MS (2009) Zinc-finger transcription factor INSM1 interrupts cyclin D1 and CDK4 binding and induces cell cycle arrest. J.Biol.Chem. 284, 5574–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimes HL, Chan TO, Zweidler-Mckay PA, Tong B, and Tsichlis PN (1996) The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol.Cell.Biol. 16, 6263–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Candal E, Alunni A, Thermes V, Jamen F, Joly JS, and Bourrat F (2007) Ol-insm1b, a SNAG family transcription factor involved in cell cycle arrest during medaka development. Dev.Biol. 309, 1–17 [DOI] [PubMed] [Google Scholar]
- 15.Welcker JE, Hernandez-Miranda LR, Paul FE, Jia S, Ivanov A, Selbach M, and Birchmeier C (2013) Insm1 controls development of pituitary endocrine cells and requires a SNAG domain for function and for recruitment of histone-modifying factors. Development 140, 4947–4958 [DOI] [PubMed] [Google Scholar]
- 16.Takagi S, Ishikawa Y, Mizutani A, Iwasaki S, Matsumoto S, Kamada Y, Nomura T, and Nakamura K (2017) LSD1 Inhibitor T-3775440 Inhibits SCLC Cell Proliferation by Disrupting LSD1 Interactions with SNAG Domain Proteins INSM1 and GFI1B. Cancer Res 77, 4652–4662 [DOI] [PubMed] [Google Scholar]
- 17.Tatemoto K, and Mutt V (1978) Chemical determination of polypeptide hormones. Proc.Natil.Acad.Sci.USA 75, 4115–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jasani B, Kreil G, Mackler BF, and Stanworth DR (1979) Further studies on the structural requirements for polypeptide-mediated histamine release from rat mast cells. Biochem.J. 181, 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Caestecker MP, Yahata T, Wang D, Parks WT, Huang S, Hill CS, Shioda T, Roberts AB, and Lechleider RJ (2000) The Smad4 activation domain (SAD) is a proline-rich, p300-dependent transcriptional activation domain. J.Biol.Chem. 275, 2115–2122 [DOI] [PubMed] [Google Scholar]
- 20.Zilfou JT, Hoffman WH, Sank M, George DL, and Murphy M (2001) The corepressor mSin3a interacts with the proline-rich domain of p53 and protects p53 from proteasome-mediated degradation. Mol.Cell.Biol. 21, 3974–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu WD, Wang HW, Muguira M, Breslin MB, and Lan MS (2006) INSM1 functions as a transcriptional repressor of the NeuroD.beta2 gene through the recruitment of cyclin D1 and histone deacetylase. Biochem.J. 397, 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang T, Chen C, Breslin MB, Song K, and Lan MS (2014) Extra-nuclear activity of INSM1 transcription factor enhances insulin receptor signaling pathway and Nkx6.1 expression through RACK1 interaction. Cell Signal. 26, 740–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang HW, Muguira M, Liu WD, Zhang T, Chen C, Aucoin R, Breslin MB, and Lan MS (2008) Identification of an INSM1 binding site in the insulin promoter: Negative regulation of the insulin gene transcription. J.Endocrinol. 198, 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osipovich AB, Long Q, Manduchi E, Gangula R, Hipkens SB, Schneider J, Okubo T, Stoeckert CJ Jr., Takada S, and Magnuson MA (2014) Insm1 promotes endocrine cell differentiation by modulating the expression of a network of genes that includes Neurog3 and Ripply3. Development 141, 2939–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavano S, Taverna E, Kalebic N, Haffner C, Namba T, Dahl A, Wilsch-Brauninger M, Paridaen J, and Huttner WB (2018) Insm1 Induces Neural Progenitor Delamination in Developing Neocortex via Downregulation of the Adherens Junction Belt-Specific Protein Plekha7. Neuron 97, 1299–1314 e1298 [DOI] [PubMed] [Google Scholar]
- 26.Xie JP, Cai T, Zhang H, Lan MS, and Notkins AL (2002) The zinc-finger transcription factor INSM1 is expressed during embryo development and interacts with the Cbl-associated protein. Genomics 80, 54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai T, Chen X, Wang R, Xu H, You Y, Zhang T, Lan MS, and Notkins AL (2011) Expression of insullinoma-associated 2 (INSM2) in pancreatic islet cells is regulated by the transcription factors Ngn3 and NeuroD1. Endocrinology 152, 1961–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Sun ZS, Xiang B, Wei CJ, Wang Y, Sun K, Chen G, Lan MS, Carmona GN, Notkins AL, and Cai T (2018) Targeted deletion of Insm2 in mice result in reduced insulin secretion and glucose intolerance. J Transl Med 16, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellitzer G, Bonne S, Luco RF, Van De Casteele M, Lenne N, Collombat P, Mansouri A, Lee J, Lan MS, Pipeleers D, Nielsen FC, Ferrer J, Gradwohl G, and Heimberg H (2006) IA-1 is Ngn3-dependent and essential for differentiation of the endocrine pancreas. EMBO J. 25, 1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duggan A, Madathany T, De Castro SCP, Gerrelli D, Guddati K, and Garcia-anoveros J (2008) Transient expression of the conserved zinc finger gene INSM1 in progenitors and nascent neurons throughout embryonic and adult neurogenesis. J.Comp.Neurol. 507, 1497–1520 [DOI] [PubMed] [Google Scholar]
- 31.Breslin MB, Zhu M, and Lan MS (2003) NeuroD1/E47 regulates the E-box element of a novel zinc-finger transcription factor, IA-1, in developing nervous system. J.Biol.Chem. 278, 38991–38997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu M, Breslin MB, and Lan MS (2002) Expression of a novel zinc-finger cDNA, IA-1, is associated with rat AR42J cells differentiation into insulin-positive cells. Pancreas 24, 139–145 [DOI] [PubMed] [Google Scholar]
- 33.Breslin MB, Wang HW, Pierce A, Aucoin R, and Lan MS (2007) Neurogenin 3 recruits CBP co-activator to facilitate histone H3/H4 acetylation in the target gene INSM1. FEBS Lett 581, 949–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang HP, Liu M, El-hodiri HM, Chu K, Jamrich M, and Tsai MJ (2000) Regulation of the pancreatic islet-specific gene beta2 (neruoD) by neurogenin 3. Mol.Cell.Biol. 20, 3292–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang T, Wang HW, Saunee NA, Breslin MB, and Lan MS (2010) Insulinoma-associated antigen-1 zinc-finger transcription factor promoters pancreatic duct cell trans-differentiation. Endocrinology 151, 2030–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang T, Saunee NA, Breslin MB, Song K, and Lan MS (2012) Functional role of an islet transcription factor, INSM1/IA-1, on pancreatic acinar cell trans-differentiation. J.Cell.Physiol. 227, 2470–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gierl MS, Karoulias N, Wende H, Strehle M, and Birchmeier C (2006) The Zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev. 20, 2465–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tao W, Zhang Y, Ma L, Deng C, Duan H, Liang X, Liao R, Lin S, Nie T, Chen W, Wang C, Birchmeier C, and Jia S (2018) Haploinsufficiency of Insm1 Impairs Postnatal Baseline beta-Cell Mass. Diabetes 67, 2615–2625 [DOI] [PubMed] [Google Scholar]
- 39.Wildner H, Gierl MS, Strehle M, Pla P, and Birchmeier C (2008) Insm1 (IA-1) is a crucial component of the transcriptional network that controls differentiation of the sympatho-adrenal lineage. Development 135, 473–481 [DOI] [PubMed] [Google Scholar]
- 40.Zhou QY, Quaife CJ, and Palmiter RD (1995) Targeted disruption of the tyrosine hydroxylase gene reveals that catecholamines are required for mouse fetal development. Nature 374, 640–643 [DOI] [PubMed] [Google Scholar]
- 41.Thompson SA, Matsumoto AM, and Palmiter RD (1995) Noradrenaline is essential for mouse fetal development. Nature 374, 643–646 [DOI] [PubMed] [Google Scholar]
- 42.Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, Matter JM, and Guillemot F (2006) Proneural bHLH and Brn proteins coregulate a neruogenic program through cooperative binding to a conserved DNA motif. Dev.Cell 11, 831–844 [DOI] [PubMed] [Google Scholar]
- 43.Farkas LM, Haffner C, Giger T, Khaitovich P, Nowick K, Birchmeier C, Paabo S, and Huttner WB (2008) Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron 60, 40–55 [DOI] [PubMed] [Google Scholar]
- 44.Monaghan CE, Nechiporuk T, Jeng S, McWeeney SK, Wang J, Rosenfeld MG, and Mandel G (2017) REST corepressors RCOR1 and RCOR2 and the repressor INSM1 regulate the proliferation-differentiation balance in the developing brain. Proc Natl Acad Sci U S A 114, E406–E415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiwatpanit T, Lorenzen SM, Cantu JA, Foo CZ, Hogan AK, Marquez F, Clancy JC, Schipma MJ, Cheatham MA, Duggan A, and Garcia-Anoveros J (2018) Trans-differentiation of outer hair cells into inner hair cells in the absence of INSM1. Nature 563, 691–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parlier D, Ariza A, Christulia F, Genco F, Vanhomwegen J, Kricha S, Souopgui J, and Bellefroid EJ (2008) Xenopus zinc finger transcription factor IA-1 (insm1) expression marks anteroventral noradrenergic neuron pregenitors in xenopus embryos. Dev.Dyn. 237, 2147–2157 [DOI] [PubMed] [Google Scholar]
- 47.Rosenbaum JN, Guo Z, Baus RM, Werner H, Rehrauer WM, and Lloyd RV (2015) INSM1: A Novel Immunohistochemical and Molecular Marker for Neuroendocrine and Neuroepithelial Neoplasms. Am.J.Clin.Pathol. 144, 579–591 [DOI] [PubMed] [Google Scholar]
- 48.Chen C, Breslin MB, and Lan MS (2016) Ectopic expression of a small cell lung cancer transcription factor, INSM1 impairs alveologenesis in lung development. BMC.Pulm.Med 16, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doxtader EE, and Mukhopadhyay S (2018) Insulinoma-associated protein 1 is a sensitive and specific marker of neuroendocrine lung neoplasms in cytology specimens. Cancer Cytopathol 126, 243–252 [DOI] [PubMed] [Google Scholar]
- 50.Rooper LM, Sharma R, Li QK, Illei P, and Westra WH (2017) INSM1 Demonstrates Superior Performance to the individual and combined use of Synaptophysin, Chromogranin, and CD56 for Diagnosing Neuroendocrine Tumors of the Thoracic Cavity. Am J Surg Pathol 41, 1561–1569 [DOI] [PubMed] [Google Scholar]
- 51.Dermawan JK, and Mukhopadhyay S (2018) Insulinoma-associated protein 1 (INSM1) differentiates carcinoid tumourlets of the lung from pulmonary meningothelial-like nodules. Histopathology 72, 1067–1069 [DOI] [PubMed] [Google Scholar]
- 52.Kuji S, Watanabe R, Sato Y, Iwata T, Hirashima Y, Takekuma M, Ito I, Abe M, Nagashio R, Omae K, Aoki D, and Kameya T (2017) A new marker, insulinoma-associated protein 1 (INSM1), for high-grade neuroendocrine carcinoma of the uterine cervix: Analysis of 37 cases. Gynecol Oncol 144, 384–390 [DOI] [PubMed] [Google Scholar]
- 53.Tanigawa M, Nakayama M, Taira T, Hattori S, Mihara Y, Kondo R, Kusano H, Nakamura K, Abe Y, Ishida Y, Okabe Y, Hisaka T, Okuda K, Fujino K, Ito T, Kawahara A, Naito Y, Yamaguchi R, Akiba J, Akagi Y, and Yano H (2018) Insulinoma-associated protein 1 (INSM1) is a useful marker for pancreatic neuroendocrine tumor. Med Mol Morphol 51, 32–40 [DOI] [PubMed] [Google Scholar]
- 54.Rooper LM, Bishop JA, and Westra WH (2018) INSM1 is a Sensitive and Specific Marker of Neuroendocrine Differentiation in Head and Neck Tumors. Am J Surg Pathol 42, 665–671 [DOI] [PubMed] [Google Scholar]
- 55.Rush PS, Rosenbaum JN, Roy M, Baus RM, Bennett DD, and Lloyd RV (2018) Insulinoma-associated 1: A novel nuclear marker in Merkel cell carcinoma (cutaneous neuroendocrine carcinoma). J Cutan Pathol 45, 129–135 [DOI] [PubMed] [Google Scholar]
- 56.Ames HM, Rooper LM, Laterra JJ, Eberhart CG, and Rodriguez FJ (2018) INSM1 Expression Is Frequent in Primary Central Nervous System Neoplasms but Not in the Adult Brain Parenchyma. J Neuropathol Exp Neurol 77, 374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lilo MT, Chen Y, and LeBlanc RE (2018) INSM1 Is More Sensitive and Interpretable than Conventional Immunohistochemical Stains Used to Diagnose Merkel Cell Carcinoma. Am J Surg Pathol 42, 1541–1548 [DOI] [PubMed] [Google Scholar]
- 58.Mukhopadhyay S, Dermawan JK, Lanigan CP, and Farver CF (2019) Insulinoma-associated protein 1 (INSM1) is a sensitive and highly specific marker of neuroendocrine differentiation in primary lung neoplasms: an immunohistochemical study of 345 cases, including 292 whole-tissue sections. Mod Pathol 32, 100–109 [DOI] [PubMed] [Google Scholar]
- 59.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, and Bishop JM (1997) Targeted expression of N-myc causes neuroblastoma in transgenic mice. EMBO J. 16, 2985–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vigneri P, Frasca F, Sciacca L, Pandini G, and Vigneri R (2009) Diabetes and cancer. Endocr.Relat.Cancer 16, 1103–1123 [DOI] [PubMed] [Google Scholar]
- 61.Pollak M (2008) Insulin and insulin-like growth factor signalling in neoplasia. Nat.Rev.Cancer 8, 915–928 [DOI] [PubMed] [Google Scholar]
- 62.Chen C, Breslin MB, and Lan MS (2015) INSM1 increases N-myc stability and oncogenesis via a positive-feedback loop in neuroblastoma. Oncotarget 6, 36700–36712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen C, Breslin MB, and Lan MS (2018) Sonic hedgehog signaling pathway promotes INSM1 transcription factor in neuroendocrine lung cancer. Cell Signal 46, 83–91 [DOI] [PubMed] [Google Scholar]
- 64.Chen C, Breslin MB, Guidry JJ, and Lan MS (2019) 5’-Iodotubercidin represses insulinoma-associated-1 expression, decreases cAMP levels, and suppresses human neuroblastoma cell growth. J Biol Chem 294, 5456–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Funa K, Steinholtz L, Nou E, and Bergh J (1987) Increased expression of N-myc in human small cell lung cancer biopsies predicts lack of response to chemotherapy and poor prognosis. Am.J.Clin.Pathol. 88, 216–220 [DOI] [PubMed] [Google Scholar]
- 66.Park KS, Martelotto LG, Peifer M, Sos ML, Karnezis AN, Mahjoub MR, Bernard K, Conklin JF, Szczepny A, Yuan J, Guo R, Ospina B, Falzon J, Bennett S, Brown TJ, Markovic A, Devereux WL, Ocasio CA, Chen JK, Stearns T, Thomas RK, Dorsch M, Buonamici S, Watkins DN, Peacock CD, and Sage J (2011) A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat.Med. 17, 1504–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Q, Notkins AL, and Lan MS (1997) Molecular characterization of the promoter region of a neuroendocrine tumor marker, IA-1. Biochem.Biophys.Res.Comm. 236, 776–781 [DOI] [PubMed] [Google Scholar]
- 68.Wang HW, Breslin MB, Chen C, Akerstrom V, Zhong Q, and Lan MS (2009) INSM1-promoter driven adenoviral HSV thymidine kinase cancer gene therapy for the treatment of primitive neuroectodermal tumors. Human Gene Ther. 20, 1308–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pedersen N, Pedersen MW, Lan MS, Breslin MB, and Poulsen HS (2006) The insulinoma-associated 1: a novel promoter for targeted cancer gene therapy for small-cell lung cancer. Cancer Gene Therapy 13, 375–384 [DOI] [PubMed] [Google Scholar]
- 70.Tseng AW, Chen C, Breslin MB, and Lan MS (2016) Tumor-specific promoter-driven adenoviral therapy for insulinoma. Cell Oncol (Dordr) 39, 279–286 [DOI] [PubMed] [Google Scholar]
- 71.Akerstrom V, Chen C, Lan MS, and Breslin MB (2012) Modifications to the INSM1 promoter to preserve specificity and activity for use in adenoviral gene therapy of neuroendocrine carcinomas. Cancer Gene Ther. 19, 828–838 [DOI] [PubMed] [Google Scholar]
- 72.Akerstrom V, Chen C, Lan MS, and Breslin MB (2013) Adenoviral insulinoma-associated protein 1 promoter-driven suicide gene therapy with enhanced selectivity for treatment of neuroendocrine cancers. Ochsner. J 13, 91–99 [PMC free article] [PubMed] [Google Scholar]
- 73.Tseng AW, Akerstrom V, Chen C, Breslin MB, and Lan MS (2016) Detection of neuroendocrine tumors using promoter-specific secreted Gaussia luciferase. Int.J.Oncol. 48, 173–180 [DOI] [PubMed] [Google Scholar]