Abstract
It is well-recognized that solid tumors are genomically, anatomically, and physiologically heterogeneous. In general, more heterogeneous tumors have poorer outcomes, likely due to the increased probability of harboring therapy-resistant cells and regions. It is hypothesized that the genomic and physiologic heterogeneity are related, because physiologically distinct regions will exert variable selection pressures leading to the outgrowth of clones with variable genomic/proteomic profiles. To investigate this, methods must be in place to interrogate and define, at the microscopic scale, the cytotypes that exist within physiologically distinct sub-regions (“habitats”) that are present at mesoscopic scales. Magnetic Resonance Imaging (MRI) provides a non-invasive approach to interrogate physiologically distinct local environments, due to the biophysical principles that govern MRI signal generation. Here, we interrogate different physiological parameters, such as perfusion, cell density, and edema, using multiparametric MRI (mpMRI). Signals from six different acquisition schema were combined voxel-by-voxel into four clusters identified using a Gaussian Mixture Model. These were compared to histological and immunohistochemical characterizations of sections that were co-registered using MRI-guided 3D printed tumor molds. Specifically, we identified a specific set of MRI parameters to classify viable-normoxic, viable-hypoxic, non-viable-hypoxic, and non-viable normoxic tissue types within orthotopic 4T1 and MDA-MB-231 breast tumors. This is the first co-registered study to show that mpMRI can be used to define physiologically distinct tumor habitats within breast tumor models.
Keywords: Magnetic Resonance Imaging, Tumor hypoxia, Tumor microenvironment, In-vivo imaging, Dynamic contrast-enhanced magnetic resonance imaging
Introduction
Solid tumors are genomically, anatomically, and physiologically heterogeneous, and this heterogeneity has prognostic significance for therapy response and emergence of resistance (1). Tumors are known to have chaotic vasculature with high permeability, low tone, and varying degrees of perfusion and oxygenation, and this has been proposed to be a major driver of the evolution of tumor heterogeneity at the genomic level (2). In a number of cancers, this regional variability of perfusion within tumors generates altered physiology, metabolism, and gene expression patterns, and predicts short time to progression and poor overall survival following therapy (3,4). Thus, methods to identify and quantify these changes would have great benefit in decision support, especially if noninvasive imaging methods could be used to identify and define physiologically distinct regions, or tumor habitats, and monitor them longitudinally (5,6).
Magnetic resonance imaging (MRI) is an excellent tool for probing sub-domain variability within tumors as it has excellent soft tissue contrast and has a myriad of contrast mechanisms that can be leveraged to provide biophysical insight. For example, the transverse relaxation time, T2, is sensitive to edema, water (hydrogen) exchange on ionizable groups across membranes, as well as macromolecular hydration layers relative to bulk water (7). The apparent diffusion coefficient (ADC), calculated from diffusion-weighted (DW) MRI sequences, is sensitive to the translational diffusion of water in tissue, which is affected by the degree of water translational restriction primarily by lipids or lipid bilayers (8). Dynamic contrast enhanced (DCE) MRI provides positive contrast in regions based on the delivery of contrast agents, which can be variable, depending the rate and extent of local perfusion. It can noninvasively report on tumor vascular characteristics, which may be highly informative in the context of spatial tumor heterogeneity (9). Solid tumors frequently show a dysfunctional vascular system, resulting in sub-regions within tumors that are variably hypoxic, nutrient deprived, and acidic, all of which can have great impact on the local selection of cellular phenotypes within these different sub-regions (2,10).
In the clinic, these image data can be combined to define spatially distinct regions with similar physiologies, thus known as “habitats” (11). However, in the clinic, there is a lack of knowledge regarding the underlying physiology and cell types that give rise to these habitats. Therefore, a critical question is whether the biophysical sensitivity of multiple MRI parameters can distinguish different tumor habitats that harbor distinct physiologies and cytotypes that can be characterized using co-registered histopathology sections. Herein, we test the hypothesis that pixel-by-pixel clusters of multiparametric MRI (mpMRI) voxels may provide insight into the existence of regions that are well vascularized or hypoxic, viable or necrotic. This is biomedically relevant as hypoxic tumors are associated with a greater propensity to metastasize and are known to be resistant to radio-and chemo-therapies (12,13). Hypoxic regions result in increased glycolytic metabolism and, because they are poorly perfused, are also known to be acidic, which is causally associated with increased invasion, metastasis, and immunosuppression (14,15). Of note, phase III clinical trials of the hypoxia activated pro-drug (HAP), evofosfamide, failed to meet their statistical endpoints, primarily because patients were not stratified according to their hypoxia status (16). Hence, a method to infer hypoxia with standard of care imaging could have significant clinical impact.
Hence, we propose that heterogeneous, physiologically distinct tumor habitats can be identified and quantified by mpMRI, and furthermore, that these regional differences represent meaningful physiological states, such as hypoxia. In this study, we used computational image analysis to cluster multiple MRI-parameter maps, on a voxel-by-voxel basis, in order to identify distinct tumor regions in vivo. These were compared to histological characteristics by using 3D-printed tumor molds to co-register MRI and histology. To date, no direct and quantitative histological validation of spatially distributed habitats, as determined by MRI, has been reported.
Materials and methods
Animal models
All procedures were approved by the Institutional Animal Care and Use Committee (IACUC), University of South Florida, under the protocols 0549 and 1160. MDA-MB-231 cancer xenograft and syngeneic 4T1 breast tumors models were developed. A detailed description is shown in Supplementary Materials and Methods.
Magnetic resonance imaging
Multiparametric MR images were acquired to study habitats. The mice were imaged on a 7T horizontal magnet (Agilent ASR 310; Santa Clara, CA) and (Bruker Biospin, Inc. BioSpec AV3HD; Billerica, MA), using a 35 mm Litzcage coil (Doty Scientific; Columbia, SC). Mice were maintained anesthetized with 2% isoflurane delivered in 1.5-liter/min oxygen ventilation. Body temperature was continuously monitored using a rectal thermometer (SAII®, SA Instruments, Stony Brook, NY) and maintained at 37°C by an external heater. Respiratory function was maintained at a range of 40–60 breaths per minute, and tumors were immobilized without pressure with tape to minimize motion artifacts.
First, anatomical T2-weighted axial images were obtained to observe the entire tumor (slice thickness of 1 mm, field of view (FOV) 30 × 30 mm2 and image size 256 × 256), which was used to delineate the tumor volume of interest (VOI) to direct the printing of the 3D-printed tumor mold. After, in order to optimize the total time of image acquisition, the slice plane was adjusted to observe the 11 central slices and used to acquire another T2-weighted image, and the T2 map, T2* map, DW-MRI, and DCE-MRI. These were combined and used to analyze the habitat imaging. T2-weighted images were obtained using a TurboRARE sequence (Repetition time (TR) = 3287 milliseconds (ms), Echo time (TE) = 72ms, slice thickness of 1 mm, FOV 30 × 30 mm2, image size 256 × 256). T2* maps were acquired with 48 gradient echoes starting at TE = 2.56 ms up to 134.63 ms with a delta TE of 2.81ms and T2 maps were with a multi echo sequence (MSME) with a train of 32 echoes starting at 7 ms up to 224 ms with a delta TE of 7 ms and a TR of 2791 ms, slice thickness of 1 mm, FOV 30 × 30 mm2, image size 256 × 256). DW-MRIs were acquired in three directions using three b-values (100, 500 and 750 s/mm2. Based on prior studies (17) these are sufficient to generate a robust ADC value with a single exponential decay. Image size was 128 × 128/interpolation to 256 × 256 for 4T1 tumors and native 256 × 256 for MDA-MB-231, TR = 700 ms, TE =15.72 ms). T1-weighted DCE-MRI were acquired for all 11 slices every 59 seconds for 22 minutes to obtain 22 time-points (TR = 231ms, TE = 6ms, FOV was 30 × 30 mm2, image size 256 × 256 and slice thickness of 1 mm) upon intravenous (using a catheter in the tail vein) injection of 0.2 mmol/kg Gadobutrol (Gadavist®; Bayer; Whippany, NJ), starting the image 3 minutes before the contrast agent injection. The total imaging time per mouse, for all sequences combined, was approximately 1 hour.
3D-printed tumor mold
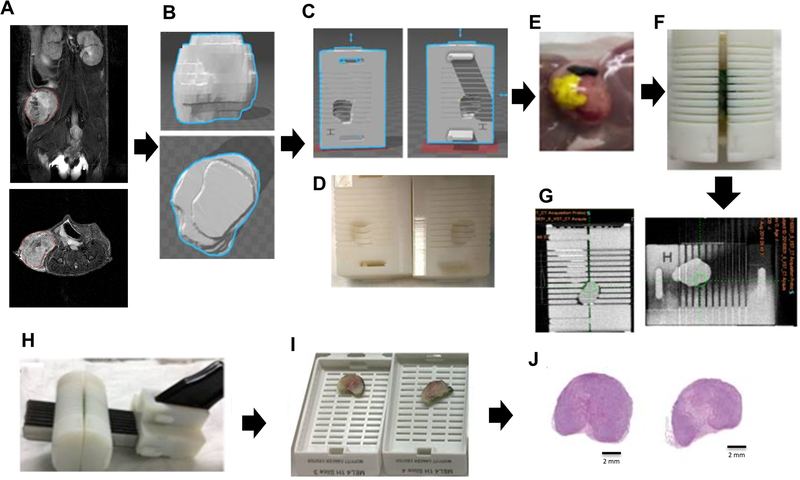
T2-weighted MR images were used to define an isosurface in order to create a 3D-printed tumor mold for each individual tumor (Figure 1A). Based on the tumor orientation in the MR images, the whole tumor was correctly positioned in a specific mold that contained slots every 2 mm (Figure 1B–G) that were aligned with the MRI slices. These slots were used to guide the slicing of the tumor to generate oriented histological slices (Figure 1H–J). The detailed protocol is described in Supplementary Materials and Methods, and the workflow is shown in Figure 1A–J.
Figure 1. Workflow for the 3D-printed tumor mold.
T2-weighted images were used to segment the tumor contours and create a 3D-printed tumor mold. (A) Tumor volume of interest (VOI) (red) in MRI T2-weighted image (upper: coronal, bottom: axial, field of view 30 × 30 mm2; image size 256 × 256); (B) 3D tumor reconstruction; (C) Mold designed in SOLIDWORKS and (D) printed in the 3D printer; (E) Tissue dyes were used to help with tumor orientation when inserting tumor into the 3D printed mold; (F) Tumor tissue inside the 3D-printed mold; (G) CT image (H) Tumor was cut in slices of 2 mm in thickness; (I) Slices were placed into individual cassettes; (J) Each tumor slice was cut into histological section of 4 μm and stained with H&E.
Histology and immunohistochemistry (IHC)
Following the MR imaging protocols, mice received an intraperitoneal injection of pimonidazole hydrochloride (60 mg/kg; Hypoxyprobe Inc; Burlington, MA) one hour prior to euthanasia. Subsequently, tumors were placed into the 3D-printed mold, fixed in Pen-Fix (Thermo Fisher Scientific, Waltham, MA), and cut in 2 mm-tumor sections, which were paraffin embedded and then subsequent serial sections of 4 μm were obtained to perform histological staining. Standard hematoxylin and eosin (H&E) and immunohistochemical staining procedures were performed for pimonidazole and cluster of differentiation 31 (CD31; also known as platelet endothelial cell adhesion molecule, PECAM-1), which were used to define the habitats in histological slides. In addition, IHC was performed for carbonic anhydrase IX (CA-IX) and peripilin-1, which were used in additional analyses. Detailed protocol is described in Supplementary Materials and Methods.
Co-registration of MRI and histological slices
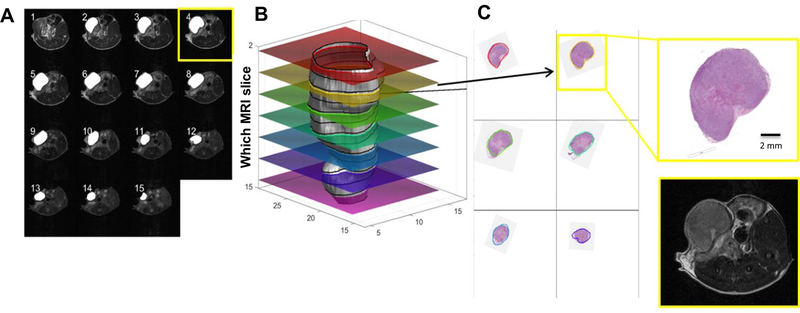
Histological slices were scanned at 20x magnification (Aperio AT2, Leica Biosystems, Buffalo Grove, IL) and loaded in Matlab (Natick, MA) for co-registration with MRI. Optimal 3D-alignment of tumor VOIs from MRI and histological slices was estimated using an optimization procedure that was designed and implemented specifically for this project (Figure 2A–C). We used an approach that we refer to as ‘non-monolithic’, in which the optimization procedure consists of outer and inner optimization problems. Namely, in each step, the outer optimization loop rotates and shifts the 3D-tumor surface reconstructed from MRI images and calculates contours obtained by its intersection with equally distant planes to mimic tumor slicing. Those contours were used by an inner optimization loop to find rotation of corresponding contours from histological slices that minimize the pixel-wise Euclidean distance between them. Both optimization loops utilized gradient-based trust-region-reflective algorithm implemented in MATLAB’s lsqnonlin function (18,19).
Figure 2. Estimation of the optimal 3D alignment of MRI and histology.
(A) T2-weighted axial slices (field of view 30 × 30 mm2; image size 256 × 256); (B) 3D representation of the tumor contour obtained by MRI (grey) and overlay of co-registered histological slices; (C) Histology images co-registered to the contours of the tumor on the surface on ‘b’.
Histological image analysis
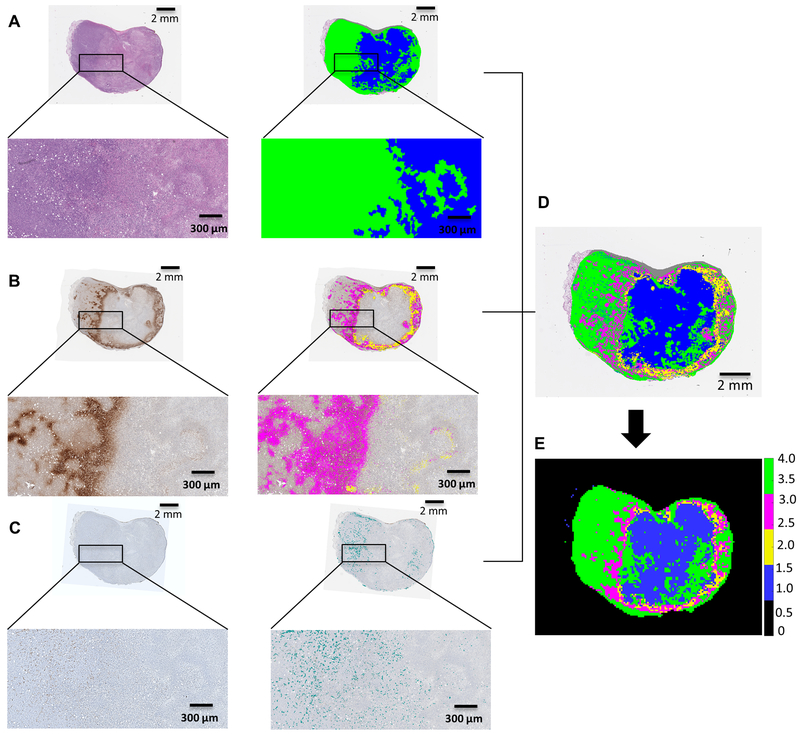
VisioPharm software (Broomfield, CO) was used to detect the positive pixels for each IHC marker (pimonidazole, CD31 and CA-IX) and to segment the H&E images into viable or non-viable tumor subregions based on cellular characteristics. Data from H&E, pimonidazole and CD31 images were imported into Matlab (Natick, MA) and used to create downsampled histological habitat maps. To do this, images were segmented into super-pixels of 117.19 μm × 117.19 μm square grid, which corresponds to one voxel size of MRI, as detailed in Supplementary material and methods (“Superpixel grid” section). Each super-pixel has multiple subpixel values. Thus, H&E was used to classify subpixels into viable or non-viable (Figure 3A). Pimonidazole positive subpixels were assigned as hypoxia (Figure 3B), and subpixels positive for CD31 (blood vessels) were assigned as viable (Figure 3C). Thus, four habitats were classified as viable-normoxic (green), viable-hypoxic (magenta), non-viable hypoxic (yellow) and non-viable normoxic (blue) (Figure 3D and E). Detailed protocols are described in Supplementary Materials and Methods, “Histological image analysis” and “Superpixel grid” sections.
Figure 3.
Segmentation of histological tumor sections. (A) Tumor section stained with hematoxylin & eosin (H&E) and respective masked representation of viable (green) and non-viable (blue) cells region; (B) Tumor section stained with pimonidazole and respective masked representation of positive pixels for pimonidazole in viable (magenta) and non-viable (yellow) regions; (C) Tumor section stained with CD-31 and respective masked representation of positive pixels for CD-31 (cyan). (D) Superimposed image classifying 4 habitats in histology, represented as viable cells in green and non-viable cells in blue, hypoxic areas in viable cells regions in magenta and hypoxia areas in non-viable cells regions in yellow. (E) Ground truth habitats map from histology, created using a super-pixel of 117.19 μm × 117.19 μm square grid to downsample the image to an equivalent MRI resolution.
Additionally, an algorithm was created in VisioPharm software (Broomfield, CO) for the identification of adipocytes, as detailed in Supplementary Materials and Methods.
Parametric maps in MRI
All parameter maps were obtained using nonlinear least squares pixel-by-pixel fit (Levenburg-Marquardt) (20,21) to the corresponding functions for T2 map, T2* map and ADC map from DW-MRI. For T2 and T2* fitting, the signal equation was of the form S(TE) = S0 exp (–TE/T2), applied to multiecho (spin or gradient) images spaced by TE. For the determination of ADC, multiple b-value images were used and fit according to: S(b) = S0 exp (–b D), where D is the ADC b is the b-value, and S0 is the intensity of the non-weighted pixel. T2 and T2* maps were computed in ParaVision (Bruker Biospin, Inc, Billerica, MA) whereas ADC maps were calculated through in-house Matlab (Natick, MA) code. DCE maps were obtained by first extracting time-series curves for each pixel, and performing nearest neighbor interpolation by a factor of 8, and then calculating the appropriate parameter through analysis of the pre- and post-contrast agent bolus time-series on a pixel-by-pixel basis also with in-house Matlab (Natick, MA) code. The slope is numerical ratio: ΔST1/Δt., where ΔS is the T1-weighted signal intensity temporal change, and Δt is the corresponding time-difference. The area under the time-series curve (AUC) is the sum of the entire curve. The time to maximum (TTM) is the time that corresponds to the maximum enhancement achieved. The slope, AUC and TTM maps were calculated from the first 20 mins following contrast injection.
Statistical clustering
For each pixel within the tumor VOI, map values of the MR relaxation, diffusion and semiquantitative DCE map parameters were associated together in a six-dimensional vector for the purposes of classification. MRI parameters were weighted equally. Before clustering, the parameters were normalized by the mean value of that parameter within the data set. Parameter map values within the tumor VOI were used to simultaneously classify four clusters, which was decided based on the number of habitats determined through histological analysis. With Matlab (Natick, MA) (Statistics and Machine Learning Toolbox), a maximum likelihood estimates of a Gaussian Mixture Model (GMM) was fit using an expectation maximization algorithm. A GMM was chosen to account for intravoxel heterogeneity, which reflects the complex microenvironment in a voxel. Pixel locations and classification information that were determined from clustering were mapped onto the corresponding pixels of T2-weighted reference images with color denoting which cluster a pixel belonged to. Clustering was performed using only the mpMRI parametric information and explicit spatial information was not used as additional constraints. Spatial information was only used to identify and track each parameter set.
Statistics
For each tumor type, statistical analyses were performed separately. One-way Analysis of variance (ANOVA) followed by Tukey post hoc was used to determine statistical significance of MR parameters values between clusters (habitats). That is, during the clustering of mpMRI data, each pixel had values of six MR parameters (T2 map, T2* map, ADC map, slope, AUC and TTM maps) associated with it. Thus, these pixel values of each MR parameters were compared between habitats (viable-normoxic, viable-hypoxic, non-viable-hypoxic and non-viable normoxic) by ANOVA using Matlab (Natick, MA). In addition, pixel values of each MR parameter were averaged, resulting in a mean value per MR parameter per sample for each habitat. These mean MR parameter values were compared between habitats by ANOVA using GraphPadPrism 7.0 (San Diego, CA).
MRI and histological habitat-maps comparison algorithm
To minimize the error in the map comparison, the MRI and histological habitat-maps were first rigidly co-registered based on their shape and intensity. The registration was restricted to rotation and translation only to avoid maps deformation. After co-registration step, each category/habitat in both maps was compared using the comparison technique based on a sliding window technique, developed in Costanza, and Kuhnert et al., (22,23). Detailed protocol is described in Supplementary Materials and Methods.
Results
A novel analysis of mpMRI data and its co-registration with histology, allowed us to successfully classify 4 different tumor habitats: viable-normoxic, viable-hypoxic, non-viable-hypoxic and non-viable normoxic, in pre-clinical breast tumor models.
3D alignment of MRI and histology
Custom code was developed to estimate the optimal 3D alignment of MRI and histological slices. The aligning algorithm tested different angles of MRI reconstructed tumor surface inclination (within -5 to +5 degrees limits) together with z-position of the initial slice to establish the alignment that minimized the total Euclidean distance between the contours from an MRI VOI and corresponding contours from the H&E (Figure 2A–C).
The quality of these fits (Q%) was expressed in terms of the Jaccard index, which was calculated as the area (in mm2) classified as tumor by both the MRI outline or aligned histology, divided by the total area encompassed by either MRI or histology. Overall, samples showed a high quality of fit, with a mean similarity index Q percentage of 83 ± 4 % for 4T1 breast tumors (n = 60 slices from 12 tumors) and 80 ± 4% for MDA-MB-231 breast tumors (n = 19 slices from 4 tumors). Individual values for each slice are shown in Table 1.
Table 1.
Quality of fit (Jaccard index; Q%) for each corresponding slice of MRI and histology.
| Sample ID | Pitch (angle in degree) | Roll (angle in degree) | Fit quality (Q) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Histological slices | ||||||||||
| MDA-MB-231 | Average (%) | 1st slice | 2nd slice | 3rd slice | 4th slice | 5th slice | 6th slice | 7th slice | ||
| MDA-1 | −2.08 | −4.94 | 84 | 83 | 77 | 90 | 85 | |||
| MDA-2 | 3.34 | 4.99 | 78 | 81 | 71 | 80 | 74 | 82 | ||
| MDA-3 | 1.87 | −4.97 | 83 | 86 | 82 | 82 | 81 | |||
| MDA-4 | −0.99 | −1.44 | 75 | 67 | 73 | 79 | 73 | 82 | 76 | |
| 4T1 | ||||||||||
| 4T1–1 | 1.55 | −0.93 | 90 | 89 | 92 | 90 | 90 | |||
| 4T1–2 | −4.92 | 1.54 | 75 | 76 | 87 | 83 | 54 | 76 | ||
| 4T1–3 | 4.99 | −4.6 | 79 | 57 | 86 | 92 | 80 | |||
| 4T1–4 | 3.36 | −4.84 | 81 | 78 | 89 | 81 | 81 | 88 | 70 | |
| 4T1–5 | −0.06 | 5.00 | 80 | 76 | 86 | 83 | 74 | |||
| 4T1–6 | 4.98 | −4.67 | 85 | 85 | 86 | 86 | 84 | |||
| 4T1–7 | 0.61 | −4.64 | 82 | 86 | 79 | 78 | 91 | 77 | ||
| 4T1–8 | −0.73 | 2.05 | 82 | 78 | 88 | 88 | 85 | 85 | 69 | |
| 4T1–9 | 0.00 | 0.00 | 85 | 74 | 85 | 91 | 91 | |||
| 4T1–10 | −3.82 | 4.63 | 88 | 89 | 93 | 89 | 90 | 87 | 82 | 86 |
| 4T1–11 | 1.40 | 4.95 | 83 | 86 | 87 | 85 | 83 | 75 | ||
| 4T1–12 | 0.49 | −4.99 | 83 | 80 | 84 | 81 | 85 | 78 | 89 | |
Distinct regions were identified by MRI and confirmed by histology
To generate habitat maps from histology; histological slices, each corresponding to an aligned MRI slice, were stained with H&E and with markers of blood vessels (CD31), hypoxia (pimonidazole), and hypoxia plus acidity (CA-IX) (24). Pimonidazole staining was observed largely in the peri-necrotic viable cell area, while CA-IX staining extended well beyond the pimonidazole regions, (Supplementary Fig. S1A–L), especially in 4T1 tumors (Supplementary Fig. S1A–F). CA-IX is a hypoxia-inducible factor (HIF) client and is thus induced by hypoxia. However, it has also been characterized as “pH-stat” which is responsible for acidifying the extracellular pH of tumors (24). This incomplete registration between CA-IX and pimonidazole staining has also been reported in other studies (25), suggesting that the CA-IX positive/pimonidazole-negative areas were “pseudo-hypoxic”, a process wherein cells express hypoxia induced proteins even in the presence of oxygen (26). Hence, only pimonidazole was used as a hypoxia marker in the further analyses.
Histological sections were segmented into four (viable-normoxic, non-viable normoxic, viable-hypoxic, or non-viable hypoxic) tumor regions, according to cellular characteristic in H&E (Figure 3A) and specific staining of pimonidazole (Figure 3B) and CD31 (Figure 3C). First, tumors were segmented into viable and non-viable tumor cells based on the staining pattern analyzed by a board-certified pathologist in the H&E sections. Non-viable cell regions were composed of necrotic cells and cells in the transition from viable to necrosis (Supplementary Fig. S2). CD31 stains vascular endothelial cells, and was observed in viable tumor regions previously classified by H&E. Hence, pixels positive for CD31 were assigned as viable, while pimonidazole positive pixels were used to classify hypoxia. Then, the output of the viable, non-viable, pimonidazole, and CD31 masks were used to create the histological habitat maps (Figure 3D), which were downsampled to the same resolution as that of the proposed habitat maps determined from MRI, using a superpixel grid (Figure 3E).
We then developed an algorithm to segment mpMRI data into different tumor regions (i.e. proposed habitats). A GMM was chosen to account for intravoxel heterogeneity, which reflects the complex microenvironment in a voxel, clearly observed in histological superpixels. Classification of tumor habitats was achieved by clustering six MRI parameter map values simultaneously: a T2 map, T2* map, ADC map, and three DCE-MRI maps (slope, AUC, and TTM). The Statistics and Machine Learning Toolbox (MATLAB Release 2018a The MathWorks, Inc., Natick, MA), was used to perform GMM clustering with diagonal covariance type, a regularization value of 0.1, centroid seeding with k-means++, and 150 repetitions. Clustering was initiated and performed in the same fashion for both tumor models. However, classification of pixels was performed independently for each model.
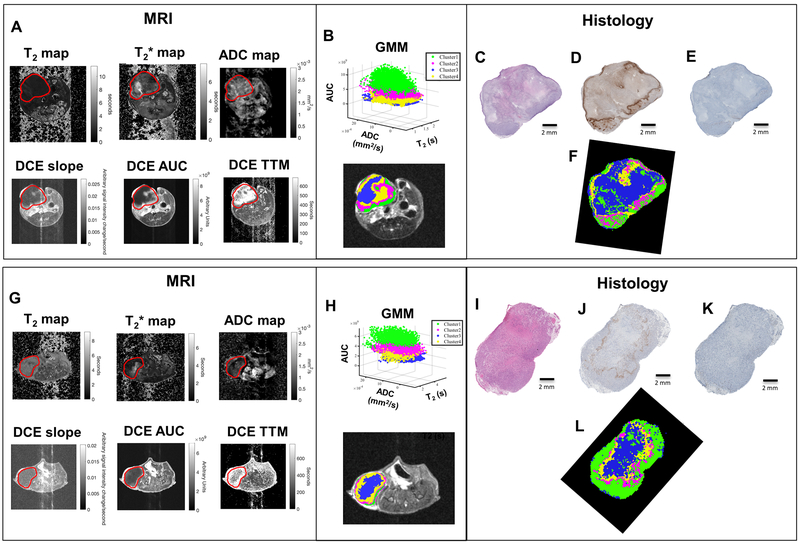
Representative MRI and histology-derived habitat maps for isogenic murine (4T1) model are shown in Figure 4A–F and supplementary Fig. S3A–F, while for the xenografted human (MDA-MB-231) breast cancer model are shown in Figure 4G–L and supplementary Fig. S3G–L. An algorithm based on a “moving/sliding window” (22,23) was used to quantify the agreement between the habitat maps from MRI and histology. A score from 0–1 was given for the agreement of each corresponding habitat and the average was calculated to obtain the total score, as shown in Supplementary Table S1.
Figure 4. Representative examples of the corresponding habitat maps from MRI and histology. 4T1 tumor is shown in (A) and MDA-MB-231 is shown in (G).
Six MRI parameter maps were obtained [T2-map, T2*-map, Apparent diffusion coefficient (ADC), Slope, Area under the curve (AUC) and time to max (TTM)]. Field of view 30 × 30 mm2; image size 256 × 256; Tumor volume of interest (VOI) is shown in red. (B and H) These six MRI parameter maps were clustered by using a Gaussian Mixture Model (GMM) to create the Habitat Maps. Cluster green shows high enhancement in DCE and cluster region blue shows low enhancement in DCE. Magenta and yellow clusters show moderate enhancement in DCE. Histological images ((C and I) H&E; (D and J) pimonidazole and (E and K) CD31) were used to create downsampled habitat-maps derived from histology (F and L) in the same resolution of MR images. The colors in histology (ground truth) habitat maps are delineated by viable cells region in green, non-viable cells region in blue and hypoxic areas in viable or non-viable regions in magenta and yellow, respectively.
Across 26 such analyses, the total moving window score ranged from 0.58 to 0.67. In addition, we calculated the scores by comparing all possibilities for incorrect classifications in the habitat maps from MRI to those of histology, in order to establish the dynamic range between correct and incorrect scores. The average for the incorrect classifications is shown in Supplementary Table S1 and the individual values for each sample are detailed in Supplementary Table S2.
As shown in Figure 4, the cluster that showed high enhancement in AUC, higher slope, and lower TTM (Figure 4A) were localized in the edge of the tumors and classified as a viable habitat (green) (Figure 4B), which were compared with the viable tumor cell regions in histology (Figure 4C–F). The core of the tumors had relatively lower DCE slope and AUC values, which were associated with the non-viable tumor cells regions in histology (blue in Figure 4F), showing the highest scores of similarities (>0.72). Clusters that showed moderate enhancement in DCE corresponded to two hypoxic areas in histology (yellow and magenta in Figure 4F). Same results were observed for the MDA-MB-231 tumor model (Figure 4G–L). These proposed habitats, hypoxic viable (H1; magenta) and hypoxic non-viable (H2; yellow) were analyzed separately or combined into one hypoxic habitat (H1+H2). When comparing the maps with H1+H2 combined, the total score for the agreement between the proposed habitat maps in MRI and histology was slightly higher.
DCE-MRI maps were the most informative to differentiate the habitats
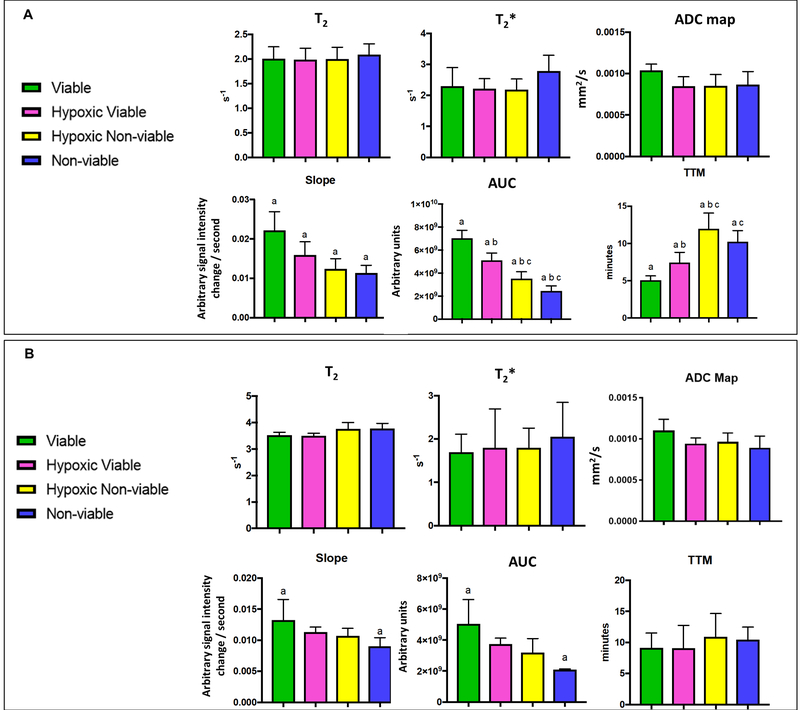
First, voxel values of each MRI-parameter, such as T2 map, T2* map, ADC map, AUC, slope and TMM were compared individually between the four MRI-defined habitats. An ANOVA multiple comparison test showed a large number of statistically significant (p <0.0001) comparisons between clusters. 216 comparisons were calculated for the 4T1 samples, and 212 were statistically significant, while 144 comparisons were calculated for the MDA-MB-231 samples, and 138 were statistically significant (Supplementary Table S3). These results indicate that the GMM algorithm was robust in classifying regions that contained distinct characteristics in each set of MRI-parameters.
Subsequently, the mean values of each MRI-parameter were calculated for each sample, and ANOVA was used to compare these mean values between the four MRI-defined clusters for 4T1 (Figure 5A) and MDA-MB-231 (Figure 5B) tumor models. These analyses indicated that only the maps derived from DCE-MRI showed a statistically significant difference between the clusters, while the T2, T2* and ADC maps were not significantly different for either tumor type. Although each individual MRI parameter is likely to contribute to the clustering, the DCE-MRI maps were the most informative to differentiate the habitats. Notably, the DCE analyses used were agnostic, and not fitted to a 2-compartment model requiring accurate knowledge of the arterial input function (AIF). In fact, histograms for slope, AUC and TTM showed different distribution in each habitat (Supplementary Fig. S4A–H). Indeed, the spatial distribution observed in the DCE-MRI maps alone (Supplementary Fig. S5A) showed a good visual correlation with pimonidazole staining in histology (Supplementary Fig. S5B and C).
Figure 5.
Mean values of each parameter from habitat maps generated by clustering six MRI-parameters [T2-map, T2*-map, Apparent diffusion coefficient (ADC), Slope, Area under the curve (AUC) and time to max (TTM)].Mean values of each parameter were compared between clusters (habitats) for the (A) 4T1 (n=6 samples) and (B) MDA-MB-231 (n=4 samples) tumor samples. Graphs represent mean and SD. p-values were obtained using one-way analysis of variance (ANOVA) followed by the Tukey test for comparison of mean values between regions (a-c indicate p<0.05 between groups).
We thus created new MRI-habitat maps without the ADC maps by clustering the other five MRI-parameters into four clusters. The MRI-cluster maps were visually similar to the MRI-cluster maps created with 6 parameters (Supplementary Fig. S6) and showed similar results in further analyses. The comparison of voxel values of each MRI-parameter between habitats (clusters) showed that 422 of 432 comparisons were statistically significant for the 4T1 samples, while 141 of 144 were statistically significant for the MDA-MB-231 samples (Supplementary Table S4). When comparing the MRI-parameter mean values for each breast tumor type, the DCE-MRI maps remained statistically different between the proposed habitats, whereas T2 did not differ significantly for both tumor types. T2* map values were higher in non-viable habitat when compared with hypoxic viable or hypoxic non-viable habitats (Supplementary Figure 7A and B), and it was statistically significant for the 4T1 tumors.
The similarity index also showed similar moving/sliding window scores when comparing the histology-habitats and MRI-cluster maps (Supplementary Table S5). Supplementary Table S6 shows the scores for all incorrect possibilities between habitats from MRI and histology.
DW-MRI did not distinguish viable and non-viable cell habitats
Surprisingly, DW-MRI did not show great value in distinguishing viable from non-viable cell habitats in these breast tumors. This was unexpected because diffusional restriction is assumed to be due to high cellularity, and thus increased ADC is commonly considered to be a marker of necrosis. Regions with non-viable cells showed both high and low ADC values (Supplementary Figure S8A and B). A close analysis of histology suggested it might be due to the mixing of non-viable cells at different stages of the necrotic process, which were classified as non-viable as detailed previously in Supplementary Figure S2.
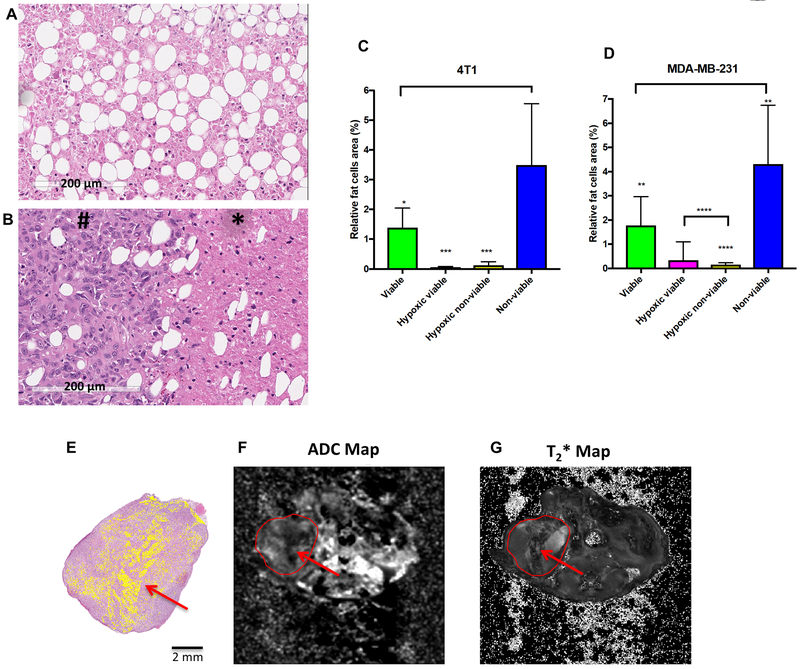
Alternatively, as these breast cancer models were orthotopic, the tumors cells were inoculated into the mammary fat pad with a high content of adipocytes. If these are in necrotic volumes, they may diffusionally restrict water and thus also contribute to the low-ADC values. To investigate this, we performed IHC for perilipin-1, a protein located on adipocyte lipid droplets, which positively stained adipocytes within the breast tumor tissue (Supplementary Figure S9A and B). Once the presence of adipocytes was confirmed, we quantified the adipocytes in each region that was previously segmented in histology (Figure 6 A and B). Results showed that the percentage of adipocytes area in the non-viable tumor cell region was significantly higher than in the viable tumor cell region (Figure 6C and D). Interestingly, it is possible to observe that a region of adipocytes identified in the H&E can be detected as a low signal region in the ADC map which is also observed in the T2* map (Figure 6 E–G).
Figure 6.
Identification of adipocytes in each segmented region (viable, hypoxic viable, hypoxic non-viable, non-viable) in histological samples. (A) Adipocytes observed in non-viable cells region; (B) Adipocytes observed in viable (#) and non-viable (*) cells region; (C and D) Relative area of adipocytes (%) in 4T1 and MDA-MB-231 histological tumor samples. Graphs represent mean and SD. p-values were obtained using one-way analysis of variance (ANOVA) followed by the Tukey test for comparison of mean values between regions (*p<0.05; ** p<0.01; ***p<0.001; ****p<0.0001 (n=6 histological samples for 4T1 and n=8 histological samples for MDA-MB-231). (E) Representative example of a MDA-MB-231 sample showing the adipocytes identified in histology (masked in yellow), which can be observed as a region with low signal in the corresponding ADC Map in (F) and is also visible in the T2* map in (G) (MR images = field of view 30 × 30 mm2; image size 256 × 256).
Discussion
It is well established that the microenvironment in solid tumors is heterogeneous, characterized by inadequate perfusion, hypoxia, and acidosis (2). Hypoxia contributes to radiation and chemotherapy resistance and consequently with aggressive tumor phenotypes and poor outcomes (12). In vivo knowledge of the spatial distribution of hypoxic and other subregions in tumors can potentially provide important prognostic information and improve treatment planning. Thus, in this study we aimed to determine whether mpMRI is capable of identifying and distinguishing hypoxic, viable, and non-viable tumor habitats. To answer this question, there were four separate problems to overcome: 1) Co-registration of MRI and histology, 2) Statistical clustering and classification of mpMRI maps into sub-regions, 3) Classification of histology for the determination of tumor habitats and 4) Measurement and scoring of agreement between imaging and histology. Based on the findings that address these questions, we have successfully demonstrated that mpMRI can identify these four tumor habitats. More importantly, our study showed that specifically semi-quantitative DCE-MRI had a great potential in identifying the hypoxic fractions in breast tumors. This is clinically relevant, given the relationship with hypoxia and resistance to therapy and the common acquisition of DCE-MRI in the workup of breast cancer patients.
In the era of targeted therapies, there is a growing interest in noninvasive techniques for monitoring therapy response. Studies have shown that functional imaging modalities, such as DW-MRI and DCE-MRI are able to longitudinally assess therapeutic responses indicating changes in cellularity and vascularization/perfusion, which precede morphological changes (27). An increase in ADC values in response to therapy has been associated with necrosis (17), while early changes in DCE-MRI parameters have been related to therapy effectiveness (17,27) and prediction of clinical outcome (28). In addition, DCE-MRI (13,29,30) as well as different mpMRI approaches (31,32), have been explored to identify hypoxia, which can be valuable in guiding therapies, for example as a tool to handle the hypoxia-induced chemoradioresistant tumor fraction, that can be integrated to treatment planning with HAPs (33) or radiation (34). Positron emission tomography (PET) based radiotracers, such as 18F-Fluoromisonidazole (18F-FMISO) and 18F-fluoroazomycin arabinoside (18F-FAZA), have also been used to identify hypoxia (35), however they suffer from low dynamic range, and are not routinely available (36), unlike DCE-MRI.
In the current study, four tumor subregions were identified by clustering mpMRI maps, and classified as viable, hypoxic viable, hypoxic non-viable and non-viable tumor cells based on the co-registered histology. MR imaging and histology co-registration is complicated by tumor shrinkage and deformations resulting from tissue fixation, differences in scale, pathology sectioning variability, and errors in determining the correspondence of slices (37). Histology is essential to characterize the in vivo tumor habitats; however, its ability to do this is highly influenced by the co-registration accuracy, as tissue deformations may influence the spatial correlation of ex vivo histology and in vivo MRI (38). Several strategies have been proposed to overcome these limitations, including fiducial markers, ex vivo MRI, as well as MRI-based 3D-printed molds (37,39). Here, by using specific 3D-printed molds we were able to co-register the slices with high accuracy, showing similarity indexes higher than 80%.
As far as the comparisons of habitats between MRI and histology, it is worth mentioning that histopathology processing can introduce errors in the direct spatial comparison of the ex vivo and in vivo habitats localization. To compare habitat maps between modalities, we used a moving/sliding window technique, which mimics human perception. Most of the existing comparison methods including Chi Square and Dice Similarity Index compare the maps on pixel-to-pixel basis, omitting the global similarity between the compared maps. Hence, the compared maps include some extent of noise and miss-registration and those methods may fail to capture the global similarity that can only be seen be human eye. Given that we have largely satisfied the criterion of MRI and histology co-registration, we demonstrated that the distinct regions, as delineated by mpMRI, are in fact habitats. This is perhaps the only realistic means to determine the biology of these MRI subregions, and sets the groundwork for future application to ensure that MRI can reliably identify tumor habitats when histological assessment is not possible.
Different analyses of mpMRI data have been used to segment tumors into distinct subregions. Notably, Henning et al. (32) were able to identify an intratumoral subregion that increased after radiotherapy, likely as a selective survival due radioresistant phenotype. Four subregions were identified using k-means clustering of DW-MRI and T2 data, while IHC differentiated two viable subregions into well-vascularized (HIF-1 negative) and hypoxic (HIF-1 positive) (32). The radioresistant hypoxic region showed intermediate ADC values and T2 lower than the well-vascularized viable region (32). More recently, a novel spatially regularized spectral clustering algorithm applied to mpMRI data accurately characterized tumor into viable, peri-necrotic (hypoxic) and necrotic subregions, which showed strong correlation with co-registered histological slides in a glioblastoma xenograft model (40). By imposing spatial constraints on a standard GMM clustering, the methodology showed greater accuracy in determining tumor subregions than other partitional clustering algorithms, such as k-means, combined fuzzy C-means (FCM) and GMM (40). It is important to note that given MRI’s spatial resolution, the presence of different habitats within single voxels is expected. By examining superpixels from histology in our analyses, it is evident that multiple habitats typically exist within each MRI voxel, thus, a mixture model, such as GMM, is preferred as it may provide insight into the mixing of biological habitats in tumors. In the current study, spatial constraints were not imposed; which suggests that the choice of MRI parameters used in this study was able to distinguish the subtle differences in tumor habitats.
Presumably, each individual MRI parameter map is sensitive to different biophysical aspects within a given voxel. Results showed that between the six parameters used, DCE-MRI maps were the most capable of distinguishing tumor subregions, as they significantly differed between clusters. Interestingly, DW-MRI did not contribute significantly to the identification of these subregions, showing similar ADC values between habitats. In addition, T2* was a significant indicator of high fat regions, which appeared to also have lower ADC values.
Historically, ADC values have been asserted to reflect tumor cellularity (41) and are generally different between viable and necrotic tumor subregions where necrotic clusters present high ADC values but differing T2 values (42–44). However, low ADC values have also been reported in regions of hypocellularity, which were suggested to be coagulative necrotic regions or areas of extreme hypoxia that has previously been reported in a diffusion-restricted necrotic region (45).
In the current work, the mean ADC values in the non-viable tumor cell regions were 0.00086 mm2/s for 4T1 and 0.00089 mm2/s for MDA-MB-231 breast tumors. Histologically, these areas were composed of mixtures of cells in the transition from viable to necrotic tissue. Cells in a final stage of necrosis will show rupture of the cell membrane and release of the cytoplasmic contents to the extracellular area. However, non-viable cell regions also contain cells where membrane rupture has not yet occurred and therefore continues to restrict the diffusion of water; and, consequently these can display continued low ADC values. In addition, other reasons may have contributed to these ADC values, such as the presence of coagulative necrosis, which is the default necrotic pattern associated with ischemia or chronic hypoxia (46), and more likely the presence of adipocytes in the tumor, as we show in Figure 6.
Thus, these results suggest that ADC itself cannot be used as a sole parameter to distinguish necrotic, hypoxic, and viable regions, at least in the case of progressing tumors used in this study, which describe necrosis in the context of progression and not in response to therapy. With regard to therapeutic response, ADC may likely increase and may presage tumor response given that relative changes in ADC are likely due to induction of cell death by an external agent (41,47). This is an important distinction and one that is not experimentally addressed herein.
Unlike DW-MRI, the DCE-maps were distinct in viable, hypoxic, and non-viable subregions. Several studies have used different DCE-MRI data analyses to delineate the spatial intratumoral vascular heterogeneity, defining tumor areas that are well-perfused, hypoxic and necrotic (48–51). Interpretations of DCE-MRI can involve qualitative analysis that require computational-based curve fitting algorithms using a bi-compartmental model with AIF. However, one of its primary challenges is the reproducibility, as the quantification is heavily influenced by the definition of an appropriate AIF (52). Semi-quantitative parameters seem to be more sensitive in the context of intratumoral heterogeneity, for example to subtle physiological differences that distinguish viable and hypoxic regions (48).
Here, we used model-free parameters to capture sensitivity to multiple aspects of microvascularity to be able to discern varying degrees of enhancement. For example, larger amounts of enhancement in the rim of the breast tumors are obvious in DCE slope maps; whereas, slower enhancement is best captured by a TTM map. In fact, the clusters showing higher enhancement in AUC, higher slope, and lower TTM were localized in the edge and corresponded to viable cells in histology. This suggests that these cells have greater access to blood supply and are in an environment rich in nutrients and oxygen (10). On the contrary, poorly perfused areas were localized in the center, and classified as non-viable.
Regions identified as hypoxic tend to have a larger TTM; hence, they are potentially identified in TTM maps, and could be missed by a standard DCE analysis that are often collected over times that are short compared to the TTM, and therefore overemphasize regions with high perfusion, high permeability and high slope. Here, we were able to identify two hypoxic subregions, both localized in the peri-necrotic area, but one extending to the viable tumor cells region and other concentrated in the non-viable tumor cells region. It is known that as a consequence of chronic hypoxia, poorly perfused tumor areas may become necrotic, or develop adaptive mechanisms that enable them to survive in acidic and hypoxic conditions, such as switching to anaerobic glycolysis as their primary energy source (53). Perhaps this population of hypoxic viable tumor cells exhibits a phenotype of therapy resistance that may be inferred by MRI. In fact, studies have shown that the spatial distribution of viable, hypoxic and necrotic tumor subregions can be determined solely based on the uptake curves from DCE-MRI (48,54).
In summary, we present a novel method for co-registration of MRI and histology, and then we showed that mpMRI permits the quantification of tumor habitats. This method holds great promise for monitoring evolutionary dynamics in tumors. If generally applicable, it may provide insight into selection pressures that occur during tumor progression, which could be used as a non-invasive imaging method for the monitoring of the Darwinian dynamics following therapy in cancer. In addition, the power of semi-quantitative DCE-MRI parameter maps in identifying hypoxia presented here is of greater interest, since DCE-MRI is already routinely used in the clinic. The ability to incorporate these analyses into routine practice for patient management and therapeutic planning is a field worth exploring in future investigations.
Supplementary Material
Statement of significance.
This study demonstrates that non-invasive imaging metrics can be used to distinguish sub-regions within heterogeneous tumors with histopathologic correlation.
Acknowledgment
This research was supported by a NIH grant awarded through the NCI (grant number 5R01CA187532; R. Gillies and G. Martinez) and in part by a Research Internship Abroad (BEPE) scholarship from FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) (grant number 2015/18541–3; B. Jardim-Perassi). This work has been supported in part by the Analytic Microscopy Core, the Tissue Core, the Image Response Assessment Team Core and the Small Animal Imaging Lab Core at the H. Lee Moffitt Cancer Center & Research Institute, a comprehensive cancer center designated by the National Cancer Institute and funded in part by Moffitt’s Cancer Center Support Grant (P30-CA076292; T. Sellers).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Swanton C Intratumor Heterogeneity: Evolution through Space and Time. Cancer Research 2012;72:4875–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillies RJ, Brown JS, Anderson ARA, Gatenby RA. Eco-evolutionary causes and consequences of temporal changes in intratumoural blood flow. Nat Rev Cancer 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Icard P, Shulman S, Farhat D, Steyaert JM, Alifano M, Lincet H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist Updat 2018;38:1–11 [DOI] [PubMed] [Google Scholar]
- 4.Ahmed F, Haass NK. Microenvironment-Driven Dynamic Heterogeneity and Phenotypic Plasticity as a Mechanism of Melanoma Therapy Resistance. Frontiers in Oncology 2018;8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connor JPB, Rose CJ, Waterton JC, Carano RAD, Parker GJM, Jackson A. Imaging Intratumor Heterogeneity: Role in Therapy Response, Resistance, and Clinical Outcome. Clinical Cancer Research 2015;21:249–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillies RJ, Beyer T. PET and MRI: Is the Whole Greater than the Sum of Its Parts? Cancer Research 2016;76:6163–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez GV. Introduction to MRI Physics. Methods Mol Biol 2018;1718:3–19 [DOI] [PubMed] [Google Scholar]
- 8.Marino MA, Helbich T, Baltzer P, Pinker-Domenig K. Multiparametric MRI of the breast: A review. J Magn Reson Imaging 2018;47:301–15 [DOI] [PubMed] [Google Scholar]
- 9.Gillies RJ, Balagurunathan Y. Perfusion MR Imaging of Breast Cancer: Insights Using “Habitat Imaging”. Radiology 2018;288:36–7 [DOI] [PubMed] [Google Scholar]
- 10.Lloyd MC, Cunningham JJ, Bui MM, Gillies RJ, Brown JS, Gatenby RA. Darwinian Dynamics of Intratumoral Heterogeneity: Not Solely Random Mutations but Also Variable Environmental Selection Forces. Cancer research 2016;76:3136–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamidreza Farhidzadeh BC, Mu Zhou, Dmitry B. Goldgof, Lawrence O. Hall, Robert A. Gatenby, Robert J. Gillies, Meera Raghavan. Prediction of treatment outcome in soft tissue sarcoma based on radiologically defined habitats. Proc SPIE 2015;9414 [Google Scholar]
- 12.Matsumoto S, Kishimoto S, Saito K, Takakusagi Y, Munasinghe JP, Devasahayam N, et al. Metabolic and physiologic imaging biomarkers of the tumor microenvironment predict treatment outcome with radiation or a hypoxia-activated prodrug in mice. Cancer Res 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellingsen C, Hompland T, Galappathi K, Mathiesen B, Rofstad EK. DCE-MRI of the hypoxic fraction, radioresponsiveness, and metastatic propensity of cervical carcinoma xenografts. Radiother Oncol 2014;110:335–41 [DOI] [PubMed] [Google Scholar]
- 14.Estrella V, Chen T, Lloyd M, Wojtkowiak J, Cornnell HH, Ibrahim-Hashim A, et al. Acidity generated by the tumor microenvironment drives local invasion. Cancer research 2013;73:1524–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pilon-Thomas S, Kodumudi KN, El-Kenawi AE, Russell S, Weber AM, Luddy K, et al. Neutralization of Tumor Acidity Improves Antitumor Responses to Immunotherapy. Cancer research 2016;76:1381–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domenyuk V, Liu X, Magee D, Gatalica Z, Stark A, Kennedy P, et al. Poly-Ligand Profiling differentiates pancreatic cancer patients according to treatment benefit from gemcitabine plus placebo versus gemcitabine plus evofosfamide and identifies candidate targets. Ann Oncol 2018;29 [Google Scholar]
- 17.Zhang XM, Wojtkowiak JW, Martinez GV, Cornnell HH, Hart CP, Baker AF, et al. MR Imaging Biomarkers to Monitor Early Response to Hypoxia-Activated Prodrug TH-302 in Pancreatic Cancer Xenografts. Plos One 2016;11:e0155289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byrd RH, Schnabel RB, Schultz GA. A trust region algorithm for nonlinearly constrained optimization. SIAM J Numer Anal 1987;24:1151–70 [Google Scholar]
- 19.Moré JJ, Sorensen DC. Computing a Trust Region Step. SIAM Journal on Scientific and Statistical Computing 1983;3:553–72 [Google Scholar]
- 20.Levenberg K A Method for the Solution of Certain Problems in Least-Squares. Quarterly Applied Mathematics 2 1944:164–8 [Google Scholar]
- 21.Marquardt D An Algorithm for Least-squares Estimation of Nonlinear Parameters. SIAM Journal Applied Mathematics 1963;11:431–41 [Google Scholar]
- 22.Kuhnert M, Voinov A, Seppelt R. Comparing raster map comparison algorithms for spatial modeling and analysis. Photogramm Eng Rem S 2005;71:975–84 [Google Scholar]
- 23.Costanza R Model goodness of fit: A multiple resolution procedure. Ecological modelling 1989;47:199–215 [Google Scholar]
- 24.Lee SH, McIntyre D, Honess D, Hulikova A, Pacheco-Torres J, Cerdan S, et al. Carbonic anhydrase IX is a pH-stat that sets an acidic tumour extracellular pH in vivo. Br J Cancer 2018;119:622–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tafreshi NK, Lloyd MC, Proemsey JB, Bui MM, Kim J, Gillies RJ, et al. Evaluation of CAIX and CAXII Expression in Breast Cancer at Varied O-2 Levels: CAIX is the Superior Surrogate Imaging Biomarker of Tumor Hypoxia. Molecular Imaging and Biology 2016;18:219–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell S, Gatenby, R.A. Gillies, R.J. Ibrahim-Hashim, A. Chapter 4 - Pseudohypoxia: Life at the Edge. Ecology and Evolution of Cancer 2017:57–68 [Google Scholar]
- 27.Chen YF, Yuan A, Cho KH, Lu YC, Kuo MYP, Chen JH, et al. Functional evaluation of therapeutic response of HCC827 lung cancer to bevacizumab and erlotinib targeted therapy using dynamic contrast-enhanced and diffusion-weighted MRI. Plos One 2017;12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen BB, Hsu CY, Yu CW, Liang PC, Hsu C, Hsu CH, et al. Early perfusion changes within 1 week of systemic treatment measured by dynamic contrast-enhanced MRI may predict survival in patients with advanced hepatocellular carcinoma. European Radiology 2017;27:3069–79 [DOI] [PubMed] [Google Scholar]
- 29.Newbold K, Castellano I, Charles-Edwards E, Mears D, Sohaib A, Leach M, et al. An Exploratory Study into the Role of Dynamic Contrast-Enhanced Magnetic Resonance Imaging or Perfusion Computed Tomography for Detection of Intratumoral Hypoxia in Head-and-Neck Cancer. International Journal of Radiation Oncology Biology Physics 2009;74:29–37 [DOI] [PubMed] [Google Scholar]
- 30.Halle C, Andersen E, Lando M, Aarnes EK, Hasvold G, Holden M, et al. Hypoxia-Induced Gene Expression in Chemoradioresistant Cervical Cancer Revealed by Dynamic Contrast-Enhanced MRI. Cancer Research 2012;72:5285–95 [DOI] [PubMed] [Google Scholar]
- 31.Featherstone AK, O’Connor JPB, Little RA, Watson Y, Cheung S, Babur M, et al. Data-driven mapping of hypoxia-related tumor heterogeneity using DCE-MRI and OE-MRI. Magn Reson Med 2018;79:2236–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henning EC, Azuma C, Sotak CH, Helmer KG. Multispectral tissue characterization in a RIF-1 tumor model: monitoring the ADC and T2 responses to single-dose radiotherapy. Part II. Magn Reson Med 2007;57:513–9 [DOI] [PubMed] [Google Scholar]
- 33.Cardenas-Rodriguez J, Li YG, Galons JP, Cornnell H, Gillies RJ, Pagel MD, et al. Imaging biomarkers to monitor response to the hypoxia-activated prodrug TH-302 in the MiaPaCa2 flank xenograft model. Magnetic Resonance Imaging 2012;30:1002–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gulliksrud K, Ovrebo KM, Mathiesen B, Rofstad EK. Differentiation between hypoxic and non-hypoxic experimental tumors by dynamic contrast-enhanced magnetic resonance imaging. Radiotherapy and Oncology 2011;98:360–4 [DOI] [PubMed] [Google Scholar]
- 35.Gaertner FC, Souvatzoglou M, Brix G, Beer AJ. Imaging of Hypoxia Using PET and MRI. Current Pharmaceutical Biotechnology 2012;13:552–70 [DOI] [PubMed] [Google Scholar]
- 36.Salem A, Asselin MC, Reymen B, Jackson A, Lambin P, West CML, et al. Targeting Hypoxia to Improve Non-Small Cell Lung Cancer Outcome. Jnci-Journal of the National Cancer Institute 2018;110 [DOI] [PubMed] [Google Scholar]
- 37.Alic L, Haeck JC, Bol K, Klein S, van Tiel ST, Wielepolski PA, et al. Facilitating Tumor Functional Assessment by Spatially Relating 3D Tumor Histology and In Vivo MRI: Image Registration Approach. Plos One 2011;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson E, Gaed M, Gomez JA, Moussa M, Pautler S, Chin JL, et al. 3D prostate histology image reconstruction: Quantifying the impact of tissue deformation and histology section location. J Pathol Inform 2013;4:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Disselhorst JA, Krueger MA, Ud-Dean SMM, Bezrukov I, Jarboui MA, Trautwein C, et al. Linking imaging to omics utilizing image-guided tissue extraction. Proceedings of the National Academy of Sciences of the United States of America 2018;115:E2980–E7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katiyar P, Divine MR, Kohlhofer U, Quintanilla-Martinez L, Scholkopf B, Pichler BJ, et al. A Novel Unsupervised Segmentation Approach Quantifies Tumor Tissue Populations Using Multiparametric MRI: First Results with Histological Validation. Mol Imaging Biol 2017;19:391–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoeny HC, Ross BD. Predicting and Monitoring Cancer Treatment Response with Diffusion-Weighted MRI. Journal of Magnetic Resonance Imaging 2010;32:2–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berry LR, Barck KH, Go MA, Ross J, Wu XM, Williams SP, et al. Quantification of viable tumor microvascular characteristics by multispectral analysis. Magnetic Resonance in Medicine 2008;60:64–72 [DOI] [PubMed] [Google Scholar]
- 43.Carano RA, Ross AL, Ross J, Williams SP, Koeppen H, Schwall RH, et al. Quantification of tumor tissue populations by multispectral analysis. Magn Reson Med 2004;51:542–51 [DOI] [PubMed] [Google Scholar]
- 44.Barck KH, Willis B, Ross J, French DM, Filvaroff EH, Carano RAD. Viable Tumor Tissue Detection in Murine Metastatic Breast Cancer by Whole-Body MRI and Multispectral Analysis. Magnetic Resonance in Medicine 2009;62:1423–30 [DOI] [PubMed] [Google Scholar]
- 45.LaViolette PS, Mickevicius NJ, Cochran EJ, Rand SD, Connelly J, Bovi JA, et al. Precise ex vivo histological validation of heightened cellularity and diffusion-restricted necrosis in regions of dark apparent diffusion coefficient in 7 cases of high-grade glioma. Neuro-Oncology 2014;16:1599–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caruso RA, Branca G, Fedele F, Irato E, Finocchiaro G, Parisi A, et al. Mechanisms of coagulative necrosis in malignant epithelial tumors (Review). Oncol Lett 2014;8:1397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theilmann RJ, Borders R, Trouard TP, Xia GW, Outwater E, Ranger-Moore J, et al. Changes in water mobility measured by diffusion MRI predict response of metastatic breast cancer to chemotherapy. Neoplasia 2004;6:831–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoyanova R, Huang K, Sandler K, Cho H, Carlin S, Zanzonico PB, et al. Mapping Tumor Hypoxia In Vivo Using Pattern Recognition of Dynamic Contrast-enhanced MRI Data. Transl Oncol 2012;5:437–U114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaudhury B, Zhou M, Goldgof DB, Hall LO, Gatenby RA, Gillies RJ, et al. Heterogeneity in intratumoral regions with rapid gadolinium washout correlates with estrogen receptor status and nodal metastasis. J Magn Reson Imaging 2015;42:1421–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang YC, Ackerstaff E, Tschudi Y, Jimenez B, Foltz W, Fisher C, et al. Delineation of Tumor Habitats based on Dynamic Contrast Enhanced MRI. Sci Rep 2017;7:9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han S, Stoyanova R, Lee H, Carlin SD, Koutcher JA, Cho H, et al. Automation of pattern recognition analysis of dynamic contrast-enhanced MRI data to characterize intratumoral vascular heterogeneity. Magnetic Resonance in Medicine 2018;79:1736–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keil VC, Madler B, Gieseke J, Fimmers R, Hattingen E, Schild HH, et al. Effects of arterial input function selection on kinetic parameters in brain dynamic contrast-enhanced MRI. Magnetic Resonance Imaging 2017;40:83–90 [DOI] [PubMed] [Google Scholar]
- 53.Kim JY, Gatenby RA. Quantitative Clinical Imaging Methods for Monitoring Intratumoral Evolution. Methods Mol Biol 2017;1513:61–81 [DOI] [PubMed] [Google Scholar]
- 54.Han SH, Ackerstaff E, Stoyanova R, Carlin S, Huang W, Koutcher JA, et al. Gaussian mixture model-based classification of dynamic contrast enhanced MRI data for identifying diverse tumor microenvironments: preliminary results. NMR in Biomedicine 2013;26:519–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.